Abstract
A comparative analysis of all published complete genomes indicated that the putative orthologs of the unannotated ychB gene of Escherichia coli follow the distribution of the dxs, dxr, and ygbP genes, which have been shown to specify enzymes of the deoxyxylulose phosphate pathway of terpenoid biosynthesis, thus suggesting that the hypothetical YchB protein also is involved in that pathway. To test this hypothesis, the E. coli ychB gene was expressed in a homologous host. The recombinant protein was purified to homogeneity and was shown to phosphorylate 4-diphosphocytidyl-2C-methyl-d-erythritol in an ATP-dependent reaction. The reaction product was identified as 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate by NMR experiments with various 13C-labeled substrate samples. A 14C-labeled specimen of this compound was converted efficiently into carotenoids by isolated chromoplasts of Capsicum annuum. The sequence of E. coli YchB protein is similar to that of the protein predicted by the tomato cDNA pTOM41 (30% identity), which had been implicated in the conversion of chloroplasts to chromoplasts.
The mevalonate pathway of terpenoid biosynthesis has been elucidated by the classical studies of Bloch, Lynen, Cornforth, and their coworkers by using yeast and animal cells (for review, see refs. 1–4). In the last few years, Rohmer, Arigoni, and their coworkers independently found that the incorporation of 13C-labeled glucose into terpenoids by certain eubacteria (5, 6) and the plant Ginkgo biloba (7) could not be explained by that pathway.
Arigoni and his coworkers (6) demonstrated the efficient conversion of 1-deoxy-d-xylulose into terpenoids in Escherichia coli, thus suggesting deoxyxylulose or its 5-phosphate as an intermediate of the alternative pathway. Subsequent work by several groups established that certain eubacteria and protozoa as well as many, if not all, plants can synthesize terpenoids by means of the 1-deoxy-d-xylulose phosphate pathway (for review, see refs. 8–10).
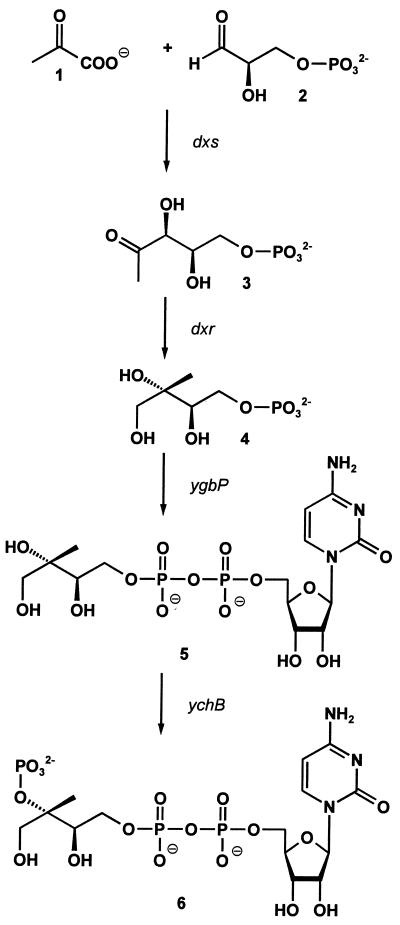
Enzymes catalyzing the formation of 1-deoxy-d-xylulose 5-phosphate (Fig. 1, 3) from glyceraldehyde 3-phosphate (Fig. 1, 2) and pyruvate (Fig. 1, 1) recently were cloned from E. coli, Capsicum annuum, and Mentha piperita (11–14). Subsequently, 1-deoxy-d-xylulose 5-phosphate was shown to be converted to the branched polyol, 2C-methyl-d-erythritol 4-phosphate (Fig. 1, 4), by a reductoisomerase specified by the dxr gene (15–18).
Figure 1.
The deoxyxylulose phosphate pathway of isoprenoid biosynthesis (for review, see refs. 8–10).
