Abstract
The engineering of transgenic crops resistant to the broad-spectrum herbicide glyphosate has greatly improved agricultural efficiency worldwide. Glyphosate-based herbicides, such as Roundup, target the shikimate pathway enzyme 5-enolpyruvylshikimate 3-phosphate (EPSP) synthase, the functionality of which is absolutely required for the survival of plants. Roundup Ready plants carry the gene coding for a glyphosate-insensitive form of this enzyme, obtained from Agrobacterium sp. strain CP4. Once incorporated into the plant genome, the gene product, CP4 EPSP synthase, confers crop resistance to glyphosate. Although widely used, the molecular basis for this glyphosate-resistance has remained obscure. We generated a synthetic gene coding for CP4 EPSP synthase and characterized the enzyme using kinetics and crystallography. The CP4 enzyme has unexpected kinetic and structural properties that render it unique among the known EPSP synthases. Glyphosate binds to the CP4 EPSP synthase in a condensed, noninhibitory conformation. Glyphosate sensitivity can be restored through a single-site mutation in the active site (Ala-100–Gly), allowing glyphosate to bind in its extended, inhibitory conformation.
Keywords: conformational change, crystal structure, genetic modification, mutation
The broad-spectrum herbicide glyphosate, the active ingredient of Roundup, inhibits 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase (EC 2.5.1.19), the enzyme catalyzing the penultimate step of the shikimate pathway toward the biosynthesis of aromatic amino acids. Roundup Ready crop lines contain a gene derived from Agrobacterium sp. strain CP4, encoding a glyphosate-tolerant enzyme, the so-called CP4 EPSP synthase (1, 2). Expression of CP4 EPSP synthase results in glyphosate-tolerant crops, enabling more effective weed control by allowing postemergent herbicide application. The substantial advantages of glyphosate-tolerant crops have resulted in rapid adoption: 87% of soybeans, 61% of cotton, and 26% of corn planted in the United States in 2005 were glyphosate-tolerant varieties (3). However, lingering concerns about the potential health and environmental effects of genetically modified organisms have limited the acceptance of such seed lines and food products, particularly in Europe and Japan.
EPSP synthase catalyzes the transfer of the enolpyruvyl moiety of phosphoenolpyruvate (PEP) to the 5-hydroxyl of shikimate-3-phosphate (S3P) (Fig. 1A). Beginning in the early 1980s, researchers sought to identify glyphosate-insensitive EPSP synthases that could be introduced into crops to provide herbicide resistance. A number of promising enzymes were identified through selective evolution, site-directed mutagenesis, and microbial screens (4–7). However, an increased tolerance for glyphosate in EPSP synthase was often accompanied by a concomitant decrease in the enzyme’s affinity for PEP, resulting in decreased catalytic efficiency. More favorable kinetic characteristics were observed in some enzymes with substitutions including Pro-101–Ser and Thr-97–Ile (numbering according to Escherichia coli EPSP synthase) (8, 9). Eventually, naturally occurring glyphosate-tolerant microbes were identified, including Agrobacterium sp. strain CP4, Achromobacter sp. strain LBAA, and Pseudomonas sp. strain PG2982 (10). The enzymes isolated from these bacteria were designated as class II EPSP synthases on the basis of their catalytic efficiency in the presence of glyphosate and their substantial sequence variation compared with EPSP synthases from plants or E. coli (1). The Agrobacterium sp. strain CP4, isolated from a waste-fed column at a glyphosate production facility, yielded a glyphosate-resistant, kinetically efficient EPSP synthase suitable for the production of transgenic, glyphosate-tolerant crops. Other class II EPSP synthases have since been described, typically from Gram-positive bacteria, including pathogenic species such as Streptococcus pneumonia (11) and Staphylococcus aureus (12).
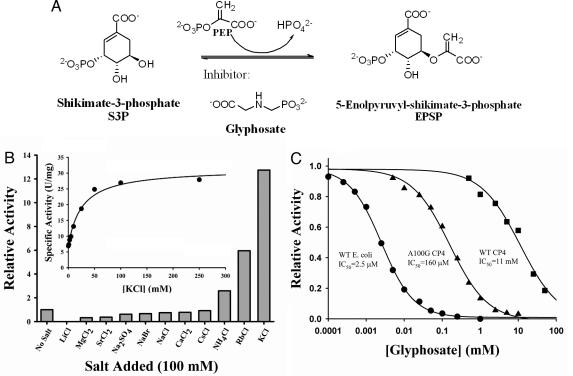
Fig. 1.
