Abstract
Colonization by the gastric pathogen Helicobacter pylori has been shown to be intricately linked to the development of gastritis, ulcers, and gastric malignancy. Little is known about mechanisms employed by the bacterium that help it adapt to the hostile environment of the human stomach. In an effort to extend our knowledge of these mechanisms, we utilized spotted-DNA microarrays to characterize the response of H. pylori to low pH. Expression of approximately 7% of the bacterial genome was reproducibly altered by shift to low pH. Analysis of the differentially expressed genes led to the discovery that acid exposure leads to profound changes in motility of H. pylori, as a larger percentage of acid-exposed bacterial cells displayed motility and moved at significantly higher speeds. In contrast to previous publications, we found that expression of the bacterial virulence gene cagA was strongly repressed by acid exposure. Furthermore, this transcriptional repression was reflected at the level of protein accumulation in the H. pylori cell.
Since its isolation from gastric biopsy samples in 1982 by Marshall and Warren (44), the small gram-negative bacterium Helicobacter pylori has been shown to infect over 50% of the world's population (45). While transmission of the bacterium between humans or from an unknown environmental reservoir is still poorly characterized, it is now understood that colonization generally results in a persistent infection that, unless specifically treated, lasts the lifetime of the infected individual (12). This has profound clinical importance, as H. pylori colonization has been shown to be associated with multiple forms of gastric disease. These range in severity from mild gastritis to duodenal and gastric ulcers and two forms of gastric malignancy, mucosa-associated lymphoid tissue lymphoma and adenocarcinoma (13, 24, 59).
In order to establish a successful infection, bacteria that colonize within the dynamic substrate of a human host must have the ability to adapt to and modify gene expression in response to changes in the host environment. A thorough understanding of the survival mechanisms employed by H. pylori during colonization of the stomach remains lacking, but one of the most obvious challenges is the constant fluctuations in pH that occur. The gastric lumen can reach a pH of 2.0 during the course of digestion, and while the mucus layer overlaying the gastric epithelium, the site of colonization of H. pylori, generally remains more neutral, the bacteria no doubt face fluctuations in pH that must be sensed and responded to (75). Because of this, mechanisms involved in acid resistance (AR) have been the focus of considerable study. AR has been shown to be intricately linked with H. pylori's ability to produce copious amounts of urease (55). The urease enzyme catalyzes the hydrolysis of urea to carbon dioxide and ammonia and helps maintain a proton motive force across the inner membrane of the bacterium (54, 64). It also leads to the extrusion of the basic ammonia molecules into the surrounding environment, thus buffering the bacterial microenvironment. Urease enzymatic activity is crucial for colonization and persistence in the stomach (6, 25, 28, 79).
In addition to urease, genes for other factors have also been shown to be important for AR. These include wbcJ, which is involved in the assembly of mature lipopolysaccharide (49); pldA, which undergoes phase variation leading to changes in the lipid composition of the outer membrane that affect AR (74); recA, which is involved in DNA damage repair (77); rocF, which is required for the arginase activity that hydrolyzes l-arginine to l-ornithine and urea (47); atpD and atpF, which encode subunits of the F1F0-ATPase (9, 48); and fur, which encodes the ferric iron uptake regulator and was recently identified as the first known transcriptional regulator required for growth in acidic conditions (10). The activities of these gene products exemplify the fact that a diverse range of factors, active at many subcompartments in the cell, are involved in the adaptive response to acid.
Microarray technology is a powerful tool for analyzing the transcriptional response of microbes to changes in environmental conditions, such as those that mimic in vivo conditions that pathogens would encounter within the context of their host. As such, two previous microarray-based studies have determined some of the transcriptional changes in H. pylori as it adapts to acid stress (3, 7). In both studies, only a single time point after exposure to acid conditions was examined. There was no overlap between the genes identified in the two studies, suggesting that neither screen was comprehensive or, alternatively, that the two sets of acid conditions examined result in changes in nonoverlapping sets of genes.
To better understand adaptive mechanisms utilized by H. pylori within the context of the host environment, we utilized spotted-DNA microarrays to characterize in a temporal manner the global changes in gene expression in response to low pH in the pathogenic H. pylori strain G27. Direct comparison to transcriptional levels of the starting culture revealed that expression of many genes was rapidly altered and remained altered for the duration of the acid exposure. Approximately 7% of the predicted open reading frames (ORFs) were significantly up or down regulated. Among these were many factors whose expression had previously been shown to be affected by low pH, as well as many factors whose pH-regulated gene expression had not previously been identified. Differential regulation of a number of the components of the flagellar regulon led us to analyze the effect of acid exposure on motility of H. pylori, and a profound increase in both the percentage and speed of motile bacteria after acid exposure was noted. In contrast to previous reports (3, 37), expression of the virulence gene cagA was repressed by acid exposure and, accordingly, a reduced level of CagA was found in the bacterial cell. Finally, mutation of the acid-repressed ansB gene, while showing no obvious effect on in vitro AR, resulted in a strain that was significantly less able to stably infect within the context of the Mongolian gerbil model of H. pylori gastric colonization.
MATERIALS AND METHODS
Bacterial strains and growth.
The H. pylori strains G27 (18) and B128 (60) were maintained as frozen stocks at −80°C in brain heart infusion media supplemented with 20% glycerol and 10% fetal bovine serum (FBS). Bacteria were grown on horse blood agar (HBA) plates containing 4% Columbia agar base (Oxoid), 5% defibrinated horse blood (HemoStat Labs, Dixon, Calif.), 0.2% β-cyclodextrin (Sigma), 10 μg of vancomycin (Sigma)/ml, 5 μg of cefsulodin (Sigma)/ml, 2.5 U of polymyxin B (Sigma)/ml, 50 μg of cycloheximide (Sigma)/ml, 5 μg of trimethoprim (Sigma)/ml, and 8 μg of amphotericin B (Sigma)/ml under microaerophilic conditions at 37°C. A microaerobic atmosphere was generated by using a CampyGen sachet (Oxoid) in a gas pack jar. For liquid culture, H. pylori was grown in brucella broth (Difco) containing 10% FBS (Gibco/BRL) with shaking in a microaerobic environment.
Acid exposure and RNA isolation.
An overnight liquid culture of G27 (optical density at 600 nm [OD600] = 0.5 to 1.0) was harvested by centrifugation and resuspended in pH 5.0 brucella broth supplemented with 10% FBS. The pH of the media was adjusted by using concentrated hydrochloric acid prior to resuspension. Bacterial samples collected prior to acid exposure represent the zero time point, and samples were harvested after 30, 60, 90, and 120 min of acid exposure. At each time point, aliquots were removed from the pH 5.0 media and were plated to determine the numbers of CFU and were harvested on a 0.45-μm-pore-size cellulose filter by vacuum filtration. Filters containing the H. pylori cells were immediately frozen in liquid nitrogen and were subsequently used for isolation of bacterial RNA.
H. pylori RNA was isolated by using TRIzol reagent (Gibco/BRL) as previously described (52). RNA concentration was quantitated by determination of the absorbance at 260 and 280 nm, and RNA integrity was verified by visualization on a 1% agarose gel.
Microarray hybridization and analysis.
Equal concentration of each test RNA (T = 30, 60, 90, and 120 min from biologically independent experiments) and the reference RNA (T = 0) were used for cDNA synthesis in a standard reverse transcriptase reaction by using Superscript II (Invitrogen) and Panorama H. pylori cDNA labeling primers (SigmaGenosys). Synthesized cDNA was purified by using Qia-Quick PCR purification columns (Qiagen) according to the manufacturer's instructions and subsequently were indirectly labeled with Cy5 and Cy3 fluorophores as previously described (62). Individual Cy5 and Cy3 reactions were combined, and unincorporated dye was removed by using a Qia-Quick PCR column according to the manufacturer's instructions. The eluate from the columns was concentrated by evaporation in a Speed Vac and was resuspended in 11 μl of Tris-EDTA. One microliter of 25-mg/ml yeast tRNA, 2.55 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.45 μl of 10% sodium dodecyl sulfate were added to each of the labeled probes. These were heated to 99°C for 2 min, cooled briefly, and then added to the H. pylori microarray for hybridization and stringency washes as has been previously described (62). The hybridized slides were scanned and analyzed by using an Axon 4000A scanner and the GENEPIX 3.0 software (Axon).
