Abstract
Various studies have demonstrated a role for E2F proteins in the control of transcription of genes involved in DNA replication, cell cycle progression, and cell fate determination. Although it is clear that the functions of the E2F proteins overlap, there is also evidence for specific roles for individual E2F proteins in the control of apoptosis and cell proliferation. Investigating protein interactions that might provide a mechanistic basis for the specificity of E2F function, we identified the E-box binding factor TFE3 as an E2F3-specific partner. We also show that this interaction is dependent on the marked box domain of E2F3. We provide evidence for a role for TFE3 in the synergistic activation of the p68 subunit gene of DNA polymerase α together with E2F3, again dependent on the E2F3 marked box domain. Chromatin immunoprecipitation assays showed that TFE3 and E2F3 were bound to the p68 promoter in vivo and that the interaction of either E2F3 or TFE3 with the promoter was facilitated by the presence of both proteins. In contrast, neither E2F1 nor E2F2 interacted with the p68 promoter under these conditions. We propose that the physical interaction of TFE3 and E2F3 facilitates transcriptional activation of the p68 gene and provides strong evidence for the specificity of E2F function.
The ability of the retinoblastoma (Rb) tumor suppressor protein to regulate cell growth is due, at least in part, to its ability to interact with and regulate the E2F family of transcription factors (8, 34). The E2F proteins have been shown to control the expression of a large number of genes involved in DNA replication, cell cycle progression, and cell fate determination. The E2F family is composed of six distinct gene products that form heterodimeric complexes with partners of the DP family. Sequence analysis reveals three distinct subfamilies of E2F genes: the E2F1, E2F2, and E2F3 genes, the E2F4 and E2F5 genes, and the E2F6 gene. This division also coincides with functional distinctions. The E2F1, E2F2, and E2F3 genes are tightly regulated by cell growth and during the cell cycle, whereas E2F4, E2F5 and E2F6 are constitutively expressed.
This cell cycle regulation of E2F1, E2F2, and E2F3 transcription is complemented by mechanisms that tightly regulate the accumulation of the proteins. An N-terminal domain unique to E2F1 to E2F3 is responsible for both ubiquitin-mediated degradation of the proteins (29) and targeting by the cyclin A/cdk2 kinase, the latter leading to inhibition of DNA binding capacity (7, 22, 23, 53). The E2F proteins also vary in their role as transcriptional regulatory activities. While E2F1 to E2F3 act as positive regulators of transcription, E2F4 and E2F5 appear to function primarily as transcriptional repressors in concert with Rb family members. E2F6 also appears to function as a transcriptional repressor but in a manner independent of Rb (4, 10, 47, 48).
Various experiments have suggested distinct functional roles for the activating E2F proteins E2F1, E2F2, and E2F3. The E2F3 protein appears to be particularly important for cell proliferation, as seen from the inhibition of E2F3 activity by antibody microinjection (25) as well as the results of deletion of the E2F3 gene (18). Moreover, the expression of a number of E2F target genes that encode key cell cycle regulatory proteins, including B-Myb, cyclin A, cdc2, cdc6, and dihydrofolate reductase, are reduced in E2F3 null fibroblasts but not in E2F1 null cells (18). In contrast, E2F1 appears to play a role in triggering an apoptotic response, either when overexpressed in the absence of survival signals (6, 21, 39, 43, 51) or in response to DNA damage (27). In addition, the ability of Myc to induce apoptosis is impaired in the absence of E2F1 function but unaltered by the absence of either E2F2 or E2F3 (26).
Given the role of the E2F1, E2F2, and E2F3 proteins as transcriptional activators, the specificity of function might best be explained by an ability of these E2F proteins to activate distinct target genes that then carry out these functions of apoptosis or proliferation. As an example, the p19ARF gene has been shown to be an E2F1 target gene (2, 6), linking the action of the Rb/E2F pathway with the p53 response leading to apoptosis. Similarly, the Apaf1 gene appears to be activated specifically by E2F1 (30). Although one potential mechanism for such specificity could be an ability of these proteins to recognize subtle differences in cis-acting promoter sequences, there is little evidence for distinct DNA sequence recognition among the E2F isoforms. More importantly, analysis of the structure of an E2F-DNA complex did not show a capacity for distinct DNA sequence recognition when the amino acid variation within the E2F family is considered (56).
An alternative mechanism for promoter specificity could involve distinct protein-protein interactions. Possibly, sequences within E2F3 allow interaction with a subset of cellular proteins that provide a basis for promoter specificity and that are distinct from the proteins that can interact with E2F1. For example, the interaction of the herpesvirus VP16 transcriptional activator protein with the cellular factor HCF-1 and the cellular Oct-1 transcription factor redirects Oct-1 to herpesvirus immediate-early promoters by virtue of expanded DNA sequence recognition (1). A further example of combinatorial specificity as a mechanism for the specificity of transcription factor function is the pancreatic islet factor STF-1, which interacts with Pbx in a cooperative fashion and targets Pbx to a subset of promoters containing STF-1 binding sites (38). Recently, we identified the YY1-binding protein RYBP as a factor which binds to E2F2 and E2F3 but not E2F1 and recruits these E2Fs to a subset of E2F target promoters containing YY1 binding sites (41).
To further explore the mechanistic basis for the specificity of E2F transcription activation, we used yeast two-hybrid screens to identify proteins that specifically interact with E2F3. In so doing, we identified the TFE3 transcription factor as a specific partner for E2F3. Recent experiments identified TFE3 as an activity that can rescue Rb-mediated growth arrest, providing a functional link between TFE3 and the Rb/E2F pathway (M. Nijman, S. Hijmans, and R. Bernards, personal communication). Moreover, previous work identified TFE3 as a fusion partner in chromosomal rearrangements in renal cell carcinomas (14, 45, 50). We further show that TFE3 and E2F3 can act synergistically to activate the p68 subunit gene of DNA polymerase α, dependent on the ability of the two proteins to physically interact, and that the two activities associate with the p68 promoter within intact cells in a mutually dependent manner.
MATERIALS AND METHODS
Plasmids and reagents.
pGAD424-TFE3 harbors a human full-length cDNA of TFE3 in the pGAD424 yeast expression vector from Clontech. p2U-DP1 was made by subcloning the full-length cDNA of mouse DP1 into the p2U yeast expression plasmid (44). The Gal4 DNA-binding domain (DBD) fusion constructs contain human E2F cDNA clones except for E2F3b, which was murine. The fusions were constructed as follows. cDNA clones encoding E2F1 (amino acids 1 to 370), E2F2 (amino acids 1 to 370), E2F3a (amino acids 1 to 398), E2F3b (amino acids 1 to 333), E2F3aΔC (amino acids 1 to 295), E2F3aΔN (amino acids 357 to 465), and E2F3aΔMB (lacking amino acids 295 to 357) proteins were inserted into pGBT9 with standard molecular biology techniques. The human placenta cDNA library was purchased from Clontech. A HindIII/BamHI fragment of the hemagglutinin (HA) tag was subcloned into pCDNA3 (Stratagene) to generate pCDNA3-HA. pCDNA3-HA-E2F1, pRC-HA-E2F2, pRC-HA-E2F3a, and pCDNA3-HA-E2F3b were made by inserting the mouse cDNAs for E2F1, E2F2, E2F3a, E2F3b, E2F3aΔMB, and E2F3aΔC into pCDNA3-HA with standard molecular biology techniques.
The E2F1 to E2F3 chimeric proteins were generated as follows. The cDNAs of pcDNA3-HAE2F1 and pRC-HA-E2F3a were mutagenized to introduce restriction sites that allow cloning of individual domain between the genes. pGEX6P1-E2F1, pGEX6P1-E2F2, pGEX6P1-E2F3a, pGEX6P1-E2F3b, and pGEX6P1-DP1 were made by inserting mouse cDNAs of E2F1, E2F2, E2F3a, E2F3b, DP1, E2F3aΔMB, E2F3aΔC, and E2F3aΔN, respectively, into pGEX6P1 (Amersham). The luciferase reporter plasmids pGV-B (Toxyo Ink.), pKL12(−164), pKL12E2FAB, and pKL12 M3-Pal1 were kindly provided by Masako Izumi (Riken, Japan) (35). The pKL12(−164) luciferase reporter plasmid contains the 5′-flanking region of the mouse p68 gene (from −164 to +97) inserted into pGV-B. The mutated p68 reporter constructs pKL12E2FAB and pKL12 M3-Pal1 were derived from pKL12(−164). pCS-2MT-TFE3 was a gift from Harvey Lodish (Massachusetts Institute of Technology). All PCR fragments were verified by sequencing according to the Sequenase kit (United States Biochemicals) instructions and by Western blot analysis.
