Abstract
Insulators are a class of elements that define independent domains of gene function. The Drosophila gypsy insulator is proposed to establish regulatory isolation by forming loop domains that constrain interactions between transcriptional control elements. This supposition is based upon the observation that insertion of a single gypsy insulator between an enhancer and promoter blocks enhancer function, while insertion of two gypsy insulators promotes enhancer bypass and activation of transcription. To investigate this model, we determined whether non-gypsy insulators interacted with each other and with the gypsy insulator. Pairs of scs or scs′ insulators blocked enhancer function. Further, an intervening scs insulator did not block gypsy insulator interactions. Taken together, these data suggest that not all Drosophila insulators interact, with this property restricted to some insulators, such as gypsy. Three gypsy insulators inserted between an enhancer and promoter blocked enhancer function, indicating that gypsy insulator interactions may be restricted to pairs. Our studies imply that formation of loop domains may represent one of many mechanisms used by insulators to impart regulatory isolation.
Keywords: enhancers/gene expression/gypsy/insulator/loop domain
Introduction
Eukaryotic genomes are organized into clusters of coordinately regulated genes. It has been estimated that 20–30% of the Drosophila genome is assembled into domains of genes with related patterns of expression (Boutanaev et al., 2002; Spellman and Rubin, 2002). A similar finding was made for the human genome, where highly expressed housekeeping genes are clustered (Lercher et al., 2002). The formation of independent domains of gene function may depend upon a class of regulatory elements, called insulators (for reviews see Gerasimova and Corces, 2001; West et al., 2002; Geyer and Clark, 2002). Insulators are defined by two properties. First, insulators block enhancer and silencer action in a position-dependent manner, preventing function when inserted between these regulatory elements and a promoter, but not when located upstream (Holdridge and Dorsett, 1991; Geyer and Corces, 1992; Kellum and Schedl, 1992; Mallin et al., 1998). Secondly, insulators protect gene expression from positive and negative chromatin effects surrounding a gene or gene locus (Kellum and Schedl, 1991; Roseman et al., 1993, 1995). Insulators do not inactivate enhancers, silencers or promoters, implying that insulators interfere with signaling between these classes of control elements (Geyer and Corces, 1992; Cai and Levine, 1995; Scott and Geyer, 1995).
The Drosophila genome contains many sequences with insulator function (for reviews see Sun and Elgin, 1999; Gerasimova and Corces, 2001; Geyer and Clark, 2002). The first identified insulators were scs and scs′ that correspond to regions of unusual chromatin structure flanking the domain of decondensation produced from transcription of two heat shock (hs) genes (Udvardy et al., 1985; Kellum and Schedl, 1991, 1992). Multiple sequences within scs and scs′ are required for insulator function (Vazquez and Schedl, 1994; Zhao et al., 1995; Gaszner et al., 1999). Two proteins bind and partially confer insulator function to scs and scs′, Zw5 and BEAF, respectively (Zhao et al., 1995; Hart et al., 1997; Gaszner et al., 1999). These proteins associate with numerous sites on the polytene chromosomes, indicating that insulators may be common in the Drosophila genome (Zhao et al., 1995; Gaszner et al., 1999).
A second Drosophila insulator is the gypsy insulator. This element was identified as the region of the gypsy retrotransposon responsible for the production of tissue-specific mutations in many genes (Geyer et al., 1988a; Peifer and Bender, 1988; Smith and Corces, 1992; Dorsett, 1993). The gypsy insulator contains 12 binding sites for the zinc finger Suppressor of Hairy-wing [Su(Hw)] protein (Parkhurst et al., 1988; Spana et al., 1988). This DNA-binding protein is essential for gypsy insulator function, as mutations in the su(Hw) gene reverse the mutagenic effects of the gypsy retrotransposon. The Su(Hw) protein recruits a second protein, Modifier of Mdg4 [Mod(mdg4)], to chromosomes (Georgiev and Gerasimova, 1989). The Su(Hw) and Mod(mdg4) proteins co-localize at hundreds of sites in the genome that do not correspond to sites of the gypsy retrotransposon (Gerasimova and Corces, 1998; Spana et al., 1988). These data indicate that, similarly to scs and scs′, gypsy-like insulators may be widely distributed.
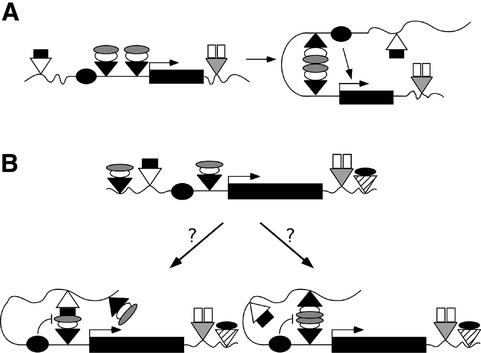
The gypsy insulator is a versatile modulator of regulatory interactions, with >20 enhancers shown to be blocked (Holdridge and Dorsett, 1991; Geyer and Corces, 1992; Dorsett, 1993; Roseman et al., 1993; Cai and Levine, 1995, 1997; Scott and Geyer, 1995; Hagstrom et al., 1996; Zhou et al., 1997; Cai and Shen, 2001; Hogga et al., 2001). Even so, this insulator does not establish an impassable barrier. Several conditions exist where enhancers overcome an intervening gypsy insulator to activate transcription (Morris et al., 1998; Scott et al., 1999; Cai and Shen, 2001; Muravyova et al., 2001). These effects are known as insulator bypass. For example, a loss of enhancer blocking occurs when two gypsy insulators are placed between an enhancer and promoter (Cai and Shen, 2001; Muravyova et al., 2001). This neutralization of gypsy insulator function was proposed to result from an interaction between gypsy insulator protein complexes to form a loop domain (Figure 1A). When two gypsy insulators are located between an enhancer and promoter, the resulting loop domain is believed to promote transcription activation by decreasing the distance between the control elements. By extension, it was suggested that a single gypsy insulator blocks enhancer-activated transcription by interacting with a second insulator upstream of the enhancer to form a loop domain that constrains the enhancer and precludes a productive interaction with the promoter (Figure 1B; Vazquez and Schedl, 1994; Gerasimova and Corces, 1998; Gerasimova et al., 2000; Cai and Shen, 2001; Muravyova et al., 2001).
