Abstract
Comparative genomics promises to rapidly accelerate the identification and functional classification of biologically important human genes. We developed the TIGR Orthologous Gene Alignment (TOGA; 〈http://www.tigr.org/tdb/toga/toga.shtml〉) database to provide a cross-reference between fully and partially sequenced eukaryotic transcribed sequences. Starting with the assembled expressed sequence tag (EST) and gene sequences that comprise the 28 TIGR Gene Indices, we used high-stringency pair-wise sequence searches and a reflexive, transitive closure process to associate sequence-specific best hits, generating 32,652 tentative ortholog groups (TOGs). This has allowed us to identify putative orthologs and paralogs for known genes, as well as those that exist only as uncharacterized ESTs and to provide links to additional information including genome sequence and mapping data. TOGA provides an important new resource for the analysis of gene function in eukaryotes. In addition, an analysis of the most widely represented sequences can begin to provide insight into eukaryotic biological processes.
The underlying goal of the Human Genome Project is the identification and functional characterization of the entire catalog of human genes. With the available complete sequences or comprehensive drafts of several eukaryotic genomes including Saccharomyces cerevisiae (Goffeau et al. 1996), Caenorhabditis elegans (The C. elegans sequencing consortium 1998), Drosophila melanogaster (Adams et al. 2000), Arabidopsis thaliana (The Arabidopsis genome initiative 2000), and human (International human genome sequencing consortium 2001; Venter et al. 2001), our ability to identify genes and analyze their functions and interactions through cross-species comparisons is improving rapidly. Nevertheless, identification and classification of gene sequences remains a significant challenge because of the lack of experimental evidence and the apparent shortcomings of the available gene prediction programs (Guigo et al. 2000). Of the estimated 35,000–60,000 human genes (Crollius 2000; Ewing and Green 2000; Liang et al. 2000a), fewer than 10,000 are represented by functionally characterized mRNA sequences in GenBank. Although many newly discovered genes might reveal their functions through disease-related studies, classifying the entire collection will require the analysis of related genes in experimentally tractable organisms. For most other eukaryotic species, the number of available gene sequences is more limited, and for many, the generation of complete genomic sequence data is not likely in the near future. However, there exist more than 7,000,000 publicly available expressed sequence tag (EST) sequences in dbEST, representing a wide diversity of eukaryotic species. Using a compact representation of those sequences within the TIGR Gene Index (TGI) databases (Liang et al. 2000b; Quackenbush et al. 2001), we created TOGA, the TIGR orthologous gene alignments, as a tool to explore genes and their relationships across species.
Cross-referencing the available genomic data has several important applications, including the identification of homologous genes in eukaryotes. Gene homologs can be separated into two classes, orthologs and paralogs (Fitch 1970; Gogarten and Olendzenski 1999; Eisen 1998). Orthologs are genes that are related by direct evolutionary descent whereas paralogs are homologous genes that are the result of a duplication event within the same lineage. The identification of orthologs is particularly important because these genes should play similar developmental or physiological roles, and consequently, their study in rodent or other models can provide insight into their functions in humans.
Although such an analysis has been performed for the completed microbial genomes and yeast (Tatusov et al. 1997, 2000), the lack of a comprehensive set of coding genes in many representative organisms has hampered the development of a similar resource for eukaryotes. For the completed C. elegans and Drosophila genomes, comparisons with the available gene sequence data revealed 2758 human–fly orthologs and 2031 human–worm orthologs, respectively, of which 1523 orthologs were common to both groups (Venter et al. 2001). The most extensive survey of orthologs in mammals is a study by Makałowski and Boguski in which they analyzed 1880 human–rodent ortholog pairs (Makałowski and Boguski 1998); 1212 rat–human pairs, 1138 mouse–human pairs, and 470 genes shared by all three species. As might be expected, both amino acid sequences and their corresponding DNA coding sequences were found to be highly conserved. More surprising is the high degree of conservation of the untranslated regions (UTRs) flanking the coding sequence: 71.0 ± 12.2% identity for mouse–human orthologs, 70.1 ± 11.4% for rat–human orthologs, and 86.3 ± 8.9% for mouse–rat orthologs.
It is this high degree of sequence conservation in the UTRs, in combination with the wealth of partial gene sequence data available through EST projects, that lead us to believe that orthologs could be identified through DNA-based sequence comparisons. Whereas more than 8,000,000 EST sequences made the necessary pair-wise comparisons a computationally and logistically daunting task, the TGI (Liang et al. 2000a; Quackenbush et al. 2001) databases, which assemble gene and EST sequences into tentative consensus (TC) sequences, make assembling a database of orthologs spanning many species feasible.
There are presently 28 species represented in the TGI (Table 1), including five mammals, 10 plants, seven eukaryotic parasites, and six other model organisms. These databases are updated every 3–6 months depending on availability of newly generated EST and gene sequence data and can be accessed at http://www.tigr.org/tdb/tgi.shtml. In total, there are 328,337 TCs, 1,211,636 singleton ESTs, and 46,511 singleton ETs (expressed transcript, or gene sequences) represented in the various TGI. It is our long-term goal to represent the full set of gene transcripts for an increasing number of organisms; these databases serve as our starting point for ortholog identification.