Data were collated with the Stanford University Microarray Database (67). Spots showing obvious abnormalities were excluded from analysis. Spot quality was further filtered by requiring that the Cy3 mean spot intensity-to-Cy5 median background intensity ratio be greater than or equal to 2.5 and the Cy5 mean spot intensity-to-Cy3 median background intensity ratio be greater than or equal to 2.5. Genes were filtered by providing that the log (base 2) of the red-to-green normalized ratio be greater than 1.8 standard deviations from the mean ratio in at least one array for each experimental set. Biologically independent experiments were analyzed separately, and the data were collapsed to reveal factors whose expression was consistently altered by acid exposure. All microarray data generated by this study are publicly available at http://genome-www.stanford.edu/microarray/.
Motility analysis.
A liquid culture of G27 was harvested by centrifugation and split into two portions: one was resuspended in pH 7.0 brucella broth plus 10% FBS and the other in pH 5.0 brucella broth plus 10% FBS. Motility was monitored by live phase-contrast microscopy. A Hamamatsu C2400 video charge-coupled device camera was used to record via an Argus-20 image processor onto S-VHS video. Resulting images were digitized into NIH Object Image for the generation of time-lapse films. Adobe Photoshop was used to assemble the figures. To trace the speed of movement of the bacteria, the TRACE function of the Argus-20 image processor was used and the individual length of the traces was calculated by using NIH Object Image.
ΔansB construction and characterization.
A deletion-insertion of the ansB coding sequence (HP0723) was constructed by allelic exchange in strain B128. The allelic exchange vector was constructed by amplification of an ansB-upstream fragment by using primers Hp0723-1 (GCGGCCGCAATCCAATCAAGCAGAAA) and Hp0723-2 (TCTAGAGAAGAGTATTGAAAGATT) and of an ansB-downstream fragment by using primers Hp0723-3 (TCTAGACATAACCTGCCCCTTCAA) and Hp0723-4 (GGTACCAACGATCGCTACAACCCCA). The primers incorporate XbaI and NotI sites and XbaI and KpnI sites, respectively (underlined), that were used for subsequent directional cloning. PCR products were subcloned into the pCR2.1-TOPO vector (Invitrogen), and fragments were then removed by restriction enzyme digestion (enzymes mentioned above) and were directionally ligated with a kanamycin resistance cassette into a pSK-derived suicide vector (36). The resulting construct was verified by restriction enzyme digestion with appropriate enzymes and was subsequently used for natural transformation of H. pylori. Allelic exchange mutants were selected by plating on HBA plates supplemented with 25 μg of kanamycin/ml, and the proper deletion-insertion was confirmed by Southern blotting.
In vitro growth curves were conducted by inoculation of brucella broth supplemented with 10% FBS to a starting OD600 of approximately 0.06 to 0.1 with an overnight-grown culture of the wild type or ansB mutant followed by subsequent determination of the OD600 at the time points indicated in Fig. 3. In vitro competition assays were conducted by mixing equal numbers of CFU of the ansB and wild-type strain in brucella broth plus 10% FBS, which was followed by growth with shaking in a microaerobic environment. Relative numbers of CFU of each strain were then determined by plating on HBA alone and HBA supplemented with kanamycin.
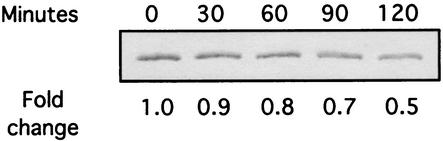
FIG. 3.
The effect of low pH on CagA accumulation. Total protein was harvested from G27 shifted to pH 5.0 brucella broth plus 10% FBS at the times indicated. An equal amount of protein from each point was separated on a 6% polyacrylamide gel, transferred to nitrocellulose membranes, and probed with an anti-CagA specific antibody. The relative change (n-fold) from the zero time point is indicated underneath each lane.
Immunoblot detection of CagA.
The relative concentration of the CagA protein was determined by collection of bacterial cells that had undergone an acid exposure as described for the microarray analysis above. Bacterial cells were lysed by boiling in standard 1× sodium dodecyl sulfate buffer, and the protein concentration was determined using a BCA kit (Pierce). An equal amount of protein from each sample was separated on a 6% polyacrylamide gel by electrophoresis, and the proteins were transferred by using a semidry transfer apparatus (E&K Scientific) to nitrocellulose membrane. The resulting blot was probed with a primary mouse monoclonal anti-CagA antibody (Austra Biological) and a secondary anti-mouse immunoglobulin G antibody conjugated to Alexa 660 (Molecular Probes). Resulting bands were visualized by scanning with an Odyssey scanner, and relative protein concentrations were determined by scanning densitometry with the same program (Licor).
Gerbil infections.
Four- to 8-week-old male Mongolian gerbils (Harlan, Indianapolis, Ind.) were fasted for 20 h prior to infection and were subsequently infected with 0.5 ml of a 1:1 mix of liquid grown wild-type B128 and ΔansB bacteria via oral gavage. This represents an inoculum of approximately 108 to 109 total bacteria. Infections were allowed to proceed for 2.5 to 3 weeks, and then animals were sacrificed, the glandular portion of the stomach was excised and homogenized with a mechanical homogenizer, and the numbers of CFU were determined by plating on HBA alone and HBA supplemented with kanamycin. The relative number of wild-type bacteria was determined by subtraction of the number of kanamycin-resistant colonies from the total number of colonies growing on the plain-HBA plates. The competitive index was determined by division of the number of mutant bacteria by the number of wild-type bacteria followed by corrections for deviations from an input ratio of 1:1.
RESULTS AND DISCUSSION
Microarray analysis of pH-regulated gene expression of G27.
To reveal dynamic changes that occur in response to low pH, it was necessary to ensure that H. pylori strain G27 would remain viable throughout the course of the experiment. This was a critical experimental parameter, as decreased viability at later time points could greatly affect the transcriptional profiles. Consequently, survival curves for G27 were determined in pH 5.0 brucella broth supplemented with 10% FBS and revealed virtually full survival of the strain throughout the 2-h exposure (data not shown). The pH of the media also remained unchanged, suggesting that these conditions would be useful for determination of expression profiles of the strain.
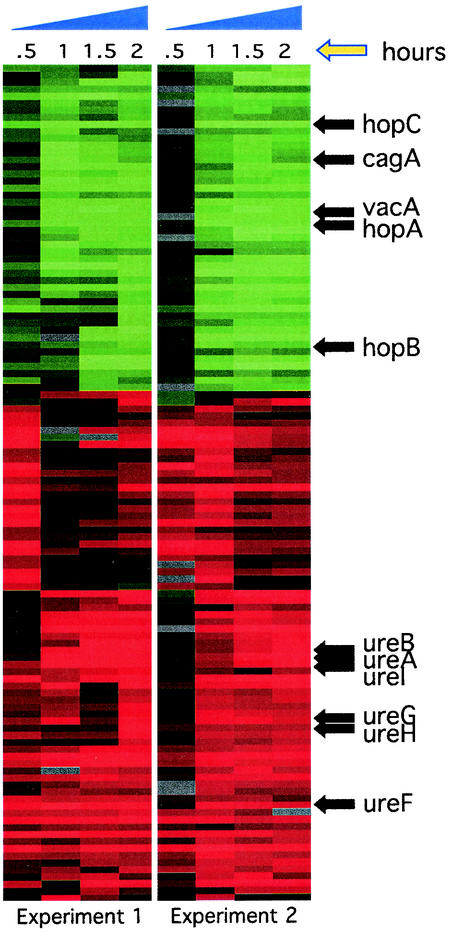
An overnight culture of G27 was harvested by centrifugation and resuspended in pH 5.0 brucella broth media as described in Materials and Methods. Samples were taken prior to resuspension and at T = 30, 60, 90, and 120 min thereafter. Two independent experiments were conducted on separate days, and total RNA was prepared from all 10 samples and utilized for transcriptome analysis. For each experiment, cDNA synthesized from RNA from the zero time point was utilized as the reference and was labeled with Cy3 while cDNA synthesized from RNA harvested after exposure to acid was labeled with Cy5. Each Cy5 and Cy3 pair was hybridized to a spotted-DNA microarray representing 98.9% of the unique ORFs found in both of the sequenced H. pylori strains (5, 78). Each DNA fragment is additionally spotted in duplicate on the array, thus allowing for technical replicates within a single hybridization (62). Ratios of each Cy5 and Cy3 fluorophore for each spot were obtained by scanning with an Axon scanner and represent the changes in total mRNA content after exposure to the acid stimulus. Data for each experiment were filtered as described in Materials and Methods, and genes showing altered patterns of expression in both experiments are schematically depicted in Fig. 1 and are listed in Table 1 and in the supplementary information at http://falkow.stanford.edu/whatwedo/supplementarydata/.
