Abstract
An earlier report showed that the US3 protein kinase blocked the apoptosis induced by the herpes simplex virus 1 (HSV-1) d120 mutant at a premitochondrial stage. Further studies revealed that the kinase also blocks programmed cell death induced by the proapoptotic protein BAD. Here we report the effects of the US3 protein kinase on the function and state of a murine BAD protein. Specifically, (i) in uninfected cells, BAD was processed by at least two proteolytic cleavages that were blocked by a general caspase inhibitor. The untreated transduced cells expressed elevated caspase 3 activity. (ii) In cells cotransduced with the US3 protein kinase, the BAD protein was not cleaved and the caspase 3 activity was not elevated. (iii) Inasmuch as the US3 protein kinase blocked the proapoptotic activity and cleavage of a mutant (BAD3S/A) in which the codons for the regulatory serines at positions 112, 136, and 155 were each replaced with alanine codons, the US3 protein kinase does not act by phosphorylation of these sites nor was the phosphorylation of these sites required for the antiapoptotic function of the US3 protein kinase. (iv) The US3 protein kinase did not enable the binding of the BAD3S/A mutant to the antiapoptotic proteins 14-3-3. Finally, (v) whereas cleavage of BAD at ASP56 and ASP61 has been reported and results in the generation of a more effective proapoptotic protein with an Mr of 15,000, in this report we also show the existence of a second caspase-dependent cleavage site most likely at the ASP156 that is predicted to inactivate the proapoptotic activity of BAD. We conclude that the primary effect of US3 was to block the caspases that cleave BAD at either residue 56 or 61 predicted to render the protein more proapoptotic or at residue 156, which would inactivate the protein.
Two diametrically opposed events take place in cells infected with herpes simplex virus 1 (HSV-1) and also in cells infected with many other viruses. On one hand, viral gene expression alters cellular homeostasis and induce the cell to initiate the process that would lead to programmed cell death. This response also occurs in infected cells besieged by the host immune system. On the other hand, apoptosis could curtail viral replication and result in the failure of the virus to express its full potential (10, 27). In the case of HSV-1, the confrontation favors the virus (10, 15, 30). Numerous publications have shown that HSV-1 blocks programmed cell death induced by heat shock (18), tumor necrosis factor alpha (10, 35), Fas ligand (10, 16, 30), osmotic shock (11, 17), and virtually every proapoptotic substance tested to date. While wild-type virus not only fails to induce apoptosis but actually blocks the cell from undergoing apoptosis induced by exogenous agents, a number of mutants have been shown to induce apoptosis even in the absence of these agents. The prevailing view is that stress induced by the function of viral gene products can induce apoptosis but that specific viral gene products block the cells from responding to metabolic injuries induced by the virus. To date, several mutants incapable of blocking the cells from undergoing apoptosis have been described (3, 10, 18, 20, 36, 38). Studies of these mutants have led to the identification of several viral proteins that block apoptosis induced by these mutants. The focus of the present study is on the US3 protein kinase shown in earlier studies to block apoptosis induced by d120, an HSV-1 mutant lacking the gene encoding the infected cell protein no. 4, the major regulatory protein of the virus (19, 21).
In noncomplementing cells infected with the d120 mutant viral gene expression is arrested at α genes and the cells exhibit cytopathogenic manifestations such as chromatin condensation, nucleosome-dependent fragmentation of cellular DNA, vacuolization and blebbing of the cytoplasm, etc. (9, 12, 18). The events leading to apoptosis are cell type dependent. In the human SK-N-SH cells, the events leading to the proapoptotic death of the cells are caspase independent. In HEp-2 and many other cell lines apoptosis induced by the d120 mutant is caspase dependent (12). Studies from this laboratory showed that ectopically expressed HSV-1 US3 protein kinase blocks apoptosis induced by the d120 mutant (19, 21). More recent studies indicated that the US3 protein kinase also blocked apoptosis induced by ectopic expression of the proapoptotic protein BAD (22). In addition, it has been reported that the US3 protein kinase contributes to HSV-mediated protection from apoptosis in vivo (2).
BAD belongs to the Bcl-2 family of proteins. Pro- and antiapoptotic members of this family dimerize through their Bcl-2 homology domains (BH1, BH2, BH3, and BH4) in order to exert their functions (4, 23, 34). BAD is a proapoptotic BH3-only member of the Bcl-2 family, since it contains only the BH3 domain. In consequence, BAD binds to antiapoptotic members of the family to mediate its proapoptotic effect (4). The activity of BAD protein is negatively regulated by phosphorylation (6). In the case of the murine BAD protein, cell survival signals, through activation of the phosphatidylinositol 3-kinase pathway, induce the Akt-mediated phosphorylation of BAD at serine 136, whereas the activated kinases RSK-1 and protein kinase A phosphorylate BAD at serine 112 (8, 13, 29, 32). Phosphorylation of both serine 112 and serine 136 enables the binding of BAD to 14-3-3 proteins. The 14-3-3 bound BAD protein becomes unavailable for interaction with Bcl-XL (14, 24, 32, 37). In addition, BAD can also be precluded from interacting with Bcl-XL by phosphorylation of serine 155 located inside the BH3 domain (7, 31, 39). In appropriately stimulated cells, BAD is dephosphorylated by calcineurin and the serine/threonine phosphatase 1-α (PP1-α) (28, 33). An Mr 15,000 product of BAD produced by cleavage at residues ASP55 and ASP61 exhibits enhanced proapoptotic activity (5).