Table 1.
Summary Statistics for Inclusion of TC and sET Sequences in TOGA for Each of the 28 Species-Specific TGI Databases Represented
| Organisms | GI | TC | sET | In TOGA | ||
|---|---|---|---|---|---|---|
| Mammal | Bos taurus (cattle) | BtGI | 16,740 | 606 | 8707 | 50.2% |
| (5) | Homo sapiens (human) | HGI | 83,892 | 6313 | 14,298 | 15.8% |
| Mus musculus (mouse) | MGI | 56,343 | 6441 | 14,152 | 22.9% | |
| Rattus norvegicus (rat) | RGI | 24,221 | 1355 | 12,052 | 47.1% | |
| Sus scrofa (pig) | SsGI | 8682 | 550 | 5490 | 59.5% | |
| Plant | Arabidopsis thaliana | AtGI | 17,163 | 9571 | 7504 | 28.1% |
| (10) | Lycopersicon esculentum (tomato) | LeGI | 11,263 | 160 | 5724 | 50.1% |
| Mesembryanthemum crystallinum | IpGI | 1381 | 0 | 979 | 70.9% | |
| Solanum tuberrosum (potato) | StGI | 3169 | 143 | 2183 | 65.9% | |
| Oryza sativa (rice) | OsGI | 8596 | 1653 | 4185 | 40.8% | |
| Glycine max (soybean) | GmGI | 14,762 | 156 | 6271 | 42.0% | |
| Zea mays (maize) | ZmGI | 9333 | 267 | 4513 | 47.0% | |
| Medicago truncatula (Medicago) | MtGI | 10,160 | 23 | 5035 | 49.4% | |
| Triticum aestivum (wheat) | TaGI | 5353 | 185 | 3231 | 58.3% | |
| Sorghum bicolor (Sorghum) | SbGI | 6252 | 71 | 3247 | 51.4% | |
| Parasite | Leishmania spp | LshGI | 381 | 694 | 266 | 24.7% |
| (7) | Trypanosoma cruzi | TcGI | 1551 | 0 | 212 | 13.7% |
| Trypanosoma brucei | TbGI | 522 | 244 | 204 | 26.6% | |
| Schistosoma mansoni | SmGI | 1525 | 57 | 393 | 24.8% | |
| Plasmodium falciparum | PfGI | 375 | 949 | 256 | 19.3% | |
| Brugia malayi | BmGI | 1735 | 26 | 521 | 29.3% | |
| Onchocerca volvulus | OvGI | 875 | 24 | 366 | 40.7% | |
| Other | Drosophila melanogaster (fly) | DGI | 10,476 | 1577 | 3538 | 29.4% |
| model | Danio rerio (zebrafish) | ZGI | 9281 | 342 | 3600 | 37.4% |
| Species | Caenorhabditis elegans | CeGI | 10,264 | 9907 | 2910 | 14.4% |
| (6) | Saccharomyces cerevisiae (yeast) | ScGI | 4028 | 1701 | 1329 | 23.2% |
| Schizosaccharomyces pombe | SpGI | 2363 | 2668 | 1462 | 29.1% | |
| Xenopus laevis (frog) | XGI | 7651 | 828 | 3785 | 44.6% | |
| Total | 28 | 328,337 | 46,511 | 116,413 | 31.1% | |
GI, Gene Index; TC, tentative consensus; sET, singleton expressed transcript; TOGA, TIGR Orthologous Gene Alignment.
RESULTS
Determination of Criteria for Ortholog Identification
Orthologs are strictly defined as genes that predate speciation and have retained their function through evolutionary history. They generally are identified using a combination of protein sequence and functional information. As our goal was to identify orthologs using DNA rather than protein sequences, we wanted to be very conservative in developing criteria for ortholog identification. Because orthologs are generally well conserved at the protein sequence level, we suspected that they should be well enough conserved at the DNA level that they could be identified by requiring reflexive, high-stringency, transitive sequence matches across three or more species; the process we used is shown schematically for three species in Figure 1.
Figure 1.
Schematic overview of the procedure used to build TIGR Orthologous Gene Alignment (TOGA). The tentative consensus (TC) sequences from the 28 TIGR Gene Index databases are searched against each other. Transitive, reflexive best matches linking three (or more) species define a tentative ortholog group (TOG). Additional nonreciprocal best matches define tentative paralogs.