Recently, we have found that 2C-methyl-d-erythritol 4-phosphate (Fig. 1, 4) is converted into 4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) in a CTP-dependent reaction by an enzyme specified by the ygbP gene of E. coli (19) (Fig. 1). This paper shows that the 2 position hydroxy group of 4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) can be phosphorylated in an ATP-dependent reaction by the enzyme specified by an unannotated ychB gene of E. coli.
Experimental Procedures
Materials.
Oligonucleotides were custom-synthesized by MWG Biotec (Ebersberg, Germany). The preparation of 13C- and 14C-labeled samples of 2C-methyl-d-erythritol 4-phosphate and 4-diphosphocytidyl-2C-methyl-d-erythritol will be reported elsewhere.
Enzymes.
The preparation of recombinant YgbP protein of E. coli has been described (19).
Construction of an Expression Plasmid for ychB of E. coli.
The ychB gene of E. coli was amplified by PCR using the primers ychBvo and ychBhi with chromosomal E. coli DNA as template (Table 1). The amplificate was used as template in a second-round PCR using the primers kEcoRI and ychBhi. The resulting PCR amplificate was digested with EcoRI and PstI and was ligated into the plasmid pNCO113 (20), which had been treated with the same enzymes. This plasmid was transformed into E. coli XL1-Blue (21), where it directed the expression of the ychB gene under the control of a T5 promoter and a lac operator.
Table 1.
Oligonucleotides used in this study
| Designation | Sequence |
|---|---|
| ychBvo | 5′-GAGGAGAAATTAACCATGCGGACACAGTGGCCC-3′ |
| ychBhi | 5′-GTCACCGAACTGCAGCTTGCCCG-3′ |
| kEcoRI | 5′-ACACAGAATTCATTAAAGAGGAGAAATTAACCATG-3′ |
Preparation and Purification of Recombinant YchB Protein from E. coli.
Crude cell extract of E. coli XL1-pNCOychB cells was prepared as described (19) and applied to a column of Q Sepharose FF (2 × 10 cm; flow rate, 3 ml/min), which then was washed with 150 ml of 50 mM Tris⋅HCl (pH 8.0) containing 1 mM DTT and 0.02% sodium azide (buffer A) and developed with a linear gradient of 0–0.5 M NaCl in buffer A (total volume, 150 ml). Fractions were combined, and ammonium sulfate was added to a final concentration of 0.5 M. The solution was applied to a column of Phenyl Sepharose 6 FF (1.6 × 10 mm; flow rate 3 ml/min), which had been equilibrated with buffer A containing 0.5 M ammonium sulfate. The column was developed with a linear gradient of 0.5–0 M ammonium sulfate in buffer A (total volume, 100 ml). Fractions were combined and concentrated to 3 ml by ultrafiltration. The solution was applied to a column of Superdex 75 HR 26/60 (flow rate, 3 ml/min), which was developed with buffer A containing 100 mM sodium chloride.
Assay of YchB Protein Activity.
Assay mixtures containing 100 mM Tris⋅HCl (pH 8.0), 5 mM MgCl2, 100 μM ATP, 5 mM DTT, 11.4 μM [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) (17.5 μCi/μmol), and 2 μg of protein in a total volume of 50 μl were incubated at 37°C for 30 min. Aliquots (40 μl) were spotted on TLC plates (Polygram SIL N-HR; Macherey & Nagel), which were developed by N-propanol/ethyl acetate/water (6:1:3) for 5 h. Radioactivity was monitored with a PhosphorImager (Storm 860; Molecular Dynamics). The Rf value of [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate (Fig. 1, 6) was 0.20.
Preparation of 4-Diphosphocytidyl-2C-Methyl-d-Erythritol 2-Phosphate (6).
