Abstract
The secreton or type II secretion machinery of gram-negative bacteria includes several type IV pilin-like proteins (the pseudopilins) that are absolutely required for secretion. We previously reported the presence of a bundled pilus composed of the pseudopilin PulG on the surface of agar-grown Escherichia coli K-12 cells expressing the Klebsiella oxytoca pullulanase (Pul) secreton genes at high levels (N. Sauvonnet, G. Vignon, A. P. Pugsley, and P. Gounon, EMBO J. 19:2221-2228, 2000). We show here that PulG is the only pseudopilin in purified pili and that the phenomenon is not restricted to the Pul secreton reconstituted in E. coli or to PulG. For example, high-level expression of the endogenous E. coli gsp secreton genes caused production of bundled pili composed of the pseudopilin GspG, and the Pul secreton was able to form pili composed of PulG-like proteins from secreton systems of other bacteria. PulG derivatives in which the C terminus was extended by the addition of eight different peptides were also assembled into pili and functioned in secretion. Three of the C-terminal peptides were shown to be exposed along the entire length of the assembled pili. Hence, the C terminus of PulG may represent a permissive site for the insertion of immunogenic epitopes or other peptide sequences. One of these PulG variants, with a six-histidine tag at its C terminus, formed nonpolar, nonbundled pili, suggesting that bundle formation and polar localization are not correlated with the ability of PulG to function in secretion. We propose that the PulG pilus is an artifactual manifestation of a periplasmic “pseudopilus” and that cycles of pseudopilus extension and retraction within the periplasm propel pullulanase through secretin channels in the outer membrane. Abnormally long pili that extend beyond the outer membrane are produced only when pilus length control and retraction are deregulated by overproduction of the major pseudopilus subunit (PulG).
The secreton or type II secretion (T2S) system permits the energy-dependent secretion of a limited number of specific proteins from the periplasm in gram-negative bacteria (33). Many of the 12 or more secreton components share extensive sequence similarity with components of the type IV piliation (T4P) system in gram-negative bacteria (24, 33). In the T2S system, these “shared” components include the pilins themselves (called pseudopilins in the secreton system) and prepilin peptidase, the enzyme that removes a short N-terminal peptide and then N-methylates type IV pilin precursors (25, 29, 35, 37) and type IV pilin-like proteins. The N-terminal prepeptide and approximately 20-residue hydrophobic domains of the type IV pilins and pseudopilins are highly conserved (24, 33). In the pullulanase (PulA)-specific secreton from Klebsiella oxytoca, there are five type IV pseudopilins (PulG, PulH, Pull, PulJ, and PulK) (31, 36, 40).
In the absence of any direct evidence for the assembly of pseudopilins into a pilus (34, 38), they were proposed to form a small, intraperiplasmic structure (the pseudopilus) that constitutes an essential element of the secreton. However, we subsequently observed that agar-grown E. coli K-12 carrying all of the 14 pul secreton genes on pBR322 (pCHAP231) produced long, bundled, predominantly polar pili that reacted with antibodies against the major pseudopilin PulG (43). Thus, the relationship between the T2S and T4P systems extends beyond their similar compositions. These long pili do not appear to interfere with pullulanase secretion (43).
In this report, we examine the production of PulG pili in greater detail. In particular, we examine whether the phenomenon is restricted to the Pul secreton and to PulG, the apparently polar localization of the PulG pilus bundles, the composition of the pilus filament, the insertion of foreign epitopes at a permissive site at the C terminus of the pilin, and their exposure on the surface of the assembled filament. We also examine how the PulG pilus spans the outer membrane and the role of minor pseudopilins in its assembly.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study were derivatives of Escherichia coli K-12 strain MC4100 [F− araD139 Δ(argF-lac)U169 psL150 relA1 flbB5301 deoC1 ptsF25]. PAP7501 (MC4100 F′Tn10 lacIq1 ΔmalE44 malG510 fimAB::kan sacB) is a derivative of PAP7460 that lacks type I pili (43). PAP7502, a derivative of PAP7500 (36), is the same as PAP7501 except that it carries the entire pul gene cluster integrated in the chromosome, and PAP9001 is the same as PAP7501 except that it lacks the F′ that carries the lacIq1 repressor gene. The Klebsiella strains used were K. planticola ATCC 15050 (20) and K. oxytoca UNF5023 (10).
Plasmids carrying pulG homologues are listed in Table 1. The exeG, espG, and xpsG genes in these plasmids were first amplified from plasmids supplied by S. Peter Howard, Michael Bagdasarian, and Nien-Tai Hu, respectively, with oligonucleotides that flank the genes and that introduce unique restriction endonuclease cleavage sites at either end. These fragments were then cloned into the appropriate vector (Table 1), and the production of the corresponding pilin was examined with antibodies against PulG, GspG (Olivera Francetic), or XpsG (Nien-Tai Hu) (cross-reactions were sufficiently strong to permit the detection of all pseudopilins with these antisera). Several independent clones of each pseudopilin gene were tested. pCHAP1380 was constructed by subcloning the xcpG (xcpT) gene from pT7.T (Alain Filloux) as a 900-bp EcoRI-HindIII fragment. (The gene homologous to pulG in the xcp gene cluster of Pseudomonas aeruginosa is generally referred to as xcpT [2]. We propose that the name be changed to xcpG, in agreement with the standard nomenclature of the T2S genes.)
TABLE 1.
Plasmids used in this study
| Plasmid | Vector/origin/resistance phenotype | Cloned genes and/or mutations | Reference and/or source of cloned fragment |
|---|---|---|---|
| pCHAP231 | pBR322:ColE1/Apr | pBR322::(pulS pulA-B pulC-O) | 8 |
| pCHAP1216 | pBR322/ColE1/Apr | pCHAP231 pulB::kan-1 ΔpulG1 | 31 |
| pCHAP1226 | pBR322/ColE1/Apr | pCHAP231 pulB::kan-1 ΔpulD | 31 |
| pCHAP1324 | pBR322/ColE1/Apr | pCHAP231 pulH::kan-2 | 31 |
| pCHAP1325 | pBR322/ColE1/Apr | pCHAP231 pulK::kan-2 | 31 |
| pCHAP1357 | pBR322/ColE1/Apr | pCHAP231 pulI::kan-2 | 31 |
| pCHAP162DC3 | pBGS19/ColE1/Kmr | lacZp-pulG(K139F) | 35 |
| pCHAP162DB3 | pBGS19/ColE1/Kmr | lacZp-pulG(K139FVLVVstop) | 35 |
| pCHAP162DB5 | pBGS19/ColE1/Kmr | lacZp-pulG(K139FATKDstop) | 35 |
| pCHAP162DC5 | pBGS19/ColE1/Kmr | lacZp-pulG(K139FVMVMVMstop) | 35 |
| pCHAP1205 | pSU18/p15/Cmr | lacZp-pulG | 35 |
| pCHAP1271 | pSU18/p15/Cmr | lacZp-pulK | 31 |
| pCHAP5236 | pSU18/p15/Cmr | lacZp-pulK* | This study |
| pCHAP1328 | pSU18/p15/Cmr | lacZp-pulJ | 31 |
| pCHAP1331 | pSU18/p15/Cmr | lacZp-pulH | 31 |
| pCHAP1351 | pSU18/p15/Cmr | lacZp-pulI | 31 |
| pCHAP1362 | pSU19/p15/Cmr | lacZp-pulG::Hiso | 35 |
| pCHAP1380 | pSU19/p15/Cmr | lacZp-xcpG(T) | This study |
| pCHAP1395 | pSU18/p15/Cmr | lacZp-epsG | This study |
| pCHAP1403 | pSU18/p15/Cmr | lacZp-exeG | This study |
| pCHAP1396 | pSU19/p15/Cmr | lacZp-xpsG | This study |
| pCCP2229 | pSU18/p15/Cmr | lacZp-outGEch | 17 |
| pCCP2245 | pSU18/p15/Cmr | lacZp-outGEch | 17 |
| pCHAP4278 | pACYC184/p15/Cmr | gspAB gspC-O | 14 |
| pCHAP4010 | pUC18/ColE1/Apr | lacZp-gspG | 14 |
| pCHAP5218 | pSU18/p15/Cmr | lacZp-gspG | This study |
| pCHAP5241 | pSU19/p15/Cmr | lacZp-pulG::blaM | This study |
| pCHAP5246 | pSU19/p15/Cmr | lacZp-pulG::c-myc | This study |
| pCHAP5247 | pSU19/p15/Cmr | lacZp-pulG::FLAG | This study |
| pCHAP5248 | pSU19/p15/Cmr | lacZp-pulG::strep-tag | This study |
| pCHAP5250 | pSU19/p15/Cmr | lacZp-pulG::LCMV (118-126) | This study |
| pCHAP5252 | pSU19/p15/Cmr | lacZp-pulG::LCMV (118-132) | This study |
Bacteria were grown at 30°C in Luria-Bertani (LB) broth or on LB agar (21) with appropriate antibiotics (ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml) and, where appropriate, 0.4% maltose to induce pul gene expression or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce genes expressed from lacZp.