Fig. 1. Models of insulator interactions. (A) The proposed mechanism for insulator bypass. Two gypsy insulators (black triangles) inserted between an enhancer (black oval) and promoter (bent arrow) permit enhancer-activated transcription because interactions between protein complexes (white, gray ovals) bound to the insulators form a loop domain that decreases the distance between the enhancer and promoter. (B) Two possible mechanisms by which a single gypsy insulator, placed between an enhancer and promoter, blocks enhancer-activated transcription. Left arrow: the gypsy insulator protein complex may interact with proteins (black rectangle) associated with a second non-gypsy insulator (white triangle) to form a loop domain that encompasses the enhancer, preventing transcriptional activation. Right arrow: formation of a loop domain by the gypsy insulator complex may require an interaction with proteins associated with a second gypsy insulator; an interaction that may occur even in the presence of an intervening non-gypsy insulator.
The loop domain model of insulator function predicts that enhancer blocking requires interactions between protein complexes associated with two insulators. Yet, previous studies have shown that a single gypsy insulator in transgenes blocks enhancer–promoter interactions at multiple, random sites throughout the genome (Geyer and Corces, 1992; Scott and Geyer, 1995; Hagstrom et al., 1996). The loop domain model may account for these effects if interactions between the gypsy insulator in the transgene and a second insulator located in the neighboring genomic DNA occur routinely. The large number of insulators within the genome may accommodate such interactions, with possible partnerships forming between gypsy and non-gypsy insulators (Figure 1B). To test these predictions, we used an insulator bypass assay to determine whether combinations of the gypsy, scs and scs′ insulators inserted between an enhancer and promoter interact. Our studies demonstrate that interactions among insulators are not a global property. Interestingly, partnerships between gypsy insulators were observed, even when separated by an intervening scs insulator. Insertion of three gypsy insulators between an enhancer and promoter restored enhancer blocking, suggesting that interactions are limited to pairs of insulators. These data indicate that gypsy insulators may be restricted to a local role in organizing regulatory interactions, perhaps by forming loop domains that preclude communication between enhancers and promoters. Further, formation of loop domains may represent one of many mechanisms of insulator action.
Results and discussion
A critical feature of the loop domain model is that at least two insulators interact. To test this model, we determined whether the repertoire of possible insulator interactions extends beyond interactions between gypsy insulators. In these studies, we employed an insulator bypass assay that used the cuticle pigmentation gene, yellow, as a reporter. Loss of yellow gene expression changes the normal cuticle color from black to yellow, while intermediate levels of expression produce intermediate levels of cuticle coloration (Geyer et al., 1988b). The yellow gene contains several independent tissue-specific enhancers; the wing and body enhancers are located 5′ of the promoter, while the bristle enhancer is located in the intron (Figure 2; Geyer and Corces, 1987). In the insulator bypass assay, the effects of two insulators inserted between the upstream wing and body enhancers and yellow promoter were determined. If two insulators interact, then the enhancer-blocking effects of the insulator should be neutralized, leading to activation of transcription and production of a dark cuticle.
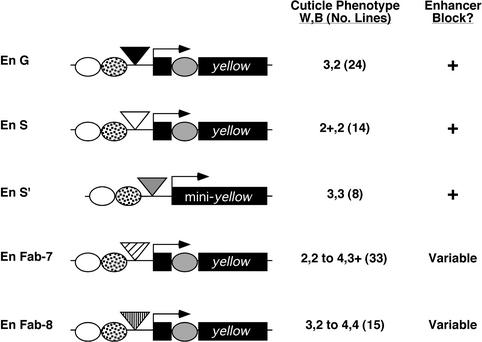
Fig. 2. Tests of enhancer blocking by several Drosophila insulators. The structure of the yellow gene is shown. Three of the yellow enhancers are diagrammed, including the wing (white oval), body (spotted oval) and bristle (gray oval) enhancers. Insulators, illustrated as triangles (black = gypsy, white = scs, gray = scs′, diagonal hatch = Fab-7, vertical hatch = Fab-8), were inserted between the wing and body enhancers and promoter. Transposons were named according to the number and arrangement of wing and body enhancers (En) and insulators (G = gypsy, S = scs, S′ = scs′). The left column summarizes average pigmentation scores assigned for each transgene, with the total number of lines analyzed shown in parentheses. The right column summarizes the enhancer blocking status, as indicated by the pigmentation score; ‘+’ refers to an enhancer block, ‘–’ refers to no enhancer block, ‘variable’ refers to a variable enhancer blocking ability among independent transgenic lines.
Identification of insulators that block the wing and body enhancers of the yellow gene
We began our studies by testing whether insertion of a single copy of scs, scs′ and the Abdominal-B insulators, Fab-7 and Fab-8, between the yellow wing and body enhancers and promoter affected gene expression (Karch et al., 1994; Hagstrom et al., 1996; Zhou et al., 1996, 1999; Barges et al., 2000). Each insulator was inserted ∼900 bp upstream of the transcription start site (Figure 2). The effects of each single, non-gypsy insulator were determined using a pigmentation assay (Figure 2, Materials and methods). A pigmentation score of 1 corresponds to levels of pigmentation associated with a complete, or nearly complete, loss of yellow gene expression, while a pigmentation score of 5 corresponds to the level of pigmentation associated with a wild-type level of gene expression (Morris et al., 1999). A pigmentation score of 2 corresponds to the level of pigmentation associated with the gypsy-induced yellow mutation, y2, and is indicative of the level of pigmentation expected for a complete block of the wing and body enhancers. Pigmentation scores that differed by a unit or more are interpreted to represent significant differences in the level of yellow gene expression (Morris et al., 1999; Materials and methods).
