Abstract
To investigate the repair of different types of DNA lesions in chromatin, we prepared mononucleosomes containing an acetylaminofluorene-guanine adduct (AAF-G), a (6-4) photoproduct, or a cyclobutane pyrimidine dimer (CPD) and measured the repair of these lesions by reconstituted 6-factor human excision nuclease. We find that incorporation into nucleosomes inhibits the repair of CPD more severely than repair of the AAF-G adduct and the (6-4) photoproduct. Equally important, we find that SWI/SNF stimulates the removal of AAF-G and (6-4) photoproduct but not of CPD from nucleosomal DNA. These results shed new light on the low rate of repair of CPDs in human cells in vivo.
Nucleotide excision repair (excision repair) removes a wide variety of structurally unrelated DNA lesions by dual incisions bracketing the lesion and spaced 24 to 32 nucleotides (nt) apart (30, 32, 41). The excision reaction is carried out by six repair factors, XPA, RPA, XPC, TFIIH, XPG, and XPF-ERCC1, in humans (5, 24, 25) and their counterparts in yeast (7). Although the excision nuclease removes all types of DNA lesions ranging from bases adducted to polyaromatic hydrocarbons (4, 12, 32) to simply methylated bases (13) and even undamaged nucleotides (3, 8), these lesions are removed with widely differing rates (3, 12, 13). In particular, the rates of removal of the two major UV photoproducts, the cyclobutane pyrimidine dimer (Pyr<>Pyr) and the (6-4) photoproduct, are of interest because, arguably, these are the two most important lesions that the excision nuclease has to deal with in nature. Numerous in vivo studies have shown that (6-4) photoproducts are repaired at 10- to 20-fold-higher rates than is Pyr<>Pyr in human cells (22, 42). Yet in vitro studies with human cell extracts (26) or 6-factor reconstituted human excision nuclease reveal only a two- to fivefold-higher rate of removal of (6-4) photoproduct than for cyclobutane thymine dimer (T<>T) (26, 27). There might be several factors contributing to the discrepancy between the in vivo and in vitro relative rates. One obvious factor is the nature of the substrates in the two systems. In vivo, the substrate is chromatin, whereas in vitro the substrate is typically naked DNA, and it has been shown recently that incorporation of a lesion into a nucleosome severely affects the rate of repair of the (6-4) photoproduct and the acetylaminofluorene-guanine (AAF-G) adduct (9, 10, 38). It was, therefore, conceivable that incorporation of the two photoproducts might have different effects on their rates of removal and the modulation of this rate by chromatin remodeling factors.
Recently, we showed that incorporation of an AAF-G adduct into nucleosomes inhibited its rate of removal by the human excision nuclease to about 25% of the rate of excision from naked DNA and that this inhibition was partly overcome by the SWI/SNF chromatin remodeling factor (10). In the present study we have used AAF-G as a reference substrate to investigate the comparative effects of incorporation into nucleosomes on the rates of excision of (6-4) photoproduct and T<>T and the modulation of these rates by SWI/SNF. Our results show that incorporation into nucleosomes has a more drastic effect on the rate of removal of T<>T than on the rate of removal of (6-4) photoproduct. Moreover, SWI/SNF stimulates the excision of (6-4) photoproduct from nucleosomes but has a negligible effect on T<>T repair. We conclude that the stimulatory effect of SWI/SNF on excision repair is dependent on the type of DNA damage and that the lack of a stimulatory effect on excision of T<>T contributes to its relatively low rate of excision in vivo.
MATERIALS AND METHODS
Enzymes and chemicals.