Epitope insertions.
We used a previously constructed mutant form of pulG carried by pCHAP162D (35) in which a DraI site was created at the end of pulG, converting a lysine residue at position 139 to phenylalanine. We designed linkers with flanking blunt (5′) and EcoRI (3′)-compatible ends and coding for c-myc, Strep-tag, lymphocytic choriomeningitis virus (LCMV) nucleoprotein, and FLAG epitopes and inserted them into the DraI and EcoRI sites of this pulG gene (the EcoRI site is downstream from pulG at the end of the 3′-truncated pulH gene in pCHAP162D). The genes were then subcloned into pSU19.
Site-directed substitution of M+5 in pulK.
pulK mutant plasmid pCHAP5236 was generated with a QuikChange site-directed mutagenesis kit (Stratagene). Oligonucleotides 5′ATCGCCCTGCTCGAGGTGCTGCTGATCCTC3′ and 5′ATCAGCAGCACCTCGAGCAGGGCGATGCC3′ were used for PCR amplification steps with a pCHAP1271 (pulK+) template. Selected clones were sequenced to confirm the presence of the correct mutation.
Immunoblotting.
Procedures for immunoblotting were essentially as used previously (43). Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in gels containing 12 or 13% acrylamide, electroblotted onto nitrocellulose membranes, and incubated first with specific antiserum [polyclonal anti-PulG at 1/6,000, anti-GspG at 1/1,000, anti-XpsG at 1/2,000, anti-XcpG(T) at 1/2,000 (Alain Filloux), anti-PulA at 1/10,000, monoclonal anti-LCMV nucleoprotein epitope at 1/1,000 (Claude Leclerc), anti-LamB at 1/5,000, monoclonal anti-His6 (Clontech), monoclonal anti-c-myc (Invitrogen), monoclonal anti-FLAG (Sigma), or polyclonal anti-Strep-tag (IBA), as recommended by the manufacturers] and then with horseradish peroxidase-coupled anti-rabbit or anti-mouse immunoglobulin G (1/1,000; Amersham). The membranes were developed by enhanced chemiluminescence (Amersham).
Pullulanase assays.
Surface-exposed pullulanase on liquid- or agar-grown cells resuspended in phosphate-buffered saline was measured in cells permeabilized in 0.5% octylpolyoxyethylene essentially as previously described (20). Secretion levels are expressed as the percentage of the enzyme activity (permeabilized cells) that could be detected in whole cells.
Detection of pili.
Bacteria were grown overnight on LB maltose agar. Immunogold labeling and transmission electron microscopy (TEM) were performed as described by Sauvonnet et al. (43), with specific antibodies diluted to 1:100 and with 10-nm gold beads on the secondary antibodies. Specimens were examined with a Philips CM12 transmission electron microscope operated under standard conditions in the 80- to 120-kV accelerating voltage range.
Purified pili were examined by TEM under low-dose conditions (10 electrons/Å2) at −700-nm defocus with a Philips CM12 transmission electron microscope operated at a 120-kV accelerating voltage.
For scanning electron microscopy (SEM) analysis, bacteria applied to poly-l-lysine-coated coverslips were immunogold labeled with anti-PulG antibodies as described above and then fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) overnight at 4°C. Cells were washed three times for 5 min (each time) in 0.2 M cacodylate buffer (pH 7.2), postfixed for 1 h in 1% (wt/vol) osmium tetroxide in 0.2 M cacodylate buffer (pH 7.2), and then rinsed with distilled water. Bacteria were dehydrated through a graded series of 25, 50, 75, and 95% ethanol solutions for 10 min (each time) and washed three times for 15 min (each time) in 100% ethanol. Dehydrated cells were consecutively immersed in 25, 50, 75, and 75% (vol/vol) hexamethyldisilazane in ethanol for 5 min (each time), immersed twice in 100% hexamethyldisilazane for 5 min (each time), and quickly air dried. Coverslips were sputter coated twice with carbon, and samples were examined with a JEOL JSM 6700F scanning electron microscope with scanning electron image (SEI) and backscatter detectors.
Immunofluorescence labeling was performed as previously described (12), with specific antibodies diluted to 1/100 and with Alexa Fluor 488 goat anti-rabbit immunoglobulin G (Molecular Probes). Bacteria were stained with propidium iodide (Molecular Probes). Samples were examined with a Zeiss confocal laser scanning microscope.
Shearing.
Bacteria were harvested from the plates and resuspended in phosphate-buffered saline to an optical density at 600 nm of 1.0 and then centrifuged twice at 13,000 × g in a microcentrifuge for 5 min (each time) to separate the bacteria (the pellet fraction) from the pilus-enriched supernatant (sheared fraction). Both fractions were precipitated with 10% trichloroacetic acid and loaded onto SDS-12% polyacrylamide gels for immunoblotting.
Purification and analysis of PulG filaments.
Plasmid pCHAP1362, encoding the PulG-His6 variant, was transformed into E. coli strain PAP9001 carrying pCHAP1216, providing all of the Pul components necessary for pilus growth and secretion in trans. Cells were grown overnight at 30°C on heavily inoculated LB-maltose agar supplemented with ampicillin and chloramphenicol. The bacterial lawn was resuspended in 1× phosphate-buffered saline with a glass spreader. Pili were sheared off by vortexing and vigorous pipetting and then centrifuged (5 min, 20,000 × g). Subsequently, the supernatant (sheared) fraction was ultracentrifuged (1 h, 150,000 × g) to concentrate the pili and remove soluble proteins. The resulting pellet was resuspended in 10 mM HEPES buffer, pH 7.5, and mixed with TALON cobalt affinity resin (Clontech) in 50 mM sodium phosphate buffer, pH 7.2, containing 300 mM NaCl. The resin was washed several times with the same buffer, and bound pili were eluted with 150 mM imidazole. The purified pili were concentrated by a second ultracentrifugation step, and the resulting pellet was suspended in small volumes of 10 mM HEPES buffer, pH 7.5, and analyzed by TEM and SDS-PAGE.
RESULTS
A threshold level of PulG is required for PulG assembly into pili.
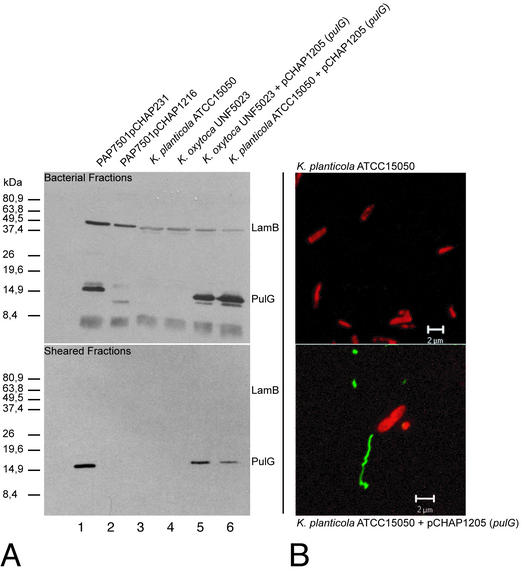
We reported previously that pseudopilin PulG assembled into bundled, apparently polar pili on the surface of E. coli expressing the pul secreton genes only when the bacteria were grown on agar plates and when pulG expression was increased (36, 43). Pili can be detected by SDS-PAGE and immunoblotting of PulG in the supernatant after shearing of bacteria harvested from plates and resuspended in liquid (43) or by direct immunovisualization with secondary antibodies coupled to gold beads (immunoelectron microscopy [immuno-TEM] [43]) or to a fluorescent dye (immunofluorescence microscopy; Fig. 1). These methods were used to examine two Klebsiella strains from which pul genes have been cloned, encapsulated K. planticola ATCC 15050 (20) and nonencapsulated K. oxytoca UNF5023 (10). PulG pili were not detected by shearing when these bacteria were grown on agar containing maltose to induce expression of the pul genes (Fig. 2). These data confirm that expression of the chromosomal pul locus is not sufficient to cause pilus production. However, in both cases, introduction of pCHAP1205, carrying the UNF5023 pulG gene, caused the appearance of PulG in the sheared fraction (Fig. 2; note that specific release of PulG was indicated by the absence of outer membrane protein LamB from the sheared fractions). Furthermore, the Klebsiella strains overexpressing pulG possessed pili (Fig. 2) that were indistinguishable from those observed in recombinant E. coli (Fig. 1). Thus, abnormally high-level pulG expression is required for PulG pilus formation in Klebsiella and the presence of the capsule does not interfere with pilus production.