Phenotypic analysis of the P[En S] lines showed light pigmentation in the wing and body tissue (scores of 2+, 2), while flies carrying the P[En S′] transgene had intermediate levels of wing and body pigmentation (scores of 3, 3). The level of pigmentation in the wing and body tissues of P[En S] and P[En S′] flies was more than one unit lower than observed for wild-type flies (scores of 5, 5) and similar to that of the flies carrying P[En G] (scores of 3, 2), suggesting that both scs and scs′ block the upstream yellow enhancers. The scs′ block appears weaker than scs, a finding that is consistent with previous studies that suggest that scs′ is a weaker insulator than scs (Vazquez and Schedl, 1994).
In contrast to scs and scs′, flies carrying the P[En Fab-7] or P[En Fab-8] transgenes showed variable levels of cuticle pigmentation (scores ranging from 2, 2 to 4, 4; Figure 2). Our results were surprising, as these insulators were shown previously to block the yellow wing and body enhancers (Zhou et al., 1999). Differences between these two studies may be the number of lines obtained or the site of insertion of the insulators in the yellow gene, which was closer to the promoter in transgenes that showed a block of enhancer function (Zhou et al., 1999). Further, our results are consistent with other studies where the degree of enhancer blocking by Fab-7 and Fab-8 was influenced by genomic context (Hagstrom et al., 1996).
Effects of pairs of scs and scs′ insulators on blocking of enhancers
Based on results from the single insulator studies, we reasoned that only pairs of the scs and scs′ insulators could be tested in our bypass assay, as these were the only insulators that provided a consistent decrease in enhancer function. Two copies of either scs or scs′ were placed between the yellow enhancers and promoter to generate P[En S λ S] and P[En 2S′], respectively. Multiple transgenic lines were established and the cuticle phenotype determined. If the scs or scs′ insulator behaved similarly to the gypsy insulator, then we predicted that insulator bypass would occur, producing transgenic flies with dark cuticle pigmentation.
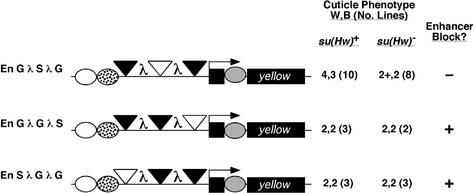
In the P[En S λ S] transgene, the pair of scs insulators was inserted between the wing and body enhancers in tandem orientation, separated by 2 kb of λ spacer DNA (Figure 3). Flies in the four P[En S λ S] transgenic lines showed low levels of body and wing pigmentation (scores of 2, 2), indicating that the yellow enhancers were unable to bypass the pair of scs insulators. We examined whether this low level of yellow gene expression might have resulted from repressive effects imposed by the λ DNA. To this end, we constructed the P[En G λ G] transposon. Seven transgenic lines carrying P[En G λ G] were established. Phenotypic analysis showed that P[En G λ G] flies had increased pigmentation in the wing and body tissues relative to P[En G], with a clear decrease in the efficiency of the enhancer block in the body tissue (scores of 3+, 3 versus 3, 2, respectively; Figures 2 and 3). Further, levels of body and wing pigmentation in P[En G λ G] flies were darker than those in P[En S λ S] flies, suggesting that the low level of pigmentation in P[En S λ S] flies cannot be attributed solely to the presence of λ DNA. Taken together, these results indicate that two scs insulators block enhancer function, implying that scs insulators do not interact.
Fig. 3. Enhancer blocking by a pair of insulators. Shown are the structures of the tested yellow transgenes. Symbols and analysis are as described in Figure 2.
In the P[En 2S′] transgene, two scs′ insulators were inserted between the wing and body enhancers as direct repeats, separated by 59 bp (Figure 3). This insulator orientation and spacing was within the range found previously to support insulator bypass of two gypsy insulators (Cai and Shen, 2001). Eight P[En 2S′] lines were obtained and pigmentation levels were assessed. We found that the level of wing and body coloration in P[En 2S′] was reduced compared with that in P[En S′] flies (scores of 3, 2 versus 3, 3; Figure 3). We conclude that two copies, if anything, produce a stronger, not weaker block of the body enhancer. These results do not support an interaction between scs′ insulators.
The location of scs and scs′ at the borders of a domain of decondensation induced by heat shock suggested that formation of a loop domain might be restricted to interactions between the heterologous insulator pair. For this reason, we constructed the P[En S λ S′] transposon, where the scs and scs′ insulators were separated by 2 kb of λ spacer DNA (Figure 3). Eight transgenic lines were obtained and phenotypic analysis completed. We found that P[En S λ S′] flies had levels of wing and body pigmentation that were lower than those observed in flies carrying the single insertion transgenes, P[En S] and P[En S′] (scores of 2, 2 versus 2+, 2 and 3, 3, respectively; Figures 2 and 3). These results indicate that the heterologous pair of scs and scs′ insulators does not interact. It is possible that our bypass assay does not reflect the behavior of scs and scs′ at the 87A heat shock locus. Interactions between endogenous scs and scs′ elements may occur but depend on sequences outside of those contained within the scs and scs′ insulators tested.
Our observations that two copies of scs or scs′ block enhancer action extend previous findings that multimerized subregions of scs or scs′ attenuate enhancer-activated transcription. Our findings demonstrate that the properties of the intact insulator are similar to those of the subregions (Vazquez and Schedl, 1994; Zhao et al., 1995; Gaszner et al., 1999). We infer from these data that pairs of scs or scs′ insulators do not interact to form loop domains. However, it is possible that these insulators do interact, but in a way that does not interfere with insulator function. Alternatively, the scs and scs′ insulators may establish loop domains through a mechanism distinct from that of the gypsy insulator, for example by an association with nuclear substructures. The recent demonstration that independent domains of gene activity result from tethering genes to the nuclear pore complex illustrates one mode whereby regulatory isolation can be conferred by an insulator without self-association (Ishii et al., 2002). The difference in the behavior of pairs of scs and scs′ insulators and the gypsy insulator implies that molecular mechanisms employed by insulators are distinct.
