Abstract
In the budding yeast Saccharomyces cerevisiae, PHO84 and PHO86 are among the genes that are most highly induced in response to phosphate starvation. They are essential for growth when phosphate is limiting, and they function in the high-affinity phosphate uptake system. PHO84 encodes a high-affinity phosphate transporter, and mutations in PHO86 cause many of the same phenotypes as mutations in PHO84, including a phosphate uptake defect and constitutive expression of the secreted acid phosphatase, Pho5p. Here, we show that the subcellular localization of Pho84p is regulated in response to extracellular phosphate levels; it is localized to the plasma membrane in low-phosphate medium but quickly endocytosed and transported to the vacuole upon addition of phosphate to the medium. Moreover, Pho84p is localized to the endoplasmic reticulum (ER) and fails to be targeted to the plasma membrane in the absence of Pho86p. Utilizing an in vitro vesicle budding assay, we demonstrate that Pho86p is required for packaging of Pho84p into COPII vesicles. Pho86p is an ER resident protein, which itself is not transported out of the ER. Interestingly, the requirement of Pho86p for ER exit is specific to Pho84p, because other members of the hexose transporter family to which Pho84 belongs are not mislocalized in the absence of Pho86p.
Yeast cells respond to phosphate starvation by up-regulating the activity of a high-affinity phosphate uptake system (1). Pho84p and Pho86p function in this phosphate transport system. Both PHO84 and PHO86 are regulated transcriptionally in response to extracellular phosphate levels by a phosphate-responsive signal transduction pathway (the PHO pathway) (2). Expression of these genes is greatly induced in low-phosphate medium (3, 4). PHO84 and PHO86 are essential for growth under low-phosphate conditions and are required for the transcriptional repression of PHO5, which encodes a secreted acid phosphatase, in high-phosphate medium.
Strains containing loss-of-function mutations in PHO84 and PHO86 exhibit similar phenotypes, e.g., phosphate uptake defects and constitutive expression of phosphate responsive genes. PHO84 was identified in a screen for mutants that express PHO5 constitutively (Phoc) (5). PHO84 encodes a member of the hexose transporter family that contains 12 transmembrane domains. Biochemical experiments demonstrate that Pho84p is a high-affinity phosphate transporter (6) that is conserved in plants and fungi (7–11). PHO86 was also identified in a genetic selection for mutants that exhibit the Phoc phenotype (12) and in a screen for mutants that confer arsenate resistance (13). PHO86 encodes a protein that associates with membranes, presumably through its two predicted transmembrane domains (4).
PHO86 is not required for transcriptional activation of PHO84 (13), raising the possibility that Pho86p is directly involved in the high-affinity phosphate uptake system. Pho86p could be a phosphate transporter that associates with Pho84p at the plasma membrane for phosphate uptake. Alternatively, Pho86p might be required for proper localization of Pho84p to the plasma membrane. Proteins destined for the plasma membrane are synthesized, processed, and folded in the endoplasmic reticulum (ER), and, once folded, they are packaged into ER-derived COPII vesicles for transport to the Golgi apparatus and then to the plasma membrane (14, 15). Accessory proteins have been identified that assist in the transport of secretory proteins through the secretory pathway. For example, Vps10p is required for the sorting of the soluble vacuolar protein carboxypeptidase Y from the Golgi to the vacuole (16), whereas Ast1p ensures efficient transport of the plasma membrane ATPase (Pma1p) from the Golgi to the plasma membrane (17). Some accessory proteins function in an early stage of the secretory pathway. One such example is Shr3p, which is required for the ER exit of the general amino acid permease (Gap1p) (18, 19). Because of the physiological importance of Pho84p in low-phosphate conditions, Pho86p may function exclusively to ensure rapid and faithful transport of the permease to the cell surface.
In this paper, we report that Pho86p is required for specific packaging of Pho84p into COPII vesicles derived from ER membranes, but itself is not packaged into COPII vesicles, indicating that Pho86p belongs to a class of “outfitters” (20), resident ER proteins that facilitate the loading of cargo into transport vesicles.
Materials and Methods
Media, Genetic Methods, and Strains.