FIG. 1.
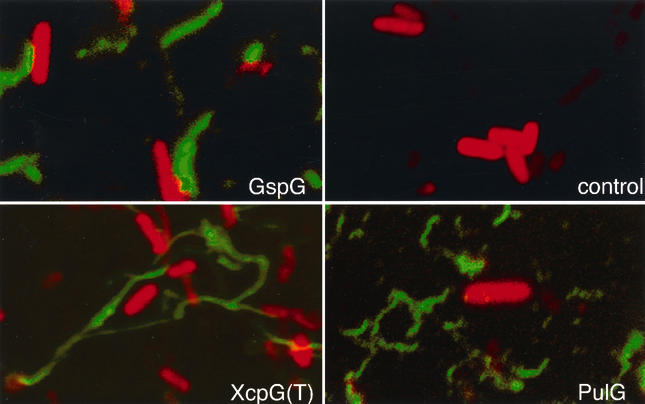
Immunofluorescence microscopy of E. coli K-12 (control) and derivatives carrying pCHAP231 {all pul genes [PulG], pCHAP1216 [pCHAP231 ΔpulG] and pCHAP4010 [gspG+] [GspG] or pCHAP1216 and pCHAP1380 [xcpG(T)+] [XcpT]} harvested from L-agar plates containing maltose to induce expression of the pul secreton genes carried by pCHAP231. The primary antibodies used were directed against the specific pseudopilins (anti-PulG was used for the control cells shown). Bacteria are stained red with propidium iodide (Molecular Probes), and bundled pili appear green because of the binding of primary antibodies, followed by Alexa Green 488-labeled secondary antibodies. Note that pili attach more tightly than bacteria to the poly-l-lysine-coated glass slides and that the bacteria wash off during processing to leave unattached pili on the slide.
FIG. 2.
Shearing (A) and immunofluorescence microscopy (B) of Klebsiella strains ATCC 15050 and UNF5023 with or without additional copies of the pulG gene on pCHAP1205. The bacteria were grown on LB agar containing maltose and IPTG (for strains with pCHAP1205). In the shearing analysis (A), sheared and cell-associated proteins were separated by SDS-PAGE and immunodetected with antibodies against PulG and the integral outer membrane protein LamB. E. coli K-12 strains carrying pCHAP231 and pCHAP1216 (pCHAP231 ΔpulG) were included as positive and negative controls, respectively. Details of the immunofluorescence study were as for Fig. 1.
Semiquantitative immunoblot analyses were used to estimate the amount of PulG present in or associated with bacteria expressing pulG at different levels (under lacZp control in plasmids with different copy numbers and with or without induction by IPTG or in the absence of the lacIq1 repressor). We estimate that the amount of PulG needed for pili to be detected by shearing, both in Klebsiella and in E. coli carrying the cloned pul genes, is three to five times that found in maltose-induced strains with chromosomal copies of the pul gene cluster.
PulG is not the only major pseudopilin that can assemble into surface pili.
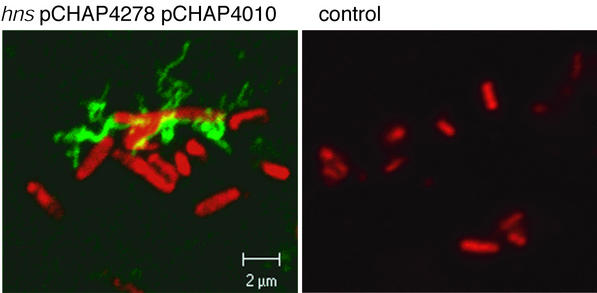
To determine whether pseudopilin assembly is unique to the Pul secreton, we examined the endogenous E. coli K-12 chitinase-specific (Gsp) secreton (14). The entire gsp gene cluster was carried by medium-copy-number pCHAP4278 in E. coli K-12 MC4100 with an hns mutation that causes derepression (14), and gspG expression was further increased by additional copies of the gene on pCHAP4010. Surface pili that could be sheared from the cells (data not shown) and that reacted with antiserum raised against the major Gsp pseudopilin, GspG, were detected when these bacteria were grown on agar (Fig. 3). Both plasmids were required for the detection of pili.
FIG. 3.
Immunofluorescence microscopy of E. coli K-12 strain MC4100 hns carrying pCHAP4278 (gspAB gspC-O+) and pCHAP4010 (gspG) of the same strain without plasmids. The primary antibodies were directed against GspG. Other details are as for Fig. 1.
To extend these analyses, we expressed pulG homologues cloned from the secreton gene clusters of several species of gram-negative bacteria in E. coli carrying pCHAP1216 (pul+ ΔpulG) and examined the recombinant strains for the ability to secrete pullulanase (complementation of ΔpulG) and pilus assembly. The outG genes from Erwinia chrysanthemi and E. carotovora were already known to complement pulG (31). This was found to be the case also for the P. aeruginosa xcpG (xcpT) gene, the E. coli K-12 gspG gene, the Vibrio cholerae epsG gene, and the Aeromonas hydrophila exeG gene (Table 2). Furthermore, all six of these pseudopilins were detected in the sheared fractions from bacteria grown on agar surfaces (Table 2). In immunofluorescence and/or immuno-TEM studies, GspG (Fig. 1), XcpG (XcpT) (Fig. 1), ExeG, and E. carotovora OutG (OutGEca; Fig. 4) were all detected in bundled, polar structures similar to those formed by PulG. However, production of E. chrysanthemi OutG (OutGEch) resulted in diffuse surface fluorescent labeling. Examination by immuno-TEM revealed that OutGEch (Fig. 4) and EpsG formed diffuse, nonbundled pili.
TABLE 2.
Complementation of ΔpulG and assembly of pili by homologues of pulG cloned and expressed in E. coli
| Gene | Origin | % Sequence similarity to PulGa
|
Complementationb | Pili detected byc:
|
|||
|---|---|---|---|---|---|---|---|
| Overall | Hydrophobic domain | Shearing | Immunofluorescence | Immuno-TEM | |||
| pulG | K. oxytoca | 100 | 100 | + | + | Bundled | Bundled |
| outG | E. chrysanthemi | 86 | 100 | + | + | Diffuse | Filaments |
| outG | E. carotovora | 86 | 100 | + | + | Bundled | Bundled |
| gspG | E. coli K-12 | 81 | 100 | + | + | Bundled | Bundled |
| exeG | A. hydrophila | 89 | 100 | + | + | NTd | Bundled |
| epsG | V. cholerae | 84 | 100 | + | + | NT | Filaments |
| XcpG(T) | P. aeruginosa | 59 | 83 | + | + | Bundled | Bundled |
| xpsG | X. campestris | 15 | 38 | − | − | NDe | ND |
Identical and conservative amino acid substitutions in the overall sequence (after cleavage of prepeptide) or in the N-terminal hydrophobic domain.
Measured in liquid cultures with an enzymatic assay for surface-exposed pullulanase.
Detected by using polyclonal antisera raised against PulG (this laboratory), GspG (this laboratory), XcpT (a gift from Alain Filloux), or XpsG (a gift from Nien-Tai Hu).
NT, not tested.
ND, not detected.
FIG. 4.
Immuno-TEM of E. coli K-12 carrying plasmids pCHAP1216 (pul+ ΔpulG) and pCCP2229 (outGEch) or pCCP2245 (outGEca) and grown on agar containing IPTG and maltose. Pili were labeled with PulG antibodies, followed by secondary antibodies tagged with 10-nm gold beads.