A solution containing 100 mM Tris⋅HCl (pH 8.0), 5 mM MgCl2, 5 mM ATP, 5 mM DTT, 5 mM [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) (117 μCi/mmol), and 100 μg of recombinant YchB protein of E. coli in a total volume of 4 ml was incubated at 37°C for 2 h. The reaction was monitored by 13C NMR. The reaction was terminated by ultrafiltration by using a Centriprep-10 device (Amicon; 5,000 rpm for 60 min). The product was purified by HPLC with a column of Nucleosil 10SB (4.6 × 250 mm; Macherey & Nagel), which was washed with 30 ml of 0.1 M ammonium formate in 40% methanol (vol/vol) and developed with a linear gradient of 0.1–1.0 M ammonium formate in 40% methanol (vol/vol).
NMR Spectroscopy.
1H NMR and 1H-decoupled 13C NMR spectra were recorded by using a AVANCE DRX 500 spectrometer from Bruker (Karlsruhe, Germany). Chemical shifts were referenced to external trimethylsilylpropane sulfonate. 31P NMR spectra were recorded by using an AC 250 spectrometer from Bruker. Chemical shifts were referenced to external 85% H3PO4.
Preparation of Chromoplasts and Incorporation Assays.
Chromoplasts of C. annuum were isolated according to published procedures (22) by using 50 mM Hepes (pH 7.6) and 1 mM DTT as suspension buffer. A solution (0.5 ml) containing 100 mM Hepes (pH 7.6), 2 mM MnCl2, 10 mM MgCl2, 2 mM NADP, 20 μM FAD, 5 mM NaF, 6 mM ATP, 1 mM NADPH, 11.4 μM [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate (Fig. 1, 6) (17.5 μCi/μmol), and chromoplasts (equivalent to 2 mg of protein) was incubated at 30°C for 15 h. The mixture was extracted twice with 1 ml of ethylacetate. The organic phase was analyzed by scintillation counting, and aliquots were spotted on TLC plates (Polygram SIL G; Macherey & Nagel), which were developed by hexane/toluene (9:1, vol/vol). Radioactivity was monitored with a PhosphorImager (Fujiifilm BAS-1500; Raytest, Straubenhardt) (23). The Rf values of β-carotene, phytoene, phytofluene, and ξ-carotene were 0.6, 0.6, 0.55, and 0.5, respectively.
Results
Genomes of organisms using the deoxyxylulose pathway for terpenoid biosynthesis must, by necessity, contain genes specifying all enzyme activities of that incompletely known pathway. If it is tentatively assumed that the deoxyxylulose phosphate pathway evolved only once, one would expect to find specific sets of orthologous genes in all complete genomes of organisms using the deoxyxylulose phosphate pathway, but not in organisms using only the mevalonate pathway. In line with this hypothesis, putative orthologs of the dxs, dxr, and ygbP genes specifying 1-deoxy-d-xylulose 5-phosphate synthase, 1-deoxy-d-xylulose 5-phosphate reductoisomerase, and 4-diphosphocytidyl-2C-methyl-d-erythritol synthase are found in a group of eubacteria using the deoxyxylulose phosphate pathway. On the other hand, putative orthologs of these genes appear to be absent in the genomes of archaea, Saccharomyces cerevisiae and Caenorhabditis elegans, which all are known to use the mevalonate pathway for isoprenoid biosynthesis (19) (Table 2).
Table 2.