FIG. 1.
Microarray analysis of the low-pH response of H. pylori. Cluster diagram showing the expression profile of the 118 genes meeting the filter criteria as explained in Materials and Methods after shift to pH 5.0. Two independent experiments are depicted, and relative expression patterns are shown for each time point. Red indicates an increase in expression, while green indicates reduced expression. Representative genes are listed and their relative location indicated by an arrow. A complete list of the regulated factors and the relative changes in expression can be found in Table 1 and as supplementary information at http://falkow.stanford.edu/whatwedo/supplementarydata/.
TABLE 1.
pH-regulated genes of H. pyloria
| Expression | Class | Gene designation
|
Gene and function | Change (n-fold) at:
|
||||
|---|---|---|---|---|---|---|---|---|
| TIGR | ASTRA | T = 30 | T = 60 | T = 90 | T = 120b | |||
| Induced | Amino acid biosynthesis | HP1282 | JHP1203 | Anthranilate synthase component, trpE | 2.4 | 2.5 | 2.5 | 2.7 |
| HP0106 | JHP0098 | Cystathionine gamma-synthase, metB | 2.3 | 1.8 | 1.8 | 3.5 | ||
| HP1050 | JHP0375 | Homoserine kinase, thrB | 1.9 | 2.3 | 3.0 | 3.1 | ||
| HP0290 | JHP0275 | Diaminopimelate decarboxylase (dap decarboxylase), lysA | 1.2 | 2.4 | 2.9 | 2.9 | ||
| HP0380 | JHP1001 | Glutamate dehydrogenase, gdhA | 2.4 | 1.2 | 1.3 | 2.0 | ||
| Biosynthesis of cofactors, pros- thetic groups, and carriers | HP0306 | JHP0291 | Glutamate-1-semialdehyde 2,1-aminomutase, hemL | 1.9 | 1.9 | 1.6 | 2.6 | |
| HP0755 | JHP0692 | Molybdopterin biosynthesis protein, moeB | 1.7 | 1.6 | 2.2 | 2.2 | ||
| Cell envelope and surface structures | HP1155 | JHP1082 | UDP-n-acetylglucosamine-n-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol n-acetylglucosamine transferase, murG | 3.4 | 1.4 | 1.3 | 1.9 | |
| HP0743 | JHP0680 | Rod shape-determining protein, mreB | 1.6 | 1.9 | 2.4 | 2.5 | ||
| HP0295 | JHP0280 | Flagellin B homolog (HAP3) | 4.2 | 1.4 | 1.9 | 2.0 | ||
| HP1119 | JHP1047 | Flagellar hook-associated protein 1 (HAP1), flgK | 3.9 | 1.7 | 1.3 | 2.1 | ||
| HP1559 | JHP1467 | Flagellar basal-body rod protein (proximal rod protein), flgB | 3.8 | NDc | 2.5 | 2.4 | ||
| HP0115 | JHP0107 | Flagellin, flaB | 3.5 | 1.0 | 1.4 | 1.4 | ||
| HP0870 | JHP0804 | Flagellar hook, flgE | 3.0 | 1.1 | 1.5 | 1.9 | ||
| HP1558 | JHP1466 | Flagellar basal-body rod protein (proximal rod protein), flgC | 2.0 | 1.1 | 1.3 | 1.6 | ||
| HP1031 | JHP0393 | Flagellar motor switch protein, fliM | 1.8 | 2.3 | 1.1 | 2.3 | ||
| HP1122 | JHP1051 | Anti-σ28 factor, flgM | 2.7 | 1.1 | 1.3 | 1.6 | ||
| Cellular processes | HP1332 | JHP1252 | Cochaperone and heat shock protein, cochaperone with DnaK, dnaJ | 4.0 | 4.0 | 3.0 | 3.1 | |
| Central intermediary metabolism | HP0069 | JHP0064 | Urease accessory protein, ureF | 3.0 | 4.0 | 3.4 | 3.5 | |
| HP0071 | JHP0066 | Urease accessory protein, ureI | 2.2 | 4.1 | 6.4 | 5.5 | ||
| HP0068 | JHP0063 | Urease accessory protein, ureG | 1.8 | 2.1 | 1.8 | 3.0 | ||
| HP0067 | JHP0062 | Urease accessory protein, ureH | 1.7 | 2.2 | 1.5 | 3.0 | ||
| HP0073 | JHP0068 | Urease alpha subunit, ureA | 1.7 | 3.8 | 7.2 | 6.9 | ||
| HP0072 | JHP0067 | Urease beta subunit, ureB | 1.2 | 3.4 | 7.1 | 7.2 | ||
| DNA metabolism, restriction and modification | HP1121 | JHP1050 | Cytosine-specific DNA methyltransferase, BSP6IM | 2.2 | 1.3 | 1.2 | 1.6 | |
| Energy metabolism | HP0294 | JHP0279 | Aliphatic amidase energy metabolism, amiF | 4.1 | 2.4 | 11 | 9.5 | |
| HP1238 | JHP1159 | Aliphatic amidase energy metabolism, amiE | 2.4 | 0.8 | 9.0 | 7.5 | ||
| JHP585 | Putative 3-hydroxyacid dehydrogenase | 1.0 | 3.2 | 4.2 | 10.1 | |||
| HP0642 | JHP0586 | NAD(P)H-flavin oxidoreductase | 1.1 | 5.7 | 8.1 | 8.1 | ||
| Fatty acid and phospholipid metabolism and biosynthesis | HP0557 | JHP0504 | Acetyl-coenzyme A carboxylase, accA | 1.4 | 1.4 | 2.3 | 1.7 | |
| Hypothetical gene | HP1233 | JHP1154 | 5.7 | 0.6 | ND | 2.8 | ||
| HP1154 | JHP1081 | 5.1 | 1.1 | 1.4 | 1.9 | |||
| HP0367 | JHP1014 | 4.6 | 1.5 | 2.3 | 2.6 | |||
| HP0119 | 4.5 | 0.9 | 4.1 | 3.7 | ||||
| HP0118 | JHP0110 | 4.3 | 1.7 | 1.7 | 3.2 | |||
| HP1327 | JHP1247 | 3.9 | 1.2 | 1.1 | 1.0 | |||
| HP1187 | JHP1113 | 3.8 | 2.6 | 3.7 | 3.6 | |||
| HP1076 | JHP0349 | 3.6 | 1.8 | 1.5 | 1.6 | |||
| HP0219 | JHP0205 | 2.9 | 3.1 | 3.4 | 3.5 | |||
| HP1173 | JHP1100 | 2.8 | 4.0 | 5.9 | 4.9 | |||
| HP1188 | 2.8 | 0.8 | 1.0 | 1.9 | ||||
| HP1022 | JHP0402 | 2.2 | 2.1 | 2.4 | 2.1 | |||
| HP1440 | JHP1333 | 2.1 | 1.1 | 1.2 | 1.3 | |||
| HP1457 | JHP1350 | 2.0 | 1.4 | 1.2 | 1.6 | |||
| HP0018 | JHP0016 | 1.7 | 2.1 | 1.7 | 2.6 | |||
| HP0554 | JHP0501 | 1.7 | 2.2 | 2.0 | 1.9 | |||
| HP0719 | JHP0657 | 0.8 | 2.0 | 2.3 | 2.8 | |||
| HP0641 | JHP0584 | 1.4 | 1.9 | 5.4 | 5.7 | |||
| HP1331 | JHP1251 | Conserved hypothetical integral membrane protein | 2.7 | 2.5 | 2.6 | 2.6 | ||
| HP0228 | JHP0213 | Conserved hypothetical integral membrane protein | 2.1 | 3.8 | 1.5 | 4.1 | ||
| HP1225 | JHP1146 | Conserved hypothetical integral membrane protein | 2.1 | 1.6 | 1.7 | 2.0 | ||
| HP0318 | JHP0301 | Conserved hypothetical protein | 2.0 | 3.2 | 1.8 | 3.8 | ||
| HP1430 | JHP1323 | Conserved hypothetical ATP-binding protein | 1.8 | ND | 2.5 | 2.2 | ||
| HP1507 | JHP1400 | Conserved hypothetical ATP-binding protein | 1.7 | 1.4 | 1.7 | 4.0 | ||
| HP0759 | JHP0696 | Conserved hypothetical integral membrane protein | 1.5 | 2.3 | 2.7 | 1.8 | ||
| HP1020 | JHP0404 | Conserved hypothetical protein | 1.3 | 3.0 | 3.1 | 2.9 | ||
| Purines, pyrimidines, nucleosides and nucleotides | HP0757 | JHP0694 | β-alanine synthetase homolog | 2.3 | 2.6 | 3.0 | 2.7 | |