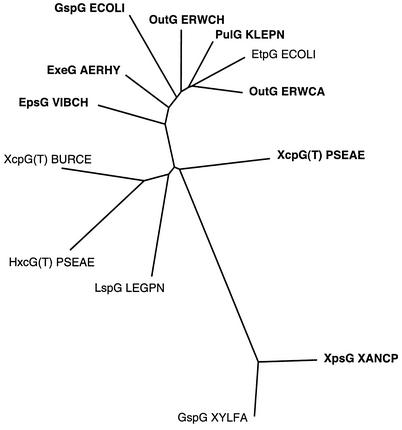
In contrast, the PulG homologue XpsG from Xanthomonas campestris could not substitute for PulG in pullulanase secretion and did not form pili in these assays (Table 2). This was not due to a failure of prepilin peptidase PulO to cleave preXpsG, which would prevent XpsG assembly (43), because XpsG in a strain with PulO migrated slightly faster than when produced without PulO, as was observed with all of the other prepseudopilins tested (data not shown). Sequence alignments revealed that XpsG is less closely related to PulG than are the other pseudopilins tested, in the usually well-conserved hydrophobic N-terminal region and especially in the downstream hydrophilic region (Table 2; Fig. 5).
FIG. 5.
Phylogenetic analysis of representative proteins in the PulG family with ClustalX. Proteins whose assembly into pili by the Pul secreton is tested here are shown in bold characters. The other proteins are EptG encoded by a plasmid found in E. coli strain O157 (44), XcpG(T) from Burkholderia cepacia, HxcG(T) from P. aeruginosa (1), LspG from Legionella pneumophila (42), and GspG from Xylella fastidiosa. This figure was prepared by Dominique Vidal-Ingigliardi.
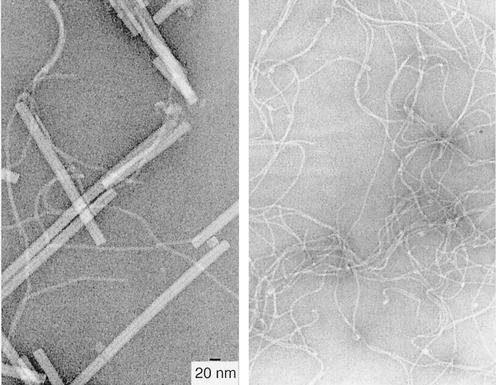
Minor pseudopilins are not detected in purified PulG pili.
To facilitate the purification of pili, we used a derivative of PulG carrying six histidine residues at its C terminus (35). When producing bacteria were examined by immuno-TEM, this PulG variant was detected as single filaments similar to those formed by OutGEch (Fig. 4) and EpsG and radiating from the entire cell surface (Fig. 6). Pili purified by cobalt affinity chromatography were found to contain three bands upon analysis by SDS-PAGE (Fig. 7). The two faster-migrating bands both reacted with antiserum against PulG and with His6 monoclonal antibodies (data not shown). N-terminal sequence analysis indicated that the faster-migrating band was mature, N-methylated PulG (35), while the upper, less intense band was cleaved but unmethylated PulG. Thus, a small amount of unmethylated PulG can be incorporated into PulG filaments, in line with the observation that unmethylated PilA pilin can be incorporated into P. aeruginosa type IV pili (29). SDS-PAGE and Coomassie blue or silver staining failed to detect any other proteins in the size range of the minor pseudopilins (14 to 35 kDa) in the purified pili, even with heavily loaded gels. Furthermore, immunoblotting with antisera against PulK and PulI failed to detect these minor pseudopilins in purified PulG-His6 pili. Therefore, PulG appears to be the only pseudopilin in the PulG filament.
FIG. 6.
Immuno-TEM of E. coli K-12 carrying plasmids pCHAP1216 (pul+ ΔpulG) and pCHAP4278 (pulG-His6). Pili were labeled with PulG antibodies, followed by secondary antibodies labeled with 10-nm gold beads.
FIG. 7.
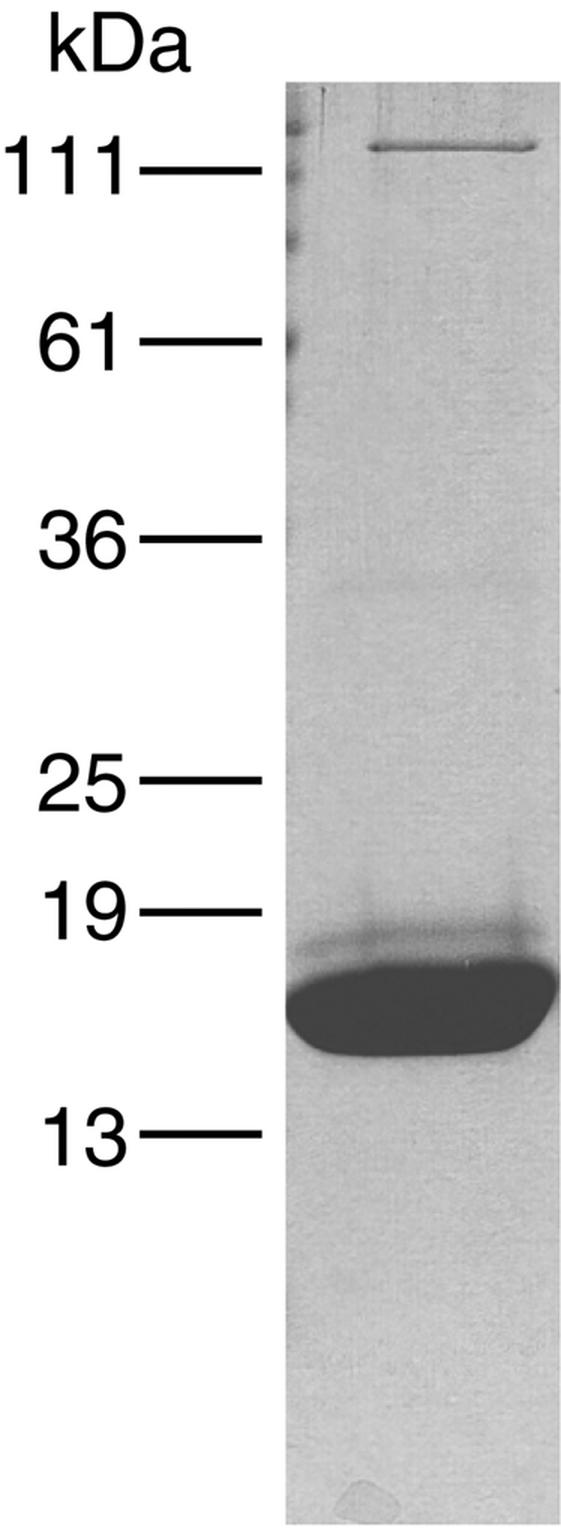
SDS-PAGE analysis of affinity-purified PulG-His6 pili. Proteins were stained with Coomassie brilliant blue.
The slow-migrating band in PulG-His6 pilus preparations comigrated with and reacted with antiserum against pullulanase (PulA). PulA could not be separated from PulG by treatment with 500 mM NaCl, followed by gel filtration on Superose 6 or cobalt affinity chromatography (data not shown). PulA did not bind to the cobalt resin in the absence of PulG-His6, and both proteins cosedimented upon immunoprecipitation with antisera against PulG (data not shown) and, thus, are tightly bound. In an attempt to determine whether PulA binds only to assembled PulG filaments, we added 0.6% octyltetraoxyethylene detergent, which dissociated the filaments, and performed a second cycle of affinity chromatography. PulG bound less well to the cobalt resin in the presence of detergent, but PulA did not bind at all, indicating that PulA adheres only to assembled PulG filaments. This observation suggests that PulG may contribute to pullulanase recognition at some stage in the secretion process. However, pilus-bound PulA represents only a small proportion of the total amount of protein present in the purified pili (Fig. 6) and of the total pool of PulA present on the cell surface. Immunogold staining of bacteria with PulG-His6-containing pili failed to demonstrate any PulA associated with the pilus, although PulA covered the cell surface, as reported previously for liquid-grown bacteria (10) (data not shown). Therefore, PulA may associate with PulG pili when the two are released from the surface by shearing (pullulanase is normally anchored to the cell surface but is released by shearing [8]).