[γ-32P]ATP (7,000 Ci/mmol) was obtained from ICN (Irvine, Calif.); EcoRI, HindIII, HinPI, and PflMI restriction enzymes and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, Mass.). T4 DNA ligase was obtained from Roche Applied Science (Indianapolis, Ind.), DNase I from Promega (Madison, Wis.), and N-acetoxy-2-acetyl-aminofluorene from Chemsyn Science Laboratories (Lenexa, Kans.). The six core human excision nuclease factors, RPA, XPA, XPC, TFIIH, XPG, and XPF-ERCC1, were purified as described previously (2, 21, 25, 31). The yeast SWI/SNF complex was a generous gift of Craig L. Peterson (University of Massachusetts, Worcester) and was prepared as described earlier (19).
Preparation of damage-containing substrates.
The substrates were prepared by ligating an oligonucleotide containing a base lesion at a predetermined site with five other oligonucleotides of partially overlapping sequences as described previously (13, 21). The 20-mer containing an AAF-G adduct was prepared by treating the oligomer with N-acetoxy-2-acetyl-aminofluorene as described earlier (18), and the modified oligomer was purified through a 20% polyacrylamide gel under nondenaturing conditions. The 8-mer oligonucleotides containing either a (6-4) photoproduct or a T<>T were prepared and purified as described elsewhere (34).
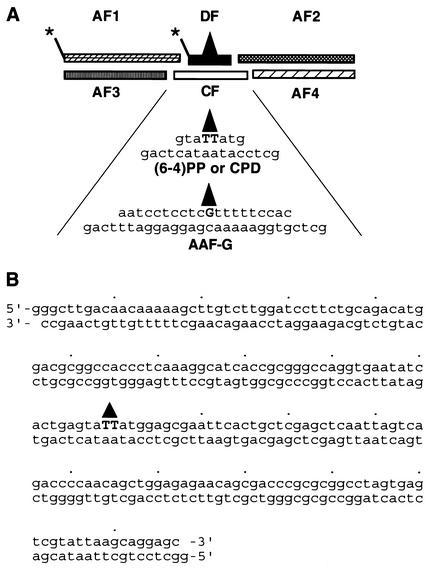
The modified oligonucleotides were labeled at the 5′ termini by using [γ-32P]ATP and polynucleotide kinase and were ligated to the other five oligomers to prepare full-length duplexes of either 209-bp (AAF-G substrate) or 197-bp [(6-4) photoproduct and T<>T substrates] as outlined in Fig. 1A (13, 21, 39). Following ligation the full-length oligomers were purified through denaturing polyacrylamide gel electrophoresis (5% acrylamide, 0.17% bisacrylamide, and 8 M urea in 2× Tris-borate-EDTA) and were annealed to obtain the corresponding duplexes. For footprinting experiments the arm oligomer 5′ to the damaged oligonucleotide was labeled with [γ-32P-]ATP before ligation with the other five oligomers that were 5′ phosphorylated with nonradioactive ATP was carried out. It must be noted that, of the three substrates, the two with the UV photoproducts have identical sequences and that the AAF-G substrate only differs in the central 20 bp. Control duplexes were prepared in the same manner, except that the central damaged oligomer was replaced by an undamaged counterpart.
FIG. 1.
Substrates. (A) Schematic of substrate construction from six oligomers by ligation. The arm fragments AF1 to AF4 are 95, 94, 90, and 91 nt in length, respectively. The length and sequences of damaged oligomers and complementary sequence (CF) for UV- and AAF-damaged substrates are shown. Asterisks indicate the positions of the radiolabel in duplexes used for footprinting and for the excision assay. (B) Sequence of the T<>T or (6-4) photoproduct-containing duplex. The triangle indicates position of the photoproduct.
Assembly and purification of nucleosomes.
The substrates were assembled into nucleosomes by using core histone proteins purified from HeLa cells as described earlier (9). Briefly, 0.5 pmol of the damage-containing duplex and 1 μg of salmon sperm DNA were incubated with core histone proteins at a 1:1 molar ratio of octamer to nucleosome unit of DNA in a high-salt buffer containing 2 M NaCl. The reaction mixture was gradually diluted to reduce the salt concentration. After assembly, the nucleosomes were purified by velocity gradient sedimentation through an 11-ml 5 to 25% sucrose gradient (10). The quality of nucleosomes was monitored by testing the fractions from the sucrose gradient on a nondenaturing polyacrylamide gel (5% polyacrylamide and 0.17% bisacrylamide in 0.5× Tris-borate-EDTA) according to methods published elsewhere (17).
