Abstract
The cDNA of LeCPK1, a calcium-dependent protein kinase, was cloned from tomato (Lycopersicon esculentum Mill.). LeCPK1 was expressed in Escherichia coli and purified from bacterial extracts. The recombinant protein was shown to be a functional protein kinase using a synthetic peptide as the substrate (syntide-2, Km = 85 μm). Autophosphorylation of LeCPK1 was observed on threonine and serine residues, one of which was identified as serine-439. Kinase activity was shown to be Ca2+ dependent and required the C-terminal, calmodulin-like domain of LeCPK1. Two classes of high- and low-affinity Ca2+-binding sites were observed, exhibiting dissociation constants of 0.6 and 55 μm, respectively. LeCPK1 was found to phosphorylate the regulatory C-terminal domain of the plasma membrane H+-ATPase in vitro. A potential role in the regulation of proton pump activity is corroborated by the apparent colocalization of the plasma membrane H+-ATPase and LeCPK1 in vivo. Upon transient expression in suspension-cultured cells, a C-terminal fusion of LeCPK1 with the green fluorescent protein was targeted to the plasma membrane. Myristoylation of the LeCPK1 N terminus was found to be required for plasma membrane targeting.
Protein kinases that are regulated by cytosolic free Ca2+ are important for signal transduction in all eukaryotes. Plants and protists have calcium-dependent protein kinases (CDPKs) that are directly activated by calcium (Harmon et al., 1987; Zhao et al., 1994). This is in contrast to calcium-stimulated protein kinases in animals and fungi that have an additional requirement for calmodulin or lipids for full activation. The cloning of the first CDPK from soybean (Glycine max; Harper et al., 1991) and the subsequent analysis of CDPKs from many other plant species (for review, see Harmon et al., 2000; Hrabak, 2000) provided an explanation for the apparent calcium sensitivity and calmodulin independence of plant CDPKs: They all share a similar primary structure consisting of four modules, i.e. an N-terminal variable region, the kinase domain followed by an auto-inhibitory domain, and a C-terminal, calmodulin-like domain. The auto-inhibitory region contains a pseudo-substrate site that, in the absence of Ca2+, binds to the catalytic center and keeps the kinase in its inactive state. Binding of Ca2+ to the calmodulin-like domain is thought to induce a conformational shift resulting in the release of the pseudo-substrate domain from the active site and kinase activation (Harmon et al., 1994, 2000; Harper et al., 1994; Hrabak, 2000).
The cytoplasmic free Ca2+ concentration under resting conditions is maintained at very low levels (10–200 nm), ensuing low CDPK activity. An increase in cytoplasmic calcium results in CDPK activation; hence, CDPKs may function as sensors of fluctuations in cytosolic Ca2+ and initiate downstream signaling events (Roberts and Harmon, 1992; Trewavas and Malhó, 1998; Harmon et al., 2000; Hrabak, 2000). A great number of both biotic and abiotic stimuli trigger an increase in the concentration of cytoplasmic free Ca2+, which then acts as a second messenger mediating a variety of cellular responses (Webb et al., 1996; Sanders et al., 1999). The specificity of the calcium signal appears to reside in characteristic spatial and temporal patterns of its concentrations (McAinsh and Hetherington, 1998; Sanders et al., 1999). CDPKs are encoded by a large gene family, and individual CDPK isoforms exhibiting different Ca2+-binding characteristics and subcellular localization may decode a subset of the calcium signals (Harmon et al., 2000). Potential downstream targets of CDPK action include soluble enzymes, transcription factors, ion channels and pumps, and cytoskeletal proteins. Very few of these proteins, however, have been identified as bona fide substrates of individual CDPKs (Harmon et al., 2000; Hrabak, 2000). We are particularly interested in the plasma membrane H+-ATPase as a potential CDPK substrate and, furthermore, in the role of proton pump regulation by calcium-dependent phosphorylation as part of the signal transduction cascade in plant defense reactions against pathogens and herbivores.
The P-type H+-ATPase is the major electrogenic pump in the plasma membrane of plant cells. It builds up and maintains an electrochemical proton gradient across the plasma membrane that drives numerous proton- and membrane potential-coupled transport processes and regulates ion channel activity (Morsomme and Boutry, 2000). Changes in proton pump activity and associated ion fluxes across the plasma membrane are among the first cellular reactions after pathogen recognition (Blumwald et al., 1998). Pathogen-derived elicitor molecules have been shown to cause either H+-ATPase activation concomitant with extracellular acidification and membrane hyper-polarization, or H+-ATPase inactivation resulting in the depolarization of the plasma membrane (Wevelsiep et al., 1993; Vera-Estrella et al., 1994; Thain et al., 1995; Hammond-Kosack et al., 1996; Xing et al., 1996; Zhou et al., 2000). A causal relationship between the proton electrochemical gradient and the activation of both wound and pathogen defense responses was established by modulation of H+-ATPase activity and by use of ionophores as elicitors of defense reactions (Doherty and Bowles, 1990; Klüsener and Weiler, 1999; Roberts and Bowles, 1999; Schaller and Oecking, 1999; Schaller et al., 2000; Engelberth et al., 2001; Schaller and Frasson, 2001).
The activity of the plasma membrane H+-ATPase is known to be regulated by reversible protein phosphorylation in a complex manner. Phosphorylation of the penultimate Thr residue in the C-terminal, autoregulatory domain of the H+-ATPase results in the 14-3-3 protein-dependent activation of the pump (Fuglsang et al., 1999; Svennelid et al., 1999; Maudoux et al., 2000). Phosphorylation at a second or additional unidentified sites, however, inhibits the H+-ATPase. The latter event appears to be calcium dependent; hence, Ca2+-dependent protein kinases have been implicated in the regulation of proton pump activity (Schaller and Sussman, 1988; Vera-Estrella et al., 1994; Kinoshita et al., 1995; Xing et al., 1996; Camoni et al., 1998; Desbrosses et al., 1998; Lino et al., 1998; De Nisi et al., 1999).
These observations point toward a role for CDPKs in plant defense as regulators of plasma membrane H+-ATPase activity. Such a function is corroborated by the well-documented rise in the cytosolic Ca2+ concentration in response to pathogen infection and wounding (Scheel, 1998). Calcium-permeable channels activated by fungal elicitors have been described in the plasma membrane of tomato (Lycopersicon esculentum Mill.) and parsley (Petroselinum crispum; Gelli et al., 1997; Zimmermann et al., 1997), and the elicitor-induced influx of Ca2+ was shown to be necessary for subsequent cellular responses (Jabs et al., 1997; Blume et al., 2000). Additional calcium channels exist in the tonoplast as well as the endoplasmic reticulum for the release of Ca2+ from internal stores (Klüsener et al., 1995; Allen and Sanders, 1997). The mobilization of Ca2+ from both intra- and extracellular stores is an early step not only in pathogen defense but also in wound signal transduction mediated by the polypeptide wound hormone systemin (Moyen et al., 1998) or by electrical signals (Vian et al., 1997; Herde et al., 1998). The influx of Ca2+ and the activity of a protein kinase are both required for the systemin-triggered depolarization of the plasma membrane and alkalinization of the extracellular space (Felix and Boller, 1995; Moyen and Johannes, 1996; Moyen et al., 1998; Schaller and Oecking, 1999; Schaller and Frasson, 2001). Hence, a calcium-stimulated protein kinase was implicated in wound signaling, but direct evidence linking a CDPK to the respective signal transduction pathway is missing (Schaller, 1999; Schaller and Oecking, 1999). In pathogen defense, on the other hand, the involvement of a CDPK was demonstrated using tobacco (Nicotiana tabacum) cells expressing the Cf9 resistance gene from tomato. In these cells, the avr9 elicitor-dependent activation of a CDPK was observed and appeared to be required for subsequent cellular responses (Romeis et al., 2000). Furthermore, correlative evidence for the involvement of a CDPK (ZmCPK10) in the elicitor-induced accumulation of pathogenesis-related proteins was presented in maize (Zea mays; Murillo et al., 2001).