US3 protein kinase blocks caspase-dependent cleavage of BAD.
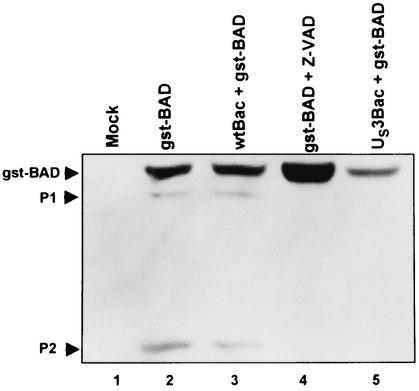
The focus of the present study concerns the role of the US3 protein kinase in blocking apoptosis induced by BAD. In an earlier report this laboratory showed that, in cells transfected with both BAD and US3, products smaller than full-size BAD disappeared and some of the full-length protein exhibited a slower electrophoretic mobility (22). To study the effect of US3 protein kinase on the BAD protein, the latter was tagged at its amino terminus with glutathione S-transferase (gstBAD) and expressed in a baculovirus vector under the cytomegalovirus immediate-early promoter as described in the legend to Fig. 1. This systems allows relatively rapid, multiplicity-dependent ectopic expression of the desired gene in virtually all cells. In this series of experiments rabbit skin cells were mock infected or were infected with either a wild-type or a recombinant baculovirus expressing the US3 protein kinase, incubated for 6 h, and then superinfected with a baculovirus expressing the full-length gstBAD fusion protein. One set of cultures was exposed to the general caspase inhibitor Z-VAD-fmk concurrent with infection with baculovirus expressing gstBAD. At 18 h after superinfection, the cells were harvested, electrophoretrically separated in denaturing gels, transferred to a nitrocellulose sheet, and reacted with the antibody to glutathione S-transferase (GST). The bands shown in Fig. 1 represent the GST protein fused to either intact BAD or amino-terminal cleavage products of the BAD protein. The results were as follows.
FIG. 1.
Photograph of electrophoretically separated cell lysates reacted with a mouse monoclonal antibody to GST. Replicate 25-cm2 flask cultures of rabbit skin cells were either mock infected or infected with 10 PFU of wt-Bac or US3-Bac per cell, incubated for 6 h, and then superinfected with 10 PFU of gstBAD-Bac per cell. One set of culture was exposed to the general caspase inhibitor Z-VAD-fmk, at a concentration of 50 μM, concurrent with gstBAD-Bac infection. Cells were scraped into their medium, pelleted by low-speed centrifugation at 4°C, rinsed twice with phosphate-buffered saline (PBS) A (0.14 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 1.5 mM KH2PO4) in the presence of three phosphatase inhibitors (10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate), and were lysed in radioimmunoprecipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in PBS A) in the presence of phosphatase inhibitors (10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate) and protease inhibitors (Complete; Roche) as recommended by the manufacturer. Lysed cells were stored on ice for 10 min before centrifugation at 14,000 rpm for 10 min (in an Eppendorf centrifuge, model 5415 C). The protein concentration of the supernatant fluids was determined with the aid of a Bio-Rad protein assay according to directions provided by the manufacturer. Protein samples denatured in disruption buffer (50 mM Tris [pH 7.0], 2.75% sucrose, 5% 2-mercaptoethanol, 2% SDS) were electrophoretically separated in a 10% denaturing polyacrylamide gel (70 μg of protein per lane), electrically transferred to a nitrocellulose sheet, blocked, and reacted with a mouse monoclonal antibody specific for GST (Santa Cruz). Protein bands were visualized through enhanced chemiluminescence detection according to the instructions of the manufacturer (Amersham Pharmacia). The upper arrowhead points to the intact chimeric protein. Arrowhead P2 points to a faint band consistent with a polypeptide originating from the cleavage of the chimeric protein at amino acid 56 or 61. Arrowhead P1 points to an additional band with an apparent Mr of 42,000. Note that both expression of the US3 protein kinase and treatment with Z-VAD-fmk blocked the appearance of the two rapidly migrating bands. The construction and the transfer plasmid encoding gstBAD were described elsewhere (22). The gstBAD-expressing baculovirus were constructed by cotransfecting the transfer plasmid gstBAD-MTS-1 with Baculogold DNA (Pharmingen) according to the manufacturer's instructions. The baculovirus encoding the US3 protein kinase under the immediate-early cytomegalovirus promoter has been described elsewhere (21).
(i) The electrophoretrically separated lysates of cells infected with the gstBAD baculovirus contained three polypeptides that reacted with the anti-GST antibody (Fig. 1, lane 2). These polypeptides had apparent Mrs of 48,000, 42,000 (P1), and 30,000 (P2) and therefore consisted of chimeric proteins containing both GST and BAD residues. The largest polypeptide corresponded to the full-length protein, whereas the smallest was consistent with a polypeptide containing GST and BAD cleaved at amino acid 56 or 61 of BAD as described elsewhere (5). We used the anti-GST antibody because the available anti-BAD antibody was directed to the carboxyl terminus and did not react with products truncated at or close to the carboxyl terminus of the protein.