Standard yeast media are as described (21) and media contained 2% glucose unless otherwise specified. No-phosphate medium is as described (12). Crosses, sporulation, and tetrad analysis were performed by standard genetic methods (22). S. cerevisiae strains used in this study were EY0664 (MATα ura3-52 leu2-3, isogenic to RSY255) and its derivatives, EY0665 (pho86Δ∷LEU2), EY0666 (pho84∷PHO84-GFP), EY0667 (pho84∷PHO84-HA), EY0668 (pho86Δ∷LEU2 pho84∷PHO84-HA), and EY0669 (pho86Δ∷LEU2 pho84∷PHO84-GFP). EY0665 was constructed by transforming EY0664 with EB1123 partially digested with KpnI and SacI, and selecting on synthetic medium lacking leucine. EY0666 and EY0667 were constructed by transforming EY0664, respectively, with EB1124 and EB1125 that were partially digested with XhoI, and selecting on minimal medium lacking uracil; the resulting strains were streaked for single colonies on 5-fluoroorotic acid plates. EY0666 was obtained by screening for Pho84p-GFP by direct fluorescence, whereas EY0667 was obtained by screening for Pho84p-HA by immunoblotting. EY0668 and EY0669 were constructed by disrupting PHO86 in EY0666 and EY0667, respectively, with EB1123 partially digested with KpnI and SacI. Other yeast strains were EY0687 (MATa end4ts his4 leu2 ura3, isogenic to RSY1580), EY0688 (MATa pma1∷HA-PMA1∷LEU2 ade2–101c his3-Δ200 leu2-Δ1 lys2–801am trp1-Δ63 ura3–52, isogenic to RSY1578), and its derivative EY0689 (pho86Δ∷TRP1). To construct EY0689, EB0477 was digested with KpnI and SacI, transformed into EY0688, and transformants were selected on synthetic medium lacking tryptophan.
Plasmids.
Plasmids pPHO84-GFP (EB0666) and pPHO86-GFP (EB0667) were constructed by fusing PCR-generated XhoI/EcoRI fragments of PHO84 and PHO86, respectively, with their native promoters in-frame to the N terminus of the green fluorescent protein (GFP) in pRS316. A disruption vector that replaces the entire ORF of PHO86 with TRP1 (EB0477) was constructed as follows. A 450-bp KpnI/EcoRI fragment derived from the 5′ noncoding region of PHO86 and a 580-bp BamHI/XbaI fragment from the 3′ noncoding region of PHO86 were generated by PCR and inserted into the Bluescript plasmid to create pTS-PHO86. An EcoRI/BglII fragment containing the TRP1 gene from plasmid pJJ248 (23) was inserted into the EcoRI/BamHI cut pTS-PHO86 to create EB0477. Another disruption vector (EB1123) for PHO86 was constructed by ligating a 4-kb PstI/BamHI fragment from EB0477 to a PstI/BamHI fragment containing the LEU2 gene from pJJ252 (23). To replace the chromosomal copy of PHO84 with PHO84-HA, we constructed an integration vector (EB1124) in three steps. First, an XhoI/SacI PCR product containing PHO84 was cloned into pRS306 digested with XhoI and SacI to create pRS306-PHO84. In this step, a polylinker containing a NotI site was added in-frame to the C-terminal coding region of PHO84 immediately preceding the stop codon. Second, a NotI fragment containing three tandem copies of the hemagglutinin epitope (HA) was inserted into the NotI site of pRS306-PHO84 to create pRS306-PHO84-HA. Third, a 0.5-kb EcoRI/SacI PCR product derived from the 3′ noncoding region of PHO84 was cloned into the EcoRI/SacI sites of pRS306-PHO84-HA to create EB1124. To integrate PHO84-GFP into the yeast genome, we constructed the integration vector EB1125 by first cloning an XhoI/SacI fragment of pPHO84-GFP into pRS306 to create pRS306-PHO84-GFP. A BamHI/SacI PCR product derived from the 3′ noncoding region of PHO84 was then cloned into pRS306-PHO84-GFP at the BamHI/SacI sites to make EB1125. Epitope-tagged PHO86 (pPHO86-HA, EB1126) was constructed as follows. A PCR-generated XhoI/EcoRI fragment of PHO86 was cloned into the XhoI/EcoRI sites of pRS316, and a polylinker containing a NotI site was added in-frame to the C-terminal coding region of PHO86 immediately preceding the stop codon. A NotI fragment containing three copies of HA was inserted into the NotI site of this construct to create EB1126. A low copy plasmid containing GAL2-GFP (URA3 marked) was a generous gift from A. Kruckeberg (University of Amsterdam, The Netherlands). The plasmid that carries ERO1-HA was a gift from J. Weissman (University of California, San Francisco).
In Vitro Vesicle Budding Assay.