With the aim to identify the CDPK(s) involved in H+-ATPase regulation and defense signaling, we have focused on kinases that are induced at the gene expression level by either wounding or treatment with a fungal toxin (fusicoccin [FC]), i.e. conditions that prompt changes in H+-ATPase activity. Such a CDPK has recently been described in tobacco (NtCDPK1; Yoon et al., 1999). We report here the cloning of the closely related LeCPK1, a CDPK from tomato plants. The LeCPK1 transcript accumulated after treatment with FC and the kinetics of transcript induction resembled those of 14-3-3 proteins, i.e. known regulators of proton pump activity. LeCPK1 was found to colocalize with the H+-ATPase at the plasma membrane in vivo, and to phosphorylate the C-terminal autoregulatory domain of the pump in vitro. A potential role for LeCPK1 as an in vivo regulator of the plasma membrane H+-ATPase is discussed.
RESULTS AND DISCUSSION
Cloning of LeCPK1
Database searches identified an expressed sequence tag (EST) clone from tomato highly similar to NtCDPK1, a CDPK from tobacco that is regulated at the level of mRNA abundance by wounding, methyl jasmonate, fungal elicitors, and chitosan (Yoon et al., 1999). Using the EST sequence information, we cloned a possible NtCDPK1 ortholog from tomato that was named LeCPK1 following the nomenclature of Hrabak et al. (1996). The 2,229-bp LeCPK1 cDNA (accession no. AJ308296) contains an open reading frame (ORF) of 1,659 bp coding for a 63-kD protein of 553 amino acids. The deduced amino acid sequence is 94% identical to that of NtCDPK1 and exhibits the modular structure typical for CDPKs (Harmon et al., 2000; Hrabak, 2000), comprising an N-terminal variable region, a kinase domain, an auto-inhibitory region, and a C-terminal, calmodulin-like domain with four EF-hand motives implicated in Ca2+ binding (Fig. 1).
Figure 1.
Comparison of the LeCPK1 and NtCDPK protein sequences. The amino acid sequences deduced from the LeCPK1 and NtCDPK cDNAs are shown. The sequences were aligned using “pileup” from the University of Wisconsin GCG program package. Identical amino acids and conservative replacements are shown with black and gray shading, respectively (box shade at http://www.ch.embnet.org/). The variable N-terminal region (>>>), the kinase domain (—), the auto-inhibitor (+++), the calmodulin-like domain (:::), and the EF hand motives (###) are indicated. Arrowheads (∧) mark the consensus sequence for N-myristoylation. The G2A mutation and the phosphorylated Ser residue are highlighted by boldface A and P, respectively.
CDPKs are encoded by a highly conserved family of genes. Therefore, to obtain a gene-specific probe for DNA and RNA gel-blot analyses, the 3′-untranslated region of the LeCPK1 cDNA was used. The specificity of the probe for the LeCPK1 gene, was demonstrated on gel blots of tomato genomic DNA, where only one hybridizing DNA fragment was detected in each lane (Fig. 2A). Using the same probe on northern blots, we found the LeCPK1 transcript to be of low abundance in leaves but more prevalent in roots and flowers (data not shown), resembling the tissue-specific expression of NtCDPK1 (Yoon et al., 1999). In leaves, however, the LeCPK1 transcript transiently accumulated after treatment with FC, a toxin from Fusicoccum amygdali that is a potent activator of the plasma membrane H+-ATPase (Marré, 1979). An initial increase in transcript abundance was observed 1.5 h after treatment of the plants with 3 μm FC and highest levels were observed after 4 h (Fig. 2B). This is in contrast to the delayed induction of pathogenesis-related protein transcripts that continue to accumulate until 8 h after FC treatment (Fig. 2B; Roberts and Bowles, 1999; Schaller and Oecking, 1999; Schaller et al., 2000). The temporal pattern of transient LeCPK1 induction is similar to that of 14-3-3 proteins (Fig. 2B; Roberts and Bowles, 1999), which are well-known regulators of H+-ATPase activity (Jahn et al., 1997; Oecking et al., 1997; Olivari et al., 1998). To further investigate the possibility of LeCPK1 itself being a regulator of proton pump activity, we expressed the protein in Escherichia coli and characterized the recombinant kinase.
Figure 2.
Northern- and Southern-blot analyses. A, DNA gel-blot analysis. Ten micrograms of tomato genomic DNA was restricted with XbaI (1), HindIII (2), EcoRI (3), and DraI (4), and was separated by agarose gel electrophoresis. DNA fragments were transferred to nitrocellulose membranes and the blot was hybridized with the radiolabeled 3′-untranslated region of the LeCPK1 cDNA. The position of DNA standards (1-kb ladder, Life Technologies/Gibco-BRL, Cleveland) is indicated and their size is given in kb. B, RNA gel-blot analyses. Five micrograms of total RNA isolated from the leaves of control plants (lane 1) and plants at 0.5, 1, 1.5, 2, 4, 6, or 8 h (lanes 2–8) after treatment with 3 μm FC was separated on formaldehyde agarose gels and subsequently transferred to nitrocellulose membranes. The blots were hybridized with the radiolabeled 3′-untranslated region of the LeCPK1 cDNA and the cDNAs of 14-3-3 and pathogenesis-related proteins, as indicated. A duplicate gel was stained with ethidium bromide as a control of RNA quantity and integrity.
Expression, Purification, and Activity of LeCPK1
The full-length LeCPK1, as well as a truncated variant lacking the C-terminal, calmodulin-like domain, were expressed as N-terminal glutathione S-transferase (GST) fusion proteins in E. coli and were designated LeCPK1-L (LeCPK1-Long) and LeCPK1-S (LeCPK1-Short), respectively. During denaturing PAGE, the recombinant proteins exhibited the molecular masses expected for the two fusion proteins. LeCPK1-L and -S were purified to near homogeneity using the GST moiety as an affinity tag (Fig. 3, A and B). Proteolytic removal of the affinity tag turned out to be impossible because the protease factor Xa cleaved the fusion proteins at a second, internal site releasing 22 amino acids from the N terminus of LeCPK1 (data not shown). Hence, all further experiments were done with the fusion proteins.
Figure 3.
Purification of recombinant LeCPK1 and characterization of the autokinase activity. A, The full-length LeCPK1 was expressed in E. coli as a GST fusion protein and the purification of recombinant LeCPK1-L was monitored by SDS-PAGE. The crude extract (1) was separated in insoluble (2) and soluble (3) fractions by centrifugation. The fusion protein was purified from the latter by affinity chromatography on a glutathione-Sepharose 4B column (Amersham Pharmacia Biotech, Dübendorf, Switzerland; 4, flow-through; 5, eluate). The size of the fusion protein is indicated in kD. In lanes 1–4 of the Coomassie Brilliant Blue-stained gel, the protein equivalent of 5 μL of E. coli culture is shown, whereas lane 5 corresponds to 0.8 μg of purified LeCPK1-L. B, A truncated form of LeCPK1 lacking the C-terminal calmodulin-like domain (LeCPK1-S) was expressed in E. coli and purified as in A. C, Recombinant LeCPK1-L and -S (L and S; 0.5 μg in each lane, two samples each) were separated by SDS-PAGE. Top, Coomassie Brilliant Blue-stained gel; lower, corresponding western blot. The blot was cut in two along the central lane (M) containing size markers (Bio-Rad Laboratories, Hercules, CA), and the two halves were assayed for autophosphorylation activity in absence (left) or presence (right) of Ca2+. The blot was autoradiographed for detection of incorporated 32P. D, LeCPK1-L autophosphorylated in presence of [γ32P]ATP was hydrolyzed in 6 n HCl. The hydrolysate was spiked with phospho-amino acid standards and separated by two-dimensional thin-layer chromatography (TLC). The plate was first sprayed with ninhydrin for detection of phospho-amino acid standards (left) and then autoradiographed (right).
LeCPK1-L and -S were tested for autokinase activity after electrophoretic separation, western blot, and renaturation on nitrocellulose membranes. LeCPK1-L incorporated labeled phosphate from [γ-32P]ATP in a Ca2+-dependent manner, whereas LeCPK1-S was inactive both in the presence or absence of calcium (Fig. 3C). This result is consistent with the proposed mechanism of activation of CDPKs, according to which the binding of Ca2+ triggers a conformational shift releasing the auto-inhibitor from the active site. In the absence of the calcium-binding domain, access to the active site is permanently blocked by the pseudo-substrate auto-inhibitor rendering the enzyme inactive (Harmon et al., 1994; Harper et al., 1994). Hydrolysis of the phosphorylated protein and TLC analysis of the amino acids in the hydrolysate revealed autophosphorylation to occur on Ser and Thr residues but not on Tyr (Fig. 3D). Hence, LeCPK1-L is a functional calcium-dependent Ser/Thr protein kinase.