| Regulatory functions | HP1021 | JHP0403 | Putative transcriptional regulator | 1.4 | 2.9 | 3.6 | 3.9 | |
| HP0278 | JHP0263 | Guanosine-5′-triphosphate-3′-diphosphate pyrophosphatase, gppA | 0.7 | 1.3 | 1.5 | 3.4 | ||
| Transcription, transcription factors and translation | HP0550 | JHP0497 | Transcription termination factor Rho, rho | 2.6 | 1.6 | 1.0 | 0.7 | |
| HP1203 | JHP1126 | Transcription termination factor NusG, nusG | 1.8 | 2.0 | 2.2 | 2.0 | ||
| HP0123 | JHP0113 | Threonyl-tRNA synthetase, thrS | 2.0 | 1.8 | 2.1 | 2.0 | ||
| HP1019 | JHP0405 | Serine protease, htrA | 2.5 | 3.4 | 4.2 | 3.9 | ||
| Transport and binding proteins | HP1171 | JHP1098 | Glutamine ABC transporter, ATP-binding protein, glnQ | 1.1 | 1.9 | 2.3 | 2.1 | |
| HP1172 | JHP1099 | Glutamine ABC transporter, periplasmic glutamine-binding protein, glnH | 1.0 | 1.8 | 2.0 | 3.6 | ||
| HP0471 | JHP0423 | Glutathione-regulated potassium efflux system, kefB | 2.1 | 1.4 | 1.2 | 1.2 | ||
| HP1339 | JHP1258 | Biopolymer transport protein, exbB | 3.0 | 1.9 | 1.8 | 2.0 | ||
| HP1340 | JHP1259 | Biopolymer transport protein, exbD | 2.3 | 1.4 | 0.9 | 2.1 | ||
| HP0876 | JHP0810 | Iron-regulated OMP, frpB | 1.4 | 3.3 | 1.9 | 1.2 | ||
| HP0715 | JHP0653 | ABC transporter, ATP-binding protein | 2.2 | 1.0 | 1.2 | 1.4 | ||
| HP1432 | JHP1321 | Putative histidine and glutamine-rich metal-binding protein | 2.0 | 2.4 | 2.8 | 3.1 | ||
| HP1465 | JHP1358 | ABC transporter, ATP-binding protein, HI1087 | 1.7 | 1.3 | 1.3 | 2.6 | ||
| Repressed | Amino acid biosynthesis | HP1038 | JHP0386 | 3-Dehydroquinase type II, aroD | 0.6 | 0.4 | 0.6 | 0.5 |
| HP0695 | JHP0633 | Hydantoin utilization protein A, hyuA | 1.0 | 0.3 | 0.4 | 0.4 | ||
| Cell envelope | HP0722 | JHP0659 | Putative OMP, hopO | 0.7 | 0.1 | 0.1 | 0.1 | |
| HP0638 | JHP0581 | OMP, omp13 (hopH) | 0.7 | 0.7 | 0.4 | 0.4 | ||
| HP0913 | JHP0849 | OMP, hopB | 0.7 | 0.5 | 0.4 | 0.4 | ||
| HP0317 | OMP, omp9 (hopU) | 0.7 | 0.4 | 0.5 | 0.4 | |||
| HP0725 | JHP0662 | Putative OMP, hopP | 0.7 | 0.7 | 0.7 | 0.4 | ||
| HP1177 | JHP1103 | OMP, omp27 (hopQ) | 0.6 | 0.1 | 0.1 | 0.1 | ||
| HP0009 | JHP0007 | OMP, hopZ | 0.6 | 0.8 | 0.4 | 0.3 | ||
| HP1469 | JHP1362 | OMP, omp31 (horJ) | 0.6 | 0.5 | 0.3 | 0.3 | ||
| HP0229 | JHP0214 | OMP, hopA | 0.5 | 0.2 | 0.2 | 0.2 | ||
| HP1501 | JHP1394 | OMP, omp32 (horK) | 0.5 | 0.1 | 0.1 | 0.1 | ||
| HP0912 | JHP0848 | OMP, hopC | 0.4 | 0.3 | 0.3 | 0.2 | ||
| HP1167 | JHP1094 | Conserved putative OMP, hofH | 0.5 | ND | 0.2 | 0.3 | ||
| Cellular processes | HP0010 | JHP0008 | Chaperone and heat shock protein, 60-kDa chaperone, groEL | 0.7 | 0.4 | 0.4 | 0.2 | |
| HP0109 | JHP0101 | Chaperone and heat shock protein, 70, 70-kDa chaperone, dnaK | 0.6 | 0.4 | 0.4 | 0.2 | ||
| HP0110 | JHP0102 | Cochaperone and heat shock protein, 24-kDa chaperone, grpE | 0.5 | 0.4 | 0.4 | 0.2 | ||
| HP0887 | JHP0819 | Vacuolating cytotoxin, vacA | 0.8 | 0.3 | 0.3 | 0.2 | ||
| HP0547 | JHP0495 | Cag pathogenicity island protein, cag26 (cagA) | 0.8 | 0.3 | 0.4 | 0.4 | ||
| Central intermediary metabolism | HP1186 | JHP1112 | Carbonic anhydrase | 1.2 | 0.5 | 0.7 | 0.3 | |
| DNA metabolism, DNA replication, recombination, and repair | HP1460 | JHP1353 | DNA polymerase III alpha subunit, dnaE | 0.6 | 0.3 | 0.4 | 0.5 | |
| Energy metabolism | HP0056 | JHP0048 | Δ-1-pyrroline-5-carboxylate dehydrogenase, putA | 0.8 | 0.4 | 0.5 | 0.5 | |
| HP0723 | JHP0661 | l-asparaginase II energy metabolism, ansB | 0.5 | 0.8 | 0.4 | 0.5 | ||
| HP1458 | JHP1351 | Thioredoxin | 0.8 | 0.4 | 0.3 | 0.4 | ||
| HP1101 | JHP1027 | Glucose-6-phosphate dehydrogenase, g6pD | 0.5 | 0.6 | 0.8 | 0.5 | ||
| Fatty acid and phospholipid metabolism | HP0871 | JHP0805 | CDP-diglyceride hydrolase, cdh | 0.6 | 0.4 | 0.5 | 0.4 | |
| HP1045 | Acetyl-coenzyme synthetase, acoE | 0.7 | 0.3 | 0.4 | 0.4 | |||
| Hypothetical gene | HP1002 | 1.4 | 0.9 | 0.4 | 0.5 | |||
| HP0637 | JHP0580 | 0.7 | 0.7 | 0.4 | 0.4 | |||
| HP0097 | JHP0089 | 0.6 | 0.4 | 0.4 | 0.3 | |||
| HP1527 | JHP1416 | 0.6 | 0.5 | 0.6 | 0.4 | |||
| HP1322 | JHP1242 | 0.6 | 0.5 | 0.5 | 0.5 | |||
| HP0111 | JHP0103 | 0.4 | 0.5 | 0.5 | 0.2 | |||
| HP1175 | JHP1102 | Conserved hypothetical integral membrane protein | 0.8 | 0.7 | 0.8 | 0.3 | ||
| HP0310 | JHP0295 | Conserved hypothetical protein | 0.7 | 0.6 | 0.6 | 0.4 | ||
| HP1285 | JHP1205 | Conserved hypothetical secreted protein | 0.6 | 0.4 | 0.4 | 0.5 | ||
| Purines, pyrimidines, nucleosides, and nucleotides | HP1178 | JHP1104 | Purine-nucleoside phosphorylase, deoD | 0.7 | 0.5 | 0.3 | 0.4 | |
| Translation | HP0382 | JHP0999 | Putative zinc-metalloprotease, yjr117w | 0.5 | 0.7 | 0.8 | 0.4 | |
| Transport and binding proteins | HP1561 | JHP1469 | Iron (III) ABC transporter, periplasmic iron-binding protein, ceuE1 | 0.7 | 0.6 | 0.3 | 0.3 | |
| HP1562 | JHP1470 | Iron (III) ABC transporter, periplasmic iron-binding protein, ceuE2 | 0.7 | 0.3 | 0.3 | 0.3 | ||
| HP1180 | JHP1106 | Pyrimidine nucleoside transport protein, nupC | 0.4 | 0.2 | 0.3 | 0.4 | ||
| HP0653 | JHP0598 | Nonheme iron-containing ferritin, pfr | 0.8 | 0.5 | 0.2 | 0.2 | ||
| HP0916 | Iron-regulated OMP, frpB2 | 0.7 | 0.3 | 0.4 | 0.3 | |||
| HP1512 | JHP1405 | Iron-regulated OMP, frpB4 | 0.5 | 0.3 | 0.3 | 0.1 | ||
| Unknown function | HP0485 | JHP0437 | Catalase-like protein | 1.1 | 0.4 | 0.5 | 0.5 | |
| HP1193 | Aldo-ketoreductase, putative | 0.7 | 0.4 | 0.6 | 0.4 | |||
Data presented are relative changes (n-fold) from the zero-time-point starting culture. Changes (n-fold) for a single experiment are given. Data for the biological replicate experiment are available as supplementary information at http://falkow.stanford.edu/whatwedo/supplementarydata/ and show a similar pattern of expression for each gene.