This assay was a modification of a previously described procedure (19), and was performed at 30°C unless otherwise specified. Yeast cultures grown in yeast extract/peptone/dextrose (YPD) or synthetic solid media were used to inoculate overnight stock cultures. Cells were inoculated into YPD or synthetic high-phosphate media until the cultures reached an optical density at 600 nm (OD600) of approximately 0.5. Yeast cells were washed three times in sterile water, inoculated into no-phosphate medium at an OD600 of 0.5, and grown for 2 h. Cells were harvested, washed twice in no-phosphate medium, and resuspended in no-phosphate medium to 2 OD600/ml, and shaken for 15 min. The cultures were labeled with 1 mCi/20 OD600/ml 35S-Promix (1,200 Ci/mmol; Amersham Pharmacia) for 3 min. Metabolic activity was stopped by the addition of NaN3 and NaF (20 mM final). Spheroplast preparation and lysis were performed as described (24). Gently lysed spheroplasts were washed once with “low-salt B88” [20 mM Hepes (pH 6.8)/50 mM KOAc/250 mM sorbitol/5 mM MgOAc], and twice with B88 [20 mM Hepes (pH 6.8)/150 mM KOAc/250 mM sorbitol/5 mM MgOAc]. The pellet was resuspended in 0.5 ml B88, and 0.5 ml “high-salt B88” [20 mM Hepes (pH 6.8)/1 M KOAc/250 mM sorbitol/5 mM MgOAc] was added. The sample was allowed to chill on ice for 10 min, and washed twice with B88. Each budding reaction contained membranes (2.5 OD600/ml lysed pheroplasts), COPII components (1–2 μg Sec13/31p, 1–2 μg Sec23/24p, and 1 μg Sar1p), or 100 μg crude cytosol (25) supplemented with 1 μg Sar1p in a total volume of 125 μl with 1× ATP regeneration system (19) and 0.1 mM GTP. Reactions were incubated at 28°C for 45 min, and 10% of the total reaction was removed for analysis (Total). The remaining reaction mixture was centrifuged at medium speed (12,000 × g) for 4 min, and 75 μl of the supernatant fraction was collected for analysis (Vesicle fraction, 60% of Total). Proteins of interest were analyzed by immunoprecipitation from Total or Vesicle fractions.
Immunoprecipitation.
Immunoprecipitations (IPs) were performed as described in ref. 19 with minor modifications. After each sample was incubated for 10 min at 55°C in 20 μl of 5% SDS, the volume was adjusted to 1 ml with IP buffer without SDS [150 mM NaCl/1% Triton X-100/50 mM TrisCl (pH 7.5)] with protease inhibitors (1 mM PMSF/0.5 μg/ml leupeptin/0.7 μg/ml pepstatin), 50 μl of 30% (vol/vol) protein G-Sepharose in IP buffer without detergents, and 2 μl (1.2 μg/μl) monoclonal anti-HA antibody (12CA5) or 2 μl polyclonal anti-Vph1p serum (a gift from T. Stevens, University of Oregon, Eugene, OR). IP reactions were washed, resolved by SDS/PAGE, and analyzed with a PhosphorImager (Molecular Dynamics).
Fluorescence Microscopy and Immunofluorescence.
Phosphate starvation and GFP direct fluorescence in live yeast cells were performed as described (26). To study Gal2p-GFP localization, we grew cells expressing Gal2p-GFP in raffinose medium to log phase, and 1% galactose was added to the culture to induce GAL2 expression for 1 h. Immunofluorescence was conducted as described (27) by using formaldehyde fixation. For Pma1p and Kar2p localization, the mouse monoclonal anti-HA antibody 16B12 (Babco, Richmond, CA) and a rabbit anti-Kar2p polyclonal antibody were used, respectively. Secondary antibodies were goat anti-mouse IgG-conjugated to BODIPY TMR-X, and goat anti-rabbit IgG-conjugated to BODIPY FL (Molecular Probes). Images of coimmunofluoresence of Pho84p-GFP or Pho86p-GFP and Kar2p were documented by using a confocal microscope (LEICA TCSNT, Wetzlar).
Results
Localization of Pho84p-GFP Is Regulated in Response to Extracellular Phosphate Levels.
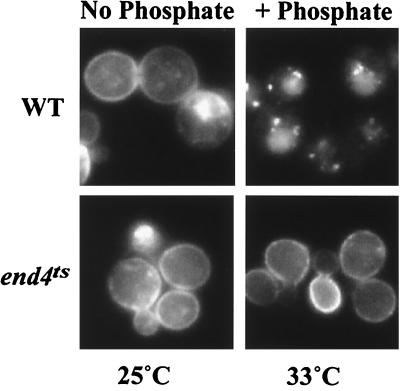
To investigate mechanisms for regulating the activity of Pho84p, we sought to study regulation of its localization in live cells by fluorescence microscopy. We constructed a fusion protein between Pho84p and GFP driven by the PHO84 promoter on a low copy plasmid (EB0666). This plasmid allowed a pho84 mutant strain to grow on low-phosphate plates and complemented the Phoc phenotype. As expected, Pho84p-GFP is localized to the plasma membrane in no-phosphate medium in wild-type cells (EY0664) (Fig. 1).
Figure 1.
Localization of Pho84p-GFP is regulated in response to extracellular phosphate levels. Wild type (EY0664, Upper) and an end4ts mutant (EY0687, Lower) harboring pPH084-GFP (EB0666) were studied. Direct fluorescence microscopy of Pho84p-GFP was performed in no-phosphate medium at 25°C (Left) and then in medium containing phosphate (final concentration 10 mM) and cycloheximide (final concentration 0.1 mg/ml) at 33°C.