Autophosphorylation of CDPKs is commonly observed; it appears to be more frequent on Ser than on Thr residues, and can have an up- or down-regulatory effect on kinase activity (Roberts and Harmon, 1992; Chaudhuri et al., 1999; Yoon et al., 1999; Hrabak, 2000). The position(s) of the phosphorylated amino acid(s), however, have not been identified previously. Using quadrupole-time of flight mass spectrometry (MS) for sequence analysis of tryptic peptides, we identified Ser-439 as one of the targets for autophosphorylation in LeCPK1-L (Fig. 4). Phosphorylation of the peptide encompassing Ser-439 was observed in two independent experiments and it appeared to be quantitative because the corresponding unphosphorylated peptide could not be detected in the tryptic digest of autophosphorylated LeCPK1-L. This autophosphorylation site is located within the calmodulin-like domain between the two EF hands closest to the kinase domain (Fig. 1). The effects of phosphorylation of Ser-439 on LeCPK1 activity, if any, remain to be identified. They may include changes in Ca2+-binding properties or the functionality of the calmodulin-like domain.
Figure 4.
Processed MS/MS spectrum of peptide m/z 642.89. MS analysis identified a tryptic peptide of autophosphorylated LeCPK1, the mass of which (m/z = 642.89; double charge; mass = 1,285 D) indicated the presence of a phosphorylated residue. The peptide was selected on basis of its mass, subjected to low-energy collision-induced decomposition, and the resulting fragments were analyzed by MS/MS. The processed (software: MaxEnt 3) MS/MS spectrum is shown. The y ion series is labeled as such and the deduced amino acid sequence is indicated. The mass difference (y9 − y8) indicates the presence of a phosphorylated Ser residue (pS), as do the ions M + H+ − [H3PO4], y10 − [H3PO4], and y9 − [H3PO4], which are derived from the peptide m/z 642.89 as a result of collision-induced decomposition of the phosphate-ester bond.
Ca2+ Binding
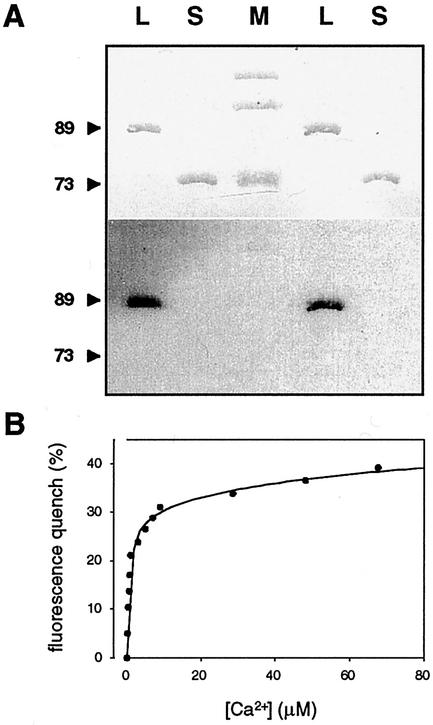
Many calcium-binding proteins are subject to a mobility shift during SDS-PAGE in the presence of Ca2+. Such a mobility shift, which is indicative of a change in protein conformation, has also been observed for a number of CDPKs (Roberts and Harmon, 1992; Zhao et al., 1994; Yoon et al., 1999; Romeis et al., 2000). In contrast, the electrophoretic mobility of LeCPK1-L was the same in the presence or absence of Ca2+ (data not shown). To obtain evidence for the direct binding of Ca2+, we therefore incubated the electroblotted LeCPK1-L and -S with 45Ca2+. Binding of the radiolabel was observed for LeCPK1-L but not for LeCPK1-S, indicating that the calmodulin-like domain is a functional calcium-binding domain (Fig. 5A).
Figure 5.
Characterization of Ca2+-binding properties. A, Recombinant LeCPK1-L and -S (L and S; 0.5 μg in each lane, two samples each) and Mr marker proteins (M, Bio-Rad) were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and stained with Ponceau S (top). The membrane was then incubated in 45Ca2+, washed, and the bound radiolabel was detected by autoradiography (lower). Positions and size (in kD) of LeCPK1-L and -S are indicated. B, LeCPK1-L (45 μg mL−1 [0.5 μm] in 50 mm HEPES, pH 7.5) was titrated with CaCl2. Two-microliter aliquots of CaCl2 (0.2 μm–2 mm in 50 mm HEPES, pH 7.5) were added and at each interval the emission spectrum was recorded using a Kontron SFM 25 fluorimeter (λex: 280 nm). The observed quench in Trp fluorescence at 325 nm was plotted against the Ca2+ concentration. Individual measurements are shown as well as the interpolated graph [SigmaPlot software, SPSS, Chicago; y = f(x) = a × x/(b + x) + c × x/(d + x), R2 = 0.993) from which high- and low-affinity Ca2+-binding constants were derived.
Furthermore, a Trp fluorescence quench was observed upon titration of LeCPK1-L with Ca2+, which was most prominent at the fluorescence maximum of 325 nm. There are four Trp residues in the catalytic domain of LeCPK1 (Trp-289, -306, -329, and -362) and the change in fluorescence reflects an altered molecular environment of these residues likely due to a change in protein conformation in response to calcium binding. The experimental data for the decrease in Trp fluorescence as a function of the increasing Ca2+ concentration are best described by a double rectangular hyperbolic function (Sigma Plot, R2 = 0.993; Fig. 5B) from which two dissociation constants of 0.6 and 55 μm can be derived. Apparently, the four Ca2+-binding EF hands fall into two classes of high and low affinity, respectively. Likewise, two Ca2+-binding sites each of low and high affinity are known to exist in calmodulin (Chin and Means, 2000). A Kd of 0.6 μm for the high-affinity Ca2+-binding site(s) in LeCPK1-L is in the range of calcium concentrations required for the activation of many CDPKs (Roberts and Harmon, 1992; Lee et al., 1998). Consistently, most of the Trp fluorescence intensity quench was observed already at very low calcium concentration (Fig. 5B), indicating that occupation of the high-affinity site(s) is sufficient to induce the conformational change required for kinase activation. This conclusion is in good agreement with the work of Zhao et al. showing that the four EF hands of Plasmodium falciparum CDPK are not functionally equivalent. Site-directed mutagenesis of the two EF hands adjacent to the catalytic domain impaired Ca2+ binding, the conformational shift, and enzyme activation, whereas mutations in the two distal EF hands had only minor effects (Zhao et al., 1994). Furthermore, it was concluded for two soybean CDPKs that binding of one or two calcium ions per molecule of enzyme is sufficient for significant activation (Lee et al., 1998).
Substrate Specificity and Subcellular Localization
The multitude of CDPK isoforms existing within a given plant species (Harmon et al., 2000; Hrabak, 2000) evokes the question of how any one isoform recognizes its substrate protein to trigger specific signaling events. The required specificity may be achieved by the substrate specificity of the kinase, and/or a specific developmental or subcellular colocalization of the two proteins.
The substrate specificities of plant CDPKs are not well defined. They do not seem to be strict because most CDPKs are able to phosphorylate histone H1 and casein (Roberts and Harmon, 1992; Hrabak, 2000). Synthetic peptides have been widely used to study the activity of CDPKs and the motif -basic-X-X-Ser/Thr- has been identified as a minimal sequence element recognized by many CDPKs (Roberts and Harmon, 1992). Consistently, we found syntide-2 (PLARTLSVAGLPGKK, recognition element underlined) to be a substrate of LeCPK1-L for which an apparent Km value of 85 μm was derived from steady-state kinetic analyses. However, two other peptides containing a similar sequence element with Lys replacing the Arg residue were not phosphorylated (data not shown).
Despite the broad substrate specificity in vitro, CDPKs are supposed to have a limited range of substrates in vivo. Although there is little experimental evidence to support this hypothesis, a higher in vivo specificity may be accomplished by temporally and spatially restricted patterns of expression during plant development (for review, see Hrabak, 2000) or by targeting of CDPK isoforms to specific subcellular compartments. CDPKs have been detected in the cytosol, in the nucleus, as well as in association with microsomal or plasma membranes (for review, see Hrabak, 2000). The primary structures of known CDPKs including that of LeCPK1 do not contain extended hydrophobic stretches that could promote membrane association. Yet, the N termini of LeCPK1 and many other CDPKs have a consensus sequence for myristoylation (MGxxxS/T, with x indicating any amino acid; Hrabak et al., 1996; Färber et al., 1997; Yalovsky et al., 1999). The covalent attachment of the 14-carbon-saturated fatty acid myristate to the amino group of the essential Gly residue in position two of the primary structure occurs cotranslationally after removal of the initiator Met. Myristoylation per se is not sufficient to anchor the protein in the lipid bilayer. Membrane association is further promoted by additional features, including palmitoylation at a Cys residue close to the myristoylated Gly and clusters of positively charged amino acids that can interact with acidic head groups of phospholipids at the cytoplasmic face of the plasma membrane (Yalovsky et al., 1999). The N terminus of LeCPK1 comprises all three features, i.e. potential acceptor sites for myristoylation and palmitoylation as well as a pair of basic residues (Fig. 1).