Key kinetic properties of CP4 EPSP synthase. (A) The reaction catalyzed by EPSP synthase. (B) The activity of CP4 EPSP synthase depends strongly on the presence of cations, such as NH4+, Rb+, and K+. (Inset) Activation by K+ is saturable with an apparent dissociation constant of 25 mM. (C) IC50 studies with wild-type CP4 EPSP synthase (■), Ala-100–Gly CP4 EPSP synthase (▴), and wild-type E. coli EPSP synthase (●) reveal that CP4 EPSP synthase is inhibited only by high millimolar concentrations of glyphosate (IC50 = 11 mM). The Ala-100–Gly mutant CP4 EPSP synthase is approximately two orders of magnitude more sensitive to glyphosate (IC50 = 160 μM). The E. coli enzyme is inhibited by even lower glyphosate concentrations (IC50 = 2.5 μM).
Despite the enormous agricultural and commercial success of genetically modified crops, the molecular basis for the glyphosate-insensitivity of the CP4 enzyme or any other class II EPSP synthase has remained obscure. Understanding glyphosate resistance in molecular detail should facilitate the optimization or replacement of existing crop lines. Additionally, this knowledge will be invaluable for the rational design of novel inhibitors of EPSP synthase to combat the emergence of glyphosate-resistant weeds. Thus, we synthesized a gene corresponding to the patented amino acid sequence of CP4 EPSP synthase (1), optimized for expression in E. coli. The wild-type CP4 enzyme and a mutant enzyme (Ala-100–Gly) were characterized by steady-state kinetics and the crystal structures, free and liganded with S3P with or without glyphosate, were determined at 1.7- to 2.1-Å resolution.
Results and Discussion
Kinetically, the most intriguing feature of CP4 EPSP synthase is the strong dependence of the catalytic efficiency on monovalent cations, namely K+, Rb+, and NH4+ (Fig. 1B). Whereas the Km for S3P appears to be independent of cations, the Km for PEP decreases from 3.5 mM to 0.2 mM in the presence of 100 mM KCl, resulting in an increase of kcat/Km by a factor of 58, from 1.9 × 103 M−1·s−1 to 1.1 × 105 M−1·s−1 (see the supporting information, which is published on the PNAS web site). The apparent dissociation constant for the interaction of K+ ions with the enzyme is ≈25 mM (Fig. 1B). It has been reported that potassium concentrations in planta are in fact sufficient to promote the enzyme’s interaction with PEP (13). In the absence of such cations, the low catalytic efficiency of CP4 EPSP synthase would render such engineered plants unsuitable. CP4 EPSP synthase maintains activity over a broad pH range and for prolonged periods at elevated temperatures (supporting information), illustrating the enzyme’s stability under harsh environmental conditions.
As expected, the CP4 EPSP synthase is insensitive to inhibition by glyphosate, exhibiting Ki and IC50 values of 6 and 11 mM, respectively (Fig. 1C; see also the supporting information). Moreover, the enzyme’s interaction with glyphosate appears to be independent of cations present: The IC50 values for glyphosate inhibition are ≈10–20 mM with or without salt added (data not shown). This finding is surprising, because PEP and glyphosate share the same binding site in EPSP synthase (14, 15). Thus, one would intuitively expect cations to modulate the binding of glyphosate; however, the effect of potassium ions on the enzyme’s activity appears to be selective toward PEP utilization. Indeed, the CP4 EPSP synthase is the prototypic class II EPSP synthase, because the catalytic efficiency remains essentially unaltered in the presence of high glyphosate concentrations. Notably, other known class II EPSP synthases display greater glyphosate sensitivity and, with the exception of the Sta. aureus enzyme, ions typically affect binding of both PEP and glyphosate (12, 16, 17).