Time is measured in minutes.
ND, no data.
Classification of acid-regulated genes.
The majority of genes that displayed altered expression upon shift to acidic pH showed a net increase in transcription; 71 of the 118 showed at least twofold up regulation at one of the time points. Sorting of the acid-regulated genes into the functional classes defined by The Institute for Genomic Research reveals that the largest class that shows altered expression is composed of hypothetical ORFs of unknown function; 36 of the 118 fall into this group. This represents approximately one-third of the identified genes and highlights our minimal understanding of many of the gene products encoded by H. pylori. Approximately 45% of the assigned ORFs of H. pylori are predicted to encode genes that are hypothetical or conserved hypothetical or of unknown function (78). This is not unique to H. pylori, however, as >50% of all of the predicted ORFs in the 87 completed genomic sequences are predicted to encode hypothetical or conserved hypothetical genes or genes of unknown function (Comprehensive Microbial Resource database at http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl). A goal of future research will be to determine the functional role of these many factors in the life cycle of H. pylori and, in particular, the roles of the genes in Table 1 in its adaptation to acid stress.
Other functional classes with the largest numbers of factors altered by exposure to acid pH include those predicted to have a role in assembly and/or maintenance of the cell envelope and its surface structures, those that encode protein transporters, and those involved in energy metabolism. Only a few pertinent genes within each of the induced and repressed classes will be discussed below.
Up regulated genes: urease.
Included on the list of up regulated genes are six genes that are part of the urease operon. Urease synthesis is directed by a seven-gene cluster, of which ureAB encode the structural components of the enzyme and ureIEFGH encode accessory genes (20, 41). Activity of the enzyme is crucial for virulence, as urease is essential for colonization and persistence in the stomach (6, 25, 28, 79). While it was previously believed that cell surface-localized urease directly created a neutral microenvironment that was conducive to bacterial survival (61), a recent study has indicated that the intracellular enzyme appears to play a more important role in AR (64). Recent work has shown that UreI acts as an inner membrane proton-gated, urea-specific channel (80). The UreI pore opens as the pH of the medium drops below 6.5 and as the cytoplasmically localized urease gains access to its urea substrate. As urease activity neutralizes the local environment, the pore closes and urea transport stops, thus providing a regulated level of enzyme activity.
Increased levels of the urease transcripts validate the ability of our microarray to identify acid-regulated components, as expression of the urease operon has previously been shown to be affected by pH (1). Akada et al. showed that levels of urease transcript were modulated by a pH-dependent posttranscriptional regulatory mechanism, as shift to an acidic pH resulted in a large increase in the levels of mRNA for each of the urease genes, even in the presence of transcriptional inhibitors. Thus, the increase shown by our microarray analysis likely reflects increased mRNA stability instead of increases in urease transcription.
Transport and binding.
Another category of genes whose level of expression is increased contains those predicted to have roles as transport and binding proteins (Table 1). Notable among these are glnQ and glnH, which encode components of a glutamine ABC transporter. It has been previously shown that glutamine can be used by H. pylori as a basic nutrient (50). The first step of this utilization is the deamination of glutamine to glutamate, which is then further dehydrogenated to yield 2-oxoglutarate and ammonia. As mentioned previously, an increased ammonia concentration could be vital to survival of H. pylori under acidic conditions, as this would facilitate the maintenance of pH homeostasis within the bacterial cell and buffer the microenvironment. In support of the importance of this metabolic pathway in acidic conditions, we observed rapid increases in transcription of gdhA, which encodes the enzyme glutamate dehydrogenase that is required for the final catalytic step prior to ammonia production. These data suggest that increased transport of glutamine into the H. pylori cell and subsequent metabolic breakdown of this to produce ammonia may be important mechanisms employed by the bacteria to facilitate survival.
Another pathway involved in the production of ammonia is also up regulated in the presence of acidic pH. The enzyme cystathionine gamma-synthase is predicted to be encoded by the metB gene of H. pylori and is induced at low pH. This enzyme's primary role is the catalysis of the committing step in methionine biosynthesis (23). However, in the absence of l-cysteine, the enzyme can catalyze the production of succinate, alpha-ketobutyrate, and ammonia via a gamma elimination reaction. It is presently unclear whether l-cysteine is limiting under the exposure conditions that we tested and whether increased levels of metB would result in the alternate reaction described, but the potential for increased availability of ammonia seems to be a recurrent theme within the adaptation process of H. pylori to acid pH.
Flagellar synthesis.
One of the most striking findings of our microarray analysis was the altered expression of a large number of genes that encode components of the flagellar apparatus (Table 1). H. pylori typically possesses a polar bundle of two to six sheathed flagella that are critical for colonization in animal models of infection (16, 26, 29, 30, 38, 58). It is believed that motility plays an essential role in that it is necessary to direct the bacterium from the extremely acidic stomach lumen to the more basic mucous layer of the gastric epithelium that is the preferred colonization site of H. pylori (71). Flagellar genes are divided into three classes according to their regulation by three different sigma factors. These classes include (i) σ54-dependent genes that are considered to provide rod and hook functions for the flagella, (ii) σ28 genes that encode proteins necessary for the flagellar filament, and (iii) σ80-dependent genes that comprise basal-body, biosynthetic, and chemotaxis genes. Analysis of the flagellar filament has shown that it is a copolymer of FlaA and FlaB (30, 42, 69, 72), where FlaA is the predominant subtype. FlaB is present as a minor constituent (39), but both FlaB and FlaA are necessary for full motility (35). flaA and flaB are regulated by σ28 and σ54, respectively. In comparison to other well-studied models, the existence of two distinct flagellin subunits, present in different amounts and under the regulation of different sigma factors, is unique. It has been suggested that H. pylori has the ability to alter the relative level of FlaA and FlaB in response to different environmental stimuli (69, 71). By doing so, the bacterium could potentially adapt to microenvironments by alteration of the filament ultrastructure to change flagellar stiffness or flexibility to better suit the particular environment being encountered.