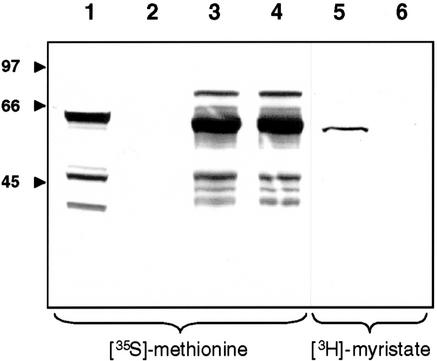
We used a coupled in vitro transcription/translation system to test for N-myristoylation of LeCPK1. When the reaction was performed in the presence of [35S]Met, wild-type LeCPK1 and a G2A-mutant, in which the myristate acceptor Gly had been substituted by Ala, were produced in similar amounts, as indicated by the apparent masses and band intensities of the predominant labeled proteins (Fig. 6). Additional weak bands correspond to less abundant proteins that may be a result of incomplete termination of translation at the first stop codon (the high-Mr protein) or additional sites of translational initiation (the lower Mr proteins). In the presence of [3H]myristate, on the other hand, only LeCPK1 but not the G2A mutant became labeled, indicating the N terminus of LeCPK1 to be a functional myristate acceptor site and Gly-2 to be essential for myristoylation (Fig. 6). The higher resolution obtained with 3H as compared with 35S during fluorography may account for the slightly different appearance of the two bands obtained for the wild-type LeCPK1 in the left and the right part of Figure 6, respectively. Alternatively, the covalent modification by N-myristoylation may result in a subtle change in electrophoretic mobility.
Figure 6.
Assay for N-myristoylation. A wheat germ coupled in vitro transcription/translation system was used to generate cDNA-encoded proteins: luciferase (lane 1, positive control), no cDNA added (lane 2, negative control), LeCPK1 (lanes 3 and 5), and G2A-CPK1 (lanes 4 and 6). In vitro translation was carried out in the presence of either [35S]Met (lanes 1–4) or [3H]myristic acid (lanes 5 and 6). The reaction products were separated by SDS-PAGE and the gel was analyzed by fluorography. The position and size (in kD) of marker proteins (Bio-Rad) are indicated.
N-terminal myristoylation has been shown previously for zucchini (Cucurbita pepo) and potato (Solanum tuberosum) CDPKs (Ellard-Ivey et al., 1999; Raíces et al., 2001), but its physiological function has not been addressed experimentally. In a study of rice (Oryza sativa) OsCPK2, myristoylation was shown to be essential for membrane association that was further enhanced by palmitoylation (Martin and Busconi, 2000). The nature of membrane to which OsCPK2 was targeted has not been identified, however. As shown below, N-myristoylation of LeCPK1 results in efficient targeting to the plasma membrane in vivo.
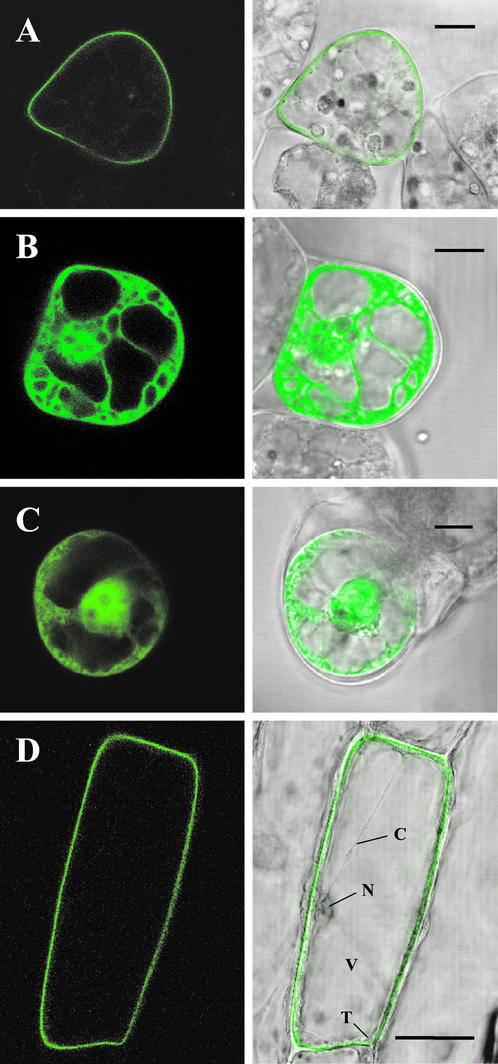
Using transient expression systems, we analyzed the subcellular localization of wild-type LeCPK1 and the G2A mutant in C-terminal fusion with green fluorescent protein (GFP) by confocal laser scanning microscopy. When expressed in suspension-cultured cells of Lycopersicon peruvianum under control of the cauliflower mosaic virus 35S promoter, the LeCPK1-GFP fusion protein was efficiently targeted to the cell periphery, likely the plasma membrane (Fig. 7A). In contrast, the G2A-GFP fusion protein localized to the cytoplasm and the nucleus (Fig. 7B) and its expression pattern was essentially indistinguishable from that of GFP alone (Fig. 7C). We obtained comparable results using the well-established transient expression system in onion (Allium cepa) epidermal cells (Scott et al., 1999; Fig. 7D; data not shown). In this system as well, we observed targeting of LeCPK1-GFP to the plasma membrane, whereas no fluorescence was found in association with the tonoplast or any other internal membrane system. The data clearly demonstrate N-terminal myristoylation to result in and to be necessary for plasma membrane targeting.
Figure 7.
Subcellular localization of GFP fusion proteins. Wild-type LeCPK1 (A) and the G2A site-directed mutant of LeCPK1 (B) were transiently expressed as C-terminal GFP-fusion proteins in suspension-cultured L. peruvianum cells. Cells in C were transformed to express GFP alone. D, Onion epidermal cells expressing the wild-type LeCPK1-GFP fusion. Letters indicate the nucleus (N), the vacuole (V), the tonoplast (T), and a cytoplasmic strand (C). The localization of GFP-fusion proteins was analyzed by confocal laser scanning microscopy (left). Merged pictures of the green fluorescence channel with the corresponding light micrographs are shown on the right. The length of the bars corresponds to 10 (A–C) and 50 (D) μm, respectively.
The nuclear localization of G2A-GFP is somewhat surprising considering the mass of the fusion protein (90 kD), which is well above the size exclusion limit of the nuclear pore complex (40 kD). Targeting to the nucleus may result from a potential nuclear localization signal (SV40 large T antigen prototype) located close to the C terminus of the LeCPK1 kinase domain (Pro-348-Arg-349-Lys-350-Arg-351). This nuclear localization signal may well be cryptic in wild-type LeCPK1 but may become active in the G2A mutant protein when plasma membrane targeting is suppressed. The physiological relevance of nuclear localization, if any, remains to be investigated.
Phosphorylation of the Plasma Membrane H+-ATPase
It has been well established that the plant plasma membrane H+-ATPase is regulated by calcium-dependent, reversible phosphorylation at multiple sites (Schaller and Sussman, 1988; Morsomme and Boutry, 2000) resulting in enhanced (Kinoshita and Shimazaki, 1999) or reduced (Lino et al., 1998; De Nisi et al., 1999) proton transport activities, respectively. Protein kinase C, Ca2+/calmodulin-dependent protein kinases, and CDPKs have been implicated in these processes (Vera-Estrella et al., 1994; Xing et al., 1996; Camoni et al., 1998; Lino et al., 1998). The positions of the phosphorylated Ser and Thr residues are largely unknown. Some of the phosphorylation sites, however, were shown to reside within the C-terminal autoregulatory domain of the H+-ATPase, which includes the highly conserved penultimate phospho-Thr residue required for the 14-3-3 and FC-mediated activation of the pump (Fuglsang et al., 1999; Svennelid et al., 1999; Maudoux et al., 2000). In vitro phosphorylation of the C-terminal domain of the plasma membrane H+-ATPase by a CDPK partially purified from maize root plasma membranes has been demonstrated but no effect on proton pump activity has been reported (Camoni et al., 1998). Therefore, we tested the H+-ATPase C-terminal domain as a substrate of LeCPK1-L.