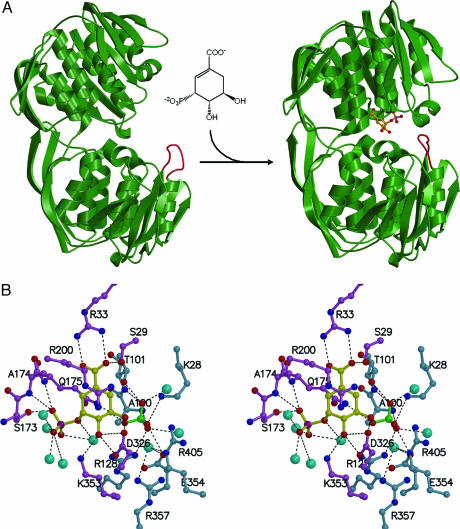
The three-dimensional structures of EPSP synthase from E. coli and Str. pneumoniae are known, but both of these enzymes are sensitive to glyphosate (15, 18, 19). As observed in the E. coli and Str. pneumoniae EPSP synthases, the CP4 EPSP synthase exists in an open, unliganded state and a closed, S3P liganded state, suggesting an induced-fit mechanism with binding of S3P as a prerequisite for the enzyme’s interaction with PEP (Fig. 2). The overall structure of the CP4 EPSP synthase is more similar to the enzyme from Str. pneumoniae than to the enzyme from E. coli. In the binary complex with S3P, the active site architecture is highly conserved, even between the CP4 and E. coli enzymes. Strictly conserved residues from both globular domains of the enzyme constitute the PEP-binding site (Figs. 2B and 3). In the unliganded state, the CP4 EPSP synthase contains highly flexible regions, particularly around the strictly conserved residue Glu-354, which is part of a 12-residue loop of the C-terminal domain (the bottom globular domain of Fig. 2A) that is barely visible in electron density maps. In the enzyme’s binary complex with S3P, this loop becomes structured and interacts with the N-terminal domain to constitute the active site. Remarkably, this loop also adopts a regular structure when the unliganded enzyme is cocrystallized with 100 mM KCl or RbCl. Several of our attempts to identify the cation binding site(s) by cocrystallization of the unliganded or S3P-liganded states of the enzyme with the more electron-dense Rb+ ion failed. It is conceivable, however, that monovalent cations may bind transiently to the enzyme, acting as chaperones in restructuring the loop, enabling CP4 EPSP synthase to interact more efficiently with PEP during a catalytic cycle (20). Once the loop has adopted a regular structure, the cation-binding site(s) may be lost.
Fig. 2.
Three-dimensional structure of CP4 EPSP synthase. (A) (Left) Unliganded CP4 EPSP synthase exists in an open conformation. (Right) Upon interaction with S3P, the enzyme undergoes a large conformational change to a closed state. Shown in orange is a loop spanning residues 347–358, which is highly flexible in the open conformation but becomes ordered in the closed conformation. This loop contains the strictly conserved EPSP synthase residues Glu-354 and Arg-357, which are involved in PEP/glyphosate binding. Monovalent cations may influence the conformation of this loop and facilitate binding of PEP. (B) Stereoview showing that, in the binary complex, S3P (yellow) binds to the enzyme residues shown in magenta through multiple hydrogen-bonding/electrostatic interactions (black dotted lines). In addition, the cyclohexene moiety of S3P is sandwiched between Arg-200 and Gln-175. Residues shown in light blue constitute the PEP/glyphosate binding site. Attracted by the accumulation of positive charges, a sulfate ion (shown in green) from the crystallization solution binds to the space occupied by the phosphate moiety of PEP or the phosphonate moiety of glyphosate in either ternary complex. Water molecules are shown as cyan spheres.
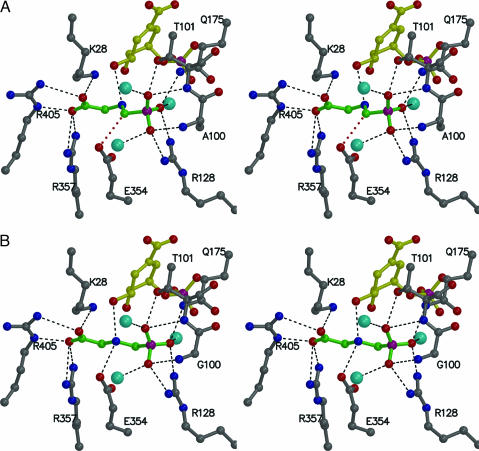
Fig. 3.
Interaction of glyphosate with CP4 EPSP synthase (stereoview). (A) Glyphosate (shown in green) binds to the active site of CP4 EPSP synthase adjacent to S3P (shown in yellow) in a condensed conformation different from that observed in E. coli or Str. pneumoniae EPSP synthase, as the result of a clash between the Ala-100 side chain and oxygen atoms of glyphosate’s phosphonate group. Notably, in this shortened conformation, the carbon atom of the phosphonate group clashes with the side chain of Glu-354 (red dotted line). (B) Replacing Ala-100 with a Gly allows glyphosate to bind in its extended conformation, positioning glyphosate’s nitrogen atom midway between the target hydroxyl of S3P and Glu-354. The view is ≈90° clockwise from that of Fig. 2B. Hydrogen bonding/electrostatic interactions are indicated by black dotted lines.