Tests of interactions between the gypsy and scs insulators
The loop domain model suggests that interactions between the gypsy insulator and other insulators within the genome occur routinely, as a consistent enhancer block is generated in transgenes that contain a single gypsy insulator. We wondered whether the gypsy insulator interacted with non-gypsy insulators to form loop domains (Figure 1B). To test this possibility, we constructed two transposons, P[En S λ G] and P[En G λ S], that contained a single gypsy and scs insulator inserted between the wing and body enhancers (Figure 3). We chose scs, as this insulator provided the strongest, most consistent block of the yellow enhancers among the Drosophila insulators tested (Figure 2). Independent transformed lines of P[En S λ G] and P[En G λ S] were established and phenotypically analyzed. In all cases, we observed that transgenic flies had a low level of pigmentation in the wing and body tissue (Figure 3). Pigmentation levels in the wing were lower than those obtained with either insulator alone, suggesting that a stronger block was conferred (scores of 2, 2 versus 2+, 2 for P[En S] and 3, 2 for P[En G]; Figures 2 and 3). We conclude that the gypsy and scs insulators do not interact to support bypass. These data imply that the formation of loop domains may be restricted to interactions between two gypsy insulators, a proposal that is inconsistent with the left part of the model shown in Figure 1B.
Effects of an intervening insulator on gypsy insulator interactions
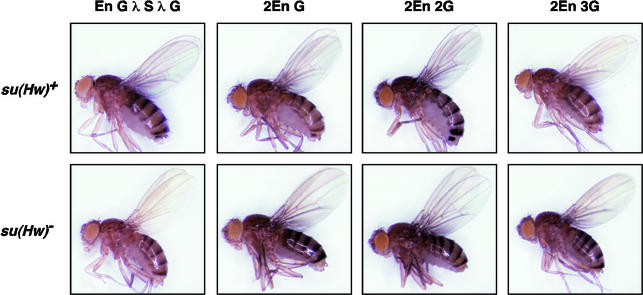
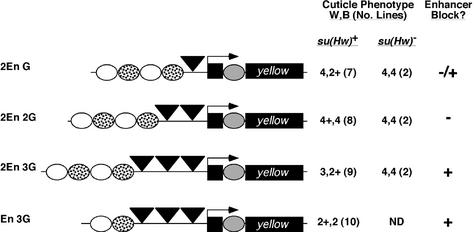
The limitation that only gypsy insulators interact imposes constraints on the loop domain model of insulator function. Routine enhancer blocking by a single gypsy insulator, as observed in transgene studies, implies that identification of another gypsy-like insulator occurs readily. This predicts that gypsy insulators interact, even in the presence of an intervening insulator (Figure 1B). To test this possibility, we constructed the transposon P[En G λ S λ G], where two gypsy insulators were separated by a 2 kb distance that included scs (Figure 4). The scs insulator was chosen, as it did not interact with the gypsy insulator (Figure 3). Phenotypic analysis of P[En G λ S λ G] flies showed high levels of wing and body pigmentation (scores of 4, 3; Figures 4 and 5). We infer from these data that the upstream wing and body enhancers bypass the insulators, as cuticle pigmentation is increased relative to P[En G] (scores of 3, 2; Figure 2). To rule out the possibility that the loss of scs insulator function in P[En G λ S λ G] flies was caused by insertion of transposons into genomic regions that were incompatible with scs enhancer blocking, we crossed lines carrying the P[En G λ S λ G] transposon into a su(Hw) mutant background. In all six lines tested, P[En G λ S λ G], su(Hw)– flies had low levels of wing and body pigmentation (scores of 2+, 2; Figures 4 and 5), demonstrating that the sites of insertion of the P[En G λ S λ G] transposon are compatible with scs insulator function.
Fig. 4. Enhancer blocking by scs in the presence of two gypsy insulators. Structures of the tested yellow transgenes are shown. In these experiments, the cuticle phenotype was determined in a su(Hw) wild-type [su(Hw)+] and mutant [su(Hw)–] background. Symbols and analysis are as described in Figure 2.
Fig. 5. Phenotypes of flies carrying insulator-containing transgenes. Shown are representative examples of su(Hw) wild-type and mutant flies carrying the indicated transgene.
The mechanism responsible for the loss of scs function in P[En G λ S λ G] flies was explored further by testing whether the large protein complexes assembled on the gypsy insulators prevented association of the scs-binding proteins. We reasoned that if this were the case, then scs function should be lost regardless of whether the scs insulator was located between or outside the two gypsy insulators. For this reason, we made the P[En S λ G λ G] and P[En G λ G λ S] transposons. Phenotypic analysis of P[En S λ G λ G] and P[En G λ G λ S] flies showed low levels of wing and body pigmentation, indicating that the scs insulator was functional in these transposons (scores of 2, 2; Figure 4). Further, the block of enhancer-activated transcription in P[En S λ G λ G] and P[En G λ G λ S] flies did not change in a su(Hw)– background (scores of 2, 2; Figure 4). Our findings demonstrate that scs function is lost only when it is located between gypsy insulators. This loss of function may result from topological constraints conferred by the gypsy insulator pair that interfere with enhancer blocking by scs, perhaps by precluding binding of the scs cognate proteins. Such constraints would be consistent with the formation of a loop domain, as shown in the right hand version of the model in Figure 1B.
Effects of three gypsy insulators placed between the upstream yellow enhancers and promoter
A second mechanism of insulator bypass involves increasing enhancer strength by increasing enhancer number (Scott et al., 1999). This effect is illustrated by the analysis of transgenic flies carrying the P[2En G] transposon that contains a duplication of the yellow wing and body enhancers (Figure 6). Phenotypic analysis indicated that in P[2En G] flies, the wing enhancer bypassed the insulator, while the body enhancer remained blocked (scores of 4, 2+; Figure 6). It may be that the threshold level of transcription required for body pigmentation is higher than in the wing, making it more difficult to observe bypass of the body enhancer. This inference is supported by the observation that bypass of two gypsy insulators inserted between the wing and body enhancers and promoter is stronger in the wing than in the body tissue (Figure 3; Muravyova et al., 2001).
Fig. 6. Enhancer-blocking effects of three gypsy insulators. Structures of the tested yellow transgenes are shown. The top three transgenes contain a duplication of the wing and body enhancers. The cuticle phenotype was determined in a su(Hw) wild-type and mutant background. Symbols and analysis used are as described in Figure 2, with the addition of –/+, referring to insulator effectiveness in the wing and body tissue, respectively. ND = not determined.