To investigate changes in the localization of Pho84p in response to changes in phosphate levels, we added phosphate to a phosphate-starved culture. Localization of Pho84p-GFP was monitored in the absence of new protein synthesis by the addition of cycloheximide. Within 30 min, most Pho84p-GFP was internalized and localized to the vacuole (Fig. 1). However, if KCl instead of KH2PO4 was added under the same conditions, most Pho84p-GFP remained at the plasma membrane (data not shown), suggesting that the changes in Pho84p-GFP localization were because of changes in the rate of internalization in response to different phosphate levels in the medium. If arsenate, a phosphate analog, was added to the phosphate-starved culture in the presence of cycloheximide, Pho84p-GFP was not internalized, indicating that the changes in Pho84p localization are specific to phosphate levels in the culture (data not shown). Because the transcription of PHO84 is regulated by the PHO pathway, we wished to determine if the change in localization of Pho84p was also regulated by this signaling pathway. To test whether this was the case, we studied Pho84p-GFP localization in other PHO mutants. In a pho85Δ strain and in a strain containing PHO81c, a hyperactive allele of PHO81, changes in localization of Pho84p-GFP are similar to wild type (data not shown), demonstrating that regulation of Pho84p localization is independent of the phosphate signaling pathway.
To test whether internalization of Pho84p-GFP was mediated by endocytosis, we examined whether internalization of Pho84p was blocked in an end4ts mutant that is defective in endocytosis at a restrictive temperature. In the end4ts mutant, grown at the permissive temperature (25°C), Pho84p-GFP was endocytosed upon addition of phosphate (data not shown). However, at the restrictive temperature (33°C), Pho84p-GFP remained at the plasma membrane even after addition of phosphate (Fig. 1), indicating that Pho84p-GFP is internalized through the endocytic pathway. We conclude that Pho84p is regulated by a posttranslational mechanism; its localization is regulated in response to extracellular phosphate levels.
PHO86 Encodes an ER Resident Protein.
Because mutations in PHO84 and PHO86 result in essentially the same phenotype, it is possible that Pho84p and Pho86p participate in the same function, uptake of inorganic phosphate. It has been proposed that Pho86p might interact with Pho84p to form a protein complex that is required for high-affinity phosphate uptake (28). If so, Pho86p would be localized to the plasma membrane in low-phosphate medium. Alternatively, Pho86p may reside in an intracellular compartment involved in the biogenesis of Pho84p.
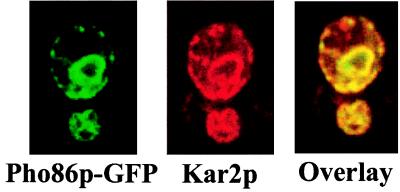
To distinguish between these two models, we studied the localization of Pho86p in low- and high-phosphate medium. We tagged the PHO86 gene at the C terminus with GFP and found that a low copy construct (EB0667) complemented the Phoc and low-phosphate lethal phenotypes of a pho86Δ strain (EY0665). In low-phosphate medium, Pho86p-GFP is localized to a perinuclear compartment coincident with Kar2p/Bip, a known ER resident protein (Fig. 2). Pho86p-GFP was also found in the ER in high-phosphate medium (data not shown), indicating that its localization is not regulated in response to extracellular phosphate levels. Thus, Pho86p is not likely to be a phosphate transporter. Instead, it may be required for the synthesis or transport of Pho84p from the ER.
Figure 2.
Pho86p-GFP is localized to the ER. Cells lacking PHO86 (EY0665) carrying pPHO86-GFP (EB0667) were starved for phosphate. Direct fluorescence of Pho86p-GFP was observed. Indirect immunofluorescence was performed by using an antibody to the ER-resident protein Kar2p/Bip. From left to right are fluorescence images corresponding to Pho86p-GFP (green), Kar2p (red), or an overlay of the two.
Pho84p-GFP Is Localized to the ER in the pho86 Mutant.
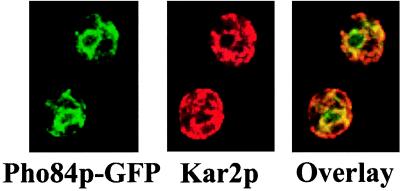
If Pho86p is involved in the transport of Pho84p, the permease may be retained in the ER in the absence of Pho86p. To test this hypothesis, we integrated PHO84-GFP (EB1125) into the yeast genome and investigated the localization of Pho84p-GFP in a strain lacking PHO86 (EY0669) by immunofluorescence in low-phosphate medium. As shown in Fig. 3, Pho84p-GFP is predominantly localized to the ER in a pho86 mutant, as indicated by its colocalization with Kar2p. By coimmunofluorescence, we also showed that Pho84p-GFP colocalizes with Ero1p-HA, another known ER resident protein (data not shown). Together with the observation that Pho84p stably accumulates in a pho86 mutant (data not shown), these results support the view that Pho86p has a role in Pho84p transport.
Figure 3.