Footprinting.
DNase I footprinting was performed as described previously (10). Briefly, 20 fmol of end-labeled naked or unfractionated nucleosomal DNA was treated with DNase I and was directly loaded onto a 5% nondenaturing polyacrylamide gel. The nucleosome band was located by autoradiography of the wet gel, the band was excised, and the DNA was purified from the gel slice and was analyzed on 5% polyacrylamide DNA-sequencing gel.
Excision assay.
The excision assay measures the release of damaged bases in 24- to 32-nt-long oligomers (14, 36). In this study, 30 pM 32P-labeled nucleosomal or naked DNA and 1.6 μg of salmon sperm DNA/ml were mixed with the six excision repair factors (42 nM RPA, 6.5 nM XPA, 2.2 nM XPC, 16 nM TFIIH, 3 nM XPG, and 6 nM XPF-ERCC1) in 12.5 μl of excision buffer containing 32 mM HEPES-KOH (pH 7.9), 64 mM KCl, 6.4 mM MgCl2, 0.24 mM EDTA, 0.8 mM dithiothreitol, 2 mM ATP, 200 μg of bovine serum albumin/ml, 5.5% glycerol, and 0.05% NP-40 and were incubated at 30°C for the indicated time. When indicated, 0.48 nM SWI/SNF was added to the reaction mixture. The reaction was stopped by phenol-chloroform extraction, and the DNA was precipitated with ethanol and was separated on an 8% polyacrylamide sequencing gel. The level of excision was determined by measuring the amount of radioactivity in the 24- to 32-nt range and the total radioactivity in the lane with a PhosphorImager and analyzing the data with the ImageQuant system (Molecular Dynamics).
RESULTS
Preparation of nucleosomes with three types of lesions.
Two duplexes 197 bp long and a third duplex of 206 bp were prepared by ligating six partially overlapping oligonucleotides, including a lesion-containing oligomer in the center where indicated. The lesions were AAF-G, (6-4) photoproduct, and T<>T. The structure of the substrate is shown schematically in Fig. 1A. The three duplexes have essentially the same sequence, with the exception of the central 20 bp, which is unique to the longer AAF-G substrate. Figure 1B shows the entire sequence of the duplexes containing the UV photoproducts. It should be noted that these substrates have no nucleosome-positioning sequences such as TG motifs or 5S ribosomal gene sequences commonly used in making uniformly phased nucleosomes in vitro (40).
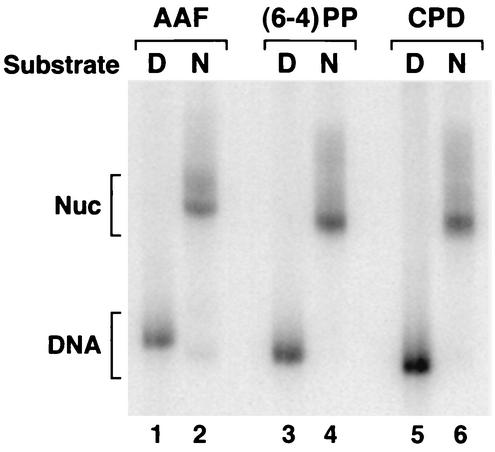
To prepare nucleosomes, the duplexes were mixed with human core histones purified from HeLa cells and were assembled into mononucleosomes by the salt dilution method (10). The nucleosome was separated from free DNA by sucrose gradient velocity sedimentation and was analyzed on a nondenaturing polyacrylamide gel. As seen in Fig. 2, the peak nucleosome fractions are essentially free of naked DNA. The trace amount of naked DNA seen in the AAF-G nucleosome most likely is due to a low level of disruption of this nucleosome during electrophoresis, as we have noticed the AAF-G nucleosome to be more susceptible to disruption by physical manipulation.