The weak action of glyphosate on CP4 EPSP synthase can be primarily attributed to an Ala residue in position 100, which is a Gly in the E. coli and Str. pneumoniae EPSP synthases. The methyl group of Ala-100 protrudes into the glyphosate-binding site, clashing with one of the oxygen atoms of glyphosate’s phosphonate group. As a result, the glyphosate molecule adopts a substantially different conformation upon interaction with the CP4 enzyme (Figs. 3 and 4). In the wild-type EPSP synthases from E. coli or Str. pneumoniae, the glyphosate molecule exists in an extended conformation (15, 18), whereas in the CP4 enzyme it adopts a shortened state, which is achieved through rotation about the N C bond adjacent to glyphosate’s carboxyl group (Fig. 4). This rotation shortens the glyphosate molecule by ≈0.6 Å. In its condensed conformation, the glyphosate molecule clashes with the side chain of Glu-354 (Fig. 3A). Ab initio energy calculations of the two distinct glyphosate conformations revealed that the condensed conformation has an ≈17 kcal/mol higher energy than the extended conformation (see Materials and Methods). Thus, only the extended, low-energy conformation of glyphosate appears to be inhibitory.
C bond adjacent to glyphosate’s carboxyl group (Fig. 4). This rotation shortens the glyphosate molecule by ≈0.6 Å. In its condensed conformation, the glyphosate molecule clashes with the side chain of Glu-354 (Fig. 3A). Ab initio energy calculations of the two distinct glyphosate conformations revealed that the condensed conformation has an ≈17 kcal/mol higher energy than the extended conformation (see Materials and Methods). Thus, only the extended, low-energy conformation of glyphosate appears to be inhibitory.
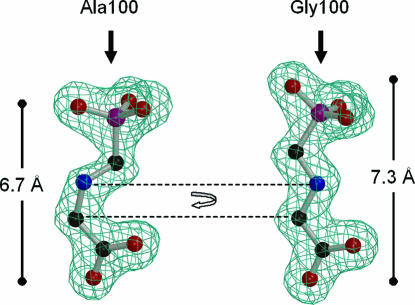
Fig. 4.
Two distinct conformations of glyphosate. Displayed are the electron densities, contoured at 3σ, derived from 1Fo − 1Fc Fourier syntheses to 1.7-Å resolution, omitting the model of glyphosate during the refinement of the ternary complexes of CP4 EPSP synthase (Left) and Ala-100–Gly CP4 EPSP synthase (Right). (Right) The conformation of glyphosate upon interaction with the Ala-100–Gly CP4 EPSP synthase is identical to the one observed in the E. coli or Str. pneumoniae enzymes. (Left) With an Ala residue in position 100, the glyphosate molecule is ≈0.6 Å shorter, mainly because of a rotation around the C N bond next to the carboxyl group.
N bond next to the carboxyl group.
We hypothesized that the mutation of this Ala-100 to Gly should restore the CP4 enzyme’s sensitivity toward glyphosate. The kinetic properties of the Ala-100–Gly CP4 EPSP synthase with respect to PEP utilization, cation dependence, and catalytic efficiency remain essentially unchanged (supporting information). However, this mutant enzyme is sensitive to inhibition by glyphosate with an IC50 value of 150 μM (Fig. 1C) and a Ki of 93 μM (supporting information). As expected, glyphosate binds to this mutant enzyme in its extended conformation (Fig. 3B). For EPSP synthase from E. coli, the residue equivalent to Ala-100 is Gly-96. Mutation of this Gly residue to Ala confers insensitivity to glyphosate but results in a large decrease in affinity for PEP (6, 7). Distinct differences in the space provided for PEP may explain the kinetic differences between the E. coli and the CP4 EPSP synthases. In particular, the clash between glyphosate’s phosphonate moiety and the Ala side chain is less pronounced in the CP4 enzyme than in the E. coli enzyme: The distance between the Cβ of Ala-100 and the nearest phosphonate oxygen of glyphosate is 2.4 Å in the CP4 enzyme versus 2.1 Å in the E. coli EPSP synthase. Although these clash distances are sufficient to hinder glyphosate from binding to either enzyme in its extended, inhibitory conformation, the active site of CP4 EPSP synthase is expected to accommodate the shorter PEP molecule more efficiently than the Gly-96–Ala E. coli EPSP synthase, rendering the CP4 enzyme more catalytically efficient. Another prominent mutation-causing glyphosate insensitivity is Pro-101–Ser in EPSP synthases from Salmonella typhimurium and goosegrass (5, 9, 21) and Pro-101–Leu in EPSP synthase from Sta. aureus (12). In the structure of the CP4 enzyme, the equivalent residue is a Leu residue in position 105, which is part of a helix in the core of the N-terminal domain. This substitution of Leu for Pro may additionally contribute to the distinct kinetic properties of the CP4 enzyme.