Yeast two-hybrid screen.
The yeast two-hybrid screen was performed as recommended in the Clontech protocol. Briefly, amino acids 1 to 397 of human E2F3a were cloned into the pGBT9 vector, and the resulting plasmid, along with DP1, was transformed into Saccharomyces cerevisiae strain pJ69a along with a human placenta library (Clontech), and the transformants were plated on selective medium plates supplemented with 5 mM 3-aminotriazole. The prey plasmid was rescued and transformed into the bacterial strain MH1066. The inserts in the recovered prey plasmids were sequenced according to the Sequenase kit (United States Biochemicals) instructions, and the interactions were reconfirmed with liquid β-galactosidase assays. Expression of the Gal4DBD-E2F fusion proteins was verified with antibodies specific to the Gal4 DBD (Clontech; 5399 to 1) and the appropriate E2Fs (Santa Cruz; sc-251, sc-633, sc-879, and sc-866). The yeast two-hybrid liquid β-galactosidase assay was performed as recommended in the Clontech protocol.
In vitro protein binding assays.
[35S]methionine-labeled TFE3 was synthesized with a coupled in vitro transcription and translation system as specified by the manufacturer (Promega). Equal volumes (25 μl of in vitro-transcribed and translated rabbit reticulocyte lysate) of [35S]TFE3 or [35S]DP1 were incubated for 24 h at 4°C with either ≈2 μg of glutathione S-transferase (GST)-Sepharose, GST-E2F1-Sepharose, GST-E2F2-Sepharose, GST-E2F3a-Sepharose, GST-E2F3b-Sepharose, GST-E2F3ΔMB-Sepharose, GST-E2F3ΔC-Sepharose, GST-E2F3ΔN-Sepharose, or GST-DP1-Sepharose in NENT-A buffer (50 mM NaCl, 20 M Tris [pH 8], 1 mM EDTA, 0.5% NP-40). Following incubation, the beads were washed in NENT-B buffer (80 mM NaCl, 20 M Tris [pH 8], 1 mM EDTA, 0.5% NP-40). Bound proteins were eluted in sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant GST fusion proteins used for the in vitro pulldown experiments were prepared as described previously (11).
Immunoprecipitations.
NIH 3T3 cells were transfected with Myc-tagged TFE3 alone or together with HA-tagged E2Fs and HA-tagged E2F mutants and chimeras. Transfected cells were lysed in lysis buffer (0.5% NP-40, 50 mM Tris [pH 8], 25% glycerol, 0.1 mM EDTA, 150 mM NaCl, and protease inhibitors [1 μg of leupeptin, 1 μg of aprotinin, and 1 μg of pepstatin per ml and 1 mM phenylmethylsulfonyl fluoride or Pefabloc (Boehringer Mannheim)]), the lysates were cleared two times with 20 μl of protein A- plus protein G-Sepharose beads (Oncogene) at 4°C for 4 h. The beads were discarded, and the lysates were incubated at 4°C for 2 h with 15 μl of anti-HA antibody immobilized on Sepharose beads (Covance). The bound proteins were resolved by SDS-PAGE. Western blots were done as described below with the indicated antibodies.
Immunoprecipitation of endogenous TFE3 and E2Fs was carried out as follows. NIH 3T3 cells were lysed in immunoprecipitation lysis buffer, and lysates were cleared as mentioned above. The cleared lysates were incubated with antibodies specific to E2F1, E2F3, or TFE3 (E2F1, sc-251; E2F3, sc-879; and TFE3, Pharmingen 15451A) immobilized onto protein A- plus protein G-agarose beads. Endogenous proteins were detected by Western blotting with specific antibodies (Santa Cruz: E2F1, sc-193; E2F3, sc-879).
Cell culture and transient transfections.
Mouse fibroblast NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Transient transfection assays were performed with Superfect reagent (Qiagen). Briefly, cells (3 × 105) were grown in a 60-mm dish for 24 h and transfected with 6 μg of total DNA with Superfect reagent. Then 0.1 μg of pCMV-β-gal was included to standardize for transfection efficiency. Transfected cells were harvested 12 h later in lysis buffer (Promega), and luciferase activity was measured with the luciferase assay system (Promega). The same extracts were used to assay for β-galactosidase activity with chlorophenol red-β-d-galactopyranoside (Sigma) as the substrate. The luciferase activity was normalized to either β-galactosidase activity or Renilla luciferase activity where indicated.
All transient transfection assays were carried out in triplicate with at least two independent assays. For transactivation by E2Fs, cells were transfected with 4 μg of reporter plasmid, 0.1 μg of pCMV-β-gal, and increasing amounts of expression plasmid for either E2F1, E2F3a, E2F3b, or E2F3aΔMB. Various amounts of pCDNA3 were added to equalize the amount of cytomegalovirus in each transfection. For transcriptional synergy by E2F and TFE3, cells were transfected with 5 μg of reporter plasmid and either 10 ng of E2F proteins in the presence or absence of 100 ng of TFE3. The control lacked both E2Fs and TFE3. Various amounts of pCDNA3 were included to equalize the amount of cytomegalovirus DNA in each transfection.
Western blot assay.
NIH 3T3 or HeLa cell lysates (nuclear or whole-cell extracts) containing equal amounts of protein were boiled for 5 min in protein sample buffer and subjected to SDS-PAGE on 8.5% polyacrylamide gels. Proteins were subsequently transferred to a polyvinylidene difluoride membrane and blocked in phosphate-buffered saline containing 5% skim milk for 2 h. Blots were then incubated with primary antibodies (1:1,000 dilution) in phosphate-buffered saline containing 5% milk overnight at 4°C, washed with phosphate-buffered saline plus 0.05% NP-40 and incubated in phosphate-buffered saline plus 0.05% NP-40 and secondary antibodies (1:5000 dilution) for 1 h at room temperature. Blots were processed with Amersham's ECL system as described by the manufacturer. Antibodies against E2F3 (sc-878), DP1 (sc-610), HA probe (sc-805), and Myc (sc-40) were purchased from Santa Cruz Biotechnology.
Chromatin immunoprecipitation assays.
We performed chromatin immunoprecipitation assays by a modification of a previously published method (46). Immunoprecipitates of wild-type and null mouse embryo fibroblasts (MEFs) and null MEFs stably expressing either E2F3a, E2F3aΔMB, or TFE3 were incubated with 1 μg each of antibodies against E2F1, E2F3a, or TFE3 (E2F1, sc-251; E2F3a, sc-879; and TFE3, Pharmingen 15451A) at 4°C overnight. We found that the polyclonal E2F1 antibody, sc-193, cross-reacted with mouse E2F3 (data not shown). Therefore, we used the monoclonal E2F1 antibody, sc-251, for addressing promoter specificity in our chromatin immunoprecipitation assays. Reversal of the cross-linking of chromatin and/or input reporter plasmid was performed as described, and samples were analyzed by semiquantitative PCR (46). Twenty-eight cycles of PCR were performed in 25 μl with 5 μl of immunoprecipitated material, 50 pmol of each primer set, 0.5 U of Taq Gold DNA polymerase (Applied Biosystems), and 0.01 μCi of [32P]dGTP or [32P]dCTP. To amplify E2F- and/or TFE3-responsive promoter regions, primer sets for p68 (accession number AB030823), positions −156 to −134 and +53 to +73, and dihydrofolate reductase (DHFR) (accession number M37144), positions −102 to −78 and +78 to +53, were used. The identification of E2F and TFE3 binding sites on each promoter was based on previously published reports (3, 9,16). PCR products were electrophoresed on 6% polyacrylamide gels. Each experiment was performed at least three independent times, and representative data are shown.
RESULTS
In considering the role of protein interactions as a basis for E2F specificity of function, we were influenced by previous work that has shown a role for an adenovirus-encoded protein in providing specificity to E2F function. The interaction of E2F with the adenovirus E2 promoter is greatly enhanced by binding of the viral E4 orf6/7 gene product (12, 13, 32, 33, 40). Whereas E2F binding to the promoter is a weak interaction, exhibiting an off-rate of less than 5 min, the interaction of E4 with E2F to form a dimeric complex on the promoter leads to significantly enhanced binding affinity, with an off-rate in excess of 60 min. The ability of E4 to elicit this effect requires the precise arrangement of E2F binding sites found in the E2 promoter. As such, the E4 protein can be seen as an E2F specificity factor, facilitating the activation of the viral E2 gene. The interaction of the E4 protein with E2F has been shown to depend on sequences within E2F proteins known as the marked box domain (20, 36). Given the fact that viral activities often mimic cellular functions, we considered the possibility that cellular proteins might function similarly to E4, facilitating the interaction of E2F with particular cellular promoters. We thus searched for proteins with yeast two-hybrid screens that bind specifically to E2F proteins dependent on the marked box domain.
