Abstract
We characterized two essential putative GTPases, Nog1p and Lsg1p, that are found associated with free 60S ribosomal subunits affinity purified with the nuclear export adapter Nmd3p. Nog1p and Lsg1p are nucleolar and cytoplasmic, respectively, and are not simultaneously on the same particle, reflecting the path of Nmd3p shuttling in and out of the nucleus. Conditional mutants of both NOG1 and LSG1 are defective in 60S subunit biogenesis and display diminished levels of 60S subunits at restrictive temperature. Mutants of both genes also accumulate the 60S ribosomal reporter Rpl25-eGFP in the nucleolus, suggesting that both proteins are needed for subunit export from the nucleolus. Since Lsg1p is cytoplasmic, its role in nuclear export is likely to be indirect. We suggest that Lsg1p is needed to recycle an export factor(s) that shuttles from the nucleus associated with the nascent 60S subunit.
In eukaryotic cells ribosomes are assembled in the nucleolus and must be transported through the nuclear pore complex to the cytoplasm, where they function in translation. The first discrete intermediate preribosomal species, the 90S particle (40), assembles around the 35S primary transcript in the nucleolus and contains integral ribosomal proteins, trans-acting rRNA processing factors, and subunit assembly factors. Recent evidence suggests that the 90S particle assembles cotranscriptionally on the primary transcript, is devoted to 40S biogenesis, and is almost entirely lacking in 60S proteins and assembly factors (5, 16). Cleavage of the transcript into 20S and 27S species in the nucleolus, the precursors of 18S and 25S, respectively, may release the pre-40S complex. The subsequent processing and assembly of the 66S particle, the nucleolar precursor of the 60S subunit (44), appears to be independent of 40S assembly (5, 16). Release of the large subunit from the nucleolus into the nucleoplasm likely coincides with the release of many of the 66S-associated biogenesis factors (9, 31), yielding a nucleoplasmic pre-60S subunit.
Several rRNA processing and protein assembly events may be associated with or required for release of the pre-60S subunit from the nucleolus into the nucleoplasm. For example, the cleavage of 27S pre-rRNA into 25S and 7S RNAs precedes release into the nucleoplasm (15). In addition, the nucleolar 66S particle contains the two proteins Rlp7p and Rlp24p that are similar to mature subunit proteins Rpl7p and Rpl24p, respectively (6, 35). Thus, it has been suggested that the exchange of Rpl7p and Rpl24p for Rlp7p and Rlp24p proteins is necessary for 60S subunit release (6, 35). Once in the nucleoplasm, the pre-60S particle undergoes further processing. For example, in yeast the 3′ trimming of 7S to 5.8S is likely a nucleoplasmic event (15) that requires the exosome, Rex1p, Rex2p, and Ngl2p (8, 29, 43). Finally, export to the cytoplasm requires the addition of the export adapter Nmd3p (11, 20) that provides the leucine-rich nuclear export sequence recognized by Crm1 in a Ran-GTP-dependent fashion. Based on genetic and physical evidence, Nmd3p may be recruited to the subunit by Rpl10p (11). The temporal sequence and cellular localization of these events is not well established, and it is possible that some of these events are not obligatorily linked with a particular compartment of the nucleus or with other events happening in parallel on the subunit.
The large and small ribosomal subunits are produced in equimolar amounts since they are derived from a common precursor. Nevertheless, they are independently matured and exported from the nucleus, with the large subunit requiring a longer transit time in the nucleoplasm (15, 24, 41). Recent advances by several research groups have provided significant insight into the nuclear export pathway of the 60S subunit. Release of the pre-60S subunit from the nucleolus requires Noc1p/Mak21p (YDR060W), Noc2p (YOR206W), and Noc3p (YLR002C) (28). Although the functions of these proteins are not known, they appear to act in a sequential fashion, with Noc1p/Mak21p and Noc2p present in the nucleolus and Noc2p and Noc3p found in the nucleoplasm.
We and others have recently shown that the nuclear export signal (NES) for the large subunit is provided by the adapter protein Nmd3p (Yhr170wp). Expression of mutant Nmd3 protein lacking the NES inhibits export of the 60S subunit (11, 20). Genetic and physical interactions between Nmd3p and Rpl10p suggest that Rpl10p may constitute at least part of the binding site for Nmd3p on the 60S subunit (11, 22). The leucine-rich NES of Nmd3p is recognized by the karyopherin Crm1p (YGR218W) (11, 20). Consequently, export of 60S subunits is blocked in crm1 (xpo1-1) conditional mutants (38) or by the Crm1p inhibitor leptomycin B (LMB) (25) in LMB-sensitive yeast (11, 20). This pathway for export of 60S subunits is conserved in metazoan cells (40a).