Conclusions
The data presented herein explain the agricultural success of Roundup Ready crops at the molecular level. In particular, a single residue in the active site (Ala-100) renders the CP4 EPSP synthase insensitive to glyphosate, whereas a highly conserved Gly residue is found at this position in known natural plant and bacterial enzymes. The continued presence of glyphosate is likely to favor mutations that reduce glyphosate sensitivity while still maintaining catalytic efficiency. It is therefore not surprising that the gene coding for CP4 EPSP synthase was isolated from a microorganism found in an extremely glyphosate-rich environment (refs. 1 and 10 and p. 632 of ref. 22). The speed at which glyphosate resistance develops depends on the organisms’ generation time and fidelity of gene replication. The current low rate of appearance of plants with naturally acquired resistance to glyphosate may be attributed to the relatively high-fidelity replication and long generation times of most plants. However, extensive use of glyphosate increases the likelihood that more glyphosate-resistant organisms will emerge on a large scale. It is conceivable that a single Ala for Gly substitution in the active site of other class II EPSP synthases will confer resistance to glyphosate.
It appears that the confined space of the active site of EPSP synthase prohibits even slight alterations of the glyphosate molecule. From our structural studies, it is evident that even conformational changes within the glyphosate molecule result in loss of inhibitory activity, which is in accordance with extensive structure–activity relationship studies on glyphosate that have been conducted in the past. More than 1,000 analogs of glyphosate have been produced and tested for inhibition of EPSP synthase, but minor structural alterations typically resulted in dramatically reduced potency, and no compound superior to glyphosate was identified (ref. 22, pp. 441–519 and 569–578). Additionally, although EPSP synthase is considered a promising target for the treatment of diseases caused by pathogenic bacteria or eukaryotic parasites (23–25), glyphosate displays little antimicrobial activity. Taken together, these findings demonstrate the pressing need for the development of entirely new inhibitors that target sites different from the PEP/glyphosate binding site of this agriculturally and medicinally important enzyme.
Materials and Methods
S3P (triethylammonium salt) was synthesized from shikimic acid by using recombinant archaeal shikimate kinase (26) and purified via anion exchange chromatography on Q-Sepharose. PEP (potassium salt) and all other chemicals were purchased from Sigma (St. Louis, MO) unless otherwise noted. Coomassie reagent (Pierce, Rockford, IL) with BSA as a standard was used to determine protein concentrations.
The amino acid sequence of CP4 EPSP synthase was obtained from U.S. Patent 5633435 (1). The sequence was initially optimized with the program package DNAWorks (27), which reverse-translated the protein sequence by using the codon frequency table for E. coli (Codon Usage Database available at www.kazusa.or.jp/codon) and divided into two segments (part A, base pairs 1–706, and part B, base pairs 701-1425) linked by the unique restriction site AsuII. The initial codon frequency threshold was set to 50%. The resulting sequence was further optimized to implement ≈30 restriction sites (unique in the vector pNCO113). Finally, flanking sequences were added for cloning of the synthetic gene sequence. Both segments (part A and part B) were synthesized by a series of consecutive PCR amplifications in analogy to Fischer et al. (28). The final PCR product corresponding to segment A (715 bp) was digested with the restriction endonucleases EcoRI and AsuII. The final PCR product corresponding to segment B (734 bp) was digested with AsuII and HindIII. Both segments were combined, ligated into the plasmid pNCO113, which had been treated with the restriction enzymes EcoRI and HindIII, and transformed into E. coli strain XL-1 Blue, resulting in the recombinant strain XL-1-pNCO-EPSPS-syn. The sequence of the synthetic gene was monitored by DNA sequencing in both directions. The synthesized gene was then ligated into pET21a (Novagen, Darmstadt, Germany), and the resulting construct was transformed into E. coli BL21(DE3) (Invitrogen, Carlsbad, CA). The Ala-100–Gly mutant CP4 EPSP synthase was produced by site-directed mutagenesis with the QuikChangeII mutagenesis kit (Stratagene, La Jolla, CA). Overexpression of soluble CP4 EPSP synthase was induced at 37°C by addition of isopropyl β-d-thiogalactoside. CP4 EPSP synthase was purified to homogeneity according to the protocol developed for Sta. aureus EPSP synthase (12).