We studied whether an increased number of insulators inserted between the duplicated set of wing and body enhancers affected enhancer blocking. Transposons were constructed which had two or three tandem copies of the gypsy insulator inserted between the duplicated set of upstream yellow enhancers and promoter, generating P[2En 2G] and P[2En 3G], respectively (Figure 6). It was expected that flies carrying either transposon would show dark cuticle pigmentation, because multiple gypsy insulators are proposed to interact (Muravyova et al., 2001). However this prediction was only partially met. In the eight P[2En 2G] lines, flies had dark pigmentation in the wing and body cuticle (scores of 4+, 4). These results suggest that the block of the body enhancers was lost, consistent with the proposal that interactions between a pair of insulators cause their neutralization. In contrast, P[2En 3G] flies had decreased pigmentation in the wing and body (scores of 3+, 2+ compared with 4+, 4 for P[2En 2G], Figures 5 and 6). These data suggest that three insulators restore insulator function.
To test whether the reduced level of pigmentation in P[2En 3G] flies resulted from repressive position effects, we crossed lines carrying the P[2En 3G] transposons into a su(Hw) mutant background. The level of cuticle pigmentation increased in the su(Hw)– flies (scores of 4, 4; Figures 5 and 6), demonstrating that position effects were not responsible. Importantly, the level of pigmentation in P[2En 2G] and P[2En 3G] flies in a su(Hw) mutant background was indistinguishable (Figures 5 and 6). These findings support the contention that three gypsy insulators confer a partial block of enhancer function.
To confirm our finding that three insulators placed between an enhancer and promoter attenuated enhancer function, we constructed the P[En 3G] transposon (Figure 6). In this construct, three insulators were inserted between a single set of wing and body enhancers and the yellow promoter. We predicted that if the third insulator restored insulator function, then the level of wing and body pigmentation would be similar to that observed in P[En G] flies. We obtained 10 P[En 3G] transgenic lines. Flies from all transgenic P[En 3G] lines had light pigmentation in the wing and body tissues (scores of 2+, 2; compare with P[En G] scores of 3, 2; Figures 2 and 6). These observations support the conclusion that three gypsy insulators prevent enhancer-activated transcription. Considering these results, we suggest that gypsy insulator interactions may be restricted to the pair of nearest neighbors, leaving the third insulator to block enhancer function. If interactions occurred between all three insulators or between the outside insulators, then insulator bypass should have occurred and dark pigmentation would have been observed; however, the opposite result was obtained.
It was unexpected that three insulators established a block of enhancer function. Previous studies indicated that interactions between three gypsy insulators promoted an interaction between the white eye enhancer and white promoter located ∼10 kb apart (Muravyova et al., 2001). However, removal of the eye enhancer or the Su(Hw) protein did not change white gene expression measurably in the majority of lines tested (www.sciencemag.org/cgi/content/full/291/5503/495/DC1). An alternative explanation for the high levels of white gene expression may be the presence of activators in the flanking DNA. Taken together, we suggest that gypsy insulator interactions may be limited to pairs, with the possibility that conditions may exist that influence the choice of gypsy insulator partnership.
Concluding perspectives
Studies described herein demonstrate that gypsy insulators show a strong propensity to interact only with each other, even in the presence of an intervening insulator (Figures 3 and 4). These data are consistent with cytological studies that show that gypsy insulators coalesce in the nucleus, in structures termed insulator bodies (Gerasimova and Corces, 1998; Gerasimova et al., 2000). However, several observations indicate that the connection between insulator bodies and enhancer blocking is not complete. First, mutations that disrupt the formation of these structures do not restore enhancer function uniformly (Georgiev and Gerasimova, 1989; Gerasimova et al., 2000). Secondly, placement of two gypsy insulators between an enhancer and promoter establishes conditions that permit enhancer action, suggesting that not all coalescence of insulators results in the loss of enhancer function. Thirdly, our data suggest that enhancer blocking may only involve interactions between pairs of gypsy insulators, which does not require formation of insulator bodies (Figure 6).
How a loop domain attenuates enhancer function is not known. One proposal is that the domain represents a unit of higher order chromatin structure that prevents interactions between proteins bound in different domains, while maximizing protein interactions within a domain (Vazquez and Schedl, 1994; Gerasimova and Corces, 1998; Gerasimova et al., 2000). This proposal connects insulators with the physical organization of chromosomal domains and implies that these elements are responsible for the global organization of chromosome structure. Studies that demonstrate that some insulators, such as scs, scs′ and HS4, localize to the boundaries of distinct chromatin domains support this proposal (Udvardy et al., 1985; Litt et al., 2001b). However, if insulator-defined loop domains correspond to a structural chromatin domain, then these domains cannot be static. The strength of an enhancer, promoter and the gypsy insulator influences the effectiveness of an enhancer block (Cai et al., 2001; Scott et al., 1999; Wei and Brennan, 2000), suggesting that the formation of loop domains by the gypsy insulator involves a competition between the promoter and insulator for enhancer interaction. Further, gypsy insulator-defined domains do not impose an impassable block to all protein–protein interactions, as FLP recombinase directs recombination between its target sites in the presence of an intervening gypsy insulator (Dunaway et al., 1997; Parnell and Geyer, 2000).
An alternative view of how a loop domain may disrupt enhancer–promoter interactions is that insulators directly impact enhancer–promoter communication (Dorsett, 1999; Geyer and Clark, 2002; West et al., 2002). One commonly held view is that enhancers exert their regulatory role by establishing contacts with the proteins bound at the promoter by looping out the intervening DNA (Ptashne, 1988). An insulator-induced loop that includes either the enhancer or promoter may interfere with productive enhancer looping, leading to a block of enhancer function. Such a model can account for our finding that scs function is lost when placed between two gypsy insulators (Figure 4). Consistent with this idea, studies have shown that topology appears to be a critical feature of insulator function, as structural changes induced by inter-chromosomal interactions reverse enhancer blocking by the gypsy insulator (Morris et al., 1998; Melnikova et al., 2002).