Pho84p-GFP is localized to the ER in the absence of PHO86. A pho86Δ strain with integrated PHO84-GFP (EY0669) was starved for phosphate and coimmunofluorescence was performed as in Fig. 2. From left to right are fluorescence images corresponding to Pho84p-GFP (green), Kar2p (red), or an overlay of the two.
Pho86p Is Not Required for Targeting of Gal2p-GFP or Pma1p-HA to the Plasma Membrane.
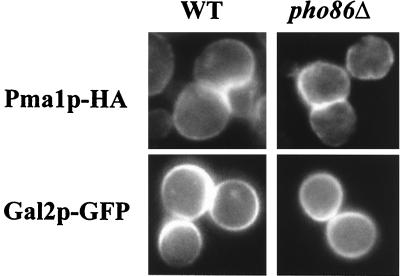
To investigate the specificity of Pho86p, we examined the subcellular localization of other plasma membrane proteins in the pho86Δ strain by indirect immunofluorescence. Pma1p-HA was localized to the plasma membrane in a pho86Δ strain (Fig. 4). However, Pho86p might still be required for the ER exit of a specific family of proteins. To determine whether deletion of PHO86 affects other transporters related in sequence to Pho84p, we investigated the localization of Gal2p-GFP in the pho86Δ strain. Gal2p is a galactose transporter in yeast, and shares 22% identity and 37% similarity with Pho84p (29). In both wild-type and pho86Δ strains, Gal2p-GFP is localized to the plasma membrane (Fig. 4). Moreover, pho86Δ cells are able to grow in a variety of conditions, including different carbon sources, and they can mate normally. From these data, we conclude that the function of Pho86p is likely to be specific to Pho84p.
Figure 4.
Pho86p is not required for proper localization of Pma1p-HA and Gal2p-GFP. (Left) Fluorescence images from wild-type cells.(Right) Images from strains lacking PHO86. Indirect immunofluorescence was performed on wild-type (EY0688) and pho86Δ (EY0689) strains expressing Pma1p-HA by using anti-HA antibodies (Upper). Direct fluorescence microscopy was performed on wild-type (EY0664) and pho86Δ (EY0665) strains expressing Gal2p-GFP grown in synthetic medium containing 2% raffinose, and 1% galactose (Lower).
Pho84p Is Packaged into COPII Transport Vesicles in Vitro.
Folded and mature secretory proteins are packaged into COPII vesicles, which bud from the ER membrane and then fuse with the Golgi apparatus. Pho86p may function in folding, cargo selection into COPII vesicles, or cargo retention in the Golgi membrane. To characterize the details of Pho84p exit from the ER, we studied its packaging into COPII vesicles derived from wild-type membranes utilizing an in vitro vesicle budding assay. This assay has been used to study packaging of Gap1p and Pma1p (19), and a subunit of the V0 complex of the vacuolar ATPase, Vph1p (P.M. and R.S., unpublished data). COPII vesicles can be generated from perforated spheroplasts by addition of ATP and GTP together with whole cytosol or purified COPII proteins that consist of a small GTPase, Sar1p, and coat proteins Sec23p, Sec24p, Sec13p, and Sec31p.
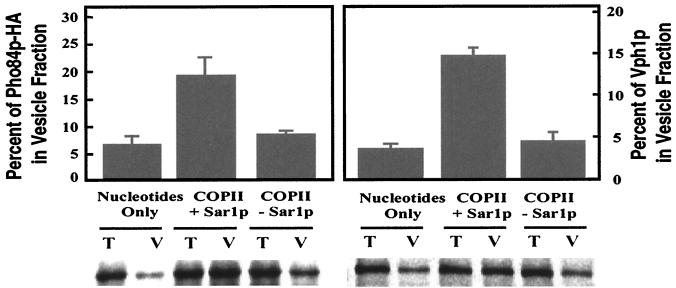
We tagged Pho84p at its C terminus with three tandem copies of the hemagglutinin antigen (HA) epitope and integrated this construct (EB1124) into the yeast genome to replace the wild-type copy of Pho84p. The resulting strain (EY0667) behaves like wild type with respect to growth in low-phosphate medium and PHO5 repression in high-phosphate medium. EY0667 cells were starved for phosphate and pulse-radiolabeled with [35S]methionine for 3 min, a short time period intended to label all newly synthesized proteins, including precursors freshly assembled in the ER. Perforated spheroplasts were prepared and incubated with purified COPII proteins to generate COPII vesicles in vitro. Vesicles were separated from membranes, and Pho84p-HA was immunoprecipitated under denaturing conditions from either the total budding reaction or the vesicle fraction and quantitatively analyzed. Pho84p-HA was packaged with approximately 20% efficiency into COPII vesicles when COPII proteins were included; the efficiency was reduced by more than 3-fold if Sar1p or any one of the purified coat proteins was omitted (data not shown), indicating that the packaging of Pho84p-HA into transport vesicles requires each of the COPII components (Fig. 5). As a control, we performed another immunoprecipitation to detect the packaging of Vph1p, from both the total and vesicle fraction of the budding reaction after Pho84p had been immunoprecipitated (Fig. 5). When perforated spheroplasts pulse-labeled for a longer time (18 min) were used in the budding reaction, the efficiency of Pho84p packaging was reduced to about 3% (data not shown), consistent with a greater proportion of Pho84p located in the compartments beyond the ER. These results demonstrate that standard COPII proteins and nucleotides are sufficient to package Pho84p-derived vesicles from wild-type membranes.