Occurrence of putative deoxyxylulose phosphate pathway genes in completely sequenced genomes
| Organism | Accession
number*
|
||||
|---|---|---|---|---|---|
| dxs | dxr | ygbP | ygbB | ychB | |
| Bacteria | |||||
| Escherichia coli K-12 MG1655 | gb AF035440 | dbj AB013300 | gb AE000358 | gb AE000358 | gb AE000219 |
| Haemophilus influenzae Rd KW20 | gb U32822 | gb U32763 | gb U32750 | gb U32750 | gb U32834 |
| Aquifex aeolicus VF5 | gb AE000712 | gb AE000688 | gb AE000734 | gb AE000715 | gb AE000713 |
| Synechocystis sp. PCC6803 | dbj D90903 | dbj D64000 | dbj D90914 | dbj D90906 | dbj D90899 |
| Bacillus subtilis 168T | dbj D84432 | emb Z99112 | emb Z99101 | emb Z99101 | emb Z99104 |
| Thermotoga maritima | gb AE001815.1 | gb AE001754.1 | gb AE001792.1 | gb AE001738.1 | gb AE001791.1 |
| Mycobacterium tuberculosis H37Rv | emb Z96072 | emb Z74024 | emb Z92774 | emb Z92774 | emb Z94752 |
| Treponema pallidum | gb AE001253 | gb AE001235 | gb AE001227 | gb AE001227 | gb AE001226 |
| Helicobacter pylori J99 | gb AE001468 | gb AE000541.1 | gb AE001474 | gb AE001474 | gb AE000644 |
| Chlamydia pneumoniae CWL029 | gb AE001686 | gb AE001618 | gb AE001642 | gb AE001639 | gb AE001675 |
| Chlamydia trachomatis D/UW-3/CX | gb AE001306 | gb AE001281 | gb AE001320 | gb AE001317 | gb AE001352 |
| Mycoplasma genitalium | — | — | — | — | — |
| Rickettsia prowazekii | — | — | — | — | — |
| Borrelia burgdorferi | — | — | — | — | — |
| Archaea | |||||
| Pyrococcus horikoshii OT3 | — | — | dbj AE000002 | — | — |
| Aeropyrum pernix K1 | — | — | — | — | — |
| Archeoglobus fulgidus | — | — | — | — | — |
| Methanobacterium thermoautotrophicum | — | — | — | — | — |
| Methanococcus jannaschii | — | — | — | — | — |
| Eukaryotes | |||||
| Caenorhabditis elegans | — | — | — | — | — |
| Saccharomyces cerevisiae | — | — | — | — | — |
gb, GenBank; dbj, Databank of Japan; emb, EMBL.
A systematic computer search for an additional gene following the distribution of dxs, dxr, ygbP, and ygbB in the complete genomes available in public domain databases retrieved the unannotated ychB gene of E. coli. This gene and its putative orthologs have the same distribution as the putative ortholog sets of dxr, dxs, and ygbP (Table 2).
The E. coli ychB gene predicts a 31-kDa peptide of 283 amino acid residues. An alignment of putative proteins with similarity to YchB from a variety of eubacteria and plants is shown in Fig. 2.
Figure 2.
Deduced amino acid sequences of ychB genes. Residues identical in more than 50% of the sequences are shown in inverse contrast. A, Chlamydia trachomatis; B, Chlamydia pneumoniae; C, Arabidopsis thaliana; D, Lysopersicon esculatum; E, Escherichia coli; F, Salmonella typhimurium; G, Haemophilus influenzae; H, Pseudomonas aeruginosa; I, Zymomonas mobilis; J, Synechocystis sp. PC6803; K, Mycobacterium tuberculosis; L, Bacillus subtilis; M, Thermotoga maritima; N, Treponema pallidum; O, Aquifex aeolicus; P, Helicobacter pylori. -, Gap; >, fragment; \\, C-terminal end; *, putative ATP-binding site.
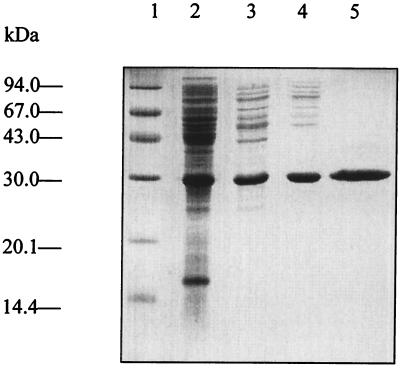
The ychB gene of E. coli was expressed in a homologous host. A recombinant E. coli strain carrying a plasmid with the ychB gene under control of a T5 promoter and lac operator was found to produce a soluble polypeptide with an apparent mass of 31 kDa (about 5% of cytoplasmic protein), which was purified to apparent homogeneity by a series of three chromatographic steps as described in Experimental Procedures (Fig. 3).
Figure 3.