FIG. 2.
Analysis of damage-containing mononucleosomes by polyacrylamide gel electrophoresis. Naked DNA (D) or nucleosomes (N) carrying the indicated lesions were separated on a 5% nondenaturing polyacrylamide gel and were visualized by autoradiography. CPD, cyclobutane pyrimidine dimer.
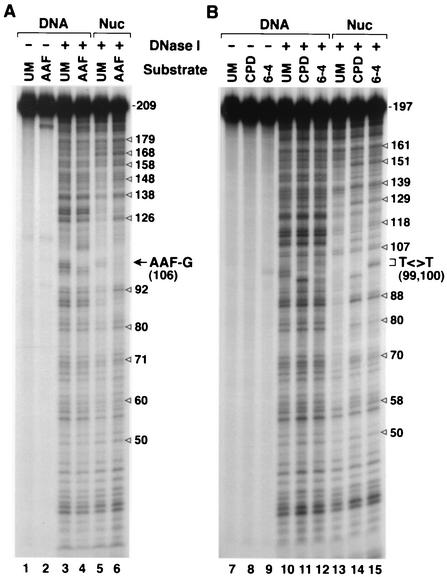
To ascertain that the DNA-protein complexes that we prepared by mixing 197- to 206-bp duplexes with human histone core proteins are indeed nucleosomes and that the damage is within the nucleosome core, we analyzed the complexes by DNase I footprinting. The results shown in Fig. 3 reveal strong 10-nt periodicity, in particular with damaged DNAs, in the cleavage pattern of all three substrates over the region of 70 to 160 bp, indicating a canonical nucleosome structure with the lesion located within the nucleosome core near the center of symmetry (Fig. 3, lanes 6, 14, and 15). With undamaged DNA the periodicity is less striking (Fig. 3, lanes 5 and 13). We have previously observed this phenomenon with the AAF-G duplex and its unmodified counterpart (10), and as suggested before, we believe that, with random sequence DNAs such as the ones used here, it is the helix deformity caused by the lesion that provides the signal for nucleosome phasing. In any event, clearly, the nucleosomes with DNA damage are strongly phased, contain the lesions within the core nucleosome, and hence are suitable to investigate the effect of nucleosomes and SWI/SNF on excision repair.
FIG. 3.
DNase I footprinting of naked DNA and nucleosomal (Nuc) substrates. (A) Footprints of the AAF-G substrate and the corresponding undamaged DNA (UM). (B) Footprints of the two UV photoproducts and of unmodified DNA of identical sequence. The nucleosome reconstitution mixtures were treated with DNase I and were directly loaded on a 5% nondenaturing polyacrylamide gel. The free and nucleosomal DNA bands were located by autoradiography, were recovered from the gel, and were then separated on 6% denaturing polyacrylamide gels that were autoradiographed. The ∼10-nt periodicity in nucleosomal DNA is indicated by open arrows, and the positions of G-AAF and the UV photoproducts are marked.
Effect of nucleosome on excision of different types of damage.