Enzymatic activity was determined by the amount of inorganic phosphate produced in a 3-min reaction using malachite green (29). CP4 EPSP synthase activity assays were conducted at 25°C in 100 μl of 50 mM Hepes-NaOH, pH 7.5/2 mM DTT with or without KCl where indicated, in parallel with E. coli EPSP synthase. Enzyme activity is expressed as micromoles of phosphate produced per minute of reaction time per milligram of enzyme (units/mg). Data evaluation was performed with SigmaPlot (SPSS Science, Chicago, IL).
The Km values for S3P and PEP were determined by fitting the data to the equation
where v is the initial velocity, Vmax is the maximum velocity, Km is the Michaelis constant, and [S] is the substrate (S3P or PEP) concentration. The IC50 values for glyphosate inhibition were obtained by fitting data to the equation
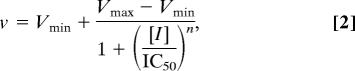 |
where v is the initial velocity, Vmax is the maximum velocity, Vmin is the minimum velocity, [I] is the concentration of glyphosate, and n is the Hill slope. The Ki was derived by determining the Km(obs) of PEP in the presence of increasing amounts of inhibitor and fitting the data to the equation
where Km(obs) is the Michaelis constant for PEP in the presence of glyphosate, [I] is the glyphosate concentration, and Km is the Michaelis constant for PEP in the absence of glyphosate.
CP4 EPSP synthase was concentrated to 40 mg/ml in 50 mM Tris·HCl, pH 8.0/2 mM DTT and crystallized by hanging drop vapor diffusion. The unliganded enzyme was crystallized in 12.5% (wt/vol) PEG 4000/50 mM KCl/2.5% DMSO/50 mM Tris·HCl, pH 8.5. The binary complex with S3P was crystallized in 1.0 M (NH4)2SO4/0.1 M KCl/1% PEG 400/50 mM Tris·HCl, pH 8.5, in the presence of 5 mM S3P. The ternary complexes of the wild-type and Ala-100–Gly enzymes were crystallized in 1.0 M (NH4)2SO4/0.1 M KCl/1% PEG 400/50 mM Hepes-Na, pH 7.5, in the presence of 5 mM S3P and 40 mM glyphosate.
Diffraction data were recorded at −180°C by using the rotation method on single flash-frozen crystals of the enzyme in its unliganded or liganded states [detector: R-axis IV++ image plate; x-rays: CuKα, focused by mirror optics; generator: Rigaku RU300 (MSC, The Woodlands, TX)]. The structure of the unliganded CP4 EPSP synthase was determined with 25% PEG 200 for cryoprotection, and 25% glycerol was used for the binary and ternary complexes. The data were reduced with XDS (30) or HKL-2000 (31). The program package CNS (32) was used for phasing and refinement; model building was performed with O (33). The structures were solved by molecular replacement. With the unliganded Str. pneumoniae enzyme (Protein Data Bank ID code 1RF5) as search model, we first determined the structure of the CP4 enzyme in its unliganded state at 2.1-Å resolution. Subsequently, the S3P-liganded state was determined at 1.64-Å resolution with both globular domains of the unliganded CP4 enzyme as search models. This binary complex served as the search model for the determination of the ternary complexes of the wild-type and Ala-100–Gly mutant enzymes. Refinement was performed with the data at the highest resolution with no sigma cut-off applied. Several rounds of minimization, simulated annealing (2,500 K starting temperature) and restrained individual B-factor refinement were carried out. Data collection and refinement statistics are summarized in Table 1. Figs. 2 and 3 were drawn with Molscript (34) and Raster3D (35); Fig. 4 was drawn with Bobscript (36) and Raster3D. Ab initio energy calculations of the two glyphosate conformations were performed with GAMESS using MP2/6–31+G* (37).
Table 1.