Our microarray analysis revealed that the overwhelming majority of pH-regulated flagellar components of H. pylori are class i genes. These include flaB, flgK, flgB, flgE, and flgC. In addition to these factors, we noted a striking increase in expression of flgM, which was recently shown to encode the H. pylori anti-σ28 factor (17). This anti-σ factor works by binding to σ28 and preventing interaction with the β-region of RNA polymerase (17), thus blocking expression of σ28-dependent genes, such as flaA. Exposure of H. pylori to acid stress thus apparently results in a transcriptional switch that results in increased expression of class I genes, which presumably affects activity of the flagellum.
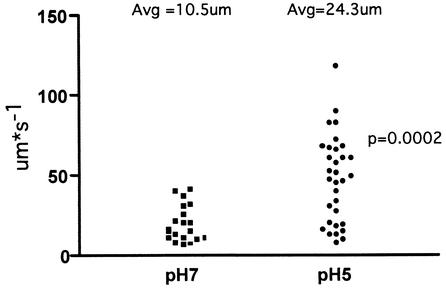
To determine whether exposure to acid would result in a phenotypic change in H. pylori motility, we conducted video microscopy of H. pylori cells exposed to neutral or acidic conditions as described in Materials and Methods. Analysis of bacteria exposed to neutral pH showed that only about 7% of these cells were motile and traveled at an average speed of 10.5 μm per s (Fig. 2). In contrast, cells that were shifted to an acidic pH showed 66% of the culture to be motile and traveled significantly faster than the pH 7.0 bacteria with an average speed of 24.3 μm per s (images are available as QuickTime movies at http://falkow.stanford.edu/whatwedo/supplementarydata/). Since the rotation of the H. pylori flagella has been shown to depend on proton motive force (56), it is likely that the bacterial cell is responding to increased concentrations of protons that are present at the acidic pH. The ability to sense the surrounding environment and alter motility could serve as a critical component of successful colonization, as it would serve the purpose of directing H. pylori to a suitable environment for infection.
FIG. 2.
The effect of acidic pH on H. pylori motility. A culture of H. pylori was split such that equal portions were suspended at pH 7.0 or 5.0. The response to the different pH conditions was monitored by video microscopy, and the speed of motile bacteria was determined as described in Materials and Methods. Each point represents one motile bacterium at the indicated pH, and the speed of that bacterium is indicated on the y axis. The average speed for each condition is indicated. The statistical significance of the differences in speed between the two sets of pH conditions was determined by using a two-tailed Mann-Whitney test, which revealed a P of 0.0002.
Response regulators.
Another factor that exhibits increased expression upon exposure to low pH is the two-component response regulator HP1021. Within the bacterial genome of H. pylori, there exists a paucity of response regulatory systems, as only six genes predicted to encode response regulators and four genes predicted to encode histidine kinases are present. Previous analysis of the response regulators has shown that at least two of the five analyzed are essential for in vitro survival (8, 46), of which HP1021 is one. Little is known about the genes that are controlled by any of the regulators, with the exception of HP0166, which was recently shown to be autoregulatory and to regulate expression of a number of genes of unknown function (21). Elucidation of regulatory networks and a greater understanding of this process would potentiate knowledge about adaptive mechanisms employed by H. pylori that allow it to colonize within the dynamic host environ. Thus, the role of HP1021 in acid survival and elucidation of the bacterial components that it controls will be of keen interest in the future.
Down regulated genes: OMPs.
Analysis of the 41 genes that reproducibly showed decreased expression between experiments also revealed that they fall into a number of different functional groups. As in the case with the induced factors, expression of a large number of hypothetical genes was affected. Somewhat surprisingly, though, this class did not represent the largest constituent of down regulated genes (Table 1). This designation instead belongs to the cell envelope classification, as 12 factors that are predicted to function as outer membrane proteins (OMPs) were identified. Comparative genomic analysis of the two sequenced H. pylori strains recently divided the 64 identified OMPs into five paralogous families based on sequence similarity and predicted secondary structure (4). The major outer membrane family is composed of two subfamilies; the Hop proteins and the Hor proteins. The Hop subfamily contains the largest number of OMPs and is characterized by the presence of a highly conserved N-terminal motif and conserved blocks of sequence at the C terminus (22, 27). Analysis of the list of acid-regulated OMPs revealed that 9 of the 12 were members of the Hop subfamily (Table 1).
The Hop subfamily of proteins contains several factors that are predicted to function as porins (22, 27) as well as adhesins to gastric mucosa (57). As a functional group, porins of gram-negative bacteria largely command the permeability properties of the outer membrane. They typically contain transmembrane diffusion channels that allow small molecules to diffuse across the outer membrane. The selectivity of many porins is typically based on size, though some exhibit a high degree of specificity for specific substrates (14, 43, 73). Little is known about the selectivity of most of the Hop porins. pH-regulated alteration of expression of the Hop proteins, specifically those acting as porins, would presumably affect permeability of the outer membrane and thus limit the accessibility of damaging agents to the bacterial periplasm and beyond. Precedence for the role of porins in survival upon exposure to acid pH can be found with the gastrointestinal pathogen Vibrio cholerae, where expression of the OmpT porin has been shown to be repressed by low pH and where the OmpU porin has been shown to be necessary for AR (51). Future experimentation to determine the selectivity of the porin proteins and the individual requirement for each in AR of H. pylori could shed valuable insight on the process of gastric colonization.
CagA.
We did not expect to identify cagA as consistently repressed by low pH in our studies, as previous reports have suggested that cagA expression is induced at low pH (3, 37). cagA is part of a pathogenicity island (PAI) that encodes a novel type IV secretion apparatus in some strains of H. pylori (2, 15). The PAI is a 40-kilobase locus that contains at least 27 genes, and H. pylori strains carrying the PAI are far more likely to be associated with serious manifestations of H. pylori infection (81). Genes contained on the PAI show homology to genes that encode secretion machinery in other pathogenic bacteria (19), and numerous studies have shown that components of the island function in the delivery of CagA to the host cell. CagA is inserted into the plasma membrane of host cells and is phosphorylated (65) by Src-like protein tyrosine kinases (66, 70). Bacterial attachment and subsequent injection of CagA into the host cell have been shown to induce striking morphological changes in the infected cell (65), presumably as a result of binding and sequestration of the phosphatase SHP-2 (32).
To determine whether levels of CagA protein were reflective of transcript levels, we performed immunoblots by using a CagA-specific antibody on total protein harvested from H. pylori cells exposed to the same conditions used for our microarray analysis. As shown in Fig. 3, relative levels of CagA decreased over time and reached a level that was approximately one-half that of the starting culture, thus showing that transcriptional repression of cagA by acid exposure is reflected at the level of CagA accumulation. It is presently unclear why our results differ from previous reports; however, direct comparisons of the results from this and the previous studies are complicated by the different media used, lengths of exposure to acidified media, sensitivity of the assay, and different strains tested. The result may suggest that there are differences in cagA expression that are intricately linked to the strain being examined. This variability could be of clinical importance due to the predicted role of CagA in disease development and progression.
Construction and characterization of an ansB mutant of H. pylori.
H. pylori is able to generate ammonium via the actions of multiple enzymes (47). These include an aliphatic amidase (68) and at least four amino acid deaminases; asparaginase, aspartase, glutaminase, and serine dehydratase (50). The role of these amino acid deaminases in protection of H. pylori from acid stress is unclear, though production of intracellular ammonium could presumably facilitate survival at low pH. Our microarray analysis showed that HP0294 and HP1238, two genes predicted to encode aliphatic amidases, were rapidly induced, while HP0723, which is predicted to encode an l-asparaginase II enzyme, AnsB, was repressed (Table 1). Two major subgroupings of l-asparaginase enzymes exist: class I enzymes are constitutive cytoplasmic enzymes, while class II enzymes are secreted to the periplasm and show regulated activity (33). The action of both classes of l-asparaginase enzymes catalyzes the hydrolysis of l-asparagine to l-aspartate and ammonium, though class II enzymes sometimes show a broader substrate specificity (33).