TFE3 is an E2F3-specific binding protein.
A screen with E2F3a as the bait yielded 11 independent clones from the approximately 6 × 105 transformants (Table 1). All 11 clones also associated with E2F3b. One each of three clones encoded fragments of CBP, YY1-binding protein, and Mga/Mad5. Because these proteins have been shown previously to interact with E2Fs, this provided a strong validation of the screen (31, 41, 37, 49). Other clones contained fragments of proteins which interacted with E2F3 and one or two other E2F family members. These included the SKI binding protein SKIP; TEF-5; and protein kinase I-β (5, 19, 55). Three out of the 11 clones were specific for E2F3a and E2F3b and failed to bind to E2F1, E2F2, or E2F4. These clones were found to contain TFE3 (amino acids 56 to 320), a fragment of retinoid X receptor binding protein, and a partial cDNA of the novel serine/threonine protein kinase WNK1 (28, 42, 52).
TABLE 1.
E2F3-specific binding partnersa
| Clone | β-Galactosidase activity
|
||||||
|---|---|---|---|---|---|---|---|
| E2F1 | E2F2 | E2F3a | E2F3b | E2F4 | E2F3ΔMB | E2F3ΔC | |
| TFE3 | − | − | + | + | − | − | − |
| RXR-BP | − | − | + | +/− | − | − | − |
| WNK1 | − | − | + | + | − | − | − |
| PKI-β | +/− | +/− | + | + | − | − | − |
| SKIP | + | − | + | + | − | − | − |
| YY1-BP | +/− | + | + | + | − | − | − |
| CBP | + | − | + | + | − | +/− | − |
| TEF-5 | + | − | + | + | − | − | − |
| B-Myb | + | + | + | + | − | +/− | − |
| Mga/Mad5 | + | − | + | + | + | +/− | − |
| SpiB | +/− | +/− | + | + | +/− | +/− | +/− |
Clones identified from the E2F3 screen that were found to be E2F3 specific. Each was assayed for interaction with the indicated E2F by cotransformation and then standard liquid β-galactosidase assays as described in the text. +, activity greater than or equal to that of the positive control; +/−, activity less than or equal to 50% of the positive control value; −, activity no greater than that of the negative control. RXR-BP, retinoid X receptor binding protein; PKI-B, protein kinase I-B; YY1-BP, YY1 binding protein.
In light of the previously defined role of the transcription factor μE3 (TFE3), a ubiquitously expressed member of the basic helix-loop-helix family of transcription factors, and the fact that several E2F-regulated promoters also contain TFE3 binding sites (E-box elements), we chose to focus on the E2F3-TFE3 interaction (15-17). Furthermore, TFE3 has recently been identified as an activity that can bypass an Rb-mediated growth arrest, demonstrating a functional relationship between TFE3 and the Rb/E2F pathway (M. Nijman, S. Hijmans, and R. Bernards, personal communication).
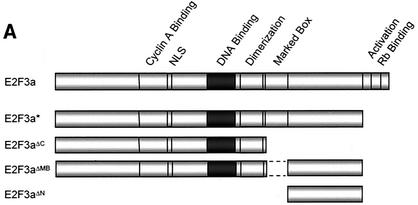
To further examine the interaction of E2F3 with full-length TFE3 in vivo, S. cerevisiae was cotransformed with full-length TFE3 fused to the Gal4 activation domain (AD) and either E2F1, E2F2, E2F3a, E2F3b, E2F4, E2F3aΔMB (lacking amino acids 295 to 357), E2F3aΔN (amino acids 357 to 465), or E2F3aΔC (amino acids 1 to 295) fused to the Gal4 DBD in the presence of DP1 (Fig. 1A). Transformants were plated on selection medium and assayed for β-galactosidase activity. TFE3 was capable of interacting with both E2F3a and E2F3b but not with the other E2F family members (E2F1, E2F2, or E2F4) (Fig. 1B). Furthermore, TFE3 failed to interact with the E2F3a mutants E2F3aΔC (amino acids 1 to 295), E2F3aΔN (amino acids 357 to 465), and, most importantly, E2F3aΔMB (lacking amino acids 295 to 357), implicating sequences within the marked box domain as being necessary for binding. Expression levels of the Gal4 DBD-E2F fusion proteins were verified with the appropriate E2F antibodies as well as an antibody specific to the Gal4 DBD (see Materials and Methods). The Gal4 DBD-E2F fusion proteins were unable to activate the lacZ reporter in the absence of cotransformed Gal4 AD-TFE3 due to deletion of the c-terminal activation domain (data not shown).
FIG. 1.
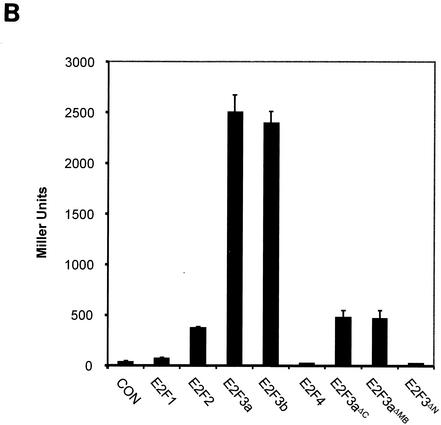
TFE3 is an E2F3-specific binding partner. (A) Schematic of E2F family members and mutants used in the yeast two-hybrid screen. Wild-type E2F1, E2F2, E2F3a, and E2F3b lacking a functional activation domain are designated with a *. E2F3aΔMB has a deletion of the marked box domain (amino acids 295 to 357). In E2F3aΔC the C terminus of E2F3 is deleted (retains amino acids 1 to 295). E2F3aΔN contains amino acids 357 to 398. NLS, nuclear localization signal. (B) A yeast liquid β-galactosidase assay was used to assess the ability of full-length TFE3 to interact with E2F family members E2F1, E2F2, E2F3a, E2F3b, and E2F4 as well as E2F3a deletion mutants. S. cerevisiae was cotransformed with Gal4 AD-TFE3, DP1, and either E2F1, E2F2, EF3a, E2F3b, E2F4, E2F3aΔMB, E2F3aΔN, or E2F3aΔC fused to the Gal4 DBD. Transformants were assayed for β-galactosidase activity, which is represented as Miller units. CON, control (Gal4 DBD).
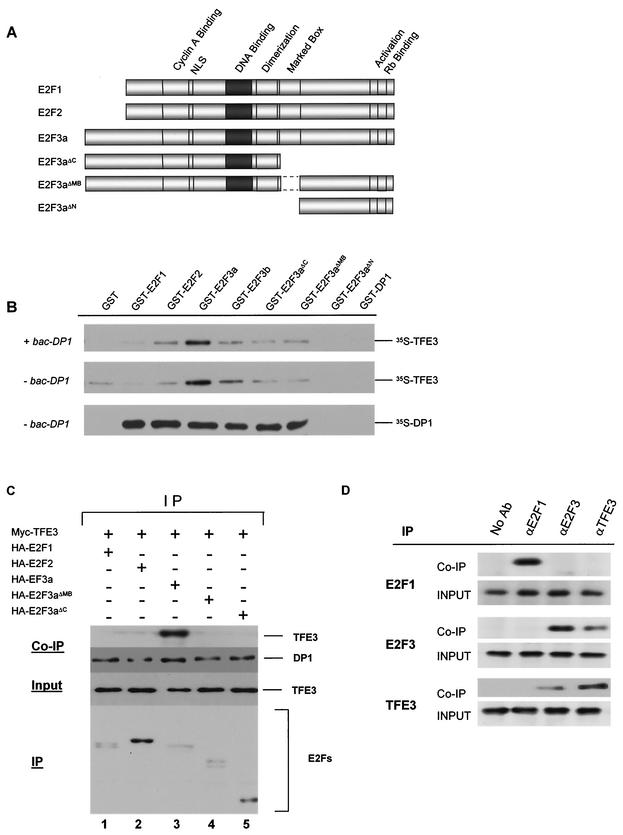
To determine whether binding of E2F3a and E2F3b to TFE3 required the heterodimerization partner DP1, we carried out in vitro binding assays with bacterially expressed GST-E2F fusions (Fig. 2A). Unlike the Gal4 DBD-E2F fusions, the GST-E2F fusions contained a functional activation domain. Equal amounts of GST-E2F fusion proteins were bound to beads and incubated in the presence of in vitro-translated 35S-labeled, Myc-tagged TFE3 in the absence and presence of bacterially expressed DP1 (bac-DP1) (Fig. 2B). TFE3 bound most strongly to E2F3a and exhibited reduced binding to E2F3b and the other E2F family members and E2F3 mutants (Fig. 2B, middle panel). Addition of bacterially expressed DP1 to the GST binding extracts did not further enhance the TFE3-E2F3 interaction (Fig. 2B, compare top and middle panels). Finally, the functional activity of the various GST-E2F fusions was tested by confirming binding to the heterodimer partner DP1 (Fig. 3A, bottom panel). As expected, DP1 did not associate with the E2F3ΔN mutant, as this mutant lacks the heterodimerization domain required for interaction with DP1. From these interaction studies, we conclude that TFE3 is an E2F3-specific binding partner, that sequences within the marked box domain facilitate binding, and that DP1 is not required for binding of E2F3 to TFE3.