The TCs and ETs from each species were compared pair wise with those from each of the 27 other species. Tentative ortholog groups (TOGs) were identified requiring transitive, reflexive best hits across at least three species with a maximum of BLASTN E-value of 10−5. This initial clustering created 87,740 TOGs, although in many instances, sequences appeared in multiple TOGs. There are several reasons this can occur. First, because of the partial sequence nature of available EST data, nonoverlapping sequence segments in some species might represent a single gene, and these segments might independently appear as best matches in a species-dependent fashion. Furthermore, by including rather primitive eukaryotes such as yeast, some orthologs and paralogs may be mixed during the clustering process. To address this potential problem, we further grouped clusters in which more than two-thirds of the sequence elements in one overlapped with another, reducing the set to 32,652 TOGs, which represent the current version of TOGA. These contain a total of 116,413 sequences from 28 species, with average pair-wise matches of 71% identity over 636 bases, and a median E-value is 6 × 10−47. Of all the 334,808 best-match pairs represented in TOGA, 297,663 (89%) are reciprocal best matches, suggesting that most sequences in each TOG are likely orthologs rather than paralogs. The total representation of each species in TOGA is listed in Table 2; the assembly process used for the creation of the TGI and TOGA is shown schematically in Figure 2.
Table 2.
Sequence Representation within Various Size Clusters for TOGA 3.0
| Cluster size (sequences) | Number of clusters | Total number of sequences | Clusters with both orthologs and paralogs |
|---|---|---|---|
| 3 | 10,943 | 32,829 | — |
| 4 | 7250 | 29,000 | — |
| 5 | 5103 | 25,515 | — |
| 6 | 2959 | 17,754 | 167 |
| 7 | 1832 | 12,824 | 193 |
| 8 | 1183 | 9464 | 191 |
| 9 | 782 | 7038 | 196 |
| 10 | 541 | 5410 | 142 |
| 11 | 388 | 4268 | 117 |
| 12 | 321 | 3852 | 95 |
| 13 | 259 | 3367 | 102 |
| 14 | 210 | 2940 | 79 |
| 15 | 150 | 2250 | 75 |
| 16 | 127 | 2032 | 65 |
| 17 | 118 | 2006 | 68 |
| 18 | 83 | 1494 | 59 |
| 19 | 56 | 1064 | 45 |
| 20 | 50 | 1000 | 43 |
| 21 | 49 | 1029 | 44 |
| 22 | 37 | 814 | 36 |
| 23 | 33 | 759 | 30 |
| 24 | 28 | 672 | 25 |
| 25 | 35 | 875 | 34 |
| 26 | 20 | 520 | 20 |
| 27 | 17 | 459 | 17 |
| 28 | 16 | 448 | 16 |
| 29 | 10 | 290 | 10 |
| 30 | 10 | 300 | 10 |
| 31 | 9 | 279 | 9 |
| 32 | 12 | 384 | 12 |
| 33 | 4 | 132 | 4 |
| 34 | 5 | 170 | 5 |
| 35 | 5 | 175 | 5 |
| 36 | 2 | 72 | 2 |
| 37 | 1 | 37 | 1 |
| 40 | 2 | 80 | 2 |
| 41 | 1 | 41 | 1 |
| 43 | 1 | 43 | 1 |
| Total | 32,652 | 116,413 | 1921 |
Clusters containing multiple sequences from a single species are considered to contain both orthologs and paralogs.
Figure 2.
Conceptual overview of information flow and the database construction process for the TIGR gene index (TGI) databases and TIGR Orthologous Gene Alignment (TOGA). For each species included in the TGI, expressed sequence tag (EST) and gene sequence data are downloaded from public sources and cleaned to remove low-quality and contaminating sequences. These are clustered at high-stringency, individual clusters assembled using CAP3 to construct the tentative consensus (TC) sequences that comprise the individual species-specific TGI databases, annotated, and released through the TGI Web site. Finally, TOGA is assembled using pair-wise comparisons and the reflexive transitive best hit process described in the text.
TOGA is a relational database that maintains the TOGs as accessionable objects that can be tracked across subsequent releases. TOGs can be searched either using a name-based search that allows users to enter a gene name and look for approximate matches or using WU-BLAST (Altschul et al. 1990; http://blast.wustl.edu) to search the data set. TOGA 3.0 can be found at http://www.tigr.org/tdb/toga/toga.shtml. TOGA reports include a graphical representation of the relationships between the component sequences, a table with summary statistics for each of the pair-wise alignments, and a multiple sequence alignment produced using CLUSTALW (Thompson et al. 1994).
Core Processes Shared by Eukaryotes
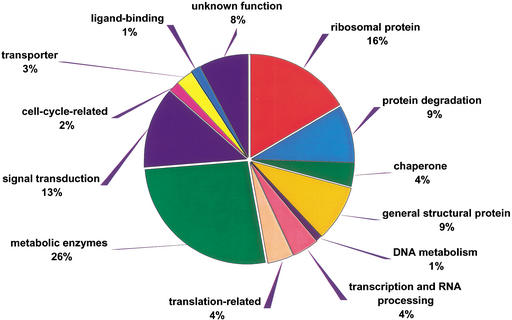
The broad representation of species within TOGA provides a unique opportunity to analyze both gene diversity and the conservation and provides a glimpse of biological processes fundamental to eukaryotes. We analyzed the 1091 TOGs containing a minimum of 14 sequences (those containing half or more of the species represented in TOGA) using the gene ontology (GO) (The Gene Ontology Consortium 2000) terms assigned to the TCs during assembly of the individual TGI (Fig. 3). Not surprisingly, metabolic enzymes represent the most extensive group of highly conserved proteins, representing 26% of the total. These include genes involved in the general carbohydrate, amino acid, nucleotide and lipid metabolism, and energy-producing ATPases. It is not surprising that ribosomal proteins (16%) and other structural proteins such as actin, tubulin, histone, cytochrome, and cyclophilin (9%) are highly represented as well.