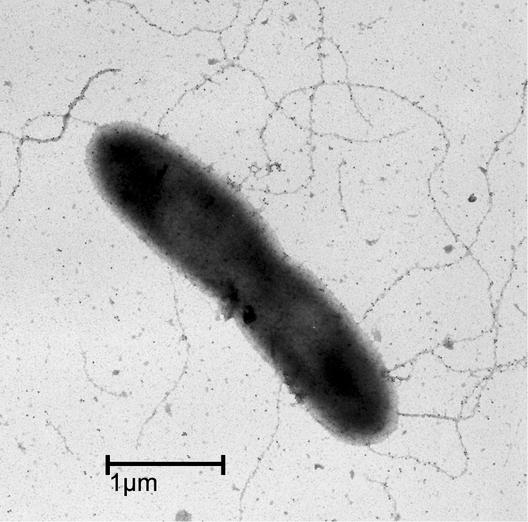
Purified PulG-His6 pilus filaments were negatively stained with 2% uranyl acetate and examined by TEM. The filaments were estimated to be 6 to 7 nm in diameter (Fig. 8, left panel). Minor kinks or blebs occasionally observed along the filaments may correspond to bound PulA (Fig. 8, right panel).
FIG. 8.
TEM of purified PulG-His6 pili after negative staining with 2% uranyl acetate. A sample of tobacco mosaic virus (18-nm diameter) was included in the sample in the left panel as a size marker.
Role of minor pseudopilins in PulA secretion and PulG assembly.
Mutations that incapacitate the minor pseudopilin genes pulI, pulJ, and pulK in derivatives of pCHAP231 all block secretion, whereas inactivation of pulH has no effect (31). The latter result was surprising because the pulH-like outH gene of E. chrysanthemi is required for secretion under some circumstances (17). To test whether pulH is required for pullulanase secretion at low copy numbers, we introduced the appropriate plasmid (pCHAP1324; pCHAP231 ΔpulH) into MC4100 carrying a pcnB::Tn10 mutation that reduces the plasmid copy number (18). In this context, the ΔpulH mutation reduced pullulanase secretion to <20% (compared to 100% with pCHAP231). Secretion was restored to 100% by a plasmid, pCHAP1331, with pulH cloned under lacZp control and a p15-derived replication origin that is not affected by pcnB. Thus, pulH is required for secretion at low pul secreton gene copy numbers. Individually, none of the other pseudopilin genes (pulG, pulI, pulJ, or pulK) could compensate for the absence of pulH at low copy numbers.
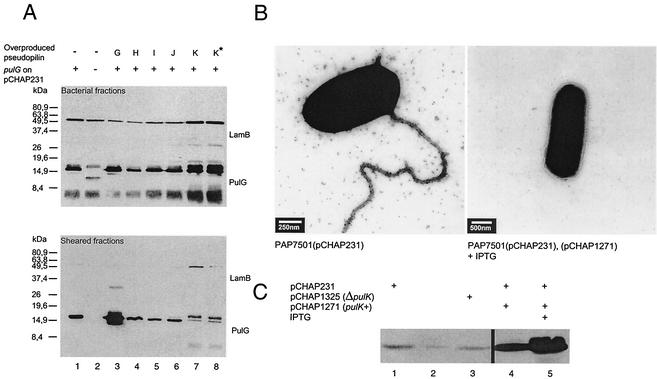
In contrast to pulI, pulH, pulJ, and pulK are not required for the formation of the PulG pilus in pcnB+ strains carrying derivatives of pCHAP231 (43) and apparently are not part of it (see above). pulI is required for efficient piliation (43) but, again, does not appear to be part of the PulG pilus (see above). To determine whether the abundance of any of the minor pilins could diminish PulG pilus production, their expression was increased by introducing plasmids bearing their respective genes under lacZp control (Table 1) into a strain already bearing pCHAP231. In all cases, the pseudopilin gene was expressed under lacZp control and, in the cases of pulH, pulI, pulJ, and pulK, approximately 50 bp of the upstream gene was fused in frame to the ATG of the lacZ gene in the vector to avoid problems of translation polarity and to ensure maximum expression (31). PulG was inefficiently released by shearing when PulK, the largest of the pseudopilins (40), was overproduced (Fig. 9A and C). Furthermore, low levels of LamB were released from these bacteria by shearing, implying that the low levels of PulG were probably released by membrane disruption rather than by shearing of pili (Fig. 9A). Indeed, immuno-TEM analysis confirmed the absence of PulG pili on the bacteria overproducing PulK (Fig. 9B). Thus, overproduction of the minor pseudopilin PulK abolished piliation although secretion was not affected, suggesting that PulK may be involved in pilus length control. Increased pulH, pulI, and pulJ expression did not affect the yield of PulG released by shearing (Fig. 9B), which may indicate that these genes are not involved in pilus length control, although the increase in expression might have been insufficient to have an observable effect. As expected, higher-level expression of pulG (pCHAP1205) increased the amount of PulG released by shearing (Fig. 9A).
FIG. 9.
Effects of increased levels of pseudopilins on the ability of E. coli K-12 to assemble PulG pili. Strain PAP7501 carrying pCHAP231 and additional plasmids bearing pseudopilin genes under lacZp control were grown on LB agar containing maltose and IPTG. Harvested cells were subjected to shearing analysis (A) or examined by immuno-TEM with primary antibodies against PulG and secondary antibodies with 10-nm gold beads (B). Proteins blotted onto nitrocellulose membranes (A) were probed with antibodies against PulG and LamB. The band migrating slightly above PulG does not react with PulG antibodies. Control cells for the shearing experiment carried pCHAP1216, the ΔpulG derivative of pCHAP231 (lane 2; ΔG). In these samples, the band migrating faster than PulG is the truncated PulG protein that is not assembled into pili. The plasmids bearing the minor pilin genes were pCHAP1205 (pulG), pCHAP1331 (pulH), pCHAP1351 (pulI), pCHAP1328 (pulJ), and pCHAP1271 (pulK). K* (lane 8) indicates that the cells carry pCHAP5236, which is the same as pCHAP1271 except that the pulK gene has a mutation converting the methionine at position +5 to glutamate. In the samples from the strain overproducing PulG (lane 3), the band migrating more slowly that PulG is the PulG dimer that is not dissociated in SDS. Panel C shows overproduction of PulK arising from the presence of the pulK gene on pCHAP1271 (lanes 4 and 5). Lane 1 shows the level of PulK in cells with pCHAP231. Lanes 2 and 3 are controls lacking the entire pul gene cluster or with the pCHAP231 derivative pCHAP1325 (pulK::kan-2), respectively. The immunoblot of total cell extracts of maltose-grown PAP7501 strains bearing the indicated plasmids was probed with antibodies against PulK that react with an unrelated protein migrating at the position of PulK in strains lacking this protein.
Certain mutations in pulG have differential effects on secretion and piliation.
We previously reported a series of insertion, substitution, and deletion mutations in pulG, some of which abolished the ability of the encoded protein to function in pullulanase secretion (35). We screened many of these mutations for their effects on pilus assembly, as determined by the shearing assay, and tested their abilities to complement a ΔpulG mutation when the bacteria are grown on agar (Table 3). In general, the data from the secretion assay were the same as those reported previously except that the blocked secretion due to the Q-2R substitution (35) could not be reproduced (Table 3) and the I+5T, M+6L double substitution abolished secretion, contrary to what was observed previously (35). In most cases, mutations that prevented secretion also blocked piliation whereas mutations that had no effect on secretion did not affect piliation (Table 3). However, two notable exceptions were the mutations replacing the E+5 residue, which abolished secretion (Table 3) (35) but did not prevent pilus formation (Table 3). In the P. aeruginosa type IV pilin PilA, E+5 is required for methylation and pilin assembly (27, 50) although methylation is not required for pilus assembly (28, 29). Thus, PulG differs from PilA in that E+5 is not required for its assembly into a pilus. This observation is intriguing because it confirms that the ability of PulG to promote secretion can be uncoupled from its ability to assemble into pili. The interesting observation that the insertion of various peptides at the C terminus of PulG affects neither secretion nor piliation (Table 3) will be pursued in greater detail below.
TABLE 3.