(ii) Infection of rabbit skin cells with wild-type baculovirus prior to infection with the virus expressing gstBAD did not affect the appearance of the rapidly migrating bands P1 and P2 (Fig. 1, lane 3). Conversely, infection of rabbit skin cells with a baculovirus expressing the US3 protein kinase blocked the appearance of the rapidly migrating bands P1 and P2 (Fig. 1, lane 5).
(iii) The caspase inhibitor, Z-VAD-fmk, blocked the appearance of the two rapidly migrating bands (P1 and P2; Fig. 1, lane 4).
We conclude from these studies that in the transduced rabbit skin cells, gstBAD was cleaved by a caspase sensitive to the general caspase inhibitor Z-VAD-fmk. Since the cleavage was also blocked by the US3 protein kinase, the results indicate that one function of US3 is to block a caspase-dependent cleavage of the BAD protein.
The role of BAD cleavage products in the induction of apoptosis.
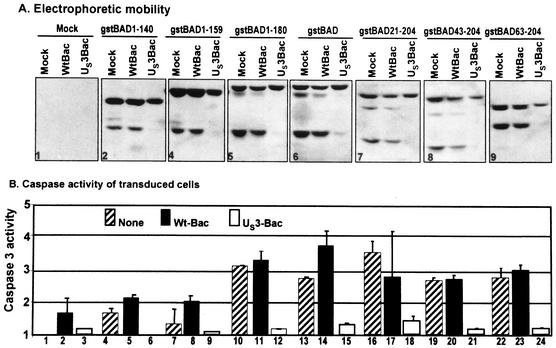
To map the cleavage site blocked by the US3 protein kinase and assess the role of the cleavage products, we constructed a series of BAD truncation mutants schematically represented in Fig. 2 according to procedures described in the legend to that figure. The truncated constructs were cloned in baculoviruses as gstBAD chimeric open reading frames. Table 1 shows the primers used for mutant construction. As illustrated in Fig. 2, the truncated mutant gstBAD1-180 lacked the carboxyl-terminal domain and the amino acids that constitute a poorly conserved BH2 domain. The gstBAD1-159 mutant contains the BH3 domain but lacked the downstream residues, whereas the gstBAD1-140 mutant lacked the entire BH3 domain and all downstream residues. Of the three amino-terminal truncations, gstBAD21-204 lacks the first 20 residues; the BAD sequence in gstBAD43-204 began at an ATG located at position 43, whereas the gstBAD63-204 mutant lacked the two known caspase 3 cleavage sites at residues 56 and 61.
FIG. 2.
Schematic diagram of the structure of the chimeric protein gstBAD (1) and the mutants used in the present study. Regulatory phosphorylation sites at positions 112, 136, and 155 are shown, along with the introduced mutations (S, serine; A, alanine). Stars indicate the two known caspase cleavage sites at residues 56 and 61. gst, GST. Amino-terminal and carboxyl-terminal truncation mutants of BAD were derived from gstBAD-MTS-1 by means of PCR with the primers listed in Table 1. The amplified truncated forms of BAD were inserted into the BamHI/NotI site of gstBAD-MTS-1, in frame with GST. The gstBAD-3S/A-MTS-1 transfer plasmid, in which the codons encoding S112, S136, and S155 were replaced with codons encoding alanines, was constructed as follows. The EcoRI/NotI fragment from gstBAD-MTS-1, encoding the gstBAD fusion protein, was subcloned into the EcoRI/NotI site of pBSK. The serine codons were then replaced with alanine codons with the aid of the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The oligonucleotides used for this purpose were S112A-sense (AGTCGCCACAGTGCGTACCCAGCGGGG), S112A-antisense (CCCCGCTGGGTACGCACTGTGGCGACT), S136A-sense (CGAGGACGCTCGCGTGCGGCTCCCCCC), S136A-antisense (GGGGGGAGCCGCAGCCGAGCGTCCTCG), S155A-sense (CTCCGAAGGATGGCCGATGAGTTTGAGGG), and S155A-antisense (CCCTCAAACTCATCGGCCATCCTTCGGAG). Mutations were confirmed by sequencing, and gstBAD-3S/A was cloned back into the EcoRI/NotI site of MTS-1 The baculoviruses were constructed and propagated as noted in the legend to Fig. 1.
TABLE 1.