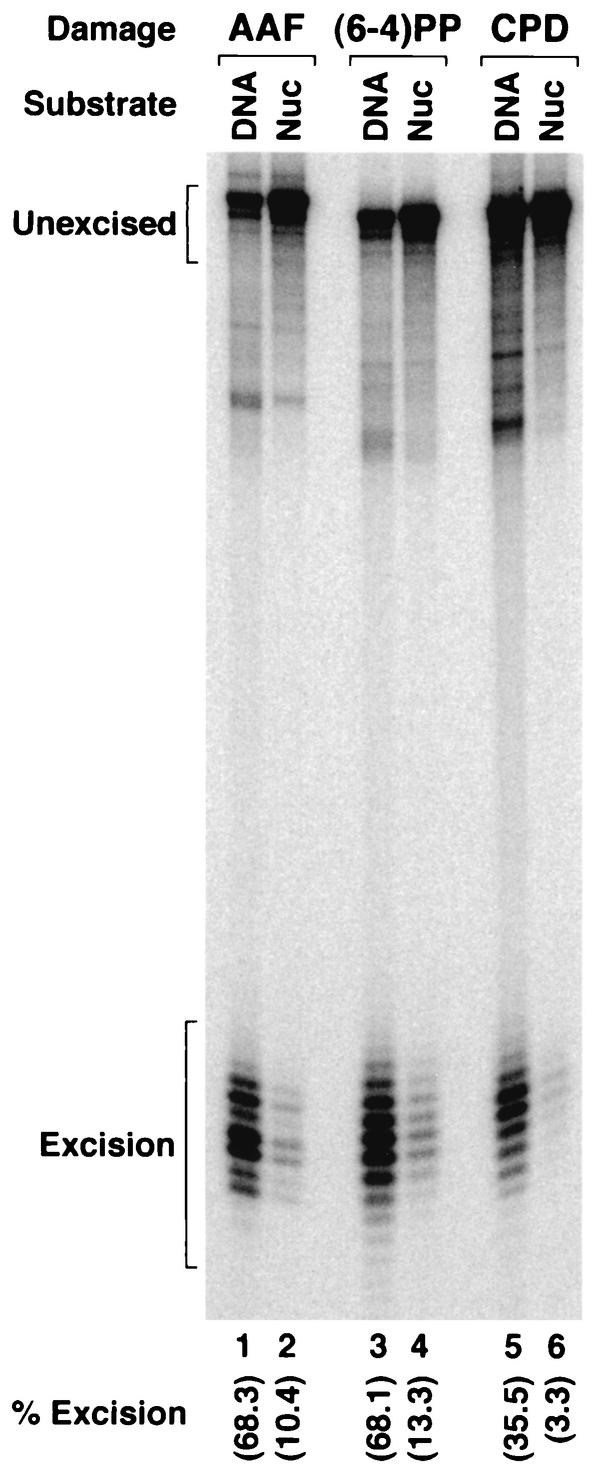
In agreement with earlier reports, AAF-G and the (6-4) photoproduct were excised more efficiently than was T<>T (23, 26) from naked DNA (Fig. 4, lanes 1, 3, and 5). In addition, as expected, incorporation of the lesions into nucleosomes suppressed excision of all three types of damage (Fig. 4, lanes 2, 4, and 6). Of significance, however, was the stronger inhibition of excision of T<>T (10-fold inhibition) than of the other two substrates (about fivefold inhibition), such that the difference between the inefficient and efficient substrates became amplified in nucleosomes. This differential inhibition of (6-4) photoproduct and T<>T may partially explain the discrepancy between the in vitro (with naked DNA) and in vivo repair rates observed for the lesions alluded to above. However, the different levels of excision that we observe for the two photoproducts [approximately fourfold-more-efficient repair of (6-4) photoproduct than of T<>T] in nucleosomes cannot account for the >10-fold difference seen in in vivo experiments. We reasoned that the repair of the two photoproducts may be affected differently by chromatin remodeling factors, which could account for the drastically slow excision of T<>T relative to that of (6-4) photoproducts in vivo. Hence, we tested the effect of SWI/SNF on excision of all three lesions from nucleosomes.
FIG. 4.
Inhibition of excision of three types of DNA lesions by nucleosomes. The indicated substrates, either as naked DNA or in nucleosomes (Nuc), were incubated with 6-factor reconstituted excision nuclease for 3 h at 30°C, and the products were separated on an 8% denaturing polyacrylamide gel. The figure shows an autoradiogram of the gel. The levels of excision were quantified by PhosphorImager and the ImageQuant system and are indicated at the bottoms of all lanes.