Effects of mutations in pulG on its ability to promote secretion and assemble into pilia
| Original plasmid | Amino acid substitution/insertion | Secretion | Piliation |
|---|---|---|---|
| pCHAP162B | G−1A | − | − |
| pCHAP162S | G−1E | − | − |
| pCHAP162F | E+5V | − | + |
| pCHAP162G | E+5A | − | + |
| pCHAP162I | F+1I | + | + |
| pCHAP162J | F+1L | + | + |
| pCHAP162KK | Q−3R | + | + |
| pCHAP162LL | Q−3P | + | + |
| pCHAP162O | I+5T, M+6L | − | − |
| pCHAP162DB3 | K133F(FVLVVstop) | + | + |
| pCHAP162DB5 | K133F(FATKDstop) | + | + |
| pCHAP162DC4 | K133F(FHHHHHHstop) | + | + |
| pCHAP162DC5 | K133F(FVMVMVMstop) | (+) | − |
pulG alleles described previously (35) were subcloned into pSU19 (Cmr) and then introduced into strain PAP7501 carrying pCHAP1216 (pCHAP231 pulG). Amino acid residues affected in PulG are indicated relative to the position of the prepilin peptidase cleavage site in pre-PulG; e.g., F+1 in processed PulG corresponds to amino acid 7 of pre-PulG. Inserted amino acids are indicated in parentheses. Bacteria were grown on L-agar plates containing maltose and assayed for pullulanase secretion and for the presence of pili, as detected by shearing, SDS-PAGE, and immunoblotting with PulG antibodies. In the secretion assay, + indicates that >80% of the pullulanase was surface exposed (secreted); −, <20% secretion; (+), 20 to 40% secretion.
PulK is unusual among pseudopilins in that it lacks the canonical glutamate at position +5. To test whether the absence of E+5 from PulK is important for its function, methionine +5 of PulK carried by pCHAP1271 was replaced with a glutamate. This substitution had no effect on the ability of the plasmid to complement the pulK mutation carried by pCHAP1325 in either pullulanase secretion or PulG pilus assembly (data not shown). Thus, the universal absence of E+5 from PulK and its homologues does not affect its processing by prepilin peptidase (4, 36) and is not necessary for its function. Overproduction of this PulK variant inhibited piliation (but not secretion) in the same way as wild-type PulK (Fig. 9).
The C terminus of PulG is a permissive insertion site.
We have already noted that addition of His6 to the C terminus of PulG did not affect its ability to promote pullulanase secretion or its assembly into pili (Table 3), although the resulting pili were unable to form bundles and were not polar (Fig. 6 and 7). Insertion of the sequences FVLVV and FATKD at the C terminus of PulG also did not affect secretion or piliation (Table 3), but the pili were bundled. However, the sequence FVMVMVM at the C terminus (pCHAP162DC5) destabilized PulG, caused the bacteria to grow slowly (35), and did not permit piliation (Table 3), although low-level pullulanase secretion was observed (35). The sequence inserted into this variant of PulG is very hydrophobic and may not be released efficiently from the plasma membrane during PulG export by the Sec machinery.
The fact that PulG-His6 filaments bind avidly to cobalt-coated beads suggests that the His6 tag is exposed along the surface of the entire filament. Unfortunately, we could not test this possibility directly because the His6 monoclonal antibodies failed to react with these filaments in immuno-TEM experiments. Since the sequences FVLVV and FATKD were equally well tolerated at the C terminus of PulG, we extended our studies to PulG derivatives bearing the following sequences: peptide 1, GSAWSHPQFEK (Strep-tag) (45), recognized by a commercial polyclonal antibody and capable of binding to streptactin-coated beads (IBA GmbH); peptide 2, EQKLISEEDL, the c-myc epitope (13) recognized by a commercial monoclonal antibody (Invitrogen); peptide 3, YPYDVPDYA, the FLAG epitope (6) recognized by a commercial monoclonal antibody (Sigma); peptide 4, RPQASGVYM, an LCMV nucleoprotein epitope (47); peptide 5, RPQASGVYMGNLTAG, a longer version of peptide 4.
In all five cases, PulG carrying the peptide was assembled into bundled pili and promoted pullulanase secretion. Thus, only the histidine tag abolished the formation of pilus bundles. Immunoblotting with PulG antibodies revealed that all five PulG derivatives were slightly larger than wild-type PulG, as expected (data not shown). However, only PulG bearing peptide 1, 2, or 3 reacted with the peptide-specific antibodies in immunoblotting experiments. Furthermore, all three peptides were detected along the entire length of the bundled pilus by immuno-TEM with the appropriate peptide-specific antibodies (see the example in Fig. 10). The failure of the antibodies directed against the LCMV peptide to react with PulG bearing peptides 4 and 5 is unexplained.
FIG. 10.
Immuno-TEM of E. coli producing PulG pili without (left) or with (right) a c-myc tag at the C terminus of the pseudopilin PulG. The primary antibodies used were specific for the c-myc epitope, which is present in PulG encoded by pCHAP5246 but absent from PulG encoded by pCHAP1362 (and therefore was not labeled; arrows).
We previously reported data suggesting that PulG with β-lactamase (BlaM) fused to its C terminus functions in secretion (34). However, although PulG-BlaM was detected in bacteria carrying pCHAP1216 (pCHAP231 ΔpulG) and expressing the pulG-blaM gene fusion from a compatible plasmid, pili were not detected, indicating that PulG-BlaM cannot be assembled.
Secretin PulD is not absolutely essential for PulG pilus assembly.
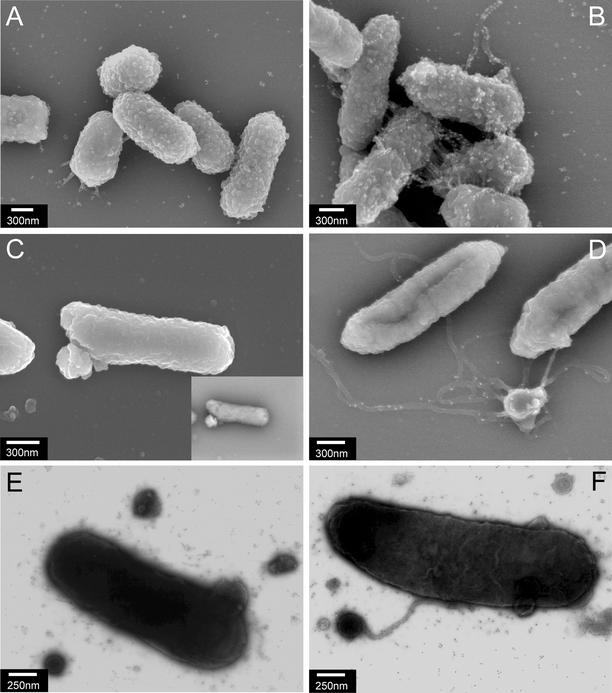
The secretin PulD, which is absolutely required for pullulanase secretion (9), forms ring-like structures (22) similar to those formed by secretin PilQ, which is required for pilus biogenesis (3, 7), leading to the idea that both pullulanase and PulG pili may traverse the outer membrane via the PulD channel (43). In Neisseria gonorrhoeae strains in which pilus retraction is abolished by a pilT mutation, the absence of PilQ does not abolish PilE pilin assembly but does prevent the extrusion of pili through the outer membrane, causing membrane blebbing and the appearance of abnormal, deformed pili on the bacterial surface (55). Overexpression of pulG results in a phenomenon (uncontrolled pilus elongation) similar to that caused by a mutation in pilT. Immuno-SEM of E. coli carrying pCHAP231 revealed the presence of bundled pili that were strongly labeled by PulG antibodies (Fig. 11B) that were not present when pulG was deleted (Fig. 11A). Deletion of pulD (pCHAP1226) also completely abolished pilus formation, as reported previously (38), but also increased membrane blebbing (Fig. 11C). When pulG expression was further increased in these bacteria by introducing pCHAP1205 (pulG+), exaggerated membrane blebbing was observed, together with stunted, deformed pili that were poorly labeled by PulG antibodies (Fig. 11D). Furthermore, immuno-TEM of these ΔpulD strains after negative staining confirmed the surface deformation seen by SEM (Fig. 11E) and revealed the presence of numerous vesicle-like structures bearing misshapen pili that were labeled, albeit inefficiently, by PulG antibodies when PulG was produced in very large amounts (Fig. 11F). The poor labeling of pili in Fig. 11D and F may indicate that PulG is covered by part of the outer membrane (55). Thus, as with PilE, a secretin is not absolutely essential for PulG pilus assembly but the pili that are produced are deformed and, apparently, are unable to traverse the outer membrane or do so by causing severe membrane perturbation.
FIG. 11.