Primers used for the construction of BAD truncation mutants
| BAD construct | Primer sequence
|
|
|---|---|---|
| Forward | Reverse | |
| gstBAD1-140 | GCCGGATCCATGGGAACCCCAAAGCAGCCC | GGGCGGCCGCTTAATTGGGGGGAGCCGAACGCGA |
| gstBAD1-159 | GCCGGATCCATGGGAACCCCAAAGCAGCCC | GGGCGGCCGCTTACTCAAACTCATCGCTCATCCT |
| gstBAD1-180 | GCCGGATCCATGGGAACCCCAAAGCAGCCC | GGGCGGCCGCTTAGGCGCTTTGTCGCATCTGTGTTGCA |
| gstBAD21-204 | CCGGATCCGATCCCGGAATCCGGAGCCTGG | CGCCGGCCGCTCACTGGGAGGGGGTGGAGCC |
| gstBAD43-204 | CCGGATCCATGTTCCAGATCCCAGAGTTTGAG | CGCCGGCCGCTCACTGGGAGGGGGTGGAGCC |
| gstBAD63-204 | CCGGATCCGGCCTGGGCCCTAGCCTCACTGAG | CGCCGGCCGCTCACTGGGAGGGGGTGGAGCC |
In this series of experiments rabbit skin cells were mock infected or were infected with wild-type or the US3-expressing baculovirus. After 6 h of incubation, the cells were superinfected with the various baculoviruses expressing the full-length or truncated forms of BAD. Cells were harvested and lysed 18 h after superinfection. The lysates were assayed for caspase 3 activity and also electrophoretically separated in denaturing gels and probed with anti-GST antibody. The electrophoretic mobility of the gstBAD chimeric proteins contained in the lysates are shown in panels 1 to 9 of Fig. 3A. The caspase 3 activities of the cell lysates shown in Fig. 3B were normalized with respect to the baseline activity of mock-infected cells (Fig. 3B, lane 1). The results may be summarized as follows.
FIG. 3.
Effect of the US3 protein kinase expression on cleavage pattern of gstBAD and derivative mutants expressed via recombinant baculovirus (A) and effect of US3 on DEVDase activity induced by gstBAD and derivative mutants (B) in rabbit skin cells. Replicate 25-cm2 flask cultures of rabbit skin cells were either mock infected or infected with 10 PFU of wt-Bac or US3-Bac per cell, incubated for 6 h, and then superinfected with 10 PFU of gstBAD-Bac or each of the truncation mutants per cell. Cells were harvested 18 h after superinfection, solubilized, electrophoretically separated on denaturing gel, electrically transferred to a nitrocellulose sheet, and reacted with mouse monoclonal antibody against gst. Caspase 3 activity in cellular extracts was assayed by using a tetrapeptide conjugated to phenylnitraniline (DEVD-pNA) (BioMol). The cells were scraped, rinsed twice with PBS A, resuspended in 150 μl of lysis solution A (0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 50 mM HEPES [pH 7.4], 1 mM dithiothreitol, 0.1 mM EDTA), and incubated on ice for 10 min. Lysates were then centrifuged at 14,000 rpm for 10 min at 4°C. Supernatant fluids were collected, and the protein content was measured with a Bio-Rad protein assay according to directions provided by the manufacturer. Equal amounts of protein were tested for DEVDase activity according to the manufacturer's instructions (BioMol). Chromophore release was quantified by measuring the absorbance at 405 nm with a spectrophotometer after 2 h. The results are expressed as the fold increase in activity compared to mock-infected cells.
(i) The GST tag did appear to interfere with the activity of BAD in activating caspase 3. Thus, the caspase 3 activity of the lysates of cells infected with the chimeric gene containing the full-length BAD (Fig. 3A, panel 6; Fig. 3B, lanes 13 and 14) was 3.5-fold higher than that of the mock-infected cells.
(ii). The caspase 3 activity of the lysates of cells infected with baculoviruses expressing gstBAD1-140 (lanes 4 and 5) or gstBAD1-159 (lanes 7 and 8) increased ∼2-fold relative to that of lysates of mock-infected cells (lane 1) and significantly less than 2-fold relative to mock-infected cells exposed to wild-type baculovirus (lane 2). We interpret these results to indicate that these constructs failed to activate caspase 3 activity and that the slight elevation of caspase 3 activity was due largely to infection with baculoviruses. In contrast, the lysates of cells infected with baculovirus gstBAD1-180 (lanes 10 and 11) did not appear to be significantly different from those of lysates of cells infected with the baculovirus expressing the full-length BAD fused to GST (lanes 13 and 14). These results are of interest from two points of view. First, whereas the residues encoding the BH3 site absent from the gstBAD1-140 were, as expected, to be essential for the induction of caspase activity, the sequences carboxyl terminal to the BH3 site (gstBAD1-180) were not essential for this activity. Since the gstBAD1-159 mutant failed to induce apoptosis, the results suggest that residues immediately carboxyl terminal to the BH3 consensus sequence may be required for the the function of the BH3 domain. Second, the truncated BAD chimeric proteins were uniformly cleaved at the 56/61 residues even though at least two of the constructs were inactive. Cleavage at the 56/61 sites was independent of the structure of the carboxyl-terminal domain of the BAD protein.
(iii) The amino-terminal truncation had a minimal effect on the activation of caspase 3 activity. It is noteworthy that the construct gstBAD63-204 was subject to the caspase cleavage even though it lacked the 56/61 cleavage sites. This observation is consistent with the conclusion that cleavage which yielded the P1 product (Fig. 1) was carboxyl terminal relative to residue 63 and therefore carboxyl terminal relative to the 56/61 cleavage sites. On the basis of the apparent molecular weights of the P1 and P2 products, we could expect that the full-length product was cleaved at residues 56/61 and again near the carboxyl terminus. Given the observation that the cleavages at the amino-terminal and carboxyl-terminal sites are independent of each other, we could expect that there exists another product of cleavages at both sites. This product would be expected to be smaller than the Mr of 15,000 reported to be present in cells transduced with baculoviruses encoding the BAD protein (5, 22).