Previous investigations of environmental conditions that regulate gene expression have, of course, revealed both induced and repressed genes. Subsequent study of these regulated factors, however, has virtually always focused on the induced genes and ignored the repressed factors as less interesting. However, a recent report showed that in some cases induced factors are not required for survival under the conditions to which they are responding (11). Another recent study also showed that activity of an acid-repressed gene was essential for low-pH survival of V. cholerae (53). In that work, expression of the recO gene, which is required for daughter strand gap repair (reviewed in reference 40), was shown to be dramatically repressed by exposure to acid. However, activity of the protein was essential for low-pH survival, as recO null mutants were 1,000-fold more sensitive to acid stress than was the wild type. In addition, vacA, which is an acid-repressed gene (Table 1), while presumably having no role in in vitro AR, was previously shown to play an important role during infection in the murine model of H. pylori infection (63). For these reasons, we chose to investigate the role of the low-pH-repressed gene ansB, predicted to encode l-asparaginase II, in H. pylori acid survival and colonization.
Deletion-insertion mutations of the ansB coding sequence were constructed by allelic exchange in the gerbil-adapted H. pylori strain B128 (60), as we were unsuccessful in establishing colonization of the G27 isolate in the Mongolian gerbil model of H. pylori. We chose to use the gerbil model, as it has several advantages over the conventional mouse model. These include a lower gastric pH and the H. pylori-inducible manifestations of typical gastritis, gastric ulceration, and carcinoma that more closely mimic disease symptoms seen in humans (34). The deletion-insertion of the ansB coding sequence was confirmed by Southern blot analysis (data not shown). The deletion-insertion mutation in ansB is unlikely to exert a polar effect, since the nearest downstream gene is convergently transcribed to ansB and since there is a predicted rho-independent terminator immediately downstream of the ansB coding sequence.
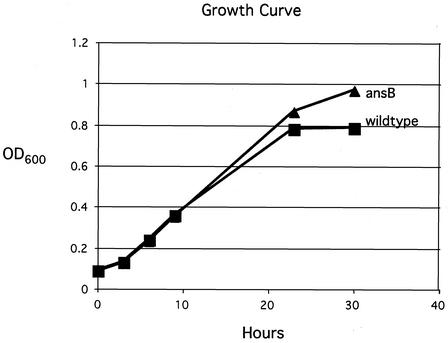
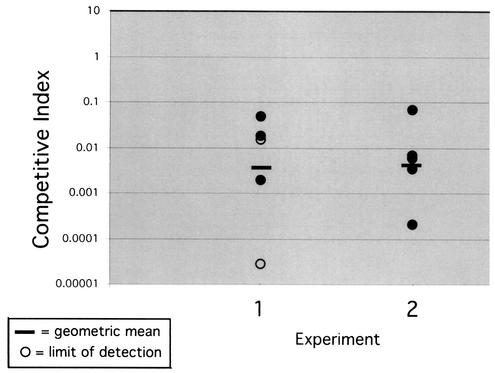
Growth curve analysis of the wild-type and ansB strains revealed that the two strains exhibited nearly identical growth curves for the duration of the experiment (Fig. 4). Acid sensitivity assays on both strains also revealed no difference in the relative levels of AR of the two strains (data not shown). Thus, ansB appears to not be required for in vitro acid survival. We next hypothesized that ansB, though subject to acid stress regulation, perhaps plays an essential role in other stressful conditions, such as during gastric colonization. To investigate the role of ansB in vivo, competition assays were conducted by coinfection of the ansB and wild-type strains in Mongolian gerbils. Five animals were infected with a 1:1 mixture of the two strains, and a replicate of the experiment was repeated on a separate day. Animals were sacrificed after 2.5 to 3 weeks, and the surviving bacteria were collected by dissection and mechanical homogenization of the stomach. Numbers of ansB and wild-type bacteria were determined, and a competitive index was calculated. Strains exhibiting equal fitness for colonization would be expected to possess a competition index at or near 1, while a mutation that rendered the strain less proficient would result in a competitive index of less than 1. As shown in Fig. 5, the ansB mutant was attenuated in its ability to compete with the wild-type strain in both experiments and showed a geometric mean competitive index of 0.004 and 0.005, respectively. This was not the case, however, when wild-type and ansB strains were grown together in vitro overnight in brucella broth, as the ansB showed a competitive index of approximately 1.0.
FIG. 4.
Growth curve analysis of wild-type B128 and its ansB derivative. Brucella broth cultures supplemented with 10% FBS were started from overnight cultures of each strain. Strains were grown in microaerophilic conditions with shaking as described in Materials and Methods, and samples were taken for OD600 measurements at the indicated times.
FIG. 5.
ansB mutants of H. pylori are defective for in vivo competition. Equal concentrations of wild-type and ansB strains were mixed and were used to infect Mongolian gerbils. Colonizing bacteria were recovered after approximately 2.5 to 3 weeks, and the number of CFU of each strain was determined by differential plating. Each circle represents an infected animal, and open circles represent animals for which no ansB derivatives were isolated. Data for two independent experiments are depicted, and the geometric means for each (0.004 and 0.005) are indicated by a black bar.
Conclusion.
Aformidable challenge in understanding how bacterial pathogens colonize and cause disease in their hosts is the development of a more thorough understanding of the adaptive mechanisms employed by these pathogens within the host. The development of microarray technology has provided a powerful tool for the simultaneous elucidation of the complete transcriptome of microbial pathogens, though the direct application of these to host-derived pathogens has been limited, with some notable exceptions (52, 82), by the inability to obtain a sufficiently large concentration of pure bacterial RNA from infected tissues. Due to this, researchers typically rely on the ability to mimic in vivo environments in vitro. The gastric pathogen H. pylori undoubtedly encounters a number of environmental stresses within the human stomach, where it colonizes. One of the most obvious factors that it must deal with is the drastic fluctuation in pH that occurs in the stomach. As such, acid-regulated gene expression is of considerable interest in understanding the process of colonization and survival of this organism.
Herein we describe a microarray-based screen to identify genes whose expression is affected upon shift to acidic pH. Our analysis revealed that 118 genes showed a twofold-or-greater change in relative expression in two independent experiments. Among these were many genes known to be regulated by low pH, such as the urease gene cluster, thus validating our ability to identify pH-regulated components. In addition, many factors whose pH regulation has not previously been recorded were identified.
Two previous array-based studies have analyzed to some extent the response of H. pylori to acid stress (3, 7). Both studies identified a number of factors that were pH regulated in the conditions that were tested, but there was no overlap between the genes identified in these two studies. Our study showed six genes whose expression patterns directly overlapped with these previous two array studies. These include HP1050, HP0743, HP0295, HP1440, HP0123 and HP0229. The low number of genes that overlap between the studies is probably due to differences in experimental design. In the study conducted by Allan et al. (3), the authors used nylon-based microarrays to analyze changes in gene expression that occur after a 30-min shift to pH 4.0 citrate buffer. In a single experiment, these authors found only 11 genes that showed differential expression. This number may be aberrantly low, because the microarray was constructed by using a library of pBluescript clones and thus probably does not contain a complete representation of all of the ORFs of the organism. In addition, nylon-based arrays and radioactive detection are of lower sensitivity than are the DNA-spotted glass slides and fluorophore-based strategies presently being used. The study by Ang et al. (7) utilized nylon-based arrays and biotinylated probes to examine acid-induced gene expression of H. pylori. Total RNA was collected from bacteria grown on acidified-agar plates for 48 h and was used to examine transcriptome profiles. The authors identified 84 genes that showed differential regulation under these conditions. Unfortunately, direct comparison of these results to ours is complicated because the bacteria were subject to a complex growth environment (colony growth), which no doubt results in very dramatic differences in the growth phases and physiological states of the bacteria examined.
It is readily accepted that a core set of genes are consistently expressed throughout the growth cycle of a bacterial cell. However, bacteria alter expression of other genes as a function of their growth phase, expressing highly divergent and specific sets of genes in the logarithmic and stationary phases. This is a well-studied paradigm in Escherichia coli, where transition from rapid growth to stationary phase results in the up regulation of genes that are considered to be part of the canonical RpoS or stationary-phase sigma factor regulon (reviewed in reference 31). These genes then function in diverse pathways involved in nutrient acquisition, stress survival, and alteration of the cell envelope. Accordingly, a recent analysis of growth-phase-dependent gene expression of H. pylori showed that, similar to what is found in other bacteria, divergent sets of genes are expressed depending on the overall growth state of the cell (76). It is possible that some of the differences in gene expression obtained by Ang et al. (7) may partially be explainable by the different growth phases of the bacteria in the two sets of conditions examined.