Nascent 60S subunits are incorporated into polysomes more slowly than are nascent 40S subunits. This likely reflects the slower processing of 60S subunits in the nucleus (39, 42), as well as a delay in the recruitment of 60S subunits into polysomes in the cytoplasm (45). The reason for this cytoplasmic delay is not clear, but it could be due to the slow addition or removal of proteins from the nascent subunit, a conformational change of the subunit, or covalent modification of the rRNA and/or r-proteins.
To identify additional proteins that bind to the free 60S subunit and that act in the biogenesis and transport of the 60S subunit, we have purified Nmd3p-bound subunits. Among the nonribosomal proteins associated with these subunits were two monomeric GTP-binding proteins, Nog1p and Lsg1p. These two proteins are both essential and required for 60S biogenesis. We show that Lsg1p is a cytoplasmic protein, whereas Nog1p has recently been described as a nucleolar protein (33).
MATERIALS AND METHODS
Strains, media, and growth conditions.
The strains used in the present study are listed in Table 1. Rich medium (yeast extract-peptone-glucose) and dropout medium (synthetic complete medium) containing 2% glucose or 1% galactose as the carbon source were as described previously (21). Yeast transformations were carried out as described previously (14).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source of reference |
|---|---|---|
| AJY272 | MATaade2 ade3 leu2 lys2-801 ura3-52 NMD3::13myc::KanMX6 | 20 |
| AJY734 | MATα ade3 leu2 lys2-801 ura3-52 nmd3-4 | 18 |
| AJY832 | MATa/α ura3-52/ura3-52 leu2Δ1/leu2 trp1Δ63/TRP1 LYS2/lys2-801 ade2 ade3 | This study |
| AJY1118 | MATα ade3 leu2 lys2-801 ura3-52 GAL1::NOG1 | This study |
| AJY1124 | MATα ade2 ade3 leu2 lys2-801 ura3-52 GAL1::NOG1 | This study |
| AJY1126 | MATaade2 ade3 leu2 lys2-801 ura3-52 GAL1::LSG1 (pAJ289) | This study |
| AJY1167 | MATα his3 leu2 lys2 ura3 lsg1Δ::KanMX4 | This Study |
| AJY1171 | MATahis3 leu2 met15Δ0 ura3 yg1099wΔ::KanMX4 | This study |
| NOP7-TAP | MATα ura3-52 trp1 lys2-801 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R nop7::NOP7-TAP | 17 |
| CH1305 | MATaade2 ade3 leu2 lys2-801 ura3-52 | 23 |
| MNY7 | MATahis3 leu2 ura3 crm1Δ::Kanr (pD2-CRM1); LMB resistant | 30 |
| MNY8 | MATahis3 leu2 ura3 crm1Δ::Kanr (pD1-CRM1T539C); LMB sensitive | 30 |
Strains.
The GAL1::NOG1 genomic fusion was made by homologous recombination as described previously (27). The oligonucleotide primers 5′-ATATAAAGAATACACTTCAGATTTAGGAGTTGCAGCGTTTATAAAGAATTCGAGCTCGTTTAAAC and 5′-ATCATTTGCTGGAGCGACAGTAGGGATATCCTTCCATGAAAGTTGCATTTTGAGATCCGGGTTTT were used in PCR to amplify plasmid pFA6a-KanMX-PGAL1. The PCR product was transformed into the wild-type diploid AJY832. Geneticin-resistant transformants were selected, and integration was confirmed by PCR. The heterozygous diploid was sporulated to give the spore clones AJY1118 and AJY1124. The GAL1::LSG1 genomic fusion was made similarly by using the oligonucleotides 5′-AATGCATTCTACTACTTAATAAAGAGGTGATGAGCTTATAAGCCAGAATTCGAGCTCGTTTAAAC and 5′-TGGCCCTTTTGGCGCCTTCCATTTCTTGGGAGCTTCTTTTGGTGGCATTTTGAGATCCGGGTTTT. The resulting heterozygous diploid was transformed with pAJ289 and sporulated to give the spore clone AJY1126.
Construction of plasmids.