LeCPK1-L phosphorylated a fusion protein of GST and CT66, the 66 C-terminal amino acids from the Nicotiana plumbaginifolia H+-ATPase PMA2, in a calcium-dependent manner (Fig. 8, A and B). Phosphorylation occurred within the PMA2-derived 66 amino acids as shown by proteolytic cleavage of the GST moiety (Fig. 8C). The phosphorylation of CT66 in vitro, and the colocalization of LeCPK1 and the H+-ATPase at the plasma membrane, are consistent with the hypothesis that the H+-ATPase is a bona fide substrate of LeCPK1 in vivo, suggesting a role for LeCPK1 in proton pump regulation.
Figure 8.
Phosphorylation of the H+-ATPase C terminus. LeCPK1-L (0.25 μg) was incubated with GST-CT66 (a fusion protein of GST and the 66 amino acids from the C terminus of the N. plumbaginifolia H+-ATPase PMA2, 1.3 μg) in the presence of [γ-32P]ATP for 1, 5, and 25 min at 25°C (lanes 1, 2, and 3, respectively). The assays shown in lanes 4, 5, and 6 were incubated for 25 min but lacked Ca2+, the substrate (CT66), or LeCPK1-L, respectively. A, Coomassie Brilliant Blue-stained SDS-PAGE gel is shown. The molecular masses of protein standards (Bio-Rad, low-Mr markers) are indicated in kD. B, Autoradiograph of the gel shown in A. C, GST-CT66 was treated with thrombin (2.5 units mg−1 GST-CT66, 3.5 h) to release the GST moiety before the phosphorylation reactions with LeCPK1-L were performed as above. The reaction products were separated electrophoretically on a Tricine/SDS gel (Schägger and Jagow, 1987). The gels were dried and analyzed by autoradiography. The positions of peptide size standards (Bio-Rad) are indicated in kD.
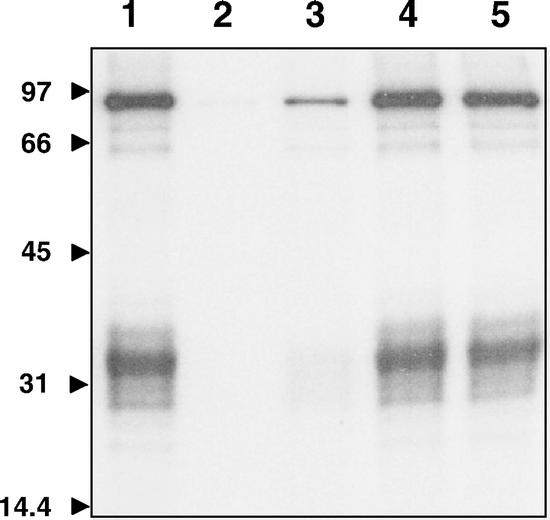
At present, we can only speculate about the physiological function of LeCPK1, but correlative evidence supports a role in plant defense reactions. We previously proposed the modulation of H+-ATPase activity by Ca2+-dependent phosphorylation to be part of the wound and pathogen defense signaling cascades (Schaller, 1999; Schaller and Oecking, 1999; Schaller et al., 2000). The polypeptide wound hormone systemin triggers a depolarization of the plasma membrane and an alkalinization of the apoplast (Felix and Boller, 1995; Moyen and Johannes, 1996), which depend on the influx of Ca2+ as well as the activity of a protein kinase: The systemin-induced alkalinization response could be mimicked by the protein phosphatase inhibitor calyculin A, whereas the protein kinase inhibitors staurosporine and K252a suppressed the systemin response (Felix and Boller, 1995; Schaller and Oecking, 1999). Likewise, staurosporine and K252a inactivated LeCPK1, inhibiting both the autokinase activity as well as the phosphorylation of CT66 (Fig. 9). Selective inhibitors of protein kinase C (bisindolylmaleimide) and Ca2+/calmodulin-dependent protein kinase II (KN-62), on the other hand, do not inhibit LeCPK1-L (Fig. 9). Likewise, these compounds did not affect the systemin-induced alkalinization response (Schaller and Oecking, 1999). Hence, the activity of systemin and LeCPK1 are affected in a similar way by various protein kinase inhibitors supporting a hypothetical role for LeCPK1 in the systemin signal transduction pathway.
Figure 9.
Effect of kinase inhibitors on the activity of LeCPK1-L. LeCPK1-L (0.13 μg) activity was assayed with GST-CT66 (2 μg) and 5 μm [γ-32P]ATP (3.5 × 107 Bq pmol−1) as the substrates (lane 1) in the presence of staurosporine (4 μm, lane 2), K252a (2 μm, lane 3), KN62 (5 μm, lane 4), and bisindolylmaleimide (0.2 μm, lane 5). The assays were performed at 25°C for 60 min. Reaction products were separated by SDS-PAGE and the gel was dried and analyzed by autoradiography.
MATERIALS AND METHODS
Cloning of LeCPK1
Database comparison revealed the sequence of a tomato (Lycopersicon esculentum Mill. cv Castlemart II) EST clone highly similar to tobacco (Nicotiana tabacum) NtCDPK1 (cLER8C16, accession no. AI773815), which formed the basis for the cloning of LeCPK1 by PCR. The 3′ end of LeCPK1 was cloned using tomato cDNA libraries from tomato shoot and flower tissues in pBluescript SK− (Stratagene, La Jolla, CA) as the template (200 ng) in the PCR with an EST-derived oligonucleotide (GGGAATTCGGTATGGGTGATGAGGCCAC) and the T7 primer as 5′ and 3′ primers, respectively. PCR products were cloned into the EcoRI and XhoI sites of pBluescript SK−. Sequence analysis of several independent clones showed that cDNAs derived from flower and shoot tissue had different polyadenylation sites but were otherwise identical and corresponded to the tomato EST clone. The main part of the LeCPK1 cDNA, including the entire ORF, was cloned by reverse transcriptase-PCR using the Smart RACE cDNA amplification system (CLONTECH, Palo Alto, CA) following the manufacturer's instructions. In a first step, single-stranded cDNA was synthesized from total tomato leaf RNA of FC-treated plants using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) and oligo(dT) as the primer. RACE-PCR was performed with a gene-specific primer derived from the cloned LeCPK1 3′ end (CGTGCTGGCTGGTGAACGTTCTCTGCTC; Microsynth, Balgach, Switzerland) and the universal primer provided with the kit. RACE-PCR products were cloned into PCR 2.1-TOPO (Invitrogen, Groningen, the Netherlands). The identity of these and all other PCR-generated clones was confirmed by sequence analysis of several independent PCR products using fluorescent dideoxy chain terminators in the cycle sequencing reaction (Perkin Elmer, Foster City, CA) and the model 373A DNA sequencer (PE-Applied Biosystems, Foster City, CA).
Northern- and Southern-Blot Analyses
Tomato plants (Ochoa Seed Co., Gilroy, CA) were grown for 14 d and were treated with 3 μm FC as described (Schaller et al., 2000). RNA was isolated from tomato leaf, cotyledon, stem, and flower tissue as well as from suspension-cultured cells using a phenol-based extraction procedure. Total RNA (5 μg) was subjected to electrophoresis on formaldehyde/agarose gels and transferred to nitrocellulose membranes according to standard protocols. For Southern-blot analysis, genomic DNA was extracted from tomato leaf tissue using the Nucleon Phytopure DNA extraction kit (Amersham Pharmacia Biotech). Ten micrograms of DNA was restricted using the enzymes indicated in the legend to Figure 2. RNA and DNA gel blots were probed with the radiolabeled 3′-untranslated region of the LeCPK1 cDNA. The blots were hybridized, washed, and evaluated as described (Schaller and Oecking, 1999).