While neutralization of gypsy insulator function is consistent with a loop domain model, other explanations are possible. For example, gypsy insulator function may depend upon the precise architecture of assembled proteins, as observed for some transcriptional regulatory elements (Merika and Thanos, 2001). Multiple Su(Hw) proteins bind to the gypsy insulator, leading to recruitment of several Mod(mdg4) proteins. It has been proposed that gypsy insulation depends upon multimerization of Mod(mdg4) proteins bound to separate insulators (Ghosh et al., 2001). The close proximity of two gypsy insulators may interfere with the establishment of an appropriate architecture of insulator proteins that promotes long-range interactions, leading to a loss of insulator action. Further, the character of protein–protein interactions depends upon many factors, such as relative affinities, distances of separation and protein levels. The close proximity of the pair of gypsy insulators may promote local interactions between gypsy insulators that lead to neutralization. However, long-range interactions may not be restricted to gypsy insulator complexes and may occur with other protein complexes, such as those assembled at enhancers or promoters (Dorsett, 1999; Geyer and Clark, 2002).
We found that interactions between pairs of insulators are not a shared property for all Drosophila insulators. It is possible that gypsy and scs/scs′ insulators represent mechanistically distinct classes of Drosophila insulators, wherein only the gypsy class of insulators function by establishing loop domains, a supposition supported by other studies (Parnell and Geyer, 2000; Hogga et al., 2001). The properties of the gypsy insulator may not be unique, as two copies of a second Drosophila insulator isolated from a retrotransposon, Idefix, appear to cause insulator neutralization (Conte et al., 2002).
Whether insulators establish loop domains in other organisms is unclear. Many vertebrate insulators contain binding sites for the CTCF protein (Bell et al., 1999; Hark et al., 2000; Kanduri et al., 2000; Filippova et al., 2001; Ishihara and Sasaki, 2002). It has been shown that CTCF sites flank the β-globin locus in chicken, mouse and human (Farrell et al., 2000; Saitoh et al., 2000). Further, CTCF sites surround the Igf2/H19 domain in human and mouse (Ishihara and Sasaki, 2002). These observations are consistent with the possibility that interactions between CTCF insulators are required for the establishment of a unique chromatin domain. However, the observation that enhancer blocking by the chicken β-globin HS4 insulator and the Igf2/H19 imprinting control element (ICR) is strengthened when the insulator is dimerized may not support this supposition (Chung et al., 1993, 1997; Kaffer et al., 2001). That CTCF-binding sites demarcate transitions of histone acetylation and methylation in the defined domains (Hebbes et al., 1994; Litt et al., 2001a,b) suggests that CTCF may locally govern chromatin structure by preventing the spread of inappropriate histone modifications (Saitoh et al., 2000).
Most eukaryotic genomes contain a diverse collection of insulators. It is likely that several mechanisms are employed by these elements to impart regulatory isolation to transcriptional control elements. Further experimentation is needed to classify these insulators. Additionally, identification of the proteins involved in insulator function will provide better insights into their mechanism of action.
Materials and methods
Cloning
The gypsy insulator contained 12 Su(Hw)-binding sites, corresponding to bp 647–1077 in the gypsy retrotransposon (Marlor et al., 1986). Scs corresponded to a 990 bp PvuII fragment, numbered 510–1503 in the scs GenBank sequence (accession No. X63731). This fragment has enhancer-blocking activity similar to the full-length scs and contains the single Zw5-binding site (Vazquez and Schedl, 1994; Gaszner et al., 1999). Scs′ corresponded to an ∼500 bp fragment, numbered 1–501 in the scs′ GenBank sequence (accession No. X63732). This fragment contains strong and weak BEAF-binding sites (Zhao et al., 1995). The Fab-7 insulator was a 1.2 kb PstI–ApaI fragment, while the Fab-8 insulator corresponded to a 0.8 kb EcoRI–HindIII fragment (bp 83 666–84 897 and 63 785–64 586 of the bithorax complex sequence, respectively; accession No. U31961). The Fab-7 and Fab-8 fragments did not include Polycomb response elements (Hagstrom et al., 1997; Mihaly et al., 1997; Barges et al., 2000).
Insulators were cloned into a 7.7 kb yellow fragment that contained 2.8 kb of 5′- and 0.13 kb of 3′-flanking DNA (Geyer and Corces, 1987). The yellow gene was modified by insertion of a NotI linker into the Eco47III site at –893 upstream of the transcription start site, between the wing and body enhancers and yellow promoter, and cloned into CaSpeRW15 (Pirrotta, 1988) to make the yw cassette (TP1291). Insulators, additional wing and body enhancers and λ DNA were cloned into the NotI site of TP1291. Each transposon was named to indicate the number of yellow wing and body enhancers included and the identity of the insulator. For example, in P[En G], the yellow gene had one set of wing and body enhancers and one gypsy insulator. In P[En G λ G], the yellow gene contained one set of wing and body enhancers and two gypsy insulators separated by 2 kb of the λ spacer DNA, corresponding to bp 23 130–25 157 (accession No. J02459). This same fragment of λ DNA was the spacer in P[En S λ G], P[En G λ S] and P[En S λ S′]. A 0.5 kb spacer, corresponding to bp 24 623–25 157, was used in P[En S λ S], P[En G λ S λ G], P[En G λ G λ S] and P[En S λ G λ G]. In some constructs, the wing and body enhancers were duplicated (–2704 to –799 relative to the transcription start site), as designated by 2En.
In the P[En Scs′] and P[En 2Scs′] transposons, the scs′ sequences, located between lox P sites, were cloned into a 5.2 kb mini-yellow gene that lacked the bristle and tarsal claw enhancers (Geyer and Corces, 1987). Insulators were inserted at –893 bp from the transcription start site. The modified mini-yellow genes were cloned into the P element vector CaSpeR 3 (Pirrotta, 1988).