Plasmids are listed in Table 2. pAJ289 (myc-LSG1) was made by PCR amplification of LSG1 from wild-type genomic yeast DNA with the primers AJO285 (5′-CATGCCATGGAACAAAAGTTGATTTCTGAAGAAGACTTGAGCTCTATGCCACCAAAAGAAGCT) and AJO286 (5′-CGTGACGTCTAATTATTTTCAATGCT). The PCR product was digested with NcoI and AatII and ligated into the same sites of pAJ151 (pRS315 with the XRN1 promoter). pAJ290 (HA-NOG1) was made similarly by using the primers AJO283 (5′-CATGCCATGGGTTACCCATACGATGTCCCAGACTACGCTGAGCTCATGCAACTTTCATGGAAG) and AJO284 (5′-GCGAAGCTTCAACGGAAATCTGTCTT). The PCR product was digested with NcoI and HindIII and ligated into the same sites of pAJ151. The lsg1-1 and lsg1-2 conditional mutants (pAJ740 and pAJ741) were made by random PCR mutagenesis of pAJ289 with the oligonucleotides AJO285 and AJO286. The PCR product was cotransformed with pAJ289 digested with BglII and XbaI to remove LSG1 coding sequence into AJY1167 (LSG1::KanMX) containing pAJ626 (myc-LSG1). Transformants were screened for temperature sensitivity on 5-fluororotic acid (5-FOA) plates. Plasmids were isolated and retransformed into AJY1167 (LSG1::KanMX). Conditional mutants nog1-1 (pAJ633) and nog1-3 (pAJ637) were constructed by a method similar to the one described for lsg1 conditional mutants, except that the oligonucleotides used for PCR amplification were AJO283 and AJO284 and pAJ290 was the template. PCR product was cotransformed with AatII- and MscI-digested pAJ290 into AJY1118 (GAL1::NOG1) containing pAJ625 (HA-NOG1). Transformants were screened for temperature sensitivity on 5-FOA plates. All lsg1 and nog1 mutants contained multiple mutations (data not shown). pAJ625 was made by ligating HA-NOG1 on an NheI-to-BamHI fragment from pAJ290 to XbaI- and BamHI-digested pRS316. pAJ626 was made by ligating myc-LSG1 on an NheI-to-BamHI fragment from pAJ289 to XbaI- and BamHI-digested pRS316. pAJ901 (LSG1-myc) was constructed by first cloning the 13myc tag from pAJ538 (19) as a SmaI-to-HindIII fragment into the same sites of pRS416. pAJ901 was then assembled in a three-part ligation: 13myc as an XbaI-to-HindIII fragment from pAJ538, the vector backbone of pAJ538 as an EagI-to-HindIII fragment, and LSG1 as a PCR fragment amplified from genomic DNA with AJO428 (5′-GCGGCGGCCGCCCATGGGACGTCTGGCCAGCATGCGGTACTTTATACGTGTGCATTATTC) and AJO415 (5′-CGCGCTAGCATTATTTTCAATGCTAAAAAC) and digested with NotI and NheI.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant markers | Source or reference |
|---|---|---|
| pAJ289 | LEU2 CEN myc-LSG1 | This study |
| pAJ290 | LEU2 CEN HA-NOG1 | This study |
| pAJ538 | LEU2 CEN NMD3-myc | 20 |
| pAJ625 | URA3 CEN HA-NOG1 | This study |
| pAJ626 | URA3 CEN myc-LSG1 | This study |
| pAJ633 | LEU2 CEN nog1-1 | This study |
| pAJ637 | LEU2 CEN nog1-3 | This study |
| pAJ740 | LEU2 CEN lsg1-1 | This study |
| pAJ741 | LEU2 CEN lsg1-2 | This study |
| pAJ901 | LEU2 CEN LSG1-myc | This study |
Indirect immunofluorescence.
Indirect immunofluorescence was performed as described previously (20). Antibodies used were the monoclonal antibody 9e10 anti-c-myc for the primary antibody (1:2,500 dilution; Covance), and the secondary antibody was Cy3-conjugated anti-mouse antibody (1:300 dilution; Amersham Pharmacia Biotech). Fluorescence was visualized on a Zeiss Axiophot microscope fitted with a ×100 objective lens and a Princeton Electronics MicroMAX charge-coupled device camera controlled with the IPLab Spectrum P software package from Signal Analytics Corp. Captured images were prepared by using Adobe Photoshop 5.0.
LMB experiments.