Figure 5.
COPII proteins and nucleotides are sufficient to package Pho84p into COPII vesicles from wild-type membranes. Permeabilized spheroplasts prepared from wild-type cells expressing Pho84p-HA (EY0667) were used in vesicle budding reactions in vitro. Reactions contained: nucleotides only (ATP and GTP), COPII + Sar1p (purified COPII proteins supplemented with nucleotides and Sar1p), and COPII − Sar1p (COPII proteins with nucleotides but without Sar1p). Pho84p-HA was immunoprecipitated from 10% of the total reactions, and the vesicle fractions were derived from 60% of the total reactions. A second immunoprecipitation was then performed to detect the amount of Vph1p in the total reactions and vesicle fractions. The immunoprecipitated proteins were resolved separately on 8% SDS/PAGE and quantified with a PhosphorImager. The percent of Pho84p-HA or Vph1p in the vesicle fraction compared with the corresponding total reaction (histogram) was calculated from the amount of radiolabeled Pho84p-HA or Vph1p (Lower), respectively. T, Total reaction. V, Vesicle fraction. Data are reported as the mean values of three independent experiments, and the error bars indicate the standard deviation.
Pho86p Is Required for Packaging Pho84p into Transport Vesicles in Vitro.
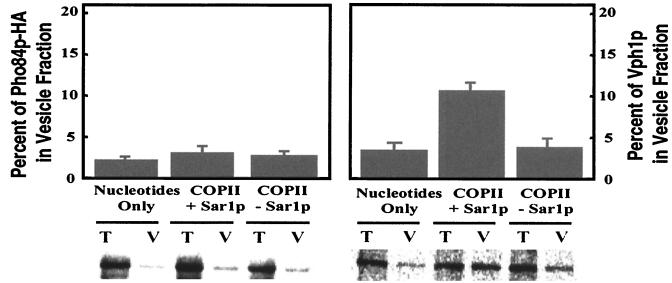
We next addressed the fate of Pho84p in membranes isolated from a pho86Δ mutant. Perforated spheroplasts were prepared from radiolabeled cells derived from a pho86Δ mutant expressing Pho84p-HA (EY0668), and the budding reactions were performed. Pho84p-HA was not detected above background levels in the vesicle fraction when purified COPII proteins were included in the budding reaction (Fig. 6). To test whether deletion of PHO86 has a general effect on cargo packaging into COPII vesicles, we investigated the packaging of Vph1p into COPII vesicles after Pho84p-HA had been immunoprecipitated. Approximately 12% of newly synthesized Vph1p was packaged by using pho86Δ mutant membranes, and this level was comparable to that achieved with wild-type membranes (see Fig. 5). These results suggest that Pho86p is not required for the general formation of COPII vesicles from the ER, or any later step in the secretory pathway. Rather, it is required at an early step for packaging of Pho84p into COPII vesicles. Thus, Pho86p belongs to a class of substrate-specific accessory proteins that have selective roles in the packaging of membrane proteins.
Figure 6.
Pho86p is required for packaging of Pho84p-HA, but not Vph1p, into COPII vesicles. Vesicle budding experiments and quantitation were performed as in Fig. 5. Permeabilized spheroplasts prepared from a pho86Δ mutant expressing Pho84p-HA (EY0668) were utilized in this experiment. Pho84p-HA was immunoprecipitated from both total and vesicle fractions by using anti-HA antibodies. Vph1p was then immunoprecipitated by using anti-Vph1 antibodies. Both proteins were resolved separately on 8% SDS/PAGE and analyzed with a PhosphorImager.
Pho86p Is Not Packaged into COPII Vesicles in Vitro.
Accessory proteins that facilitate cargo transport from ER to Golgi can be divided into three classes: “outfitters,” “escorts,” or “guides” (20). A fundamental feature of an “outfitter” is that it is not expected to accompany its cargo to the Golgi, and thus would not be packaged into COPII vesicles. Even though Pho86p is localized to the ER when yeast cells are grown in high- or low-phosphate medium, we could not rule out the possibility that Pho86p shuttles between the ER and the Golgi, as is expected of “escort” and “guide” proteins.