(A to D) Immuno-SEM of E. coli strain PAP9001 immunogold labeled with anti-PulG antibodies. Except for the main image in panel C (see details), all SEM pictures were taken with SEI and backscatter detectors. (A) Bacteria expressing all of the pul genes except pulG (PAP9001/pCHAP1216). Pili are not detected, and only background labeling is observed. (B) Bacteria expressing all of the pul genes, including pulG (pCHAP231). Specific labeling is observed on pili that interconnect the bacteria. (C) Bacteria expressing all of the pul genes except pulD (encoding the secretin; pCHAP1226). The main image was obtained with an SEI detector alone for clearer resolution of the bacterial cell surface. The image of the same bacterium in the top inset was taken with backscatter detectors. (D) Bacteria expressing all of the pul genes except pulD (pCHAP1226) and overexpressing pulG (pCHAP1205). Note the reappearance of surface pili because of higher-level pulG expression, despite the absence of secretin (compare panels C and D). (E and F) Immuno-TEM of bacteria similar to those in panels C and D.
DISCUSSION
Pseudopili or real pili?
The data presented here confirm and extend our previous observation that PulG can be assembled into pili provided that pulG expression is increased compared to the level maximally achieved by maltose-induced induction of the chromosomal pulC-O operon, of which it is part. We believe that PulG pili are aberrant manifestations of another structure, probably the elusive “pseudopilus” (34, 38), that has more physiological relevance. One idea that we find particularly attractive is that the pseudopilus acts as a piston that pushes secreted proteins into the large compartment in the lumen of the secretin channel (22, 23), allowing release of the proteins into the medium and subsequent pilus retraction. This “piston” model of T2S, first proposed by Hobbs and Mattick (16), is based on the well-documented phenomenon of type IV pilus elongation and retraction that produces twitching motility and related patterns of bacterial movement across solid surfaces (19, 49). Although the mechanisms involved remain unclear, it seems to be accepted that pilus elongation is promoted by an ATPase (PilB in P. aeruginosa, PilF in Neisseria) and that pilus retraction results from disassembly promoted by another, related ATPase (PilT in both bacteria) that operates in reverse (53, 54). The factors controlling pilus length and the switch from elongation to retraction are unknown.
The piston model is supported by the observation that the absence of secretin PulD, the portal via which pullulanase is proposed to cross the outer membrane, blocks both pullulanase secretion and the appearance of long PulG pili on the cell surface without preventing PulG assembly into filaments (Fig. 11D). However, unlike the T4P system, the secreton has only one ATPase (PulE in the Pul secreton), which is related both to PilB/F and to PilT. How, then, could retraction occur? We propose that elongation and retraction of PulG pseudopili are both stimulated by PulE or that the electrochemical potential across the plasma membrane that is required for secretion (30) is directly or indirectly responsible for PulG pilus disassembly. We further propose that elongation may be arrested and disassembly (retraction) may be triggered by incorporation into the base of the filament of a minor pseudopilin, possible PulK (Fig. 9), or by the release of pullulanase into the lumen of the PulD secretin channel. In this scenario, pilus growth beyond the outer membrane would result from the increased levels of PulG, which titrate the hypothetical “stop-assembly” signal (PulK?) or stimulate rapid and uncontrolled assembly of PulG pili. In this context, the observation that a small amount of pullulanase copurifies with PulG pili is particularly interesting since it is the only interaction between pullulanase and a secreton component that we have been able to detect so far. This may suggest that the two proteins interact as part of the secretion cascade.
Although we find this model attractive, it is not fully supported by all of the observations reported here and elsewhere. For example, the fact that very high-level expression of pulG in a strain lacking the secretin PulD causes membrane deformation and production of aberrant pili (Fig. 11D) similar to those observed in an N. gonorrhoeae pilT pilQ mutant (55) suggests that PulG pili span the outer membrane via the lumen of the PulD secretin channel. Thus, these pili would be expected to prevent secretion by blocking the channel, but this does not appear to be the case (43). This enigma was discussed previously (43), when we proposed the possible coexistence of open (functional) channels and channels plugged by PulG pili in the same cell. Another possibility is that the surface-associated pullulanase that we detected in agar-grown, PulG pilus-producing bacteria was secreted before the secretin channels were plugged by PulG. Unfortunately, it is not possible to perform kinetic experiments on pullulanase secretion with bacteria grown on agar.
The fact that several different PulG homologues from bacteria whose secretons are not interchangeable can be assembled into pili by the Pul secreton and can replace PulG in secretion indicates that pseudopilins are not the main determinants of secretion specificity, a role probably played by PulC and/or PulD (5, 17, 31, 48). However, the type IV pilin PpdD can be assembled but cannot substitute for PulG in secretion (43). Thus, a pilus-like structure is not itself sufficient to act as a piston to permit secretion. PpdD pilus growth may not be controlled by components of the Pul secreton (e.g., PulK) so that it cannot form the short pseudopili that stimulate secretion.
Bundling and polar localization of PulG pili.
Two PulG homologues (OutGEch and EpsG) and PulG-His6 all formed nonbundled pili but still promoted pullulanase secretion, indicating that functionality in pullulanase secretion does not correlate with bundle formation. The mechanism of bundle formation by type IV pili is not understood. PulG pilus bundles appear to be located at a cell pole, raising the possibility that the entire secreton is located at the pole and, hence, that secretion occurs here. Indeed, recent studies indicate that components of the V. cholerae Eps secreton are located at the cell pole, where secretion was observed (46). However, nonbundled PulG-His6 pili were evenly distributed over the cell, suggesting that the ability of (pseudo)pili to form polar bundles is not correlated with the ability of the protein to function in secretion. These data do not necessarily cast doubt on the reported polar localization of the secreton in V. cholerae, since our studies were carried out in a heterologous system (K. pneumoniae pul secreton genes expressed in E. coli) at higher levels of major pseudopilin production and under growth conditions different from those used in the study of the V. cholerae Eps secreton. Furthermore, we suspect that the elongated pseudopili that we observed upon increased pulG expression are redundant structures that do not function in secretion; hence, their location does not necessarily reflect the site at which secretion occurs.
The failure of PulG-His6 to form bundles is probably due to the polyhistidine tag, since replacement of His6 with other short peptides did not prevent bundling. Structural analyses of purified PulG monomers and of PulG filaments, currently under way, should reveal more information on the packing of the subunits in the pili and on surface-exposed residues. The sequences of the OutGEch and OutGEca proteins and of EpsG and ExeG are so closely related (91% overall identity in both cases) that it may be possible to identify bundle-inducing amino acids by site-directed mutagenesis. All four proteins are sufficiently related to PulG that it may be possible to predict their structures from that of PulG, providing more information on the bundling phenomenon.
Polyvalent foreign epitope presentation by PulG pili.
The fortuitous finding that PulG can form pili even when its C terminus is extended by at least 15 amino acids suggests that these pili may be useful as carriers of polyvalent immunogenic peptides. One of the potential advantages of such a system is that repeated exposure of one or more epitopes along the length of the pilus should increase immunogenicity. The possible use of pili for such purposes is not a new idea (11, 39, 41, 51), but many previous studies failed because insertions prevented polymerization of the pilus filament because of the unpredictable structural constraints. It will be interesting to determine the length and number of the foreign peptides that can be tagged onto PulG, whether there are other permissive sites for epitope insertion, and whether inserted peptides are immunogenic when delivered either on whole bacteria or in purified pili. Bacteria producing pili bearing appropriate peptides exposed along their entire length may also bind avidly to specific ligands or receptors.
However, there are at least three potential drawbacks to the use of PulG pili for these purposes: their fragility (they were not designed to withstand shear forces), their failure to assemble in liquid-grown cultures (43), and the unsuitability of E. coli K-12 for use as a live vaccine strain. Some of these problems can be easily overcome. For example, the entire pul gene cluster can be expressed in strains of Salmonella enterica serovar Typhimurium that are better suited for use as live vaccines. Problems related to the extreme fragility and lack of production of PulG pili in liquid cultures are more difficult to resolve. Interestingly, we have repeatedly observed that E. coli K-12 expressing maltose-induced pul secreton genes carried by the pBR322 derivative pCHAP231 or K. oxytoca UNF5023 expressing chromosomal pul genes and plasmid-encoded pulG exhibit enhanced biofilm formation in a simple microtiter dish assay (15, 32) (data not shown). The significance of this observation is difficult to ascertain, although it presumably indicates that PulG pili are produced under these circumstances. Although type IV pili have been shown to play a role in biofilm formation (26, 52), we note that PulG pili adhere very tightly to glass and plastic and form dense networks that link bacteria together (Fig. 11B). Therefore, the adherent bacterial masses that result may not correspond to the organized structures that are generally considered to be biofilms.