FIG. 2.
E2F3 interaction with TFE3. (A) Schematic of E2F family members and mutants used for the in vitro binding assays. Full-length E2F1, E2F2, E2F3a, E2F3b, and E2F3aΔMB have a deletion of the marked box domain (amino acids 295 to 357). In E2F3aΔC the C terminus of E2F3 is deleted (retains amino acids 1 to 295). E2F3aΔN contains amino acids 357 to 398. (B) GST-E2F fusion proteins bound to beads were incubated in the presence of in vitro-translated 35S-labeled Myc-TFE3 or in vitro-translated 35S-labeled DP1 as a control, in either the presence or absence of bacterially expressed DP1. GST alone was used as a negative control in these experiments. (C) NIH 3T3 cells were transfected with Myc-TFE3 and HA-tagged E2F1, E2F2, E2F3a, E2F3aΔMB, or E2F3aΔC (lanes 1 to 5, respectively). Unlike the Gal4 DBD-E2F fusions, the HA-E2F fusions contained a functional activation domain. Transfected cells were lysed in immunoprecipitation lysis buffer at 24 h posttransfection. HA antibody immobilized on Sepharose beads (Covance) was used to immunoprecipitate the HA-tagged E2Fs. Myc-TFE3 and DP1 that coimmunoprecipitated with the E2F proteins were detected by Western blotting with specific antibodies (Co-IP). The level of TFE3 expressed in the transfected cells was measured by Western blotting of aliquots before the immunoprecipitation (input). Finally, the amounts of the E2Fs that were recovered in the immunoprecipitations (IP) was determined by Western blotting with HA antibody. (D) NIH 3T3 cells were lysed in immunoprecipitation lysis buffer, and lysates were incubated with antibodies to E2F1, E2F3, or TFE3 immobilized on protein A- plus protein G-agarose beads (Oncogene). The levels of endogenous E2F1, E2F3, and TFE3 were measured by Western blotting of aliquots before the immunoprecipitation (input). Endogenous TFE3 that coimmunoprecipitated with the E2F proteins was detected by Western blotting with a TFE3-specific antibody. Similarly, endogenous E2F proteins that coimmunoprecipitated with TFE3 were detected by Western blotting with antibodies specific to E2F1 and E2F3. No Ab, no antibody.
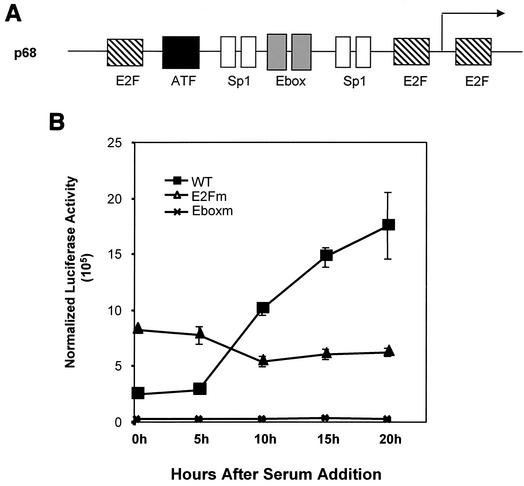
FIG. 3.
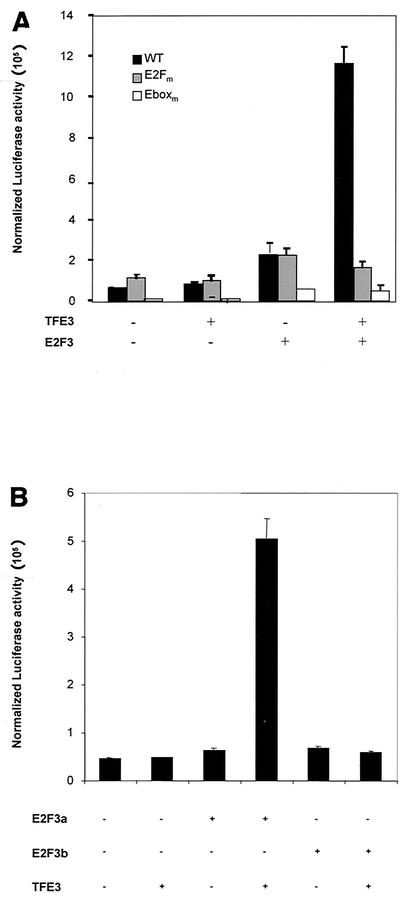
Role of TFE3 and E2F3a in p68 promoter activity. (A) Schematic depiction of the DNA polymerase α p68 subunit promoter. Binding sites for known transcription factors are indicated in relation to the start site of transcription. (B) NIH 3T3 cells were transfected with reporter plasmids and starved for 48 h. After starvation, cells were stimulated to grow by the addition of serum to 20%. Samples were taken at the indicated times, and luciferase activity was measured. Luciferase activity was normalized to β-galactosidase activity (normalized luciferase activity). WT, wild-type promoter; E2Fm, E2F mutant promoter; Eboxm, E-box mutant promoter.
We next assessed whether TFE3 and E2F3a can interact in mammalian cells. Full-length Myc-TFE3 protein was expressed in NIH 3T3 cells by transient transfection together with HA-E2F1, HA-E2F2, HA-E2F3a, HA-E2F3aΔMB, or HA-E2F3aΔC (Fig. 2C). Like the GST-E2F fusions, the HA-E2F fusions contained a functional activation domain. The resulting complexes were recovered and analyzed by SDS-PAGE with antibodies directed against the HA and Myc tags. Consistent with the binding results from S. cerevisiae, TFE3 coimmunoprecipitated with HA-E2F3a (Fig. 2C, lane 3) but not HA-E2F1, HA-E2F2, or HA-E2F3a mutants lacking the marked box domain. To verify that the various HA-E2F fusions were equally functional, we determined the levels of endogenous DP1 coimmunoprecipitated with the HA-E2F fusions (Fig. 2C, middle panel). From this analysis, we conclude that TFE3 and E2F3a associate with one another in mammalian cells and that binding is dependent on the marked box domain.
We next verified that endogenous TFE3 and E2F3a interact in vivo by immunoprecipitation of endogenous E2F1, E2F3a, or TFE3 from NIH 3T3 cell lysates. Consistent with the binding data in Fig. 2C, TFE3 coimmunoprecipitated with E2F3a but not with E2F1 (Fig. 2D, bottom panel). Similarly, E2F3a but not E2F1 coimmunoprecipitated with TFE3 (Fig. 2D, top and middle panels). Endogenous protein levels of E2F1, E2F3a, and TFE3 were measured by Western blotting of aliquots before the immunoprecipitation (input) as an internal control. This analysis further confirmed that TFE3 and E2F3a associate with one another in vivo.
Role for TFE3 and E2F3a in control of DNA polymerase α p68 promoter.
Analysis of the promoter regions of numerous E2F target genes identified a subset of genes that contain both E2F binding sites and E-box elements (binding sites for TFE3). Among these is the promoter of the DNA polymerase α p68 subunit gene (Fig. 3A). Analysis of promoter mutations that alter either the E2F elements or the E-box elements demonstrated the importance of both elements in the function of the p68 promoter following stimulation of cell proliferation (Fig. 3B), a result consistent with previous work (35). In particular, mutation of the E-box elements was seen to completely abolish promoter activity, while mutation of the E2F elements resulted in both derepression in quiescent cells and reduced activation at G1/S.
Given the evidence that TFE3 associates with E2F3a, we investigated the functional significance of this interaction in the activation of the p68 promoter. NIH 3T3 cells were transfected with the p68 luciferase reporter constructs (wild-type, E2Fm, and E-boxm) together with plasmids encoding TFE3 alone, E2F3a alone, or both transcription factors together, and harvested for determination of luciferase and β-galactosidase activity at 12 h posttransfection (Fig. 4A). At the concentration of plasmid employed, neither E2F3a nor TFE3 alone led to a significant increase, but the two together resulted in a nearly 10-fold increase, indicating that TFE3 and E2F3a can synergistically activate the p68 promoter. Mutation of either the E2F-binding sites (E2Fm) or the E-box like elements (E-boxm) abrogated the TFE3-E2F3a synergistic effect, confirming that the two factors act synergistically and indicating that the presence of one binding site alone is insufficient for recruiting both binding partners (Fig. 4A).