Summary of data collection and structure refinement
| Data set | CP4 unliganded | CP4:S3P | CP4:S3P:glyphosate | CP4(A100G):S3P:glyphosate |
|---|---|---|---|---|
| Sapce group | P212121 | P21 | P21 | P21 |
| Unit cell dimension, Å | a = 55.5, b = 76.5, c = 88.9, α = β = γ = 90° | a = 63.2, b = 44.9, c = 77.2, α = γ = 90°, β = 106.3° | a = 63.1, b = 45.0, c = 77.3, α = γ = 90°, β = 106.5° | a = 63.2, b = 44.7, c = 77.0, α = γ = 90°, β = 106.0° |
| Protein atoms | 3,247 | 3,260 | 3,260 | 3,260 |
| Ligand atoms | 0 | 16 | 26 | 26 |
| Solvent molecules | 280 | 510 | 542 | 491 |
| rmsd bonds, Å | 0.01 | 0.011 | 0.01 | 0.011 |
| rmsd angles, ° | 1.7 | 1.63 | 1.63 | 1.69 |
| Resolution range | 15–2.1 (2.2–2.1) | 15–1.64 (1.7–1.64) | 15–1.7 (1.74–1.7) | 15–1.7 (1.76–1.7) |
| Unique reflections | 23,463 (2,967) | 50,983 (4,995) | 45,806 (2,965) | 44,077 (4.254) |
| Completeness, % | 96.4 (95.1) | 99.7 (98.9) | 99.3 (97.3) | 96.1 (93.2) |
| I/σI | 20.9 (7.3) | 34.9 (3.8) | 29.3 (4.6) | 24.3 (6.8) |
| Rmerge,† % | 5.3 (15.7) | 6.4 (38.0) | 7.0 (37.7) | 7.8 (26.1) |
| Rcryst,‡ % | 17.9 | 15.8 | 15.8 | 14.8 |
| Rfree,§ % | 23.2 | 18.9 | 18.4 | 17.4 |
Values in parentheses refer to the highest resolution shell. rmsd calculations are from ideal values.
†Rmerge = 100 × ΣhΣi ∣Ihi − Ih∣/ΣhiIhi, where h are unique reflection indices.
‡Rcryst = 100 × Σ∣Fobs− Fmodel∣/ΣFobs, where Fobs and Fmodel are observed and calculated structure factor amplitudes, respectively.
§Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement [1,174 for unliganded CP4, 1,419 for CP4:S3P, 1,290 for CP4:S3P:glyphosate, and 1,259 for CP4(A100G):S3P:glyphosate].
Supplementary Material
Acknowledgments
We thank Drs. Bernhard Trinczek and Andreas Becker (University of Kansas) for discussions, Jennifer Biery (University of Kansas) for research assistance, and Sang-Kil Son (University of Kansas) for the energy calculations of the glyphosate molecule. This work was supported by National Institutes of Health Grant 1R01 GM70633-01.
Abbreviations:
- EPSP
5-enolpyruvylshikimate-3-phosphate
- PEP
phosphoenolpyruvate
- S3P
shikimate-3-phosphate.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2GG4 (unliganded CP4 EPSP synthase), 2GG6 (S3P-liganded CP4 EPSP synthase), 2GGA (S3P:glyphosate-liganded CP4 EPSP synthase), and 2GGD (Ala-100–Gly CP4 EPSP synthase-liganded with S3P and glyphosate)].
References
- 1.Barry G. F., Kishore G. M., Padgette S. R., Stallings W. C. 5633435. U.S. Patent. 1997
- 2.Padgette S. R., Kolacz K. H., Delannay X., Re D. B., LaVallee B. J., Tinius C. N., Rhodes W. K., Otero Y. I., Barry G. F., Eichholz D. A., et al. Crop Sci. 1995;35:1451–1461. [Google Scholar]
- 3.Johanns M., Wiyatt S. D., editors. National Agriculture Statistics Service. Acreage. Washington, DC: U.S. Dept. of Agriculture; 2005. Jun 30, [Google Scholar]
- 4.Comai L., Sen L. C., Stalker D. M. Science. 1983;221:370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- 5.Padgette S. R., Re D. B., Gasser C. S., Eichholtz D. A., Frazier R. B., Hironaka C. M., Levine E. B., Shah D. M., Fraley R. T., Kishore G. M. J. Biol. Chem. 1991;266:22364–22369. [PubMed] [Google Scholar]
- 6.Eschenburg S., Healy M. L., Priestman M. A., Lushington G. H., Schönbrunn E. Planta. 2002;216:129–135. doi: 10.1007/s00425-002-0908-0. [DOI] [PubMed] [Google Scholar]
- 7.Sost D., Amrhein N. Arch. Biochem. Biophys. 1990;282:433–436. doi: 10.1016/0003-9861(90)90140-t. [DOI] [PubMed] [Google Scholar]
- 8.Spencer M., Mumm R., Gwyn J. 6040497. U.S. Patent. 2000
- 9.Stalker D. M., Hiatt W. R., Comai L. J. Biol. Chem. 1985;260:4724–4728. [PubMed] [Google Scholar]