Expression and Purification of Recombinant LeCPK1-L and -S
Two constructs were generated for the expression of LeCPK1 in Escherichia coli as GST fusion proteins, i.e. LeCPK1-L corresponding to the entire ORF and LeCPK1-S, coding for a truncated protein lacking the C-terminal calmodulin-like domain (compare with Fig. 1). The respective regions of the LeCPK1 cDNA were amplified by PCR using Pwo DNA polymerase (Roche Diagnostics, Rotkreuz, Switzerland) and synthetic oligonucleotide primers (forward primer, 5′-ATGGGTGGTTGTTTTAGCAAGAAGT-3′; reverse primers for LeCPK1-L and -S, 5′-GGGGTACCCTAGAAAAGCTTTTGTTGTGGTTG-3′ and 5′-GGGGTACCCTATTACAGAGGGTTGATTTCTTCTTCA-3′, respectively). The PCR products were cloned into the StuI/KpnI sites of pGEX-G, a derivative of pGEX-3x (Amersham Pharmacia Biotech) for the expression of N-terminal GST fusion proteins under the control of the isopropylthio-β-galactoside (IPTG)-inducible tac promoter. The constructs were transformed into E. coli BL21 codon plus (DE3)-RIL (Stratagene). The fusion proteins were purified from IPTG-induced cultures by affinity chromatography on immobilized glutathione and analyzed by SDS-PAGE as described (Hauser et al., 2001).
Autophosphorylation and Phospho-Amino Acid Analysis
Autophosphorylation of LeCPK1-L and -S was analyzed on western blots. The immobilized proteins were denatured for 1 h in 50 mm Tris/HCl, 7 m guanidine/HCl, 50 mm dithiothreitol, 2 mm EDTA, and 0.25% (w/v) dry milk. Proteins were then renatured in the same buffer with 100 mm NaCl replacing the guanidine/HCl. To assay Ca2+-dependent autophosphorylation, the membranes were then incubated for 30 min at 25°C in either kinase buffer A (50 mm HEPES, pH 7.5; 10 mm MgCl2; 1 mm EGTA; 2 mm dithiothreitol; and 1.1 mm CaCl2) with 0.012 μm [γ-32P]ATP (3 × 1010 Bq μmol−1), or the same buffer with 4 mm EGTA replacing CaCl2 (kinase buffer B). The membranes were washed repeatedly in 30 mm HEPES, pH 7.5, and analyzed on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Phospho-amino acids were analyzed essentially as described by Shi et al. (1999). In brief, LeCPK1-L (5 μg) was autophosphorylated in 0.5 mL of buffer A containing 0.05 μm [γ-32P]ATP (7.4 × 1010 Bq μmol−1), the protein was precipitated, and then hydrolyzed in 6 n HCl during 1 h at 110°C. The hydrolysate was dried in vacuo, spiked with phospho-amino acid standards (12 μg each of l-phospho-Ser, -Thr, and -Tyr), and analyzed by two-dimensional TLC on cellulose plates (first dimension, iso-butyric acid:0.5 m NH4OH = 5:3 [v/v]; second dimension, propionic acid:1 m NH4OH:iso-propanol = 45:17.5:17.5 [v/v]). Phospho-amino acid standards were visualized by spraying with ninhydrin (0.25% [w/v] in acetone), and the labeled amino acids were detected by autoradiography.
Determination of Autophosphorylation Sites
LeCPK1-L (100 μg) was allowed to autophosphorylate in 100 μL of kinase buffer A containing 10 μm [γ-32P]ATP (3.5 × 107 Bq pmol−1) during 1 h at 25°C. The protein was then digested with factor Xa (factor Xa cleavage and removal kit, Roche Diagnostics) resulting in the release of the GST moiety as well as the removal of the N-terminal 22 amino acids of LeCPK1 due to the presence of an internal processing site. The proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. In-gel digestion with trypsin (Promega, Mannheim, Germany) was done as described by Jensen et al. (1998). Phosphopeptides were purified on three ZipTip MC tips (Millipore, Eschborn, Germany) charged with Fe3+, Ni2+, and Cu2+, respectively. Part of the radiolabel was retained on each of the three tips. The eluates were combined, reduced to dryness in vacuo, and resuspended in 60% (v/v) acetonitrile and 0.1% (v/v) trifluoroacetic acid. MS spectra were recorded on a Q-TOF2 instrument (Micromass, Manchester, UK) using nano-electrospray ionization. Peptides corresponding in mass to possible phosphopeptides of LeCPK1 were fragmented by collision-induced decomposition and amino acid sequences were derived from the resulting MS/MS spectra using the software MassLynx 3.5 including the MaxEnt 3 module (Micromass).
Ca2+-Binding Activity
The Ca2+-binding capacities of LeCPK1-L and LeCPK1-S were compared after SDS-PAGE and subsequent electrophoretic transfer of the proteins onto nitrocellulose membranes. The membranes were washed three times for 20 min in 60 mm KCl, 5 mm MgCl2, and 10 mm imidazol/HCl, pH 6.8. They were then incubated for 10 min in 20 mL of the same buffer containing 20 μCi 45CaCl2 (3.5 × 108 Bq mg−1), rinsed in a large excess of water, and autoradiographed.
The Ca2+-binding constants were determined for LeCPK1-L (45 μg mL−1 in 50 mm HEPES, pH 7.5) by fluorescence spectroscopy using an SFM 25 fluorimeter (λex: 280 nm, Kontron, Neufahrn, Germany). Two-microliter aliquots of CaCl2 stock solutions in 50 mm HEPES, pH 7.5, were added to result in a final Ca2+ concentration ranging from 0.2 μm to 2 mm. At each interval, the emission spectrum was recorded. The maximum change of Trp fluorescence was observed at 325 nm and was plotted against the Ca2+ concentration in the assay. The software SigmaPlot was used for the evaluation of the data.
Kinase Activity Assay
Assays of LeCPK1-L activity with syntide-2 (PLARTLSVAGLPGKK, Sigma, St. Louis) as the substrate were performed as described by Harmon et al. (1994) with minor modifications. The 50-μL reaction mixture contained 30 nm recombinant LeCPK1-L and varying concentrations of syntide-2 in kinase buffer A with 0.1 mg mL−1 bovine serum albumin. After 5 min at 30°C, the reaction was started by addition of 60 μm [γ-32P]ATP (3,000 Bq pmol−1). The reaction was terminated after 15 min by spotting 10-μL aliquots onto phosphocellulose paper (Whatman P81). The paper was first washed in a large excess of 150 mm H3PO4, then in acetone, air dried, and counted in a liquid scintillation counter.
N-Myristoylation Assay
Myristoylation assays were performed in a cell-free wheat germ transcription/translation system (Promega) essentially as described (Ellard-Ivey et al., 1999). PCR was employed to amplify the ORF of LeCPK1 as well as a mutant cDNA that codes for a Gly substituting Ala in the second amino acid position (G2A-CPK1) using synthetic oligonucleotide primers (CPK1–5′, CCTCTAGAATGGGTGGTTGTTTTAGCAA; G2A-CPK1–5′, CCTCTAGAATGGCTGGTTGTTTTAGCAAGAAGT; and 3′, GGTCTAGAGGGAAAAGCTTTTGTTGTGGTTGT) and Pwo DNA polymerase. The PCR products were cloned into the XbaI site of pBluescript SK− and clones containing the insert in the correct orientation with respect to the T7 promoter were identified by restriction analysis. The plasmids (0.6 and 1 μg, respectively) were linearized with NotI and used as templates in the TNT-coupled transcription/translation system according to the manufacturer's instructions (Promega). Translation products were radiolabeled in the presence of either 10 μm [35S]l-Met (3.7 × 1013 Bq mmol−1; Hartmann Analytic, Braunschweig, Germany) or 40 μm [9,10-3H]myristic acid (1.9 × 1012 Bq mmol−1; Moravek Biochemicals, Brea, CA). The reaction products were separated by SDS-PAGE and analyzed by fluorography.
Transient Expression of LeCPK1/GFP Fusion Proteins
The ORFs of wild-type LeCPK1 and the G2A mutant (see above) were cloned into pCL60 (gift of Dr. Claudio Lupi, Swiss Federal Institute of Technology, Zurich), a derivative of pBluescript SK− that allows the transient expression of proteins in C-terminal fusion with enhanced GFP (CLONTECH) under control of the cauliflower mosaic virus 35S promoter. The constructs (35S-CPK1-GFP and 35S-G2A-GFP) were delivered into onion (Allium cepa) epidermal cells (Scott et al., 1999) and suspension-cultured cells of Lycopersicon peruvianum (Felix and Boller, 1995) using a particle inflow gun (Vain et al., 1993); 4 μg of plasmid DNA and 4 μg of gold particles per shot). Cells were allowed to recover for 24 to 48 h before they were analyzed by confocal laser scanning microscopy using an ArKr laser at 488 nm (Leica TCS SP, Leica DM IRBE; Leica Microsystems, Wetzlar, Germany).