FIG. 4.
Synergistic activation of p68 promoter by TFE3 and E2F3a (A) NIH 3T3 cells were transfected with either the wild-type p68 promoter (WT), a promoter containing E2F site mutations (E2Fm), or one with mutations in the E-box elements (Eboxm), together with a plasmid expressing TFE3 alone, E2F3a alone, or TFE3 and E2F3a. Cells were harvested 12 h posttransfection and assayed for luciferase activity and β-galactosidase activity. Luciferase activity was normalized to β-galactosidase activity. (B) NIH 3T3 cells were transfected with the p68 promoter and 10 ng of either empty vector, an E2F3a-expressing plasmid, or an E2F3b-expressing plasmid alone or in combination with 100 ng of the TFE3-expressing plasmid. TFE3 alone was used as a control.
Since E2F3b is also capable of binding to TFE3, we asked whether E2F3b, like E2F3a, could activate p68 gene transcription in synergy with TFE3. NIH 3T3 cells were transfected as above with the wild-type p68 luciferase reporter together with plasmids encoding E2F3a, E2F3b, TFE3, or combinations of E2F proteins with TFE3. Whereas E2F3a was able to synergistically activate transcription with TFE3, E2F3b was unable to activate transcription in the presence of TFE3 (Fig. 4B). Furthermore, when overexpressed, E2F3b antagonized the ability of E2F3a and TFE3 to synergistically activate p68 gene transcription (data not shown), suggesting that the E2F3b isoform might function as a dominant negative inhibitor of E2F3a. Indeed, overexpression of E2F3b was observed to inhibit transactivation by E2F3a, again suggesting a potential role for E2F3b as a dominant negative inhibitor of E2F3a (data not shown).
Synergistic activation of transcription by E2F3a and TFE3, dependent on the E2F3 marked box domain.
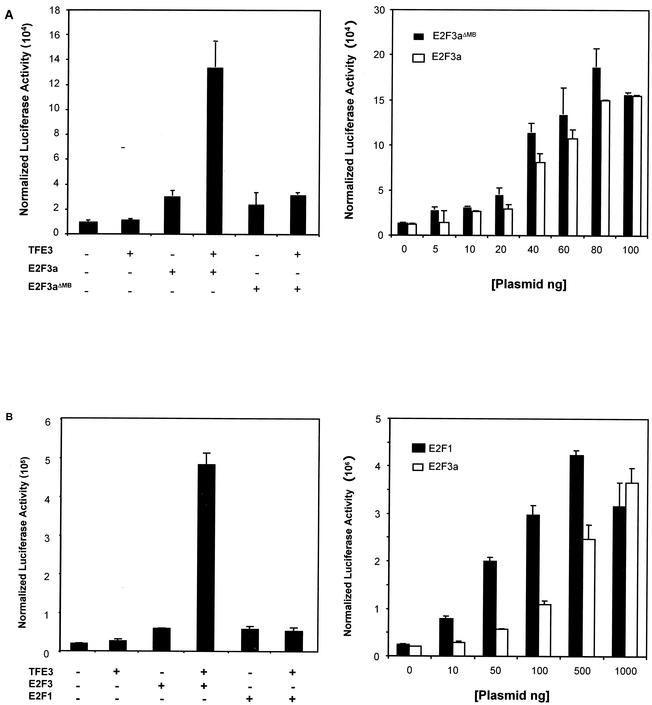
The binding data suggest that the E2F3a marked box domain is necessary for binding to TFE3 (Fig. 1 and 2). We next addressed the role of the marked box domain with respect to E2F3a-TFE3 synergy of the p68 promoter. NIH 3T3 cells were transfected with the wild-type p68 reporter construct together with plasmids encoding either E2F3a, E2F3aΔMB, TFE3, or combinations of the E2F3a proteins with TFE3. As seen in previous assays, TFE3 synergistically activated transcription with wild-type E2F3a (Fig. 5A, left panel). In contrast, E2F3aΔMB, unlike full-length E2F3a, was unable to synergistically activate transcription of the p68 gene in the presence of TFE3. Although the E2F3a marked box mutant was not able to synergize with TFE3 to activate the p68 promoter, this mutant was still capable of transactivation when overexpressed. Indeed, transfection of NIH 3T3 cells with the p68 reporter and increasing amounts of E2F3aΔMB led to a dose-dependent activation of the reporter which was comparable to that displayed by wild-type E2F3a (Fig. 5A, right panel). This finding confirms that E2F3aΔMB, when overexpressed, is capable of binding to the E2F elements within the p68 promoter and activating transcription; thus, the failure to synergize with TFE3 is not due to a general defect of the mutant E2F3 protein.
FIG. 5.
Synergistic activation of the p68 promoter by E2F and TFE3 is marked box-dependent and specific to E2F3a. (A) Left panel. NIH 3T3 cells were transfected with the p68 promoter and 10 ng of either empty vector, an E2F3a-expressing plasmid, or an E2F3aΔMB-expressing plasmid alone or in combination with 100 ng of the TFE3-expressing plasmid. TFE3 alone was used as a control. Right panel. NIH 3T3 cells were transfected with the p68 promoter and increasing amounts of E2F3a or E2F3aΔMB expression plasmids. Cells were then harvested for determination of luciferase and β-galactosidase activity. (B) Left panel. NIH 3T3 cells were transfected either with 10 ng of plasmids expressing E2F3a or E2F1 or with TFE3 in combination with 10 ng of E2F3a or E2F1. TFE3 alone was used as a control. Cells were harvested as above. Right panel. NIH 3T3 cells were transfected with the wild-type p68 promoter and increasing amounts of expression vector for either E2F1 or E2F3a and harvested for determination of luciferase and β-galactosidase activity as above.
The binding data also show that TFE3 is an E2F3-specific binding partner. To assess whether TFE3-mediated transcriptional synergy of the p68 gene is also E2F3 specific, we assayed the ability of TFE3 to synergize with E2F1. NIH 3T3 cells were transfected with the wild-type p68 reporter together with plasmids encoding E2F1 alone, E2F3a alone, or E2F1 or E2F3a in combination with TFE3. As shown in Fig. 5B, left panel, and consistent with the results in Fig. 4 and 5A, E2F3a was able to synergistically activate the p68 promoter together with TFE3. In contrast, the combination of E2F1 and TFE3 did not lead to a synergistic activation of the p68 promoter, even though overexpressed E2F1 is capable of activating the p68 gene (Fig. 5B, right panel). These data correlate with the binding data and suggest that physical association with TFE3 is required for cooperative transcription by E2Fs. Furthermore, these data suggest that while E2F1 can activate transcription of the p68 gene when expressed at high levels, only E2F3a can activate in synergy with TFE3.
E2F3 marked box domain mediates TFE3 specificity.
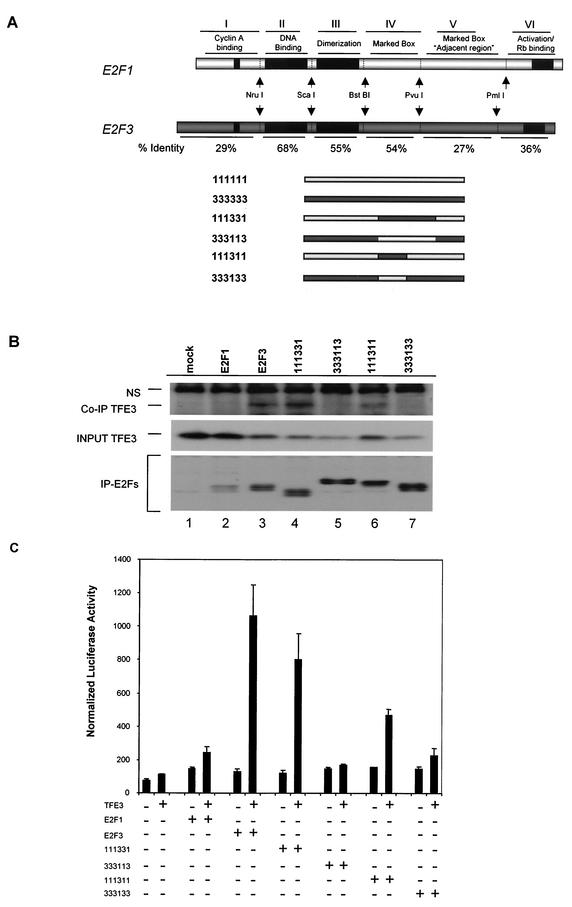
The data so far suggest that the marked box domain of E2F3 is necessary for binding to TFE3 as well as for allowing E2F3 and TFE3 to synergistically activate transcription of the p68 gene. To further investigate the role of the marked box in E2F function and to determine whether this domain of E2F3 is both necessary and sufficient for binding to TFE3, we created a series of chimeric proteins which contained the marked box and adjacent sequences of either E2F1 or E2F3 (Fig. 6A). Briefly, the cDNAs of HA-E2F1 and HA-E2F3 were mutagenized to introduce restriction sites that allowed the cloning of individual domains between the genes. Full-length Myc-TFE3 protein was expressed in NIH 3T3 cells by transient transfection together with HA-E2F1, HA-E2F3, and the various chimeras. Consistent with the binding data in Fig. 2C, TFE3 coimmunoprecipitated with HA-E2F3 but not HA-E2F1 (compare lanes 2 and 3). Assays for the interaction of TFE3 with the chimeras revealed that the E2F3 marked box domain was in fact sufficient to allow an interaction with TFE3 (Fig. 2C, lane 6). It does appear that the sequence adjacent to the marked box domain also contributed to the interaction, since the 111331 chimera was more efficient in TFE3 binding than the 111311 chimera (Fig. 2C, compare lanes 4 and 6). In contrast, the E2F1 marked box domain was not able to confer an interaction with TFE3 (Fig. 2C, lanes 5 and 7).