- 10.Barry G. F., Kishore G. M., Padgette S. R. 92/04449. Int. Patent. 1992
- 11.Du W., Wallis N. G., Mazzulla M. J., Chalker A. F., Zhang L., Liu W. S., Kallender H., Payne D. J. Eur. J. Biochem. 2000;267:222–227. doi: 10.1046/j.1432-1327.2000.00994.x. [DOI] [PubMed] [Google Scholar]
- 12.Priestman M. A., Funke T., Singh I. M., Crupper S. S., Schönbrunn E. FEBS Lett. 2005;579:728–732. doi: 10.1016/j.febslet.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder J. I., Ward J. M., Gassmann W. Annu. Rev. Biophys. Biomol. Struct. 1994;23:441–471. doi: 10.1146/annurev.bb.23.060194.002301. [DOI] [PubMed] [Google Scholar]
- 14.Eschenburg S., Kabsch W., Healy M. L., Schönbrunn E. J. Biol. Chem. 2003;278:49215–49222. doi: 10.1074/jbc.M309741200. [DOI] [PubMed] [Google Scholar]
- 15.Schönbrunn E., Eschenburg S., Shuttleworth W. A., Schloss J. V., Amrhein N., Evans J. N., Kabsch W. Proc. Natl. Acad. Sci. USA. 2001;98:1376–1380. doi: 10.1073/pnas.98.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du W., Liu W. S., Payne D. J., Doyle M. L. Biochemistry. 2000;39:10140–10146. doi: 10.1021/bi000890v. [DOI] [PubMed] [Google Scholar]
- 17.Fischer R. S., Rubin J. L., Gaines C. G., Jensen R. A. Arch. Biochem. Biophys. 1987;256:325–334. doi: 10.1016/0003-9861(87)90453-x. [DOI] [PubMed] [Google Scholar]
- 18.Park H., Hilsenbeck J. L., Kim H. J., Shuttleworth W. A., Park Y. H., Evans J. N., Kang C. Mol. Microbiol. 2004;51:963–971. doi: 10.1046/j.1365-2958.2003.03885.x. [DOI] [PubMed] [Google Scholar]
- 19.Stallings W. C., Abdel-Meguid S. S., Lim L. W., Shieh H. S., Dayringer H. E., Leimgruber N. K., Stegeman R. A., Anderson K. S., Sikorski J. A., Padgette S. R., Kishore G. M. Proc. Natl. Acad. Sci. USA. 1991;88:5046–5050. doi: 10.1073/pnas.88.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priestman M. A., Healy M. L., Becker A., Alberg D. G., Bartlett P. A., Lushington G. H., Schönbrunn E. Biochemistry. 2005;44:3241–3248. doi: 10.1021/bi048198d. [DOI] [PubMed] [Google Scholar]
- 21.Baerson S. R., Rodriguez D. J., Tran M., Feng Y., Biest N. A., Dill G. M. Plant Physiol. 2002;129:1265–1275. doi: 10.1104/pp.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franz J. E., Mao M. K., Sikorski J. A. Glyphosate: A Unique Global Herbicide. Washington, DC: Am. Chem. Soc.; 1997. [Google Scholar]
- 23.Alibhai M. F., Stallings W. C. Proc. Natl. Acad. Sci. USA. 2001;98:2944–2946. doi: 10.1073/pnas.061025898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConkey G. A. Antimicrob. Agents Chemother. 1999;43:175–177. doi: 10.1128/aac.43.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDevitt D., Payne D. J., Holmes D. J., Rosenberg M. J. Appl. Microbiol. 2002;92:28S–34S. [PubMed] [Google Scholar]
- 26.Daugherty M., Vonstein V., Overbeek R., Osterman A. J. Bacteriol. 2001;183:292–300. doi: 10.1128/JB.183.1.292-300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoover D. M., Lubkowski J. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer M., Romisch W., Saller S., Illarionov B., Richter G., Rohdich F., Eisenreich W., Bacher A. J. Biol. Chem. 2004;279:36299–36308. doi: 10.1074/jbc.M404406200. [DOI] [PubMed] [Google Scholar]
- 29.Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 31.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. B. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. B. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 35.Merrit E. A., Bacon D. J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 36.Esnouf R. M. J. Mol. Graphics. Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S., et al. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.