Phosphorylation of the C-Terminal Domain of the H+-ATPase
A PCR-fragment (forward primer, TATGAATTCCATGGGCTGCAAGTTCCT; and reverse primer, TATGTCGACTCAAACAGTGTATGATTG) coding for the 66 C-terminal amino acids of the Nicotiana plumbaginifolia plasma membrane H+-ATPase PMA2 was cloned into pGEX-2T (Amersham Pharmacia Biotech) for expression in E. coli of GST-CT66, an N-terminal fusion of GST with the carboxy terminus of the proton pump (Jelich-Ottmann et al., 2001). The fusion protein was purified from IPTG-induced cultures by affinity chromatography on immobilized glutathione. When applicable, the GST moiety was released proteolytically using 1 unit of thrombin (Amersham Pharmacia Biotech) per 2.5 μg of fusion protein during 3 h at 25°C.
Two micrograms of GST-CT66 was used directly, or 5 μg of the thrombin-digested fusion protein was tested as substrates of LeCPK1-L in 15 μL of kinase buffers A or B containing 5 μm [γ-32P]ATP (3.5 × 107 Bq pmol−1). Protein kinase inhibitors (4 μm staurosporine, 2 μm K-252a, 5 μm KN-62, and 0.2 μm bisindolylmaleimide; Alexis Corporation, San Diego) were added as indicated. The reaction products were separated by SDS-PAGE using the Laemmli-buffer system, or on Tricine/SDS gels according to Schägger and Jagow (1987). The gels were dried and analyzed by autoradiography.
ACKNOWLEDGMENTS
We thank Claudio Lupi for the plasmid pCL60, Birgit Klüsener (Ruhr-Universität Bochum) for valuable discussions, and Nikolaus Amrhein (Swiss Federal Institute of Technology, Zurich) for his support and critical reading of the manuscript.
Footnotes
The work was supported by the Swiss National Science Foundation (grant no. 31–56855.99 to A.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.000869.
LITERATURE CITED
- Allen GJ, Sanders D. Vacuolar ion channels of higher plants. Adv Bot Res. 1997;25:217–251. [Google Scholar]
- Blume B, Nürnberger T, Nass N, Scheel D. Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell. 2000;12:1425–1440. doi: 10.1105/tpc.12.8.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Lam BC-H. Early signal transduction pathways in plant-pathogen interactions. Trends Plant Sci. 1998;3:342–346. [Google Scholar]
- Camoni L, Fullone MR, Marra M, Aducci P. The plasma membrane H+-ATPase from maize roots is phosphorylated in the C-terminal domain by a calcium-dependent protein kinase. Physiol Plant. 1998;104:549–555. [Google Scholar]
- Chaudhuri S, Seal A, DasGupta M. Autophosphorylation-dependent activation of a calcium-dependent protein kinase from groundnut. Plant Physiology. 1999;120:859–866. doi: 10.1104/pp.120.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- De Nisi P, Dell'Orto M, Pirovano L, Zocchi G. Calcium-dependent phosphorylation regulates the plasma-membrane H+-ATPase activity of maize (Zea mays L.) roots. Planta. 1999;209:187–194. doi: 10.1007/s004250050621. [DOI] [PubMed] [Google Scholar]
- Desbrosses G, Stelling J, Renaudin JP. Dephosphorylation activates the purified plant plasma membrane H+-ATPase: possible function of phosphothreonine residues in a mechanism not involving the regulatory C-terminal domain of the enzyme. Eur J Biochem. 1998;251:496–503. doi: 10.1046/j.1432-1327.1998.2510496.x. [DOI] [PubMed] [Google Scholar]
- Doherty HM, Bowles DJ. The role of pH and ion transport in oligosaccharide-induced proteinase inhibitor accumulation in tomato plants. Plant Cell Environ. 1990;13:851–855. [Google Scholar]
- Ellard-Ivey M, Hopkins RB, White TJ, Lomax TL. Cloning, expression and N-terminal myristoylation of CpCPK1, a calcium-dependent protein kinase from zucchini (Cucurbita pepo L.) Plant Mol Biol. 1999;39:199–208. doi: 10.1023/a:1006125918023. [DOI] [PubMed] [Google Scholar]
- Engelberth J, Koch TGS, Bachmann N, Rechtenbach J, Boland W. Ion channel-forming alamethicin is a potent elicitor of volatile biosynthesis and tendril coiling. Cross talk between jasmonate and salicylate signaling in lima bean. Plant Physiol. 2001;125:369–377. doi: 10.1104/pp.125.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Färber P, Graeser R, Franklin RM, Kappes B. Molecular cloning and characterization of a second calcium-dependent protein kinase of Plasmodium falciparum. Mol Biochem Parasitol. 1997;87:211–216. doi: 10.1016/s0166-6851(97)00052-2. [DOI] [PubMed] [Google Scholar]
- Felix G, Boller T. Systemin induces rapid ion fluxes and ethylene biosynthesis in Lycopersicon peruvianum cells. Plant J. 1995;7:381–389. [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG. Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J Biol Chem. 1999;274:36774–36780. doi: 10.1074/jbc.274.51.36774. [DOI] [PubMed] [Google Scholar]
- Gelli A, Higgins VJ, Blumwald E. Activation of plant plasma membrane Ca2+-permeable channels by race-specific fungal elicitors. Plant Physiol. 1997;113:269–279. doi: 10.1104/pp.113.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Silverman P, Raskin I, Jones JD. Race-specific elicitors of Cladosporium fulvum induce changes in cell morphology and the synthesis of ethylene and salicylic acid in tomato plants carrying the corresponding Cf disease resistance gene. Plant Physiol. 1996;110:1381–1394. doi: 10.1104/pp.110.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF. CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5:154–159. doi: 10.1016/s1360-1385(00)01577-6. [DOI] [PubMed] [Google Scholar]
- Harmon AC, Putnam-Evans C, Cormier MJ. A calcium-dependent but calmodulin-independent protein kinase from soybean. Plant Physiol. 1987;83:830–837. doi: 10.1104/pp.83.4.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Yoo B-C, McCaffery C. Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33:7278–7287. doi: 10.1021/bi00189a032. [DOI] [PubMed] [Google Scholar]
- Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science. 1991;252:951–954. doi: 10.1126/science.1852075. [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang J-F, Lloyd SJ. Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33:7267–7277. doi: 10.1021/bi00189a031. [DOI] [PubMed] [Google Scholar]
- Hauser F, Strassner J, Schaller A. Cloning, expression, and characterization of tomato (Lycopersicon esculentum) aminopeptidase P. J Biol Chem. 2001;276:31732–31737. doi: 10.1074/jbc.M103179200. [DOI] [PubMed] [Google Scholar]
- Herde O, Peña-Cortés H, Willmitzer L, Fisahn J. Remote stimulation by heat induces characteristic membrane-potential responses in the veins of wild-type and abscisic acid-deficient tomato plants. Planta. 1998;206:146–153. [Google Scholar]
- Hrabak EM. Calcium-dependent protein kinases and their relatives. Adv Bot Res. 2000;32:185–223. [Google Scholar]
- Hrabak EM, Dickmann LJ, Satterlee JS, Sussman MR. Characterization of eight new members of the calmodulin-like domain protein kinase gene family from Arabidopsis thaliana. Plant Mol Biol. 1996;31:405–412. doi: 10.1007/BF00021802. [DOI] [PubMed] [Google Scholar]
- Jabs T, Tschöpe M, Colling C, Hahlbrock K, Scheel D. Elicitor-stimulated ion fluxes and O2− from the oxidative burst are essential components in triggering defense gene activation and phytoalexin biosynthesis in parsley. Proc Natl Acad Sci USA. 1997;94:4800–4805. doi: 10.1073/pnas.94.9.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9:1805–1814. doi: 10.1105/tpc.9.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelich-Ottmann C, Weiler EW, Oecking C. Binding of regulatory 14-3-3 proteins to the C-terminus of the plant plasma membrane H+-ATPase involves part of its autoinhibitory region. J Biol Chem. 2001;276:39852–39857. doi: 10.1074/jbc.M106746200. [DOI] [PubMed] [Google Scholar]
- Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. In: Link AJ, editor. Methods in Molecular Biology, 112: Proteome Analysis Protocols. Totowa, NJ: Humana Press; 1998. pp. 513–530. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Nishimura M, Shimazaki K-I. Cytosolic concentration of Ca2+ regulates the plasma membrane H+-ATPase in guard cells of fava bean. Plant Cell. 1995;7:1333–1342. doi: 10.1105/tpc.7.8.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K. Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J. 1999;18:5548–5558. doi: 10.1093/emboj/18.20.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klüsener B, Boheim G, Liss H, Engelberth J, Weiler EW. Gadolinium-sensitive, voltage-dependent calcium release channels in the endoplasmic reticulum of a higher plant mechanoreceptor organ. EMBO J. 1995;14:2708–2714. doi: 10.1002/j.1460-2075.1995.tb07271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klüsener B, Weiler EW. Pore-forming properties of elicitors of plant defense reactions and cellulolytic enzymes. FEBS Lett. 1999;459:263–266. doi: 10.1016/s0014-5793(99)01261-2. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yoo B-C, Harmon AC. Kinetic and calcium-binding properties of three calcium-dependent protein kinase isozymes from soybean. Biochemistry. 1998;37:6801–6809. doi: 10.1021/bi980062q. [DOI] [PubMed] [Google Scholar]
- Lino B, Baizabal-Aguirre VM, Gonzáles de la Vara LE. The plasma-membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta. 1998;204:352–359. doi: 10.1007/s004250050266. [DOI] [PubMed] [Google Scholar]
- Marré E. Fusicoccin: a tool in plant physiology. Annu Rev Plant Physiol. 1979;30:273–288. [Google Scholar]
- Martin ML, Busconi L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000;24:429–435. doi: 10.1046/j.1365-313x.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P. A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J Biol Chem. 2000;275:17762–17770. doi: 10.1074/jbc.M909690199. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Hetherington AM. Encoding specificity in Ca2+ signalling systems. Trends Plant Sci. 1998;3:32–36. [Google Scholar]
- Morsomme P, Boutry M. The plant plasma membrane H+-ATPase: structure, function and regulation. Biochim Biophys Acta. 2000;1465:1–16. doi: 10.1016/s0005-2736(00)00128-0. [DOI] [PubMed] [Google Scholar]
- Moyen C, Hammond-Kosack KE, Jones J, Knight MR, Johannes E. Systemin triggers an increase of cytoplasmic calcium in tomato mesophyll cells: Ca2+ mobilization from intra- and extracellular compartments. Plant Cell Environ. 1998;21:1101–1111. [Google Scholar]
- Moyen C, Johannes E. Systemin transiently depolarizes the tomato mesophyll cell membrane and antagonizes fusicoccin-induced extracellular acidification of mesophyll tissue. Plant Cell Environ. 1996;19:464–470. [Google Scholar]
- Murillo I, Jaeck EC, M J, San Segundo B. Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitors and infection. Plant Mol Biol. 2001;45:145–158. doi: 10.1023/a:1006430707075. [DOI] [PubMed] [Google Scholar]
- Oecking C, Piotrowski M, Hagemeier J, Hagemann K. Topology and target interaction of the fusicoccin-binding 14-3-3 homologs of Commelina communis. Plant J. 1997;12:441–453. [Google Scholar]
- Olivari C, Meanti C, De Michelis MI, Rasi-Caldogno F. Fusicoccin binding to its plasma membrane receptor and the activation of the plasma membrane H+-ATPase. Plant Physiol. 1998;116:529–537. doi: 10.1104/pp.116.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raíces M, Chico JM, Téllez-Iñón MT, Ulloa RM. Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development. Plant Mol Biol. 2001;46:591–601. doi: 10.1023/a:1010661304980. [DOI] [PubMed] [Google Scholar]
- Roberts DM, Harmon AC. Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:375–414. [Google Scholar]
- Roberts MR, Bowles DJ. Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiol. 1999;119:1243–1250. doi: 10.1104/pp.119.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JDG. Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell. 2000;12:803–815. doi: 10.1105/tpc.12.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF. Communicating with calcium. Plant Cell. 1999;11:691–706. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schaller A. Oligopeptide signalling and the action of systemin. Plant Mol Biol. 1999;40:763–769. doi: 10.1023/a:1006279409687. [DOI] [PubMed] [Google Scholar]
- Schaller A, Frasson D. Induction of wound response gene expression in tomato leaves by ionophores. Planta. 2001;212:431–435. doi: 10.1007/s004250000413. [DOI] [PubMed] [Google Scholar]
- Schaller A, Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell. 1999;11:263–272. doi: 10.1105/tpc.11.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Roy P, Amrhein N. Salicylic acid-independent induction of pathogenesis-related gene expression by fusicoccin. Planta. 2000;210:599–606. doi: 10.1007/s004250050049. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Sussman MR. Phosphorylation of the plasma membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta. 1988;173:509–518. doi: 10.1007/BF00958964. [DOI] [PubMed] [Google Scholar]
- Scheel D. Resistance response physiology and signal transduction. Curr Opin Plant Biol. 1998;1:305–310. doi: 10.1016/1369-5266(88)80051-7. [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou P-L, Robertson D, Allen NS. Model system for plant cell biology: GFP imaging in living onion epidermal cells. BioTechniques. 1999;26:1125–1132. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KS, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell. 1999;11:2392–2406. doi: 10.1105/tpc.11.12.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell. 1999;11:2379–2391. doi: 10.1105/tpc.11.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain JF, Gubb IR, Wildon DC. Depolarization of tomato leaf cells by oligogalacturonide elicitors. Plant Cell Environ. 1995;18:211–214. [Google Scholar]
- Trewavas A, Malhó R. Ca2+ signalling in plant cells: the big network! Curr Opin Plant Biol. 1998;1:428–433. doi: 10.1016/s1369-5266(98)80268-9. [DOI] [PubMed] [Google Scholar]
- Vain P, Keen N, Murillo J, Rathus C, Nemes C, Finer JJ. Development of the particle inflow gun. Plant Cell Tissue Organ Cult. 1993;33:237–246. [Google Scholar]
- Vera-Estrella R, Barkla BJ, Higgins VJ, Blumwald E. Plant defense to fungal pathogens. Activation of host-plasma membrane H+-ATPase by elicitor-induced enzyme dephosphorylation. Plant Physiol. 1994;104:209–215. doi: 10.1104/pp.104.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian A, Henry-Vian C, Schantz R, Schantz M-L, Davies E, Ledoigt G, Desbiez M-O. Effect of calcium and calcium-counteracting drugs on the response of Bidens pilosa L. to wounding. Plant Cell Physiol. 1997;38:751–753. [Google Scholar]
- Webb AAR, McAinsh MR, Taylor JE, Hetherington AM. Calcium ions as intracellular messengers in higher plants. Adv Bot Res. 1996;22:45–96. [Google Scholar]
- Wevelsiep L, Rüpping E, Knogge W. Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol. 1993;101:297–301. doi: 10.1104/pp.101.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing T, Higgins VJ, Blumwald E. Regulation of plant defense response to fungal pathogens: two types of protein kinases in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell. 1996;8:555–564. doi: 10.1105/tpc.8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodríguez-Conceptión M, Gruissem W. Lipid modifications of proteins—slipping in and out of membranes. Trends Plant Sci. 1999;4:439–445. doi: 10.1016/s1360-1385(99)01492-2. [DOI] [PubMed] [Google Scholar]
- Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HS. Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol. 1999;39:991–1001. doi: 10.1023/a:1006170512542. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Pokutta S, Maurer P, Lindt M, Franklin RM, Kappes B. Calcium-binding properties of a calcium-dependent protein kinase from Plasmodium falciparum and the significance of individual calcium-binding sites for kinase activation. Biochemistry. 1994;33:3714–3721. doi: 10.1021/bi00178a031. [DOI] [PubMed] [Google Scholar]
- Zhou F, Anderson CH, Burhenne K, Hertz Fischer P, Collinge DB, Thordal-Christensen H. Proton extrusion is an essential signalling component in the HR of epidermal single cells in the barley-powdery mildew interaction. Plant J. 2000;23:245–254. doi: 10.1046/j.1365-313x.2000.00777.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Nürnberger T, Frachisse J-M, Wirtz W, Guern J, Hedrich R, Scheel D. Receptor-mediated activation of a plant Ca2+-permeable ion channel involved in pathogen defense. Proc Natl Acad Sci USA. 1997;94:2751–2755. doi: 10.1073/pnas.94.6.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]