(iv) The US3 protein kinase blocked activation of caspase 3. The inhibition of caspase 3 activity is evident in virtually all nine panels shown in Fig. 3. The conclusion to be drawn from the experiments shown in Fig. 3 is that the primary effect of US3 is to block the caspase-dependent cleavage of BAD.
A central question arising from these studies is whether US3 protein kinase acts by phosphorylation of the serines known to inactivate the proapoptotic activity of the BAD protein.
The US3 protein kinase blocks BAD-induced apoptosis independently of the phosphorylation of Ser112, Ser136, or Ser155.
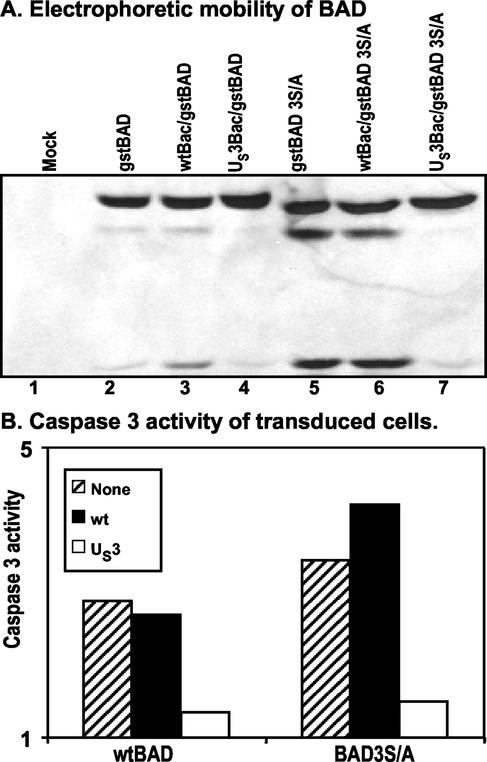
The BAD protein is rendered inactive by phosphorylation of the three regulatory serines at positions 112, 136, and 155 (6). One hypothesis to explain the effect of the US3 protein kinase is that it induces the phosphorylation of the regulatory serines, thereby precluding the sequence of events that lead to the activation of BAD. To test this hypothesis, we constructed a mutant (gstBAD-3S/A) in which the codons Ser112, Ser136, and Ser155 were each replaced with alanine codons as described in the legend to Fig. 2. Rabbit skin cells were mock infected or were infected with either wild-type baculovirus or a US3-expressing baculovirus, incubated for 6 h, and then superinfected with either gstBAD or gstBAD-3S/A baculovirus. The lysates of cells harvested 18 h after the superinfection were tested for the presence of intact and cleaved gstBAD fusion proteins and for the induction of caspase 3 activity as described in the legend to Fig. 3. The results (Fig. 4) were that the caspase 3 activity of cells infected with the baculovirus encoding the wild-type or mutated gstBAD fusion protein was induced at levels >2-fold greater than those of mock-infected cells. The activity of the construct containing the mutant BAD appeared to be higher than that of the construct containing the wild-type BAD. Of particular interest was the observation that US3 protein kinase blocked the activation of caspase 3 and the cleavage of both wild-type and mutated BAD constructs.
FIG. 4.
Effect of the US3 protein kinase expression on cleavage pattern of gstBAD and the gstBAD-3S/A mutant expressed via recombinant baculovirus in rabbit skin cells (A) and effect of US3 on DEVDase activity induced by gstBAD and gstBAD-3S/A (B). Replicate 25-cm2 flask cultures of rabbit skin cells were either mock infected or infected with 10 PFU of wt-Bac or US3-Bac per cell, incubated for 6 h, and then superinfected with 10 PFU of gstBAD-Bac or gstBAD-3S/A-Bac per cell. Cells were harvested and electrophoretically separated or assayed for DEVDase activity as described in the legend to Fig. 3.
US3 does not recruit BAD to the 14-3-3 proteins.
BAD protein phosphorylated at serines 112 and 136 is bound by 14-3-3 proteins and sequestered in the cytoplasm. This translocation precludes the interaction of BAD with Bcl-XL and therefore protects the cell from apoptosis. One hypothesis that could account for the effect of US3 protein kinase on BAD is that a modification of BAD mediated by the US3 protein kinase enables the interaction with the 14-3-3 proteins even in the absence of phosphorylation of the regulatory serines.