SDS/PAGE. Lane 1, molecular mass markers; lane 2, crude cell extract of E. coli XL1-pNCOychB; lane 3, YchB protein after Sepharose Q FF chromatography; lane 4, YchB protein after Phenyl Sepharose chromatography; lane 5, YchB protein after Superdex chromatography.
Reaction mixtures containing recombinant YchB protein, [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) and ATP afforded a product that could be separated from the substrate by TLC (Rf = 0.2). Product formation was shown to depend on the presence of ATP as second substrate. These data suggested tentatively that YchB protein catalyzes an ATP-dependent phosphorylation of 4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5). The enzyme was specific with regard to the diphosphocytidyl compound as substrate.
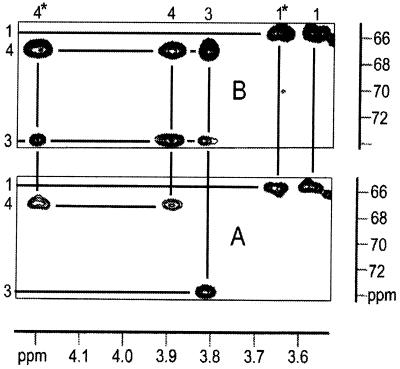
NMR experiments then were performed to determine the structure of the enzyme product. To enhance the sensitivity and selectivity of these experiments, we used [1,2,2-methyl,3,4-13C5]-, [1,3,4-13C1]-, respectively [2,2-methyl-13C2]4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) as substrates. 1H, 13C, and 31P NMR spectra were obtained with the crude incubation mixtures or after HPLC purification of the respective enzyme products. The 1H1H, 1H13C, and 13C13C spin networks were gleaned by two-dimensional DQF-COSY (double quantum filtered–correlated spectroscopy), HMQC (heteronuclear multiple quantum correlation), HMQC-TOCSY (HMQC–total correlation spectroscopy), HMBC (heteronuclear multiple quantum multiple bond correlation), and INADEQUATE experiments (Table 3) establishing 2C-methylerythritol and cytidine as structural motifs. As an example, Fig. 4 shows HMQC and HMQC-TOCSY spectra of the crude incubation mixture using [1,2,2-methyl,3,4-13C5] 4-diphosphocytidyl-2C-methyl-d-erythritol as substrate. The HMQC spectrum (Fig. 4A) displays 13C1H correlations of C-1, C-3, and C-4 with their directly attached hydrogen atoms. In the HMQC-TOCSY spectrum (Fig. 4B), extended 1H spin systems connected by 1H TOCSY transfer are correlated to the respective 13C atoms. Thus, C-3 shows correlations to the directly attached H-3 as well as to the coupled diastereotopic hydrogen atoms at C-4.
Table 3.
NMR data of 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate
| Position | Chemical
shifts, ppm
|
Coupling constants,
Hz
|
Correlation
pattern
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1H† | 13C‡ | 31P§ | JHH | JPC | JPP | JCC | DOF-COSY | HMBC¶ | INADEQUATE†† | |
| 1 | 3.58(m,1) | 65.78 | 3.8∥(P-2) | 39.8††(2) | 1* | 2 | ||||
| 1* | 3.64(m,1) | 1 | ||||||||
| 2 | 81.91 | 7.4¶(P-2) | 38.9¶(2-me) | 2-methyl,1,3 | 1,2-methyl,3 | |||||
| 2-methyl | 1.26(s,3) | 17.92 | 1.9¶(P-2) | 38.9¶(2) | 1,3 | 2 | ||||
| 3 | 3.81(m,1) | 73.96 | 7.3∥(P-2,P-4) | 4,4* | 2,4 | |||||
| 4 | 3.89(m,1) | 67.16 | 5.7|(P-4) | 42.9††(3) | 3,4* | 3 | ||||
| 4* | 4.19(m,1) | 3,4 | ||||||||
| 1′ | 5.87(d,1) | 90.21 | 4.4 | 2′ | Cyt-6 | |||||
| 2′ | 4.21(t,1) | 75.24 | 4.9 | 1′,3′ | ||||||
| 3′ | 4.25(t,1) | 70.22 | 4.9 | 2′,4′ | ||||||
| 4′ | 4.17(m,1) | 83.85 | 3′,5′ | |||||||
| 5′ | 4.09(ddd,1) | 65.49 | 12.2,5.4 | 4′,5′* | ||||||
| 5′* | 4.17(m,1) | 5′ | ||||||||
| Cyt-2 | 156.50 | Cyt-6 | ||||||||
| Cyt-4 | 165.49 | Cyt-5,Cyt-6 | ||||||||
| Cyt-5 | 6.07(d,1) | 97.35 | 7.6 | Cyt-6 | Cyt-6 | |||||
| Cyt-6 | 7.92(d,1) | 143.03 | 7.6 | Cyt-5 | Cyt-5 | |||||
| P(2) | 0.49 | 1.7¶(2-me),7.6¶(2) | ||||||||
| P(4) | −7.28 | 20.8 | ||||||||
| P(5′) | −8.00 | 20.8 | ||||||||
Indicates downfield shifted 1H NMR signals of diastereotopic hydrogen pairs.