Germline transformation
P transposons were injected at a concentration of 400 µg/ml, with the ‘wings clipped’ helper plasmid pπ25.7 at a concentration of 200 µg/ml into the host strain y1 w67c23 (Rubin and Spradling, 1982; Karess and Rubin, 1984). Southern analysis determined the number of inserts and verified the structural integrity of the yellow gene for each transgenic line. Only lines with single insertions were analyzed. Selected lines were crossed into a su(Hw)v/su(Hw)f mutant background (Roseman et al., 1995) to determine the contribution of the gypsy insulator to the yellow phenotype observed.
Analysis of yellow phenotypes
Flies were raised at 25°C, 70% humidity on standard corn meal and agar medium. Phenotypic analysis involved crossing transgenic males to y1 w67c23 females and assessment of wing and body pigmentation in females, as described previously (Morris et al., 1999). ‘Wing’ refers to the wing blade, and ‘body’ refers only to pigmentation in the abdominal stripes, not the interstripe abdominal cuticle or the thoracic cuticle. Pigmentation was scored in 3- to 4-day-old female progeny by comparison with a series of five parallel controls. A score of 1 represents null or nearly null, and a score of 5 represents wild-type. A score of 2 is the phenotype of the y2 gypsy-induced allele. Intermediate phenotypes of 3 and 4 corresponded to females carrying y82f29/y18 and y2/y18, respectively. For each cross, 20–30 females were scored independently by at least two people. There can be small differences in pigmentation score between flies of a given genotype scored at a particular time. In the figures, the average phenotype obtained from at least two independent crosses is listed. A plus sign indicates that the average level of pigmentation was slightly greater than that of the corresponding control. We only consider differences in pigmentation to be significant if the scores differ by a unit or more (Morris et al., 1999).
Acknowledgments
Acknowledgements
We thank Jennifer Leff, Katie O’Malley and Joanna Sparano for technical support, Craig Hart for plasmids containing the scs and scs′ insulators, and Lori Wallrath and members of the Geyer laboratory for their critical reading of this manuscript and their thoughtful and insightful comments on these experiments. This work was supported by a National Institute of Health grant to P.K.G. (GM42539).
References
- Barges S., Mihaly,J., Galloni,M., Hagstrom,K., Muller,M., Shanower,G., Schedl,P., Gyurkovics,H. and Karch,F. (2000) The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development, 127, 779–790. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–396. [DOI] [PubMed] [Google Scholar]
- Boutanaev A.M., Kalmykova,A.I., Shevelyov,Y.Y. and Nurminsky,D.I. (2002) Large clusters of co-expressed genes in the Drosophila genome. Nature, 420, 666–669. [DOI] [PubMed] [Google Scholar]
- Cai H. and Levine,M. (1995) Modulation of enhancer–promoter interactions by insulators in the Drosophila embryo. Nature, 376, 533–536. [DOI] [PubMed] [Google Scholar]
- Cai H.N. and Levine,M. (1997) The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J., 16, 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H.N. and Shen,P. (2001) Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science, 291, 493–495. [DOI] [PubMed] [Google Scholar]
- Cai H.N., Zhang,Z., Adams,J.R. and Shen,P. (2001) Genomic context modulates insulator activity through promoter competition. Development, 128, 4339–4347. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Bell,A.C. and Felsenfeld,G. (1997) Characterization of the chicken β-globin insulator. Proc. Natl Acad. Sci. USA, 94, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte C., Dastugue,B. and Vaury,C. (2002) Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol., 22, 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. (1993) Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics, 134, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. (1999) Distant liaisons: long-range enhancer–promoter interactions in Drosophila. Curr. Opin. Genet. Dev., 9, 505–514. [DOI] [PubMed] [Google Scholar]
- Dunaway M., Hwang,J.Y., Xiong,M. and Yuen,H.L. (1997) The activity of the scs and scs′ insulator elements is not dependent on chromosomal context. Mol. Cell. Biol., 17, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell C.M., Grinberg,A., Huang,S.P., Chen,D., Pichel,J.G., Westphal,H. and Felsenfeld,G. (2000) A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse β-globin locus. Proc. Natl Acad. Sci. USA, 97, 14554–14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G.N., Thienes,C.P., Penn,B.H., Cho,D.H., Hu,Y.J., Moore,J.M., Klesert,T.R., Lobanenkov,V.V. and Tapscott,S.J. (2001) CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet., 28, 335–343. [DOI] [PubMed] [Google Scholar]
- Gaszner M., Vazquez,J. and Schedl,P. (1999) The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer–promoter interaction. Genes Dev., 13, 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P.G. and Gerasimova,T.I. (1989) Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet., 220, 121–126. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell, 92, 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet., 35, 193–208. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Byrd,K. and Corces,V.G. (2000) A chromatin insulator determines the nuclear localization of DNA. Mol. Cell, 6, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Clark,I. (2002) Protecting against promiscuity: the regulatory role of insulators. Cell. Mol. Life Sci., 59, 2112–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P.K. and Corces,V.G. (1987) Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev., 1, 996–1004. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Corces,V.G. (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev., 6, 1865–1873. [DOI] [PubMed] [Google Scholar]
- Geyer P.K., Green,M.M. and Corces,V.G. (1988a) Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc. Natl Acad. Sci. USA, 85, 8593–8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P.K., Richardson,K.L., Corces,V.G. and Green,M.M. (1988b) Genetic instability in Drosophila melanogaster: P-element mutagenesis by gene conversion. Proc. Natl Acad. Sci. USA, 85, 6455–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D., Gerasimova,T.I. and Corces,V.G. (2001) Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J., 20, 2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller,M. and Schedl,P. (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev., 10, 3202–3215. [DOI] [PubMed] [Google Scholar]
- Hagstrom K., Muller,M. and Schedl,P. (1997) A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics, 146, 1365–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr,C.J., Katz,D.J., Ingram,R.S., Levorse,J.M. and Tilghman,S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature, 405, 486–489. [DOI] [PubMed] [Google Scholar]
- Hart C.M., Zhao,K. and Laemmli,U.K. (1997) The scs′ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol., 17, 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga I., Mihaly,J., Barges,S. and Karch,F. (2001) Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell, 8, 1145–1151. [DOI] [PubMed] [Google Scholar]