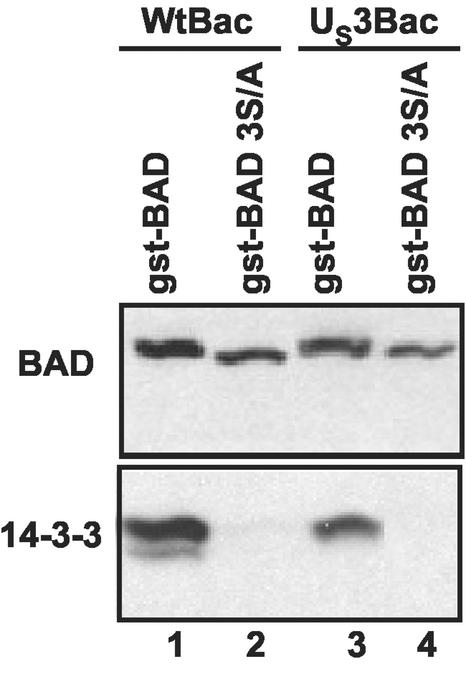
To test this hypothesis, rabbit skin cells were infected with a wild-type or recombinant baculovirus expressing the US3 protein kinase. After 6 h of incubation, the cells were superinfected with either baculoviruses encoding GST fused to wild-type or 3S/A mutant BAD. At 18 h after infection, the cells were harvested, lysed, and reacted with glutathione-agarose beads. After an extensive rinse the bound proteins were subjected to chromatography on denaturing polyacrylamide gels and reacted with antibody to BAD and the 14-3-3 proteins. As shown in Fig. 5, the eluate obtained from glutathione-agarose beads bound both wild-type and mutant gstBAD chimeric proteins. However, only the chimeric protein containing wild-type BAD sequences bound the 14-3-3 proteins.
FIG. 5.
Photograph of immunoblot of affinity purified gstBAD or gstBAD-3S/A and coprecipitated 14-3-3 proteins. Replicate 25-cm2 flask cultures of rabbit skin cells were infected with 10 PFU of wt-Bac or US3-Bac per cell, incubated for 6 h, and then superinfected with 10 PFU of either gstBAD-Bac or gstBAD-3S/A-Bac per cell. Cells were harvested 18 h after the superinfection, lysed in 500 μl of lysis buffer B (142.5 mM KCl, 5 mM MgCl2, 10 mM HEPES, 0.25% Nonidet P-40) in the presence of phosphatase and protease inhibitors, and incubated on ice for 10 min. Lysates were pelleted at 14,000 rpm for 10 min at 4°C. The supernatant fluids were collected and reacted overnight with constant agitation at 4°C with 50 μl of a 50% slurry of glutathione beads (Pharmacia Biotech). The beads were pelleted by low-speed centrifugation and rinsed three times with lysis buffer B. The proteins complexed with the pulled-down gstBAD fusion proteins were denatured in disruption buffer (50 mM Tris [pH 7.0], 2.75% sucrose, 5% 2-mercaptoethanol, 2% SDS) and electrophoretically separated in an 11% denaturing polyacrylamide gel. Immunoblotting was done as described in the legend to Fig. 1. The electrophoretically separated proteins were reacted with a rabbit polyclonal antibody specific for BAD (Santa Cruz) or with a rabbit antibody specific for the 14-3-3 proteins (Santa Cruz) and then visualized by enhanced chemiluminescence detection.
The role of US3 in blocking apoptosis induced by BAD.
These results indicate that the mechanism by which the US3 protein kinase blocks activation of the BAD protein does not depend on the phosphorylation of the regulatory serines or the interaction of BAD with the 14-3-3 proteins.
In an earlier article this laboratory reported that the US3 protein kinase blocked the apoptosis induced by the HSV-1 d120 mutant at a premitochondrial stage (21). Further studies revealed that the kinase also blocks programmed cell death induced by the proapoptotic protein BAD (22). In the studies reported here, we investigated the effects of the US3 protein kinase on the function and state of the BAD protein. We report several key findings below.
(i) In uninfected cells BAD was processed by at least two proteolytic cleavages that are blocked by caspase inhibitors. The untreated transduced cells expressed elevated caspase 3 activity.
(ii) In cells cotransduced with the US3 protein kinase, the BAD protein was not cleaved and the caspase 3 activity was not elevated.
(iii) Inasmuch as the US3 protein kinase blocked the proapoptotic activity and cleavage of a mutant (BAD3S/A) in which the codon for the regulatory serine at positions 112, 136, and 155 was replaced with an alanine codon, the US3 protein kinase does not act by phosphorylation of these sites nor is the phosphorylation of these sites required for the antiapoptotic function of the US3 protein kinase.
(iv) The US3 protein kinase does not enable the binding of the BAD3S/A mutant to the antiapoptotic family of proteins 14-3-3.
(v) Finally, although the cleavage of BAD at ASP56 and ASP61 has been reported and results in the generation of a more effective proapoptotic protein with an Mr of 15,000, we show here the existence of a second caspase-dependent cleavage site most likely at ASP156.
Relevant to the interpretation of these results are the following findings.
(i) A feature of BAD-induced apoptosis is the establishment of a positive feedback loop. Thus, activated BAD titrates out Bcl-XL and then acts to release cytochrome c from mitochondria. Assembly of the apoptosome complex and activation of procaspase 9 follow. This in turn results in activation of caspase 3, one of the most important apoptotic effectors. Caspase 3 cleaves numerous cellular proteins, among which is BAD itself. The carboxyl-terminal product of the BAD cleavage exhibits stronger mitochondrial localization and higher apoptotic capability than intact BAD, thus closing the feedback loop (5). The key finding relevant to the biology of BAD is that the US3 protein kinase abolishes caspase activity in all cells, including those that were transduced by wild-type or mutant BAD constructs and those that were not exposed to BAD. Therefore, US3 appears to affect caspase activation independently of BAD. The central question of whether US3 acts solely on downstream effectors of BAD in blocking apoptosis or whether it acts also on BAD was not unambiguously answered. The data currently available suggest that US3 may act solely on effector caspases. Thus, the BAD protein lacks the consensus site for direct phosphorylation of BAD by the US3 protein kinase (25, 26). The hypothesis that US3 induces the phosphorylation of one or more of the regulatory serines known to block activation of BAD has proven to be untenable inasmuch as a US3 protein kinase blocked apoptosis induced by the BAD3S/A mutant. The experimental design used in the present study precluded a more thorough analysis of the apparent change in the electrophoretic mobility of the BAD protein reported in the earlier study (22).