† Referenced to external trimethylsilylpropane sulfonate. The multiplicities and the relative integral values of 1H NMR signals of an unlabeled sample are indicated in parentheses.
‡ Referenced to external trimethylsilylpropane sulfonate.
§ Referenced to external phosphoric acid (85%, vol/vol).
¶ From the spectrum of [2,2-methyl-13C2]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate.
∥ From the spectrum of [1,3,4-13C1]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate.
†† From the spectrum of [1,2,2-methyl,3,4-13C5]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate.
Figure 4.
1H13C correlation NMR spectra of [1,2,2-methyl,3,4-13C5]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate. (A) Part of HMQC spectrum. (B) Part of HMQC-TOCSY spectrum. Mixing time for 1H1H-TOCSY transfer, 60 ms.
The 1H-decoupled 31P NMR spectrum of the enzyme product obtained from [2,2-methyl-13C2]4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5) displayed two doublets at −7.28 ppm respectively −8.00 ppm reflecting the presence of a pyrophosphate motif (31P31P coupling constant, 20.8 Hz) and a double-doublet at 0.49 ppm reflecting a phosphomonoester motif (31P13C coupling constants, 7.6 Hz and 1.7 Hz). Without 1H-decoupling, the 31P NMR signals at −7.28 and −8.00 ppm were broadened, whereas the multiplicity and the linewidth of the signal at 0.49 ppm were not affected.
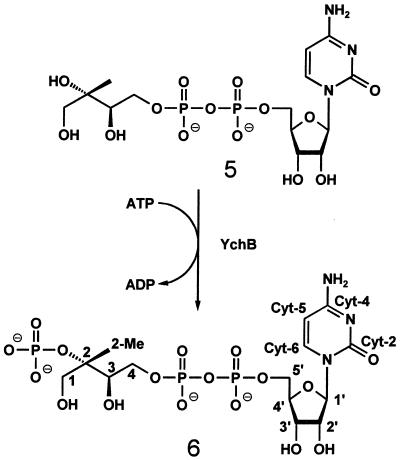
The absence of scalar 1H31P coupling in case of the orthophosphate moiety is in line with the product structure shown in Fig. 5. Moreover, the location of the orthophosphate group at C-2 is confirmed by a significant downfield shift of the 13C NMR chemical shift of C-2 (81.9 ppm) with respect to the chemical shift of substrate C-2 (73.8 ppm) (19) as well as by the detected 31P13C coupling pattern (Table 3). In the case of a hypothetical 1- or 3-phosphate, no 31P13C coupling to the methyl carbon atom is expected (4JPC). The 31P13C coupling pattern detected with the sample obtained from [1,3,4-13C1]4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 5, 5) (Table 3) further confirmed the structure of the product as 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate (Fig. 5, 6).
Figure 5.
Enzyme reaction catalyzed by YchB protein of E. coli.