FIG. 6.
Marked box domain of E2F3 is both necessary and sufficient for binding to TFE3. (A) Schematic of the chimeric E2Fs described in this study. The nomenclature describes the identity of individual domains in the chimeras: first digit, amino terminus/cyclin A binding; second digit, DNA binding domain; third digit, DP1 dimerization domain; fourth digit, marked box; fifth digit, marked box adjacent region; sixth digit, transactivation/Rb binding domain. The resultant chimeric E2F cDNAs were cloned into a cytomegalovirus-HA expression vector. (B) NIH 3T3 cells were transfected with Myc-TFE3 and HA-tagged E2F1 to E2F3 chimeric proteins. Transfected cells were lysed in immunoprecipitation lysis buffer 24 h posttransfection. HA antibody immobilized on Sepharose beads (Covance) was used to immunoprecipitate the HA-tagged E2Fs. Myc-TFE3 that coimmunoprecipitated with the E2F proteins was detected by Western blotting with specific antibodies (Co-IP TFE3). The level of TFE3 expressed in the transfected cells was measured by Western blotting of aliquots before the immunoprecipitation (input TFE3). Finally, the amounts of the E2Fs that were recovered in the immunoprecipitations (IP-E2Fs) were determined by Western blotting with HA antibody. NS, nonspecific. (C) NIH 3T3 cells were transfected with the p68 promoter and 10 ng of either empty vector, expression plasmids for E2F1 or E2F3, or the E2F1 to E2F3 chimeric proteins alone or in combination with 100 ng of the TFE3 expression plasmid. TFE3 alone was used as a control. Cells were then harvested for determination of Renilla luciferase activity.
We next addressed the ability of the chimeras to mediate transcriptional synergy of the p68 promoter in the presence of TFE3. NIH 3T3 cells were transfected as above with the wild-type p68 luciferase reporter together with plasmids encoding HA-E2F1, HA-E2F3, Myc-TFE3, and the various chimeras. Consistent with our previous results, E2F3 but not E2F1 was able to synergistically activate the p68 promoter together with TFE3. In contrast, the combination of E2F1 and TFE3 or E2F3 chimeras containing the marked box region of E2F1 together with TFE3 did not lead to activation of the p68 promoter (Fig. 6C). Assays of the chimeras revealed an ability to synergize with TFE3 that coincided with the physical interaction with TFE3. Specifically, the chimera containing the E2F3 marked box domain and adjacent sequence was equal to the wild-type E2F3 protein in synergizing with TFE3. The chimera containing only the E2F3 marked box domain was capable of synergistic activation with TFE3 but reduced compared to the chimera containing the adjacent sequences. In contrast, the chimera containing the E2F1 marked box and adjacent sequence was not able to synergize with TFE3. These data thus provide strong evidence for a role of the marked box domain of E2F3 in mediating a physical interaction with TFE3 that coincides with the ability to mediate a cooperative transcription activation.
Mutually dependent interaction of TFE3 and E2F3 with the p68 promoter.
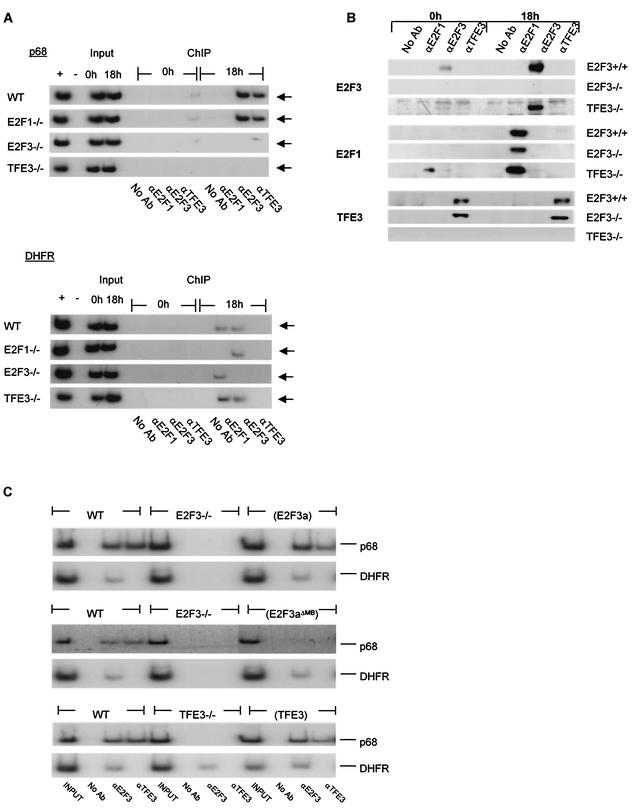
The promoter activation assays provide evidence for a role for E2F3 and TFE3 in activation of the p68 gene that coincides with the physical interaction of these two proteins. To connect these two sets of assays and provide evidence for an interaction of TFE3 and E2F3a in the control of p68 expression in vivo, we used chromatin immunoprecipitation assays to examine the interaction of these proteins with the p68 promoter in intact cells (Fig. 7A). Wild-type, E2F3−/−, TFE3−/−, and E2F1−/− mouse embryo fibroblast cultures were brought to quiescence and then stimulated to grow by the addition of serum to a 20% final concentration. Cells were harvested either at quiescence or 18 h following growth stimulation and then cross-linked with formaldehyde. Extracts were prepared, DNA was fragmented, and chromatin material was immunoprecipitated with antibodies specific for E2F3a and TFE3. DNA was released from the immunoprecipitates and then used for PCR assays to measure the presence of the p68 promoter sequences.
FIG. 7.
Interaction of E2F3 and TFE3 with the p68 promoter following growth stimulation. (A) Chromatin immunoprecipitation (ChIP) assays were used to examine the interaction of E2F3 and TFE3 with the p68 promoter in intact cells. Mouse embryo fibroblasts (wild type [WT], E2F1−/−, E2F3−/−, and TFE3−/−) were harvested either at quiescence or 18 h following serum stimulation and cross-linked with the addition of formaldehyde as described in the text. Chromatin was immunoprecipitated with antibodies to either E2F1, E2F3, or TFE3. After reversing the cross-link, DNA released from the immunoprecipitates was used for PCR analysis to measure the presence of p68 promoter sequences. The arrows indicate the position of the p68 promoter PCR product. No Ab, no antibody. (B) Aliquots of the immunoprecipitates from panel A were analyzed in an SDS-acrylamide gel and assayed for the presence of E2F1, E2F3, and TFE3 by Western blotting with specific antibodies. (C) Chromatin immunoprecipitation assays were used to examine the interaction of E2F1, E2F3, and TFE3 with the DHFR promoter in intact cells as described above. (D) Chromatin immunoprecipitation assay for interaction of E2F3 and TFE3 with the endogenous p68 and DHFR promoters in wild-type, TFE3−/−, and TFE3−/− MEFs stably expressing TFE3 protein (TFE3).
As shown in Fig. 7A (top panel), the p68 promoter was detected in the E2F3a immunoprecipitates from wild-type cells isolated at 18 h following serum stimulation but not in the quiescent-cell sample, consistent with the accumulation of E2F3a protein as cells are stimulated to grow (compare the Western blot analysis for E2F3a in the 0- and 18-h samples in Fig. 7B). Similarly, the p68 promoter was not detected in either the E2F3−/− or the TFE3−/− cells, implicating both factors as being important in the formation of the stable promoter complex.