Figure 3.
Functional role assignments based on gene ontology (GO) terms for the 1091 most highly represented proteins in TIGR Orthologous Gene Alignment (TOGA), each of which contains sequence from 14 or more species.
Proteins involved in signal transduction (Reith 2001), including GTP-binding proteins, protein kinase and phosphatase, and other protein-modifying enzymes, as well as 14-3-3 protein (Burbelo and Hall 1995), are more highly represented (13% of the total) than are receptors. This suggests, as one might expect, that much of the core signal processing machinery is maintained through evolution whereas the genes involved in receiving signals have evolved to meet each organism's particular needs.
Processing of genetic information is also well conserved, with genes involved in transcription and translation (Hampsey 1998; Ibba and Soll 1999; Squires and Zaporojets 2000) each representing 4% of the total. Representative genes include DNA helicase, RNA helicase, the general transcription factors such as TBP, RNA polymerase, poly(A)-binding protein, small nuclear ribonucleoprotein, splicing factors, translation initiation factors, and elongation factors; elongation factor-1α is the most highly conserved sequence, with 43 sequences from the 28 species appearing in a single ortholog group (TOG14405).
Surprisingly, genes encoding components of the protein degradation pathway (Hochstrasser 1995) are among the most highly conserved (9% of the total), including proteasome subunits containing both ATPase and non-ATPase regulatory subunits, various peptidase, and ubiquitin, polyubiquitin, ubiquitin fusion proteins, and ubiquitin conjugating enzymes. Although protein degradation is rarely considered as important as protein synthesis, the high degree of sequence conservation across diverse species suggest that protein degradation is fundamental to the survival of eukaryotic cells.
Molecular chaperones (Smith et al. 1998), which are involved in the stress response and eventual defense of the living cells, represent another group of highly conserved proteins (4%). These include heat shock proteins (HSP70–90), T-complex proteins (TCPs), DNA J, and other stress-induced proteins. The other comparably smaller groups of conserved proteins include proteins involved in cell cycle, ligand-binding proteins, and transporters.
Finally, many conserved sequences in eukaryotes are annotated as unknown proteins, including those found only through anonymous EST sequencing projects and those predicted by purely computational methods. Although the existence of these genes and their functional classification would require additional laboratory analysis, conservation across multiple, often distantly related species of these expressed sequences in TOGA (9%) provides strong evidence that these do, in fact, represent real protein coding genes and imply that they may, in fact, represent genes playing crucial cellular roles. Thus, TOGA, as well as similar ortholog analysis, can provide a means of prioritizing newly discovered genes for further study.
Orthologous Genes in the Biochemical Pathways
Regardless of biological function, genes and their protein products do not operate in isolation, but rather as components of complex biological and biochemical pathways (Kanehisa and Goto 2000). The study of these pathways in a variety of organisms has contributed significantly to understanding the roles played by each of the participating genes. As an initial effort to use TOGA to reconstruct orthologous pathways, we identified the TOGs representing genes important in cell cycle control, focusing on p53 and the Rb tumor suppressor (Fig. 4). Our analysis is based on a simplified model of cell cycle control representing the core biochemical processes involved; more complete analyses of the genes involved in the process can be found elsewhere (Kohn 1999; Sherr 2000; Harbour and Dean 2000).
Figure 4.
Representation of the cell cycle control pathway within TIGR Orthologous Gene Alignment (TOGA). The regulatory molecules in the cell cycle are shown in green boxes together with the associated TOGA accession numbers (in italics). When there are multiple proteins for a single element of the pathway, the corresponding TOGA accessions are listed in the adjacent blue boxes (linked by pink bidirectional arrows). Genes associated with human diseases are shown in red.
In general, cell cycle progression is controlled by cyclin–CDK complex containing the regulatory cyclin and catalytic kinase, which processes the mitogenic signals through RAS signal transduction pathway. In response to extracellular mitogens, cyclin D–dependent kinase (CDK4 and CDK6) initiate phosphorylation of Rb. This leads to the end of Rb-mediated repression and activated expression of the S-phase genes including genes for DNA replication, DNA synthesis, and cell cycle regulators through the pathway of E2F/DP (Dyson 1998; Harbour and Dean 2000); phosphorylation of Rb is completed by cyclin E–CDK2. Completion of S-phase progression also requires the activity of cyclin A–CDK2 and cyclin B1-CDC2; CDK1 activity is required for G2/M transition. Two types of inhibitors inhibit CDK activity: INK4A-D and Cip/Kip. Whereas INK4A-D inhibits cyclin D–CDK4/6, Cip/Kip inhibits both cyclin E–CDK2 and cyclin A–CDK2 (Sherr and Roberts 1999). Recent work indicates that Cip/Kip may act as the positive regulator for cyclin D–CDK activity (Sherr 2000). The normal cell cycle is disrupted and growth arrest occurs when DNA damage is signaled through the ATM/ABL pathway by the tumor suppressor p53 (Levine 1997; Taylor and Stark 2001). When the DNA damage is severe, apoptosis is induced by p53-mediated activation of BAX and BCL (Vousden 2000); the CDK inhibitor p21Cip1 and GADD45 are transcriptionally up-regulated by p53 (el-Deiry et al. 1993; Kastan et al. 1992).