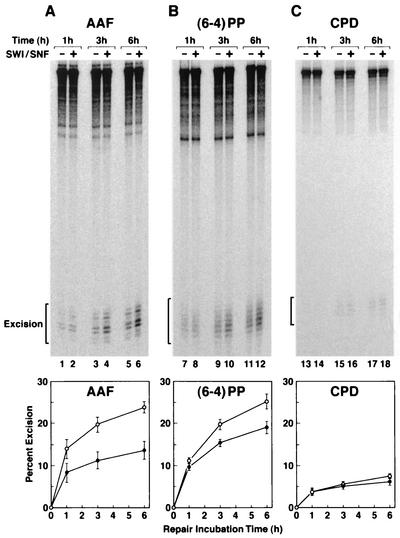
Differential effect of SWI/SNF on excision of different types of damage.
Inclusion of SWI/SNF in the reaction mixture stimulated the rate and extent of excision of AAF-G by a factor of about 2 as we reported previously (10) (Fig. 5A). Similarly, SWI/SNF stimulated excision of the (6-4) photoproduct by about 1.5-fold (Fig. 5B). In contrast, this chromatin remodeling factor had no effect on either the rate or the extent of T<>T excision under our assay conditions (Fig. 5C). Thus, recognizing the limitations of our experimental system, we conclude that the stronger inhibition of excision of T<>T than of (6-4) photoproduct by the nucleosome and the lack of a stimulatory effect by at least one remodeling factor on T<>T excision from a nucleosome may be the two major determinants of the very low rate of excision of Pyr<>Pyr in vivo, compared to the moderate rate observed with naked DNA and the reconstituted human excision nuclease in vitro (24; this work).
FIG. 5.
Effect of SWI/SNF on repair of three different lesions in mononucleosomes. Nucleosomal DNAs (30 pM) were incubated with reconstituted excision nuclease in the absence or presence of SWI/SNF (0.48 nM) for the indicated times, and the reaction products were separated on 8% polyacrylamide gels. The top panels show representative autoradiograms of excision gels. In the bottom panel the level of excision as percentage of input substrates is plotted from three independent experiments; the bars indicate standard errors.
DISCUSSION
We have made three observations of relevance to DNA repair in chromatin in this study: the effects of DNA lesions on nucleosome phasing, the dependence of repair inhibition by nucleosome on the type of DNA damage, and finally the dependence of stimulation of excision nuclease by SWI/SNF on the type of DNA lesion.
Effect of damage on nucleosome phasing.
As a common practice, nucleosome reconstitution experiments are performed with special DNA sequences, such as TG motifs or 5S ribosomal gene sequences, to obtain nucleosomes with uniform phasing (11, 33). We previously observed that, with a 200-bp duplex with an AAF-G adduct in the center, a uniform nucleosome phasing could be achieved without the need for a special sequence (10). In this study we show that the same high-quality phasing is conferred by (6-4) and T<>T photoproducts as well. It is likely that the DNA bending and kinking caused by these lesions help position them in the center of the dyad symmetry of the nucleosome core. While the crystal structures of AAF-G or (6-4) photoproduct containing DNAs are not available at present, the structure of a T<>T-containing decamer shows that the photoproduct bends the DNA by 30° (29) in agreement with an earlier solution circularization study (15). It is known that DNA bends such as these, induced by whatever mechanism, help in nucleosome phasing (11).
Inhibition of repair by nucleosomes.
Recent studies with defined nucleosomal substrates have revealed that both human and Xenopus nucleotide excision repair (10, 11, 17, 38) as well as human base excision repair (28) are inhibited by nucleosomal structure (see reference 6). In this study we show that the repair of AAF-G and (6-4) photoproduct, which are presumed to cause more drastic structural changes in DNA than T<>T (which nevertheless kinks the duplex by 30°), is affected less severely than that of T<>T repair. It is conceivable that the gentler bending caused by T<>T in DNA makes the structural deformity of this lesion less obvious and hence less accessible to damage recognition proteins when the T<>T is in nucleosomes. While this is a plausible explanation, a more definitive answer should come from the crystal structures of mononucleosomes containing each of these lesions.