(ii) The evidence that BAD is cleaved by a caspase at a second site near the carboxyl terminus of the protein came as a surprise since such a cleavage has not been reported previously. If we assume that the cleavage is effected by the same enzyme that cleaves the amino-terminal sites, the predicted cleavage site would be at residue ASP156. This site is in the sequence of the BH3 domain. We should note that not all changes of the BH3 domain affect proapoptotic activity of BAD. Thus, Adachi et al. (1) reported that the substitution of the residue corresponding to ASP156 of the human homologue of BAD with GLY disrupted the interaction of BAD with Bcl-2, but the mutation did not affect the interaction of BAD with Bcl-XL or its targeting to the mitochondria. These authors also reported that the proapoptotic activity of the mutant was comparable to that of wild-type BAD. Nevertheless, since the gstBAD1-159 was inactive, we could expect that the cleavage would also inactivate BAD. Our results would therefore suggest that effector caspases both activate and inactivate the BAD protein.
(iii) In an earlier study this laboratory reported a decrease in the amounts of BAD protein accumulating in cells transduced by both BAD and US3 protein kinase (22). We also noted a decrease in the accumulation of BAD protein in cells doubly transduced in the present study. We should note, however, that the downregulation of total BAD in cells coexpressing the US3 protein kinase appears to be cell type dependent since in SK-N-SH cells the amounts of BAD protein in doubly transduced cells were larger than the amounts detected in cells transduced with wild-type BAD only (data not shown). The decrease in the amount accumulating in doubly infected cells is not due to proteasome-dependent degradation of BAD since the treatment of cells expressing gstBAD with the proteasome inhibitor MG132 did not result in an increase in the amount of total BAD (data not shown). We conclude that either BAD protein synthesis is induced de novo in transduced rabbit skin cells or that BAD is subject to degradation by a non-proteasome-dependent pathway. It is of interest that the US3 protein kinase is not an universal blocker of all proapoptotic injuries to the cell caused by HSV infection. For example, cells infected with HSV-1 mutant lacking the glycoprotein D gene cause apoptosis that is blocked by glycoprotein D, glycoprotein J, or by ectopic overexpression of the mannose 5-phosphate receptor but not by US3 (data not shown). The results presented here suggest that US3 targets caspases. The range and relative effectiveness of US3 in blocking the action of these enzymes remain to be determined.
Acknowledgments
These studies were carried out at the University of Chicago and were aided by grants from the National Cancer Institute (CA87661, CA83939, CA71933, CA78766, and CA88860) and the U.S. Public Health Service.
REFERENCES
- 1.Adachi, M., and K. Imai. 2002. The proapoptotic BH3-only protein BAD transduces cell death signals independently of its interaction with Bcl-2. Cell Death Differ. 9:1240-1247. [DOI] [PubMed] [Google Scholar]
- 2.Asano, S., T. Honda, F. Goshima, D. Watanabe, Y. Miyake, Y. Sugiura, and Y. Nishiyama. 1999. US3 protein kinase of herpes simplex virus type 2 plays a role in protecting cornel epithelial cells from apoptosis in infected mice. J. Gen. Virol. 80:51-56. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, M., J. O'Toole, and J. A. Blaho. 1999. Induction and prevention of apoptosis in human Hep-2 cells by herpes simplex virus type 1. J. Virol. 73:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chittenden, T., C. Flemington, A. B. Houghton, R. G. Ebb, G. J. Gallo, B. Elangovan, G. Chinnadurai, and R. J. Lutz. 1995. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 14:5589-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condorelli, F., P. Salomoni, S. Cotteret, V. Cesi, S. M. Srinivasula, E. S. Alnemri, and B. Calabretta. 2001. Caspase cleavage enhances the apoptosis-inducing effects of BAD. Mol. Cell. Biol. 21:3025-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Goth, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231-241. [DOI] [PubMed] [Google Scholar]
- 7.Datta, S. R., A. Katsov, L. Hu, A. Petros, S. W. Fesik, M. B. Yaffe, and M. E. Greenberg. 2000. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6:41-51. [PubMed] [Google Scholar]
- 8.del Peso, L., M. Gonzales-Garcia, C. Page, R. Herrera, and G. Nunez. 1997. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278:687-689. [DOI] [PubMed] [Google Scholar]