Most of these cell cycle regulators are present in TOGA, reflecting the high degree of conservation of the general cell cycle regulation pathway. Although the cyclins and CDKs are well conserved across plants, animals, and fungi, many of the cell cycle inhibitors (Cip/Kip, CDK4A-D) are detected only within animal lineage. These inhibitors function downstream of p53 or are induced by TGF-β, and these pathways previously have been detected only in animal systems. This suggests that plants and fungi possess different controlling mechanisms for these aspects of the cell cycle checkpoint pathway.
Consistent with the previous literature, TOGA only has orthologs of p53 represented within the vertebrates and the Rb tumor suppressor protein within the mammals. Although it has been asserted that Drosophila homologs of p53 exist (Ollmann et al. 2000), the nucleotide similarity is clearly above the 10−5 E-value threshold used to construct TOGA. Homologs for Rb tumor suppressor protein have been claimed in plants (Durfee et al. 2000). However, we were able to identify only human, mouse, rat Rb orthologs, implying the sequence similarity at the nucleotide level is not sufficient for TOGA to identify any orthologs of Rb.
TOGA also was able to identify E2F/DP family proteins, which play multiple roles in cell cycle control, including regulation of S-phase progression, apoptosis induction, and even tumor suppression. Although homologs of these have been reported previously in plants (Magyar et al. 2000), our analysis suggests that many of the downstream targets are absent and that therefore the biological processes in plants are clearly distinct. An ortholog of the newly discovered protein from β-transcripts of INK4A, p14-ARF, which connects the Rb and p53 pathways, was previously found only in mouse (as p19-ARF; Sherr 2000), and its failure to be identified by TOGA suggests that it is not yet represented in mammalian EST libraries.
In summary, even though the cyclin–CDK complexes are well conserved and function as the cell cycle driver in animals, plants, and fungi, the other major regulators, specially the tumor suppressor proteins, are only shared within the animals, or more strictly within the vertebrate and mammalian lineages.
Homologs of Human Disease Gene in Eukaryotes
One of the goals of the human genome project has been to identify and characterize the full catalog of human disease genes. One component of that effort has been the search for orthologous genes in model organisms (Bassett et al. 1996; Mushegian et al. 1997; Rubin et al. 2000; Fortini et al. 2001), in which they can be characterized more easily through classic functional analysis. We attempted to identify homologs of human disease genes in eukaryotic organisms represented in TOGA using a slightly modified list of 288 human disease genes previously compiled for a survey of the predicted gene in the Drosophila genome (Rubin et al. 2000; Fortini et al. 2001). The TOGs representing these genes were identified through a combination of TBLASTN and gene name searches against the TIGR Human Gene Index and manual analysis and curation of the identified sequences using the corresponding OMIM (http://www.ncbi.nlm.nih.gov/omim) and GenBank records.
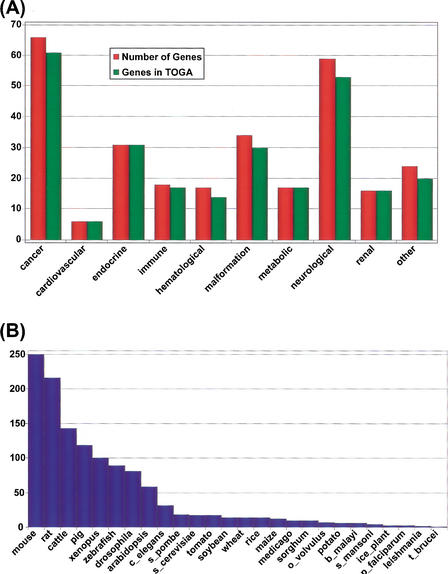
Of the 288 disease genes analyzed for Drosophila, we were able to identify 265 with orthologs in TOGA (Fig. 5A). In many instances, this is the first time the homologs of these genes have been identified in many of the organisms represented in TOGA (Fig. 5B). As expected, mouse and rat, which have been used extensively as models of human disease, have the greatest representation of human disease genes, with 250 and 216, respectively, and at least one homolog from both mouse and rat were identified for 203 human disease genes. Representation in other species is influenced by two factors: their evolutionary distance from human and the degree of sequence sampling by EST and genome sequencing projects. As one might expect, the greatest number of nonhuman homologs was identified from the evolutionarily close species; vertebrates have significantly more homologs than other species, and most animals are more highly represented than plants. However, sequence sampling also plays a role in gene identification. For example, within the mammals, more human homologs were identified in mouse and rat than in the more closely related cattle and pig largely because of the more limited EST sequencing projects in these nonrodent species. Furthermore, Arabidopsis, the only plant with a complete genome sequence, has more human disease homologs than any other plant.