Differential effects of SWI/SNF on repair of UV photoproducts.
In addition to the stronger inhibition of T<>T repair than of (6-4) photoproduct repair by nucleosomes, the repair of T<>T is not stimulated by SWI/SNF. This likely contributes to its very low rate of repair in vivo. At present we do not have a definitive explanation for why T<>T repair is not stimulated by SWI/SNF. Based on structural considerations alone, it is safe to assume that the (6-4) photoproduct distorts the duplex more severely than does T<>T and hence would be more accessible to any protein approaching the nucleosome (20). However, there is no evidence that SWI/SNF has a higher affinity for damaged DNA. It is likelier that the damage recognition factors RPA, XPA, and XPC detect the (6-4) photoproduct more readily than T<>T and in doing so help recruit SWI/SNF to the damage site (10), which in turn remodels the nucleosome and facilitates the assembly of the entire excision nuclease and eventual damage removal. It must be noted, however, that in addition to SWI/SNF there are other chromatin remodeling factors that alter DNA-histone interactions by either SWI/SNF-like ATP-dependent action or by covalent modification (1, 16, 35, 37) and that the effects of these on repair of various lesions must be tested in vitro to provide a more comprehensive understanding of the different rates of removal of the two major UV photoproducts in vivo.
Acknowledgments
We thank C. L. Peterson for SWI/SNF and J. T. Reardon for comments on the manuscript.
This work was supported by NIH grant GM32833.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Bessho, T., A. Sancar, L. H. Thompson, and M. P. Thelen. 1997. Reconstitution of human excision nuclease with recombinant XPF·ERCC1 complex. J. Biol. Chem. 272:3833-3837. [DOI] [PubMed] [Google Scholar]
- 3.Branum, M. E., J. T. Reardon, and A. Sancar. 2001. DNA repair excision nuclease attacks undamaged DNA: a potential source of spontaneous mutations. J. Biol. Chem. 276:25421-25426. [DOI] [PubMed] [Google Scholar]
- 4.Buschta-Hedayat, N., T. Buterin, M. T. Hess, M. Missura, and H. Naegeli. 1999. Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA. Proc. Natl. Acad. Sci. USA 96:6090-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans, E., J. G. Moggs, J. R. Hwang, J. M. Egly, and R. D. Wood. 1997. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green, C. M., and G. Almouzni. 2002. When repair meets chromatin. EMBO Rep. 3:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1995. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270:12973-12976. [DOI] [PubMed] [Google Scholar]
- 8.Hanawalt, P. C. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 21:8949-8956. [DOI] [PubMed] [Google Scholar]
- 9.Hara, R., J. Mo, and A. Sancar. 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20:9173-9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara, R., and A. Sancar. 2002. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 22:6779-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes, J. J., and A. P. Wolffe. 1992. Transcription factor interaction with nucleosomal DNA. Bioessays 14:597-603. [DOI] [PubMed] [Google Scholar]
- 12.Hess, M. T., U. Schwitter, M. Petretta, B. Giese, and H. Naegeli. 1997. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl. Acad. Sci. USA 94:6664-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, J. C., D. S. Hsu, A. Kazantsev, and A. Sancar. 1994. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. USA 91:12213-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, J. C., D. L. Svoboda, J. T. Reardon, and A. Sancar. 1992. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl. Acad. Sci. USA 89:3664-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain, I., J. Griffith, and A. Sancar. 1988. Thymine dimers bend DNA. Proc. Natl. Acad. Sci. USA 85:2558-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kingston, R. E., and G. L. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 17.Kosmoski, J. V., E. J. Ackerman, and M. J. Smerdon. 2001. DNA repair of a single UV photoproduct in a designed nucleosome. Proc. Natl. Acad. Sci. USA 98:10113-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindsley, J. E., and R. P. P. Fuchs. 1994. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry 33:764-772. [DOI] [PubMed] [Google Scholar]