Isolated chromoplasts of C. annum were shown to incorporate [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate into carotenoids. Specifically, a mixture of phytoene and β-carotene obtained from the incubation mixtures by solvent extraction and TLC was shown to contain 9.4% of the proffered radioactivity; 0.3% respectively 0.5% of proffered radioactivity were diverted to phytofluene and ξ-carotene. The diversion of radioactivity from [2-14C]4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate to the terpenoid fraction was not diminished by the addition of unlabeled 2C-methyl-d-erythritol 4-phosphate (Fig. 1, 4) to reaction mixtures (data not shown).
Discussion
We have shown previously that YgbP protein of E. coli catalyzes the formation of 4-diphosphocytidyl-2C-methyl-d-erythritol (Fig. 1, 5). The present data show that this compound can be phosphorylated at the 2 position by the YchB gene product of E. coli. The resulting 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate (Fig. 1, 6) can be incorporated into carotenoids by isolated chromoplasts. These data leave no doubt that 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate (Fig. 1, 6) serves as an intermediate in the deoxyxylulose phosphate pathway of terpenoids.
The translation product of a tomato cDNA designated pTOM41 (Table 4) (24) initially was identified as a chromoplast-targeted protein. The protein sequence of pTOM41 is similar (30% identical residues) to the YchB protein of E. coli and contains a putative plastid leader sequence. The transcript of the cognate cDNA is up-regulated during the chloroplast/chromoplast transition. The translation product was incorporated into the soluble fraction of the tomato chromoplasts (24). These observations are all in line with the metabolic role of ychB orthologs as established by the present work.
Table 4.
Orthologs of ychB in incompletely sequenced genomes
| Organism | Accession or Contig number* |
|---|---|
| Bacteria | |
| Actinobacillus actinomycetemcomitans | Contig5I0 |
| Bordelella bronchiseptica | Contig2244 |
| Bordetella pertussis | Contig408 |
| Campylobacter jejuni | Cj.seq |
| Caulobacter crescentus | gcc_I346 |
| Chlorobium tepidum | gct_5 |
| Clostridium acetobutylicum | AE00I437 |
| Clostridium difficile | ContigI239 |
| Deinococcus radiodurans | 8896 |
| Enterococcus faecalis | gef_6342 |
| Klebsiella pneumoniae | Contig3I |
| Mycobacterium avium | M.avium_24 |
| Mycobacterium bovis | Contig750 |
| Mycobacterium leprae | ContigI080 |
| Neisseria gonorrhoeae | ContigI2I |
| Neisseria meningitidis serogroup A | NM.seq |
| Pasteurella multocida | Contig264 |
| Porphyromonas gingivalis | II94 |
| Pseudomonas aeruginosa | Contig54 |
| Salmonella typhimurium LT2 | gb M77236 |
| Salmonella typhi | Contig334 |
| Salmonella paratyphi | SPA.0.2446 |
| Shewanella putrefaciens | 4279 |
| Sinrhizobium meliloti | 423II4AI2.xI |
| Staphyloccoccus aureus | Contig856 |
| Streptococcus mutans | Contig435 |
| Streptococcus pyogenes | Contig7 |
| Thiobacillus ferrooxidans | 203I |
| Vibrio cholerae | 666_I752 |
| Yersinia pestis | Contig648 |
| Zymomonas mobilis | gb AF088896.I |
| Plants | |
| Arabidopsis thaliana | gb AC005I68 |
| Solanum lycopersicum | gb U62773 |
Contig numbers as of October 19, 1999.
The alignment of deduced amino acid sequences of putative ychB orthologs shows a glycine-rich conserved motif extending from amino acid residue position 90–107 (E. coli) (Fig. 2). Post et al. (25) tentatively assigned this region as an ATP-binding site. The amino acid sequence of the YchB protein shows some similarity to other kinases, such as homoserine or mevalonate kinases. In Deinococcus radiodurans (Table 4), a putative ychB ortholog is tightly downstream of the putative ygbP ortholog and the ubiA gene specifying 4-hydroxybenzoate octaprenyl transferase. All three ORFs are likely to be transcriptionally coupled. Our study illustrates the power of comparative genome analysis for elucidation of metabolic pathway.