Figure 5.
Human disease genes in TIGR Orthologous Gene Alignment (TOGA). A list of 288 human disease genes45,46, with the redundant entry INK4 removed, were mapped to the TIGR Human Gene Index, which was used to identify the corresponding TOGA accession. (A) Representation of these genes within each of 10 disease categories, and their corresponding representation within TOGA. (B) Representation of these same disease genes by orthologs in various model organisms.
Some of the homologs for the human disease genes, specifically those involved in the cell cycle control including ras, myc, cyclin D, CDK4, Rb, INK4A, INK4C, Bcl, ATM, Abl, and p53 are shown in Figure 4. Most of these genes are either oncogenes or tumor suppressor genes. Studies on these homologs in the model species would greatly accelerate our understanding of the related disease.
Disease genes not found in TOGA include five cancer-related genes, one involved in hematological disease, three related to immune disorders, four involved in malformation syndromes, six neurological disorder genes, and four others. It is not our intention to conclude that these genes do not have the homologs in other eukaryotic organisms. Rather, many of these genes simply could not be identified because of the stringency imposed by requiring transitive, reciprocal best matches across three or more species, and it is likely that the remainder will be identified as more sequence data become available. Indeed, we were able to find a single nonhuman reciprocal best match for all the missing human disease genes in TOGA (generally in mouse, which has the greatest EST sequence representation). Finally, we identified only 81 disease gene homologs in Drosophila, fewer than half of that found in the analysis of fly genome (Rubin et al. 2000; Fortini et al. 2001). Although many of the identified genes do have good reciprocal best matches with human genes, they failed to meet the stringent criteria used to assemble TOGA, suggesting that they may have been misidentified previously or that our DNA-based criteria for finding orthologs fails for such highly diverged sequences and that protein-based approaches would be more appropriate in this case. Details of this analysis including the OMIM entries, gene name, gi number, HGI accession number, TOG accession number, and the identified TCs from each species are available at http://www.tigr.org/tdb/toga/human_disease.shtml.
Phylogenetic Analysis of Orthologous Eukaryotic Genes
Molecular sequences have surpassed morphological and structural characters for use in phylogenetic studies. TOGA represents the most extensive collection of eukaryotic gene sequences and provides a unique resource for a detailed investigation of evolutionary relationships. Indeed, for each TOG, one can use the sequence alignments produced by CLUSTALW as the basis for constructing a phylogenetic tree. However, trees based on individual genes may well be biased and multiple sequences are required for a more complete analysis of the genetic relationships between organisms (Nei et al. 2001).
Using TOGA, we selected TOGs representing multiple species and performed such an analysis. Rather than generating separate trees and averaging over them, we collected TOGs for the species of interest. Within each group, the sequences were individually aligned using CLUSTALW and trimmed to include only the overlapping region defined as that between the first and last consensus site across all represented species. Trimmed sequences were concatenated and used as input for PAUP, and trees were constructed using the distance, maximum parsimony, and maximum likelihood methods (Eisen 1998).
Figure 6A shows the results for the 21 nonparasitic eukaryotic species using 52 orthologous sequences spanning nearly 21,500 nucleotides. As one would expect, there are three main branches, differentiating the plants (10 species) from the fungi (two species) and the metazoa (nine species). Within the metazoa, the vertebrates form a distinct subgroup, with mammals further differentiated from others. Within the plants, the monocots are clearly separated from the dicots, which show greater sequence divergence. To validate these results, we performed a further analysis for the plants using 236 sequences spanning more than 122 kb and metazoa with 242 sequences spanning 118 kb (Figs. 6B,C). These results provide confirmation of our underlying assumption in the construction of the TGI: that EST sequences can be accurately assembled to produce TC sequences representing actual genes and that these TCs can provide valuable data for functional annotation and analysis in species that are not yet well characterized.
Figure 6.
Phylogenetic analysis of conserved genes in TOGA. Sequences were collected from TIGR Orthologous Gene Alignment (TOGA) representing (A) plants, fungi, and metazoa, (B) plants only, and (C) metazoa only, trimmed to their matching regions, concatenated, aligned, and used to construct phylogenetic trees. As expected, the alignments derived from TOGA allow a faithful reconstruction of the known evolutionary relationships between these species.
DISCUSSION
Efforts to catalog the collection of eukaryotic genes are progressing rapidly. Although both public and private efforts have provided a nearly complete draft human sequence and greatly accelerated the pace of genomic sequencing projects in other model species, the annotation of the genome, including the identification of gene sequences, remains a significant challenge. Comparative genomics will play a crucial role in the further analysis of genes and gene function, as well as in the identification of noncoding regulatory regions. The genes encoded within each genome provide a natural index that will allow those genomes to be cross-referenced. Although such analysis is clearly best conducted using the sequences of completed genomes and comprehensive lists of the genes and proteins they encode, for many species such data may not be available for quite some time, if ever. However, there exists an extensive body of EST and gene sequence data that can be used to search for orthologous genes.