- 19.Logie, C., and C. L. Peterson. 1999. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 304:726-741. [DOI] [PubMed] [Google Scholar]
- 20.Mann, D. B., D. L. Springer, and M. J. Smerdon. 1997. DNA damage can alter the stability of nucleosome: effects are dependent on damage type. Proc. Natl. Acad. Sci. USA 94:2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsunaga, T., C. H. Park, T. Bessho, D. Mu, and A. Sancar. 1996. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 271:11047-11050. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, D. L., and R. S. Nairn. 1989. The biology of the (6-4) photoproducts. Photochem. Photobiol. 49:805-819. [DOI] [PubMed] [Google Scholar]
- 23.Mu, D., E. Bertrand-Burggraf, J. C. Huang, R. P. P. Fuchs, and A. Sancar. 1994. Human and E. coli excinucleases are affected differently by the sequence context of acetylaminofluorine-guanine adduct. Nucleic Acids Res. 22:4869-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mu, D., D. S. Hsu, and A. Sancar. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271:8285-8294. [DOI] [PubMed] [Google Scholar]
- 25.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 26.Mu, D., M. Tursun, D. R. Duckett, J. T. Drummond, P. Modrich, and A. Sancar. 1997. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell. Biol. 17:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu, D., M. Wakasugi, D. S. Hsu, and A. Sancar. 1997. Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272:28971-28979. [DOI] [PubMed] [Google Scholar]
- 28.Nilsen, H., T. Lindahl, and A. Verreault. 2002. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 21:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, H., K. Zhang, Y. Ren, S. Nadji, N. Sinha, J. S. Taylor, and C. Kang. 2002. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. USA 99:15965-15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 31.Reardon, J. T., D. Mu, and A. Sancar. 1996. Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease. J. Biol. Chem. 271:19451-19456. [DOI] [PubMed] [Google Scholar]
- 32.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 33.Shrader, T. E., and D. M. Crothers. 1989. Artifical nucleosome positioning sequences. Proc. Natl. Acad. Sci. USA 86:7418-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, C. A., and J. S. Taylor. 1993. Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn, trans-syn-I, (6-4), and Dewar photoproducts of thymidylyl(3′→5′)-thymidine. J. Biol. Chem. 268:11143-11151. [PubMed] [Google Scholar]
- 35.Strahl, B. D., and D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 36.Svoboda, D. L., J. S. Taylor, J. E. Hearst, and A. Sancar. 1993. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J. Biol. Chem. 268:1931-1936. [PubMed] [Google Scholar]
- 37.Thoma, F. 1999. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 18:6585-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ura, K., M. Araki, H. Saeki, C. Masutani, T. Ito, S. Iwai, T. Mizukoshi, Y. Kaneda, and F. Hanaoka. 2001. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 20:2004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakasugi, M., and A. Sancar. 1999. Order of assembly of human DNA excision repair nuclease. J. Biol. Chem. 274:18759-18768. [DOI] [PubMed] [Google Scholar]
- 40.Wolffe, A. P., and J. J. Hayes. 1993. Transcription factor interactions with model nucleosomal templates. Methods Mol. Genet. 2:314-329. [Google Scholar]
- 41.Wood, R. D. 1997. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272:23465-23468. [DOI] [PubMed] [Google Scholar]
- 42.Ye, N., M. S. Bianchi, N. O. Bianchi, and G. P. Holmquist. 1999. Adaptive enhancement and kinetics of nucleotide excision repair in humans. Mutat. Res. 435:43-61. [DOI] [PubMed] [Google Scholar]