Acknowledgments
We thank A. Werner and F. Wendling for help with the preparation of the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 369), the Fonds der Chemischen Industrie, and the Hans-Fischer-Gesellschaft.
Abbreviations
- COSY
correlated spectroscopy
- HMQC
heteronuclear multiple quantum correlation
- TOCSY
total correlation spectroscopy
References
- 1.Qureshi N, Porter J W. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 47–94. [Google Scholar]
- 2.Bloch K. Steroids. 1992;57:378–382. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]
- 3.Bach T J. Lipids. 1995;30:191–202. doi: 10.1007/BF02537822. [DOI] [PubMed] [Google Scholar]
- 4.Bochar D A, Friesen J A, Stauffacher C V, Rodwell V W. In: Comprehensive Natural Products Chemistry. Barton D, Nakanishi K, editors. Vol. 2. Oxford: Pergamon; 1999. pp. 15–44. [Google Scholar]
- 5.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broers S T J. Thesis No. 10978. Zürich: Eidgenössische Technische Hochschule Zürich; 1994. [Google Scholar]
- 7.Schwarz M K. Thesis No. 10951. Zürich: Eidgenössische Technische Hochschule Zürich; 1994. [Google Scholar]
- 8.Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk M, Bacher A. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 9.Rohmer M. Prog Drug Res. 1998;50:135–154. doi: 10.1007/978-3-0348-8833-2_3. [DOI] [PubMed] [Google Scholar]
- 10.Rohmer M. In: Comprehensive Natural Products Chemistry. Barton D, Nakanishi K, editors. Vol. 2. Oxford: Pergamon; 1999. pp. 45–68. [Google Scholar]
- 11.Sprenger G A, Schörken U, Wiegert T, Grolle S, deGraaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lois L M, Campos N, Putra S R, Danielsen K, Rohmer M, Boronat A. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange B M, Wieding M R, McCaskill D, Croteau R. Proc Natl Acad Sci USA. 1998;95:2100–2104. doi: 10.1073/pnas.95.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouvier F, d'Harlingue A, Suire C, Backhaus R A, Camara B. Plant Physiol. 1998;117:1423–1431. doi: 10.1104/pp.117.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi S, Kuzuyama T, Watanabe H, Seto H. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange B M, Croteau R. Arch Biochem Biophys. 1999;365:170–174. doi: 10.1006/abbi.1999.1168. [DOI] [PubMed] [Google Scholar]
- 17.Schwender J, Müller C, Zeidler J, Lichtenthaler H K. FEBS Lett. 1999;455:140–144. doi: 10.1016/s0014-5793(99)00849-2. [DOI] [PubMed] [Google Scholar]
- 18.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Türbachova I, Eberl M, Zeidler J, Lichtenthaler H K, et al. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 19.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk M H. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stüber D, Matile H, Garotta G. In: Immunological Methods. Lefkovits F, Pernis P, editors. IV. San Diego: Academic; 1990. pp. 121–152. [Google Scholar]
- 21.Bullock W O, Fernandez J M, Short J M. BioTechniques. 1987;5:376–379. [Google Scholar]
- 22.Camara B. Methods Enzymol. 1993;214:352–265. [Google Scholar]
- 23.Fellermeier M, Kis K, Sagner S, Maier U, Bacher A, Zenk M H. Tetrahedron Lett. 1999. , 2743–2746. [Google Scholar]
- 24.Lawrence S D, Cline K, Moore G A. Plant Mol Biol. 1997;33:483–492. doi: 10.1023/a:1005785321165. [DOI] [PubMed] [Google Scholar]
- 25.Post D A, Hove-Jensen B, Switzer R L. J Gen Microbiol. 1993;193:259–266. doi: 10.1099/00221287-139-2-259. [DOI] [PubMed] [Google Scholar]