The TGIs use the available EST and gene sequence data in GenBank and reduce the more than 8,000,000 sequences to significantly fewer, high-quality consensus sequences. Through comparison of the TC sequences comprising Gene Index databases constructed for 28 species, we have been able to identify more than 32,000 tentative orthologs containing sequences from three or more species. These have been organized in the TIGR Orthologous Gene Alignment database. TOGA represents TOGs as accessionable objects in a database that allows rapid navigation between the TGIs, and through them, to mapping and genome sequence data and other data. As more genomic and EST sequence data become available, we will expand our catalog of orthologs using both DNA sequences and the proteins they encode. TOGA provides a framework in which the data can be organized and TOGA will continue to develop and expand to meet the challenges presented by this increased data.
Identification of orthologs and paralogs using automated methods and only DNA sequences is clearly a difficult task. Indeed, in our attempts to provide a broad representation of eukaryotes and to be comprehensive in our analysis, we have clearly mixed some orthologs and paralogs into TOGs. This is due to several factors, including the incomplete sampling of the genes that are provided by existing data and our use of S. cerevisiae and S. pombe and other eukaryotes that predate many gene duplication events in higher animals and plants. Even so, our we believe that TOGA represents the best representation of orthologs given the available data.
Our analysis of the TOGs presented here represents an attempt to show some potential applications of the TOGA database. Ultimately, one should consider the relationships revealed through this analysis as a set of hypotheses that can be tested. Furthermore, one should be cautious of negative results or concluding that there are missing genes in some species. Clearly, much of what we observe reflects the degree of coverage of the complete genomes and transcriptomes of the organisms surveyed. Nevertheless, our analysis provides some measure of confidence in the relationships that TOGA represents and suggests that many of the TOGs contain true orthologs.
METHODS
Assembly of TIGR Gene Index (TGI) Databases
The TIGR Gene Indices (TGIs) including all 28 used in this study were separately assembled using methods described previously (Liang et al. 2000a; Quackenbush et al. 2001). Briefly, EST sequences and coding gene sequences were downloaded from dbEST and GenBank records. Sequences were trimmed to remove vector, poly-A/T tails, adaptor sequences, and contaminating bacterial sequences. We also included additional curated expressed transcript (ET) sequences from the EGAD, a curated database of nonredundant transcript sequences maintained at TIGR (http://www.tigr.org/tdb/egad/egad.html). Sequences were compared using BLAST; those sharing ≥95% identity over regions at least 40 base pairs in length, with unmatched overhangs less than 20 base pairs were placed into clusters. The sequences comprising each cluster were assembled using CAP3 (Huang and Madan 1999) to produce TCs (Tentative Human Consensus sequences [THCs] in humans). A TC containing a known gene was assigned the function of that gene; TCs without assigned functions were searched using DPS (Huang et al. 1997) against a nonredundant protein database; high-scoring hits were used to assign a putative function.
Identification of Tentative Orthologue Groups (TOGs)
Tentative Consensus sequences (TCs) and the singleton Expressed Transcripts (sETs) from each of the TIGR Gene Indices were separately searched against the TCs and sETs comprising the other Gene Indices using WU-BLAST (Altschul et al. 1990; http://blast.wustl.edu), and the best hit for each sequence was recorded. Matches meeting or exceeding an established maximum BLASTN E-value of 10−5 were stored in TOGA, a relational database, implemented in SYBASE, designed to capture relationships between orthologous and paralogous genes. The results of these searches were used to identify reciprocal best hit pairs. A reciprocal best hit pair is defined as a pair of TCs in separate species in which the first member of the pair has as its best hit in the second species, the second member and the second member has as its best hit in the first species the first member. Tentative Orthologue Groups (TOGs) were constructed by selecting reciprocal best hit pairs that link TCs in three or more species. Tentative paralogs to these groups were identified as TCs that were not a member of a reciprocal best hit pair, but whose best hit was a TC contained within an existing TOG. Multiple alignments of each TOG, both with and without the paralog sequences, were performed using CLUSTALW (Thompson et al. 1994) and are displayed at http://www.tigr.org/tdb/toga/toga.shtml with links to the individual TC reports; alignments also can be viewed using JALVIEW (M. Camp, unpubl., see http://www2.ebi.ac.uk/∼michele/jalview).
Analysis of Human Disease Genes
Based on the previously published list of human disease-related genes (Rubin et al. 2000; Fortini et al. 2001), the associated GenPept and OMIM records were assembled. TBLASTN was used to search the available protein sequences against the human TCs represented in TOGA, and the results were manually curated. When sequence-based matches could not be found, the OMIM records were reviewed, and new DNA or protein sequences were selected to provide the broadest possible representation of this gene set; orthologs were identified in TOGA using the human TC accessions.