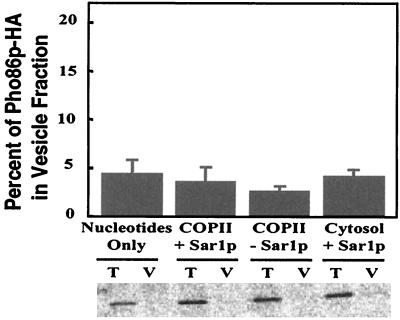
To follow the fate of Pho86p, we constructed a low copy plasmid (ARS/CEN) in which three copies of the HA epitope were introduced at the C terminus of Pho86p under the control of its native promoter (EB1126). The epitope-tagged Pho86p complemented the Phoc phenotype of a pho86Δ strain (EY0665) and allowed the strain to grow in low-phosphate medium. Indirect immunofluorescence indicated that Pho86p-HA was localized to the ER both in high- and low-phosphate medium (data not shown), confirming the previous observations made with Pho86p-GFP. To study the packaging of Pho86p into COPII vesicles, we transformed Pho86p-HA into a pho86Δ strain, pulse-labeled the cells for 3 min, and prepared perforated spheroplasts. We performed the budding reactions with either whole cytosol or purified COPII proteins supplemented with nucleotides. As shown in Fig. 7, Pho86p-HA did not appear above background levels in the vesicle fractions. As a control, Vph1p was packaged into the transport vesicles under the same conditions (data not shown). These data indicate that Pho86p is an ER resident protein that is likely to function as an “outfitter,” and thus, its role must precede the incorporation of Pho84p into COPII vesicles.
Figure 7.
Pho86 is not packaged into COPII vesicles. Cells lacking PHO86 (EY0665) and carrying plasmid pPHO86-HA (EB1126) were pulse-labeled, and perforated spheroplasts were prepared. Vesicle budding experiments were performed as in Fig. 5, except cytosol + Sar1p (cytosol supplemented with Sar1p and nucleotides) were also included in the reactions. Pho86p-HA was analyzed by immunoprecipitation and quantitation on a PhosphorImager.
Discussion
We have shown that Pho86p functions in the secretory pathway—it is required for packaging of Pho84p, a high-affinity phosphate transporter, into COPII vesicles. Pho86p is likely to belong to a class of “outfitters,” resident ER proteins that facilitate the export of cargo molecules from the ER (20). Pho86p might be required for folding, maturation, or oligomerization of Pho84p in the ER so that it can assume a conformation competent for incorporation into COPII vesicles. Another possibility is that Pho86p specifically recruits COPII proteins to the ER and then is displaced by have been reported Pho84p during the budding reaction.
Recently, many examples have been reported of accessory proteins that facilitate the ER exit of cargoes. Three such proteins in S. cerevisiae, Vma21p, Chs7p, and Gsf2p, are required for the ER exit of Vph1p (30), chitin synthase, Chs3p (31), and a glucose transporter, Hxt1p (32), respectively. Though these three accessory proteins are localized to the ER in steady state, it is not known if they function prior to the incorporation of their respective cargoes into COPII vesicles or if these proteins themselves are transported out of the ER. However, both Vma21p and Gsf2p possess C-terminal, cytoplasmic retrieval (-KKXX) sequences, which suggests that these molecules cycle between the ER and the Golgi apparatus. In contrast, Pho86p appears to function in a manner similar to that of Shr3p. Neither Pho86p nor Shr3p has a recognizable retrieval signal. Shr3p is required for packaging of a family of amino acid permeases into COPII vesicles, but itself is not included in these vesicles (19). Interestingly, other than the C-terminal retrieval signals on two of these proteins, these accessory proteins do not share homology, and they facilitate exit of different classes of cargoes from the ER. It is unclear whether these proteins function as part of the quality control procedure, or part of the cargo selection process involving COPII coat proteins and other sorting receptors.
Our data suggest that the function of Pho86p is specific to Pho84p. Pho86p is not required for the ER exit of other plasma membrane proteins that were investigated. Interestingly, it is not required for the plasma membrane targeting of Gal2p, which belongs to the same 12-transmembrane-domain protein family. In this respect, Pho86p apparently functions in a manner different from Shr3p and Gsf2p, which are required for the ER exit of a family of transporters; Shr3p is required for Gap1p and Hip1p (19), and Gsf2p for Hxt1p and Gal2p (32). It is not known whether Pho86p directly interacts with Pho84p. A direct interaction between these two proteins could explain the specificity of Pho86p in substrate selection. The nature of this interaction could be explored by using Pho84p homologues from other species expressed in wild-type and pho86 mutants, or by the use of Pho84p-Gal2p chimeric proteins.
Among more than 6,000 gene products in yeast, it is estimated that at least 10% of them go through the secretory pathway (14). When phosphate is limiting, the major components of the secretory pathway are very different from those that are present in phosphate-rich conditions. It is likely that there exist mechanisms to ensure efficient ER exit of different cargoes under certain physiological conditions and that the sorting itself may be regulated by nutritional conditions. Nitrogen regulation of sorting and stability of the general amino acid permease, Gap1p, in the Golgi apparatus is an example of nutritionally regulated transport in yeast (33). Pho86 may play such a role because its transcription is highly induced under low-phosphate conditions.