Acknowledgments
This work was supported, in part, by EC research contract HPRN-CT-2000-00075 and by grants to G.V. from the Fondation pour la Recherche Médicale and the Caisse Nationale d'Assurance Maladie et Maternité des Travailleurs Non Salariés des Professions Non Agricoles.
We are grateful to Michael Bagdasarian, Alan Collmer, Alain Filloux, Olivera Francetic, Peter Howard, and Nien-Tai Hu for cloned copies of pseudopilin genes and for antisera, to Olivera Francetic for help with experiments on GspG, to Dominique Vidal-Ingigliardi for computer analyses, to Christine Schmidt for help with the electron microscopy, to Frank Ebel for help with fluorescence microscopy and for help in preparing antisera against PulK and PulI, to Claude Leclerc for helpful discussions concerning the potential use of PulG in epitope delivery and for antibodies, to Jacques D'Alayer for microsequencing, and to all of the members of the Unité de Génétique Moléculaire for support.
REFERENCES
- 1.Ball, G., E. Durand, A. Lazdunski, and A. Filloux. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475-485. [DOI] [PubMed]
- 2.Bally, M., A. Filloux, M. Akrim, G. Ball, A. Lazdunski, and J. Tommassen. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121-1131. [DOI] [PubMed] [Google Scholar]
- 3.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 4.Bleves, S., A. Lazdunski, J. Tommassen, and A. Filloux. 1998. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX. Mol. Microbiol. 27:31-40. [DOI] [PubMed] [Google Scholar]
- 5.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J. Mol. Biol. 27:205-219. [DOI] [PubMed] [Google Scholar]
- 6.Brizzard, B. L., R. G. Chubert, and D. L. Vizard. 1994. Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and a peptide elution. BioTechniques 16:730-735. [PubMed] [Google Scholar]
- 7.Collins, R. F., L. Davidsen, J. P. Derrick, R. C. Ford, and T. Tonjum. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d'Enfert, C., C. Chapon, and A. P. Pugsley. 1987. Export and secretion of the lipoprotein pullulanase by Klebsiella pneumoniae. Mol. Microbiol. 1:107-116. [DOI] [PubMed] [Google Scholar]
- 9.d'Enfert, C., I. Reyss, C. Wandersman, and A. P. Pugsley. 1989. Protein secretion by gram-negative bacteria: characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J. Biol. Chem. 264:17462-17468. [PubMed] [Google Scholar]
- 10.d'Enfert, C., A. Ryter, and A. P. Pugsley. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Der Vartarian, M., M. C. Mechin, B. Jaffeux, Y. Bertin, I. Felix, and B. Gaillard-Martinie. 1994. Permissible peptide insertions surrounding the signal peptide-mature protein junction of the ClpG prepilin: CS31A fimbriae of Escherichia coli as carriers of foreign sequences. Gene 148:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebel, F., T. Podzadel, M. Rohde, A. U. Kresse, S. Krämer, C. Deibel, C. A. Guzman, and T. Chakraborty. 1998. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30:147-161. [DOI] [PubMed] [Google Scholar]
- 13.Evans, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francetic, O., D. Belin, C. Badaut, and A. P. Pugsley. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 142:27-30. [DOI] [PubMed] [Google Scholar]
- 16.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 17.Lindeberg, M., G. P. C. Salmond, and A. Collmer. 1996. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologs reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol. Microbiol. 20:175-190. [DOI] [PubMed] [Google Scholar]
- 18.Lopilato, J., S. Bortner, and J. Beckwith. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205:285-290. [DOI] [PubMed] [Google Scholar]
- 19.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-101. [DOI] [PubMed] [Google Scholar]
- 20.Michaelis, S., C. Chapon, C. d'Enfert, A. P. Pugsley, and M. Schwartz. 1985. Characterization and expression of the structural gene for pullulanase, a maltose-inducible secreted protein of Klebsiella pneumoniae. J. Bacteriol. 164:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot protein PulS, structure and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 25.Nunn, D. N., and S. Lory. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 4375-4382. [DOI] [PMC free article] [PubMed]
- 26.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 27.Pasloske, B. L., and W. Paranchych. 1988. The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol. Microbiol. 2:489-495. [DOI] [PubMed] [Google Scholar]
- 28.Pasloske, B. L., D. G. Scraba, and W. Paranchych. 1989. Assembly of mutant pilins in Pseudomonas aeruginosa: formation of pili composed of heterologous subunits. J. Bacteriol. 171:2142-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepe, J. C., and S. Lory. 1998. Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa: effect on leader peptidase and N-methyltransferase activities in vitro and in vivo. J. Biol. Chem. 273:19120-19129. [DOI] [PubMed] [Google Scholar]
- 30.Possot, O., L. Letellier, and A. P. Pugsley. 1997. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol. Microbiol. 24:457-464. [DOI] [PubMed] [Google Scholar]
- 31.Possot, O., G. Vignon, N. Bomchil, F. Ebel, and A. P. Pugsley. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 182:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratt, L. A., and K. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 33.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pugsley, A. P. 1996. Multimers of the precursor of a type IV pilin-like component of the general secretory pathway are unrelated to pili. Mol. Microbiol. 20:1235-1245. [DOI] [PubMed] [Google Scholar]
- 35.Pugsley, A. P. 1993. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol. Microbiol. 9:295-308. [DOI] [PubMed] [Google Scholar]
- 36.Pugsley, A. P., N. Bayan, and N. Sauvonnet. 2001. Disulphide bond formation in secreton component PulK provides a possible explanation for the role of DsbA in pullulanase secretion. J. Bacteriol. 183:1312-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pugsley, A. P., and B. Dupuy. 1992. An enzyme with type IV prepilin peptidase activity is required to process a component of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol. Microbiol. 6:751-760. [DOI] [PubMed] [Google Scholar]
- 38.Pugsley, A. P., and O. Possot. 1993. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PulG protein into a multiprotein complex. Mol. Microbiol. 10:665-674. [DOI] [PubMed] [Google Scholar]
- 39.Rani, D., M. Bayer, and D. Schifferli. 1999. Polymeric display of immunogenic epitopes from herpes simplex virus and transmissible gastroenteritis virus surface proteins on an enteroadherent fimbria. Clin. Diagn. Lab. Immunol. 6:30-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyss, I., and A. P. Pugsley. 1990. Five additional genes in the pulC-O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023 that are required for pullulanase secretion. Mol. Gen. Genet. 222:176-184. [DOI] [PubMed] [Google Scholar]
- 41.Rondot, S., K. G. Anthony, S. Dübel, N. Ida, S. Wiemann, K. Beyreuther, L. S. Frost, M. Little, and F. Breitling. 1998. Epitopes to F-pilin are incorporated into functional recombinant pili. J. Mol. Biol. 279:589-603. [DOI] [PubMed] [Google Scholar]
- 42.Rossier, O., and N. P. Cianciotto. 2001. Type II secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauvonnet, N., G. Vignon, A. P. Pugsley, and P. Gounon. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, H., B. Henkel, and H. Karch. 1997. A gene cluster related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol. Lett. 148:265-272. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, T. G., J. Koepke, R. Frank, and A. Skerra. 1996. Molecular interactions between the Strep-tag affinity peptide and its cognate target, streptavidin. J. Mol. Biol. 255:753-766. [DOI] [PubMed] [Google Scholar]
- 46.Scott, M., Z. Dossani, and M. Sandkvist. 2001. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc. Natl. Acad. Sci. USA 94:13978-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebo, P., C. Fayolle, O. d'Andria, D. Ladant, C. Leclerc, and A. Ullmann. 1995. Cell-invasive activity of epitope-tagged adenylate cyclase of Bordetella pertussis allows in vitro presentation of a foreign epitope to CD8+ cytotoxic T cells. Infect. Immun. 63:3851-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shevchik, V. E., J. Robert-Badouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J 16:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strom, M. S., and S. Lory. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa: effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656-1664. [PubMed] [Google Scholar]
- 51.Thiry, G., A. Clippe, T. Scarcez, and J. Petre. 1989. Cloning of DNA sequences encoding foreign peptides and their expression in the K88 pili. Appl. Environ. Microbiol. 55:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch, C. B., M. Hobbs, S. P. Livingstone, V. Krishnapillai, and J. S. Mattick. 1991. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in bacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 54.Wolfgang, M., H.-S. Park, S. F. Hayes, J. P. M. van Putten, and M. Koomey. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 95:14973-14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfgang, M., J. P. M. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]