- 9.DeLuca, N. A., M. McCarth, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galvan, V., and B. Roizman. 1998. Herpes simplex virus type 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvan, V., R. Brandimarti, and B. Roizman. 1999. Herpes simplex virus 1 blocks caspase-3-independent and caspase-dependent pathways to cell death. J. Virol. 73:3219-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvan, V., R. Brandimarti, J. Munger, and B. Roizman. 2000. Bcl-2 blocks a caspase-dependent pathway of apoptosis activated by herpes simplex virus 1 infection in Hep-2 cells. J. Virol. 74:1931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, S. Y., A. Karpia, L. Zhu, and A. J. Hsueh. 1997. Interference of BAD (Bcl-xL/Bcl-2-associated death promoter)-induced apoptosis in mammalian cells by 14-3-3 isoforms and P11. Mol. Endocrinol. 11:1858-1867. [DOI] [PubMed] [Google Scholar]
- 15.Jerome, K. R., J. F. Tait, D. M. Koelle, and L. Corey. 1998. Herpes simplex virus type 1 renders infected cells resistant to cytotoxic-T-lymphocyte-induced apoptosis. J. Virol. 72:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerome, K. R., Z. Chen, R. Lang, M. R. Torres, J. Hofmeister, S. Smith, R. Fox, C. J. Froelich, and L. Corey. 2001. HSV and glycoprotein J inhibit caspase activation and apoptosis induced by granzyme B or Fas. J. Immunol. 167:3928-3935. [DOI] [PubMed] [Google Scholar]
- 17.Koyama, A. H., and Y. Miwa. 1997. Suppression of apoptotic DNA fragmentation in herpes implex virus type 1-infected cells. J. Virol. 71:2567-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leopardi, R., and B. Roizman. 1996. The herpes simplex major regulatory protein ICP4 blocks apoptosis induced by the virus or hyperthermia. Proc. Natl. Acad. Sci. USA 93:9583-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase Us3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindberg, A., and J. P. Krevi. 2002. Splicing inhibiton a the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology 294:189-198. [DOI] [PubMed] [Google Scholar]
- 21.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munger, J., and B. Roizman. 2001. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc. Natl. Acad. Sci. USA 98:10410-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oltvai, Z. N., C. L. Milliman, and S. J. Korsmeyer. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74:609-619. [DOI] [PubMed] [Google Scholar]
- 24.Pastorino, J. G., M. Tafani, and J. L. Farber. 1998. Tumor necrosis factor induces phosphorylation and translocation of BAD through a phosphatidylinositide-3-OH kinase-dependent pathway. J. Biol. Chem. 274:19411-19416. [DOI] [PubMed] [Google Scholar]
- 25.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purves, F. C., A. D. Deana, F. Marchiori, D. P. Leader, and L. A. Pinna. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889:208-215. [DOI] [PubMed] [Google Scholar]
- 27.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 28.Salomoni, P., F. Condorelli, S. M. Sweeney, and B. Calabretta. 2000. Versatility of BCR/ABL-expressing leukemic cells in circumventing proapoptotic BAD effects. Blood 96:676-684. [PubMed] [Google Scholar]
- 29.Scheid, M. P., and V. Duronio. 1998. Dissociation of cytokine-induced phosphorylation of Bad and activation of PKB/Akt: involvement of MEK upstream of Bad phosphorylation. Proc. Natl. Acad. Sci. USA 95:7439-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sieg, S., Z. Yildirim, D. Smith, N. Kayagaki, H. Yagita, Y. Huang, and D. Kaplan. 1996. Herpes simplex virus type 2 inhibition of Fas ligand expression. J. Virol. 70:8747-8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan, Y., M. R. Demeter, H. Ruan, and M. J. Comb. 2000. BAD ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J. Biol. Chem. 275:25865-25869. [DOI] [PubMed] [Google Scholar]
- 32.Tan, Y., H. Ruan, M. R. Demeter, and M. J. Comb. 1999. p90RSK blocks Bad-mediated cell death via a protein kinase C-dependent pathway. J. Biol. Chem. 274:34859-34867. [DOI] [PubMed] [Google Scholar]
- 33.Wang, H.-G., N. Pathan, I. M. Ethel, S. Krajewski, Y. Yamaguchi, F. Shibasaki, F. McKeon, T. Bobo, T. F. Franke, and J. C. Reed. 1999. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 284:339-343. [DOI] [PubMed] [Google Scholar]
- 34.Yin, X. M., Z. N. Oltvai, and S. J. Korsmeyer. 1994. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 369:272-273. [DOI] [PubMed] [Google Scholar]
- 35.Yu, K., Y., B. Kwon, J. Ni, Y. Zhai, R. Ebner, and B. S. Kwon. 1999. A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J. Biol. Chem. 274:13733-13736. [DOI] [PubMed] [Google Scholar]
- 36.Zachos, G., M. Koffa, C. M. Preston, J. B. Clements, and J. Conner. 2001. Herpes simplex virus type 1 blocks the apoptotic host cell defense mechanisms that target Bcl-2 and manipulates activation of p38 mitogen-activated protein kinase to improve viral replication. J. Virol. 75:2710-2728. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Zha, J., H. Harada, E. Yang, J. Jockel, and S. J. Korsmeyer. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not Bcl-XL. Cell 87:589-592. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, X. M., Y. Liu, G. Payne, R. J. Lutz, and T. Chittenden. 2000. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J. Biol. Chem. 275:25046-25051. [DOI] [PubMed] [Google Scholar]