Future experiments to identify the role of each of the identified factors in adaptation to acidic environments should provide valuable information concerning the mechanisms employed by H. pylori during colonization within the gastric environment of the human host. As AR is a crucial part of H. pylori's ability to survive in its human host and cause disease, study of some of these factors may facilitate elucidation of novel targets for vaccine development and antimicrobial therapy.
Acknowledgments
We are grateful to D. Israel and R. Peek for providing the B128 gerbil-colonizing strain and for helpful discussion of animal experiments. A. Camilli, E. Joyce, L. Thompson, M. Amieva, and A. Mueller provided invaluable discussion and critical review of the manuscript.
D.S.M. is supported by NIH training grant AI07502-06, the Damon Runyon Cancer Research Fund, and Stanford Digestive Disease Center grant DK56339. Research in the laboratory of S.F. and L.S.T. is supported by grants CA92229 and AI38459 from the NIH.
Editor: V. J. DiRita
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 3.Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. [DOI] [PubMed] [Google Scholar]
- 4.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 6.Andrutis, K. A., J. G. Fox, D. B. Schauer, R. P. Marini, J. C. Murphy, L. Yan, and J. V. Solnick. 1995. Inability of an isogenic urease-negative mutant stain of Helicobacter mustelae to colonize the ferret stomach. Infect. Immun. 63:3722-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ang, S., C.-Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J.-T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bijlsma, J. J., A. L. M. Lie, I. C. Nootenboom, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 10.Bijlsma, J. J., B. Waidner, A. H. Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birrell, G. W., J. A. Brown, H. I. Wu, G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99:8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaser, M. J. 1990. Epidemiology and pathophysiology of Campylobacter pylori infections. Rev. Infect. Dis. 12(Suppl. 1):S99-S106. [DOI] [PubMed] [Google Scholar]
- 13.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. BMJ 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 15.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clyne, M., T. Ocroinin, S. Suerbaum, C. Josenhans, and B. Drumm. 2000. Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect. Immun. 68:4335-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colland, F., J. C. Rain, P. Gounon, A. Labigne, P. Legrain, and H. De Reuse. 2001. Identification of the Helicobacter pylori anti-sigma28 factor. Mol. Microbiol. 41:477-487. [DOI] [PubMed] [Google Scholar]
- 18.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 20.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doig, P., M. M. Exner, R. E. Hancock, and T. J. Trust. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 177:5447-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duchange, N., M. M. Zakin, P. Ferrara, I. Saint-Girons, I. Park, S. V. Tran, M. C. Py, and G. N. Cohen. 1983. Structure of the metJBLF cluster in Escherichia coli K12. Sequence of the metB structural gene and of the 5′- and 3′-flanking regions of the metBL operon. J. Biol. Chem. 258:14868-14871. [PubMed] [Google Scholar]
- 24.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Exner, M. M., P. Doig, T. J. Trust, and R. E. Hancock. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrero, R. L., V. Cussac, P. Courcoux, and A. Labigne. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 174:4212-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foynes, S., N. Dorrell, S. J. Ward, Z. W. Zhang, A. A. McColm, M. J. Farthing, and B. W. Wren. 1999. Functional analysis of the roles of FliQ and FlhB in flagellar expression in Helicobacter pylori. FEMS Microbiol. Lett. 174:33-39. [DOI] [PubMed] [Google Scholar]
- 30.Haas, R., T. F. Meyer, and J. P. van Putten. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8:753-760. [DOI] [PubMed] [Google Scholar]
- 31.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress response, 1st ed. ASM Press, Washington, D.C.
- 32.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 33.Huser, A., U. Kloppner, and K. H. Rohm. 1999. Cloning, sequence analysis, and expression of ansB from Pseudomonas fluorescens, encoding periplasmic glutaminase/asparaginase. FEMS Microbiol. Lett. 178:327-335. [DOI] [PubMed] [Google Scholar]
- 34.Ikeno, T., H. Ota, A. Sugiyama, K. Ishida, T. Katsuyama, R. M. Genta, and S. Kawasaki. 1999. Helicobacter pylori-induced chronic active gastritis, intestinal metaplasia, and gastric ulcer in Mongolian gerbils. Am. J. Pathol. 154:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karita, M., M. K. Tummuru, H. P. Wirth, and M. J. Blaser. 1996. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect. Immun 64:4501-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostrzynska, M., J. D. Betts, J. W. Austin, and T. J. Trust. 1991. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labigne, A., V. Cussac, and P. Courcoux. 1991. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J. Bacteriol. 173:1920-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 43.Maier, C., E. Bremer, A. Schmid, and R. Benz. 1988. Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli. Demonstration of a nucleoside-specific binding site. J. Biol. Chem. 263:2493-2499. [PubMed] [Google Scholar]
- 44.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed]
- 45.Matysiak-Budnik, T., and F. Megraud. 1997. Epidemiology of Helicobacter pylori infection with special reference to professional risk. J. Physiol. Pharmacol. 48(Suppl. 4):3-17. [PubMed] [Google Scholar]
- 46.McDaniel, T. K., K. C. Dewalt, N. R. Salama, and S. Falkow. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 6:15-23. [DOI] [PubMed] [Google Scholar]
- 47.McGee, D. J., F. J. Radcliff, G. L. Mendz, R. L. Ferrero, and H. L. Mobley. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGowan, C. C., T. L. Cover, and M. J. Blaser. 1997. Analysis of F1F0-ATPase from Helicobacter pylori. Infect. Immun. 65:2640-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGowan, C. C., A. Necheva, S. A. Thompson, T. L. Cover, and M. J. Blaser. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 30:19-31. [DOI] [PubMed] [Google Scholar]
- 50.Mendz, G. L., and S. L. Hazell. 1995. Aminoacid utilization by Helicobacter pylori. Int. J. Biochem. Cell Biol. 27:1085-1093. [DOI] [PubMed] [Google Scholar]
- 51.Merrell, D. S., C. Bailey, J. B. Kaper, and A. Camilli. 2001. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 183:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 54.Meyer-Rosberg, K., D. R. Scott, D. Rex, K. Melchers, and G. Sachs. 1996. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology 111:886-900. [DOI] [PubMed] [Google Scholar]
- 55.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakamura, H., H. Yoshiyama, H. Takeuchi, T. Mizote, K. Okita, and T. Nakazawa. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect. Immun. 66:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537-1548. [DOI] [PubMed] [Google Scholar]
- 58.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 60.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Physicians 110:531-544. [PubMed] [Google Scholar]
- 61.Phadnis, S. H., M. H. Parlow, M. Levy, D. Ilver, C. M. Caulkins, J. B. Connors, and B. E. Dunn. 1996. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 64:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 65.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Selbach, M., S. Moese, C. R. Hauck, T. F. Meyer, and S. Backert. 2002. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277:6775-6778. [DOI] [PubMed] [Google Scholar]
- 67.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25:989-998. [DOI] [PubMed] [Google Scholar]
- 69.Spohn, G., and V. Scarlato. 2001. Motility, chemotaxis, and flagella, p. 239-248. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 70.Stein, M., F. Bagnoli, R. Halenbeck, R. Rappuoli, W. J. Fantl, and A. Covacci. 2002. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43:971-980. [DOI] [PubMed] [Google Scholar]
- 71.Suerbaum, S. 1995. The complex flagella of gastric Helicobacter species. Trends Microbiol. 3:168-171. [DOI] [PubMed] [Google Scholar]
- 72.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szmelcman, S., M. Schwartz, T. J. Silhavy, and W. Boos. 1976. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and lambda-resistant mutants as measured by fluorescence quenching. Eur. J. Biochem. 65:13-19. [DOI] [PubMed] [Google Scholar]
- 74.Tannaes, T., N. Dekker, G. Bukholm, J. J. Bijlsma, and B. J. Appelmelk. 2001. Phase variation in the Helicobacter pylori phospholipase A gene and its role in acid adaptation. Infect. Immun 69:7334-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor, D. N., and J. Parsonnet. 1995. Epidemiology and natural history of H. pylori infections, p. 551-564. In M. J. Blaser, P. F. Smith, J. Ravdin, H. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York City, N.Y.
- 76.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed]
- 77.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, D. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 79.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 81.Xiang, Z., S. Censini, P. F. Bayeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]