The TFE3 immunoprecipitate also contained p68 promoter sequence, and again, this was substantially increased in the growth-stimulated sample. Unlike E2F3a, the TFE3 protein was present in both quiescent and growing cells (Fig. 7B), suggesting that the ability of TFE3 to interact with the p68 promoter might be dependent on the presence of the E2F3a protein. In support of this model, the assay of TFE3 chromatin immunoprecipitates from extracts of E2F3 null cells showed that binding of TFE3 to the p68 promoter was markedly reduced despite the presence of normal TFE3 protein levels in the E2F3 null cells (Fig. 7B). Conversely, an assay of E2F3a chromatin immunoprecipitates from TFE3 null extracts revealed that binding of E2F3a to the p68 promoter was dependent on TFE3 (Fig. 7A). In contrast, the absence of E2F1 (in the E2F1−/− cells) did not alter the interaction of either E2F3 or TFE3 with the p68 promoter. Together, these results demonstrate that the interaction of TFE3 and E2F3a with the p68 promoter is facilitated by the presence of both proteins.
Additional assays were performed as controls for the specificity of the chromatin interactions. Since an analysis of the DHFR promoter sequence revealed the presence of multiple E2F elements but an absence of canonical TFE3 binding sequences, we reasoned that E2F interactions with this promoter should be independent of TFE3. As shown in Fig. 7C, both E2F1 and E2F3a interacted with the DHFR promoter and appeared to do so independently, since the absence of either protein did not seem to affect the binding of the other protein (see E2F1−/− and E2F3−/− cells). Consistent with the absence of potential E-box elements within the DHFR promoter region, we saw no evidence for an interaction of TFE3 with the DHFR promoter in the in vivo chromatin immunoprecipitation assays (Fig. 7C), demonstrating a promoter specificity to the E2F3-TFE3 interaction.
Finally, to demonstrate that the interaction of E2F3 and TFE3 with the p68 promoter was facilitated by the presence of both proteins, we carried out chromatin immunoprecipitation assays in wild-type and E2F3 or TFE3 null MEFs stably expressing either E2F3a, E2F3aΔMB, or TFE3 (Fig. 7D). Cells were brought to quiescence and then stimulated to grow by the addition of serum to a 20% final concentration. Cells were harvested at 18 h following growth stimulation and then cross-linked with formaldehyde as described above. As shown in Fig. 7D, the p68 promoter was detected in both the E2F3 and TFE3 immunoprecipitates of wild-type, E2F3−/− MEFs stably expressing E2F3a and TFE3−/− MEFs stably expressing TFE3 but not in E2F3−/−, E2F3−/− MEFs stably expressing E2F3aΔMB, or TFE3−/− cells. These data confirm that the interaction of either E2F3a or TFE3 with DNA is facilitated by the presence of both proteins and is dependent on the marked box of E2F3.
DISCUSSION
Although there is considerable evidence for a role of the E2F family members in processes such as proliferation, apoptosis, differentiation, oncogenesis, and DNA repair, relatively little is known about the mechanisms that discriminate between the selective roles of the individual family members. We and others have proposed that these proteins regulate a distinct subset of target genes and thus can exhibit diverse roles in the cell. The work we describe here provides a mechanistic basis for understanding the action of these proteins as transcriptional regulatory activities. In particular, we propose that the ability of E2F3a and TFE3 to physically interact provides a mechanism for functional synergy involving these two proteins. In this model, specificity of function is achieved by the ability of transcription factors to interact, coupled with the joint presence of binding sites for the factors in a given promoter. It is the potential for multiple such interactions that provides the basis for the concept of combinatorial specificity, in which a limited number of transcription factors can achieve the regulation of a large number of target genes (54).
We expect that E2F3a makes use of other partners besides TFE3 to achieve specificity of function. For instance, the analysis of DHFR promoter interactions demonstrates that the binding of E2F3 is independent of TFE3. Moreover, other recent experiments have demonstrated an interaction between E2F3a and RYBP that provides a bridge between E2F3 and the YY1 transcription factor to facilitate binding to the cdc6 promoter and thus providing another basis for specific promoter interaction (41).
Role for marked box as a protein interaction domain.
The E2F marked box domain was originally identified by sequence analysis as a region of sequence conservation within the E2F family (24). Less conserved than the DNA binding domain sequences, the marked box region exhibits approximately 55% homology across E2F1 to E2F3 and slightly less with the other E2F family members. This conservation suggests a functional role for this domain, and the work that we present here suggests a role for the domain in facilitating protein interactions that provide promoter specificity.
Prior work has demonstrated a role for the marked box domain in mediating the interaction of the adenovirus E4 orf6/7 protein with E2F (20) that then allows the formation of a stable complex on the viral E2 promoter (12, 32, 33, 40). In the absence of E4, E2F can bind to the E2 promoter but does so with weak affinity and does not effectively stimulate transcription. The ability of this E2F-E4 complex to bind with high affinity to the E2 promoter is dependent on the architecture of the promoter, whereby the two E2F binding elements must be properly spaced and aligned to allow the formation of the complex. Given the fact that this arrangement of binding sites is largely unique to the E2 promoter, the effect of E4 is to specifically activate E2 transcription by directing E2F to the promoter.
In the case of the p68 promoter, and possibly other promoters that have E-box and E2F elements, it is the joint presence of these elements that allows binding of TFE3 and E2F3 and thus provides the specificity. The fact that the chromatin interaction of E2F3 is facilitated by the presence of TFE3 and vice versa suggests that the consequence of the E2F3-TFE3 is similar to that of the E2F-E4 interaction, a stabilization of an otherwise weak interaction. However, it is also clear from the inability of E2F3b to synergistically activate the p68 promoter in the presence of TFE3 that a stable interaction with DNA is not sufficient to form a transcriptionally competent complex. It is likely that the E2F3a-TFE3 interaction may be the basis for recruiting additional factors to DNA which are necessary for transcriptional activation and which may not be efficiently recruited by E2F3b/TFE3.
Basis for specificity of E2F function within the E2F family.
The fact that E2F3 can interact with TFE3, resulting in a synergistic activation of the p68 promoter, whereas E2F1 does not interact with TFE3 and thus does not synergize in transcriptional activation provides a basis for the E2F3-specific activation of transcription. In this particular instance, a promoter jointly containing a TFE3 site (E-box element) together with an E2F site would be a potential E2F3 target. It is important to note that this model does not limit the action of E2F3 to TFE3, nor does it limit the action of TFE3 to E2F3. Indeed, other work has shown that TFE3 can act synergistically with Smad proteins in the activation of transcription (15, 16). Rather, this represents an example of combinatorial specificity achieved by the joint action of promoter-specific transcription factors and provides one basis for the specificity that distinguishes the action of E2F3 from that of other E2Fs.
These results predict that a promoter containing an E-box element paired with an E2F element is a potential E2F3-specific target. Presumably, further studies will lead to identification of additional E2F3 partners as well as partners for other E2F proteins so as to expand the understanding of promoter sequences that specify activation of transcription. As this information develops, including an understanding of the constraints on the spatial organization of the sites, it should be possible to expand an understanding of gene-specific control through computational approaches that have the ability to recognize the promoter-specific code of combinations of cis-acting elements that specify and direct the joint interactions of transcription factors.
Acknowledgments
We thank Masako Izumi and Fumio Hanaoka (Riken, Japan), Harvey Lodish (MIT) for gifts of reagents, and Nancy Jenkins and Neal Copeland for providing the TFE3 null mice. We also thank Kaye Culler for help with preparation of the manuscript and Nina Laakso and Lazlo Jakoi for technical assistance.