Phylogenetic Analysis
We began phylogenetic analysis by first dividing the species represented in the TGI into five groups: plants, consisting of the 10 plant species; mammals, containing human, mouse, rat, cattle, and pig; vertebrates, including the mammals plus frog and zebrafish; higher animals, consisting of the vertebrates plus fly and nematode; higher eukaryotes, including plants, higher animals, and the two yeast species. TOGs were classified by their representation of all of the species within each group; when more than one sequence from a single species was represented in a TOG, a single representative from that species was selected using a simple voting scheme based on the number of the best reciprocal hits within that cluster. Sequences from each of the represented species were extracted from the appropriate TGI database, and CLUSTALW was used to align the relevant sequences. Regions matching across all species were identified and extracted from these alignments, and all aligned sequences for a particular group were concatenated and loaded into PAUP, which was used to construct phylogenetic trees using the distance, maximum likelihood, maximum parsimony, and bootstrapping methods with default settings.
WEB SITE REFERENCES
http://blast.wustl.edu, WU-BLAST Home Page.
http://www.ncbi.nlm.nih.gov/omim, Online Mendelian Inheritance in Man.
http://www.tigr.org/tdb/egad/egad.html, The TIGR EGAD Database.
http://www.tigr.org/tdb/tgi.shtml, The TIGR Gene Index Databases.
http://www.tigr.org/tdb/toga/human_disease.shtml, Disease Gene Assignments in TOGA.
http://www.tigr.org/tdb/toga/toga.shtml, The TOGA Database.
http://www2.ebi.ac.uk/∼michele/jalview, Jalview Sequence Alignment Viewer.
Acknowledgments
The authors are indebted to Michael Heaney and Susan Lo for database support, and Vadim Sapiro, Billy Lee, Sonja Gregory, Rajeev Karamchedu, Jeff Shao, Corey Irwin, Jacqueline Neubrech, and Eddy Arnold for computer system support, and Norman Lee and Jonathan Eisen for thoughtful comments and suggestions. This work was supported by grants from the U.S. Department of Energy and the National Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: johnq@tigr.org; FAX: (301) 838-0208.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.212002.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bassett DE, Boguski MS, Hieter P. Yeast genes and human disease. Nature. 1996;379:589–590. doi: 10.1038/379589a0. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Hall A. 14–3–3 Proteins, hot numbers in signal transduction. Curr Biol. 1995;5:95–96. doi: 10.1016/s0960-9822(95)00022-4. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Crollius HR. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat Genet. 2000;25:235–237. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Durfee T, Feiler HS, Gruissem W. Retinoblastoma-related proteins in plants: Homologues or orthologues of their metazoan counterparts? Plant Mol Biol. 2000;43:635–642. doi: 10.1023/a:1006426808185. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: Improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;25:232–234. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Fortini ME, Skupski MP, Bokuski MS, Hariharan IK. A survey of human disease gene counterparts in the Drosophila genome. J Cell Biol. 2001;150:F23–F29. doi: 10.1083/jcb.150.2.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–31. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Olendzenski L. Orthologs, paralogs and genome comparisons. Curr Opin Genet Dev. 1999;9:630–636. doi: 10.1016/s0959-437x(99)00029-5. [DOI] [PubMed] [Google Scholar]
- Guigo R, Agarwal P, Abril JF, Burset M, Fickett JW. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 2000;10:1631–1642. doi: 10.1101/gr.122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR. A tool for analyzing and annotating genomic sequences. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kohn KW. Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol Biol Cell. 1999;10:2703–2734. doi: 10.1091/mbc.10.8.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J. An optimized protocol for analysis of EST sequences. Nucleic Acids Res. 2000a;28:3657–3665. doi: 10.1093/nar/28.18.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J. Gene index analysis of the human genome estimates approximately 120,000 genes. Nat Genet. 2000b;25:239–240. doi: 10.1038/76126. ; Erratum, Nature Genet. 26: 501. [DOI] [PubMed] [Google Scholar]
- Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inze D. Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett. 2000;486:79–87. doi: 10.1016/s0014-5793(00)02238-9. [DOI] [PubMed] [Google Scholar]
- Makałowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Bassett DE, Jr, Bokuski MS, Bork P, Koonin EV. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Prot Natl Acad Sci. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc Natl Acad Sci. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. Drosophila p53 is a structure and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR gene indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acid Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith AD. Protein kinase-mediated signaling networks: Regulation and functional characterization. Methods Mol Biol. 2001;124:1–20. doi: 10.1385/1-59259-059-4:1. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The Pezcoller lecture: Cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Smith DF, Whitesell L, Katsanis E. Molecular chaperones: Biology and prospects for pharmacological intervention. Pharmacol Rev. 1998;50:493–513. [PubMed] [Google Scholar]
- Squires CL, Zaporojets D. Proteins shared by the transcription and translation machine. Annu Rev Microbiol. 2000;54:775–798. doi: 10.1146/annurev.micro.54.1.775. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vousden KH. p53: Death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]