Acknowledgments
We thank A. Kruckeberg for the plasmid expressing Gal2p-GFP, T. Stevens for anti-Vph1p serum, J. Weissman for the plasmid expressing Ero1p-HA, and B. Lesch for the preparation of purified COPII components. We also thank T. Shroyer for making the disruption vector for PHO86 (EB0477). We thank the members of the O'Shea and Schekman laboratories for their technical assistance and constructive comments. We are especially grateful to A. Spang and L. Jan for their critical comments on the manuscript. This work was supported by Grant GM51377 from the National Institutes of Health (to E.K.O.), and by the Howard Hughes Medical Institute (to R.S.). W.-T.W.L. was supported by a Howard Hughes Medical Institute predoctoral fellowship.
Abbreviations
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
References
- 1.Nieuwenhuis B J, Borst-Pauwels G W. Biochim Biophys Acta. 1984;770:40–46. doi: 10.1016/0005-2736(84)90071-3. [DOI] [PubMed] [Google Scholar]
- 2.Oshima Y. In: The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 159–180. [Google Scholar]
- 3.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yompakdee C, Bun-ya M, Shikata K, Ogawa N, Harashima S, Oshima Y. Gene. 1996;171:41–47. doi: 10.1016/0378-1119(96)00079-0. [DOI] [PubMed] [Google Scholar]
- 5.Ueda Y, Oshima Y. Mol Gen Genet. 1975;136:255–259. doi: 10.1007/BF00334020. [DOI] [PubMed] [Google Scholar]
- 6.Berhe A, Friestedt U, Persson B L. Eur J Biochem. 1995;227:566–572. doi: 10.1111/j.1432-1033.1995.tb20426.x. [DOI] [PubMed] [Google Scholar]
- 7.Harrison M J, van Buuren M L. Nature (London) 1995;378:626–629. doi: 10.1038/378626a0. [DOI] [PubMed] [Google Scholar]
- 8.Muchhal U S, Pardo J M, Raghothama K G. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leggewie G, Willmitzer L, Riesmeier J W. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. Proc Natl Acad Sci USA. 1997;94:7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Trieu A T, Blaylock L A, Harrison M J. Mol Plant–Microbe Interact. 1998;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- 12.Lau W-T W, Schneider K R, O'Shea E K. Genetics. 1998;150:1349–1359. doi: 10.1093/genetics/150.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bun-ya M, Shikata K, Nakade S, Yompakdee C, Harashima S, Oshima Y. Curr Genet. 1996;29:344–351. [PubMed] [Google Scholar]
- 14.Kaiser C A, Gimeno R E, Shaywitz D A. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Pringle J R, Broach J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 91–228. [Google Scholar]
- 15.Barlowe C. Biochim Biophys Acta. 1998;1404:67–76. doi: 10.1016/s0167-4889(98)00047-0. [DOI] [PubMed] [Google Scholar]
- 16.Marcusson E G, Horazdovsky B F, Cereghino J L, Gharakhanian E, Emr S D. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 17.Chang A, Fink G R. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ljungdahl P O, Gimeno C J, Styles C A, Fink G R. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- 19.Kuehn M J, Schekman R, Ljungdahl P O. J Cell Biol. 1996;135:585–595. doi: 10.1083/jcb.135.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann J M, Malkus P, Schekman R. Trends Cell Biol. 1999;9:5–7. doi: 10.1016/s0962-8924(98)01414-7. [DOI] [PubMed] [Google Scholar]
- 21.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1993. [Google Scholar]
- 22.Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1978. [Google Scholar]
- 23.Jones J S, Prakash L. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 24.Rexach M F, Latterich M, Schekman R W. J Cell Biol. 1994;126:1133–1148. doi: 10.1083/jcb.126.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spang A, Schekman R. J Cell Biol. 1998;143:589–599. doi: 10.1083/jcb.143.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaffman A, Rank N M, O'Shea E K. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwappach B, Stobrawa S, Hechenberger M, Steinmeyer K, Jentsch T J. J Biol Chem. 1998;273:15110–15118. doi: 10.1074/jbc.273.24.15110. [DOI] [PubMed] [Google Scholar]
- 28.Yompakdee C, Ogawa N, Harashima S, Oshima Y. Mol Gen Genet. 1996a;251:580–590. doi: 10.1007/BF02173648. [DOI] [PubMed] [Google Scholar]
- 29.Hodges P E, McKee A H Z, Davis B P, Payne W E, Garrels J I. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham L A, Hill K J, Stevens T H. J Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trilla J A, Duran A, Roncero C. J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherwood P W, Carlson M. Proc Natl Acad Sci USA. 1999;96:7415–7420. doi: 10.1073/pnas.96.13.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberg K J, Rowley N, Kaiser C A. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]