- Holdridge C. and Dorsett,D. (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol., 11, 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K. and Sasaki,H. (2002) An evolutionarily conserved putative insulator element near the 3′ boundary of the imprinted Igf2/H19 domain. Hum. Mol. Genet., 11, 1627–1636. [DOI] [PubMed] [Google Scholar]
- Ishii K., Arib,G., Lin,C., Van Houwe,G. and Laemmli,U.K. (2002) Chromatin boundaries in budding yeast: the nuclear pore connection. Cell, 109, 551–562. [DOI] [PubMed] [Google Scholar]
- Kaffer C.R., Grinberg,A. and Pfeifer,K. (2001) Regulatory mechanisms at the mouse igf2/h19 locus. Mol. Cell. Biol., 21, 8189–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C., Pant,V., Loukinov,D., Pugacheva,E., Qi,C.F., Wolffe,A., Ohlsson,R. and Lobanenkov,V.V. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol., 10, 853–856. [DOI] [PubMed] [Google Scholar]
- Karch F., Galloni,M., Sipos,L., Gausz,J., Gyurkovics,H. and Schedl,P. (1994) Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res., 22, 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R.E. and Rubin,G.M. (1984) Analysis of P transposable element functions in Drosophila. Cell, 38, 135–146. [DOI] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell, 64, 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol., 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M.J., Urrutia,A.O. and Hurst,L.D. (2002) Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet., 31, 180–183. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson,M., Gaszner,M., Allis,C.D. and Felsenfeld,G. (2001a) Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science, 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson,M., Recillas-Targa,F., Prioleau,M.N. and Felsenfeld,G. (2001b) Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J., 20, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallin D.R., Myung,J.S., Patton,J.S. and Geyer,P.K. (1998) Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics, 148, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor R.L., Parkhurst,S.M. and Corces,V.G. (1986) The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol., 6, 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L., Gause,M. and Georgiev,P. (2002) The gypsy insulators flanking yellow enhancers do not form a separate transcriptional domain in Drosophila melanogaster. The enhancers can activate an isolated yellow promoter. Genetics, 160, 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M. and Thanos,D. (2001) Enhanceosomes. Curr. Opin. Genet. Dev., 11, 205–208. [DOI] [PubMed] [Google Scholar]
- Mihaly J., Hogga,I., Gausz,J., Gyurkovics,H. and Karch,F. (1997) In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development, 124, 1809–1820. [DOI] [PubMed] [Google Scholar]
- Morris J.R., Chen,J.L., Geyer,P.K. and Wu,C.T. (1998) Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl Acad. Sci. USA, 95, 10740–10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.R., Chen,J., Filandrinos,S.T., Dunn,R.C., Fisk,R., Geyer,P.K. and Wu,C. (1999) An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics, 151, 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin,A., Gracheva,E., Parshikov,A., Belenkaya,T., Pirrotta,V. and Georgiev,P. (2001) Loss of insulator activity by paired Su(Hw) chromatin insulators. Science, 291, 495–498. [DOI] [PubMed] [Google Scholar]
- Parkhurst S.M., Harrison,D.A., Remington,M.P., Spana,C., Kelley,R.L., Coyne,R.S. and Corces,V.G. (1988) The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev., 2, 1205–1215. [DOI] [PubMed] [Google Scholar]
- Parnell T.J. and Geyer,P.K. (2000) Differences in insulator properties revealed by enhancer blocking assays on episomes. EMBO J., 19, 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M. and Bender,W. (1988) Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc. Natl Acad. Sci. USA, 85, 9650–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. (1988) Vectors for P-mediated Transformation in Drosophila. Butterworths, Boston, MA. [DOI] [PubMed]
- Ptashne M. (1988) How eukaryotic transcriptional activators work. Nature, 335, 683–689. [DOI] [PubMed] [Google Scholar]
- Roseman R.R., Pirrotta,V. and Geyer,P.K. (1993) The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J., 12, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R.R., Johnson,E.A., Rodesch,C.K., Bjerke,M., Nagoshi,R.N. and Geyer,P.K. (1995) A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics, 141, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Bell,A.C., Recillas-Targa,F., West,A.G., Simpson,M., Pikaart,M. and Felsenfeld,G. (2000) Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J., 19, 2315–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.S. and Geyer,P.K. (1995) Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J., 14, 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.C., Taubman,A.D. and Geyer,P.K. (1999) Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics, 153, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.A. and Corces,V.G. (1992) The suppressor of Hairy-wing binding region is required for gypsy mutagenesis. Mol. Gen. Genet., 233, 65–70. [DOI] [PubMed] [Google Scholar]
- Spana C., Harrison,D.A. and Corces,V.G. (1988) The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev., 2, 1414–1423. [DOI] [PubMed] [Google Scholar]
- Spellman P.T. and Rubin,G.M. (2002) Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol., 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F.L. and Elgin,S.C. (1999) Putting boundaries on silence. Cell, 99, 459–462. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Maine,E. and Schedl,P. (1985) The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol., 185, 341–358. [DOI] [PubMed] [Google Scholar]
- Vazquez J. and Schedl,P. (1994) Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J., 13, 5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. and Brennan,M.D. (2000) Polarity of transcriptional enhancement revealed by an insulator element. Proc. Natl Acad. Sci. USA, 97, 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Gaszner,M. and Felsenfeld,G. (2002) Insulators: many functions, many mechanisms. Genes Dev., 16, 271–288. [DOI] [PubMed] [Google Scholar]
- Zhao K., Hart,C.M. and Laemmli,U.K. (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell, 81, 879–889. [DOI] [PubMed] [Google Scholar]
- Zhou J., Barolo,S., Szymanski,P. and Levine,M. (1996) The Fab-7 element of the bithorax complex attenuates enhancer–promoter interactions in the Drosophila embryo. Genes Dev., 10, 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zhou J., Cai,H.N., Ohtsuki,S. and Levine,M. (1997) The regulation of enhancer–promoter interactions in the Drosophila embryo. Cold Spring Harbor Symp. Quant. Biol., 62, 307–312. [PubMed] [Google Scholar]
- Zhou J., Ashe,H., Burks,C. and Levine,M. (1999) Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development, 126, 3057–3065. [DOI] [PubMed] [Google Scholar]