J.R.N. is an Investigator of the Howard Hughes Medical Institute. P.H.G. was supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Babb, R., C. C. Huang, D. J. Aufiero, and W. Herr. 2001. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA-binding structure. Mol. Cell. Biol. 21:4700-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 3.Brodin, G., A. Hgren, C.-H. Dijke, and R. Heuchel. 2000. Efficient TGF-β induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J. Biol. Chem. 275:29023-29030. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright, P., H. Muller, C. Wagener, K. Holm, and K. Helin. 1998. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17:611-623. [DOI] [PubMed] [Google Scholar]
- 5.Dahl, R., B. Wani, and M. J. Hayman. 1998. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene 26:1579-1586. [DOI] [PubMed] [Google Scholar]
- 6.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dynlacht, B. D., K. Moberg, J. A. Lees, E. Harlow, and L. Zhu. 1997. Specific regulation of E2F family members by cyclin-dependent kinases. Mol. Cell. Biol. 17:3867-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 9.Fry, C. J., A. Pearson, E. Malinowski, S. M. Bartley, J. Greenblatt, and P. J. Farnham. 1999. Activation of the murine dihydrofolate reductase promoter by E2F1. J. Biol. Chem. 274:15883-15891. [DOI] [PubMed] [Google Scholar]
- 10.Gaubatz, S., J. G. Wood, and D. M. Livingston. 1998. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. USA 95:9190-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giangrande, P. H., E. A. Kimbrel, D. P. Edwards, and D. P. McDonnel. 2000. The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol. Cell. Biol. 20:3102-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, S., D. A. Engel, and T. Shenk. 1989. An adenovirus early region 4 gene product is required for induction of the infection-specific form of cellular E2F activity. Genes Dev. 3:1062-1074. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, S., and T. Shenk. 1989. E2F from adenovirus-infected cells binds cooperatively to DNA containing two properly oriented and spaced recognition sites. Mol. Cell. Biol. 9:4495-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heimann, P., H. El Housni, G. Ogur, M. A. Weterman, E. M. Petty, and G. Vassart. 2001. Fusion of a novel gene, RCC17, to the TFE3 gene in t(X;17) (p11.2;q25.3)-bearing papillary renal cell carcinomas. Cancer Res. 61:4130-4135. [PubMed] [Google Scholar]
- 15.Hua, X., X. Liu, D. O. Ansari, and H. F. Lodish. 1998. Synergistic cooperation of TFE3 and Smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 12:3084-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua, X., Z. A. Miller, H. Benchabane, J. L. Wrana, and H. F. Lodish. 2000. Synergism between transcription factors TFE3 and Smad3 in transforming growth factor β-induced transcription of the Smad7 gene. J. Biol. Chem. 275:33205-33208. [DOI] [PubMed] [Google Scholar]
- 17.Hua, X., Z. A. Miller, G. Wu, Y. Shi, and H. F. Lodish. 1997. Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: Interactions of promoter DNA, transcription factor mE3, and Smad proteins. Proc. Natl. Acad. Sci. USA 96:13130-13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690-703. [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquemin, P., J. A. Martial, and I. Davidson. 1997. Human TEF-5 is prentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-β enhancer. J. Biol. Chem. 272:12928-12937. [DOI] [PubMed] [Google Scholar]
- 20.Jost, C. A., D. Ginsbreg, and W. G. Kaelin, Jr. 1996. A conserved region of unknown function participates in the recognition of E2F family members by the adenovirus E4 ORF 6/7 protein. Virology 220:78-90. [DOI] [PubMed] [Google Scholar]
- 21.Kowalik, T. F., J. DeGregori, J. K. Schwarz, and J. R. Nevins. 1995. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J. Virol. 69:2491-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek, W., M. E. Ewen, S. Shirodkar, Z. Arany, W. G. Kaelin, and D. M. Livingston. 1994. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78:161-172. [DOI] [PubMed] [Google Scholar]
- 23.Krek, W., G. Xu, and D. M. Livingston. 1995. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell 83:1149-1158. [DOI] [PubMed] [Google Scholar]
- 24.Lees, J. A., M. Saito, M. Valentine, T. Look, E. Harlow, N. Dyson, and K. Helin. 1993. The retinoblastoma protein binds to a family of E2F transcription factors. Mol. Cell. Biol. 13:7813-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leone, G., J. DeGregori, Z. Yan, L. Jakoi, S. Ishida, R. S. Williams, and J. R. Nevins. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 12:2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C.-H. Park, P. Giangrande, L. Wu, H. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105-113. [DOI] [PubMed] [Google Scholar]
- 27.Lin, W.-C., F.-T. Lin, and J. R. Nevins. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15:1833-1845. [PMC free article] [PubMed] [Google Scholar]
- 28.Macchi, P., L. Notarangelo, S. Giliani, D. Strina, M. Repetto, M. G. Sacco, P. Vezzoni, and A. Villa. 1995. The genomic organization of the human transcription factor 3 (TFE3) gene. Genomics 28:491-494. [DOI] [PubMed] [Google Scholar]
- 29.Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. 1999. Interaction between SCFSKP2 ubiquitin protein ligase and E2F-1 underlies regulation of E2F-1 degradation. Nat. Cell Biol. 1:14-19. [DOI] [PubMed] [Google Scholar]
- 30.Moroni, M. C., E. S. Hickman, E. L. Denchi, G. Caprara, E. Colli, F. Cecconi, H. Muller, and K. Helin. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3:552-558. [DOI] [PubMed] [Google Scholar]
- 31.Morris, L., K. E. Allen, and N. B. La Thangue. 2000. Regulation of E2F transcription by cyclin E-cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2:232-239. [DOI] [PubMed] [Google Scholar]
- 32.Neill, S. D., C. Hemstrom, A. Virtanen, and J. R. Nevins. 1990. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc. Natl. Acad. Sci. USA 87:2008-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neill, S. D., and J. R. Nevins. 1991. Genetic analysis of the adenovirus E4 6/7 transactivator: interaction with E2F and induction of a stable DNA-protein complex are critical for activity. J. Virol. 65:5364-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9:585-593. [PubMed] [Google Scholar]
- 35.Nishikawa, N. S., M. Izuni, H. Uchida, M. Yokoi, H. Miyazawa, and F. Hanaoka. 2000. Cloning and characterization of the 5′-upstream sequence governing the cell cycle-dependent transcription of mouse DNA polymerase α 68 kDa subunit gene. Nucleic Acids Res. 28:1525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor, R. J., and P. Hearing. 1994. Mutually exclusive interaction of the adenovirus E4-6/7 protein and the retinoblastoma gene product with internal domains of E2F-1 and DP-1. J. Virol. 68:6848-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F and Myc responsive genes in G0. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 38.Peers, B., S. Sharma, T. Johnson, M. Kamps, and M. Montminy. 1995. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: importance of the FPWMK motif and of the homeodomain. Mol. Cell. Biol. 15:7091-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin, X.-Q., D. M. Livingston, W. G. Kaelin, and P. D. Adams. 1994. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA 91:10918-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raychaudhuri, P., S. Bagchi, S. D. Neill, and J. R. Nevins. 1990. Activation of the E2F transcription factor in adenovirus-infected cells involves E1A-dependent stimulation of DNA-binding activity and induction of cooperative binding mediated by an E4 gene product. J. Virol. 64:2702-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlisio, S., T. Halperin, M. Vidal, and J. R. Nevins. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21:5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seol, W., H. S. Choi, and D. D. Moore. 1995. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol. Endocrinol. 9:72-85. [DOI] [PubMed] [Google Scholar]
- 43.Shan, B., and W.-H. Lee. 1994. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol. Cell. Biol. 14:8166-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikorski, R. S., and P. A. Hieter. 1989. System of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skalsky, Y. M., P. M. Ajuh, C. Parker, A. I. Lamond, G. Goodwin, and C. S. Cooper. 2001. PRCC, the commonest TFE3 fusion partner in papillary renal carcinoma is associated with pre-mRNA splicing factors. Oncogene 20:178-187. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and Rb families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 47.Trimarchi, J. M., B. Fairchild, R. Verona, K. Moberg, N. Andon, and J. A. Lees. 1998. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl. Acad. Sci. USA 95:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trimarchi, J. M., B. Fairchild, J. Wen, and J. A. Lees. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98:1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trouche, D., and T. Kouzarides. 1996. E2F1 and E1A12S have homologous activation domains regulated by Rb and CBP. Proc. Natl. Acad. Sci. USA 93:1439-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weterman, M. A., M. Wilbrink, and A. Geurts van Kessel. 1996. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1) (p11;q21)-positive papillary renal cell carcinomas. Proc. Natl. Acad. Sci. USA 93:15294-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, X., and A. J. Levine. 1994. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. USA 91:3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, B., J. M. English, J. L. Wilsbacher, S. Stippec, E. J. Goldsmith, and M. H. Cobb. 2000. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem. 275:16795-16801. [DOI] [PubMed] [Google Scholar]
- 53.Xu, M., K.-A. Sheppard, C.-Y. Peng, A. S. Yee, and H. Piwnica-Worms. 1994. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol. Cell. Biol. 14:8420-8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto, K. R., B. D. Darimont, R. L. Wagner, and J. A. Iniguez-Lluhi. 1998. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harb. Symp. Quant. Biol. 63:587-598. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, L., L. Yu, Q. Tu, M. Zhang, H. He, W. Chen, J. Gao, J. Yu, Q. Wu, and S. Zhao. 2000. Cloning and mapping of human PKIB and PKIG, a comparison of tissue expression patterns of three members of the protein kinase inhibitor family, including PKIA. Biochemistry 349:403-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, N., E. Fraenkel, C. O. Pabo, and N. P. Pavletich. 1999. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13:666-674. [DOI] [PMC free article] [PubMed] [Google Scholar]