Overnight cultures were diluted twofold into fresh medium and cultured for 1 h. Cells were then concentrated 20-fold in fresh medium, LMB (M. Yoshida) was added at a final concentration of 0.1 μg/ml, and cultures were incubated for an additional 30 min. Cells were fixed for 1 h with a 3.7% final concentration of formaldehyde and treated as described above by indirect immunofluorescence.
Rpl25-eGFP localization.
Overnight cultures were diluted twofold into fresh medium and grown for 30 min at 30°C. Cultures were divided in half, with one half shifted to 37°C and the remainder grown at 30°C for 3 h. Hoechst 34442 (1 μg/ml [final concentration]) was added to cultures during the last 30 min of growth. Green fluorescent protein (GFP) signal was visualized as described for indirect immunofluorescence.
Pulse-chase labeling and immunoprecipitation of ribosomal subunits.
For 35S immunoprecipitation pulse-chase experiments, 250-ml cultures of CH1305 expressing pAJ901 (Lsg1-myc) or NOP7-TAP (J. Woolford) were grown to mid-log phase in medium lacking methionine. Cultures were concentrated to 9 ml in fresh medium. Proteins were labeled by adding 0.5 mCi (0.85 nmol) of l-[35S]methionine for 5 min, followed by the addition of excess unlabeled l-methionine (5,200 nmol). At the indicated times, 1.5-ml samples were collected, and cells were pelleted and flash frozen. Immunoprecipitations were performed on labeled extracts as described previously (19). Immunoprecipitated proteins were analyzed on 12% polyacrylamide gels, which were either dried and exposed to autoradiography or analyzed by Western blotting.
For the Lsg1-60S competition assay, l-[35S]methionine-labeled protein extracts were prepared and incubated with increasing amounts of unlabeled wild-type yeast extract or purified unlabeled 60S subunits for 1.5 h at 4°C. Immunoprecipitations were then carried out as described previously (18). Immunoprecipitated proteins were analyzed on 12% polyacrylamide gels. Proteins were visualized by Western blotting or by autoradiography of the gels after drying.
Northern blotting.
RNA was extracted as described previously (36) and separated by using 1.2% agarose-formaldehyde gels for the large rRNA species and 6% polyacrylamide-7 M urea gels for the small rRNA species. RNA was transferred to nylon membranes (Zetaprobe; Bio-Rad) and probed with 32P-end-labeled oligonucleotide probes as previously described (46). The probes were AJO130 (23S, 20S; 5′-TCTTGCCCAGTAAAAGCTCTCATGC), AJO190 (18S; 5′-GTCTGGACCTGGTGAGTTTCCC), AJO191 (5.8S; 5′-CGCTGCGTTCTTCATCGATGCG), AJO192 (25S; 5′-CCCGCCGTTTACCCGCGCTTGG), AJO214 (27SA,B; 5′-GTTCGCCTAGACGCTCTCTTC), AJO249 (5S; 5′-TCTGGTAGATATGGCCGCAACC), AJO282 (7S) 5′-GGCCAGCAATTTCAAGTTA), and AJO313 (35S, 27SA; 5′-TCCAGTTACGAAAATTCTTGTTTTTGACAA).
Other methods.
Polysome analysis and rRNA pulse-chase experiments were performed as described previously (18). For the mass spectrometric identification of proteins, Nmd3-13myc and associated proteins were immunoprecipitated from extracts of AJY272 as described previously (19). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% or 4 to 20% gradient polyacrylamide gels and then stained with Coomassie blue. Protein bands were excised, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF), and then protein identification was done by using ProFound (W. M. Keck Facility, Yale University).
RESULTS
Nog1p and Lsg1p are coimmunoprecipitated with Nmd3p-60S complexes.
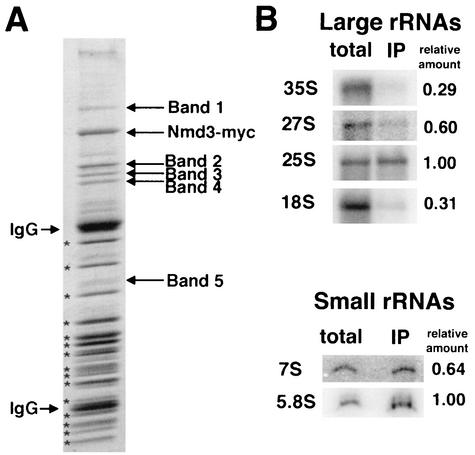
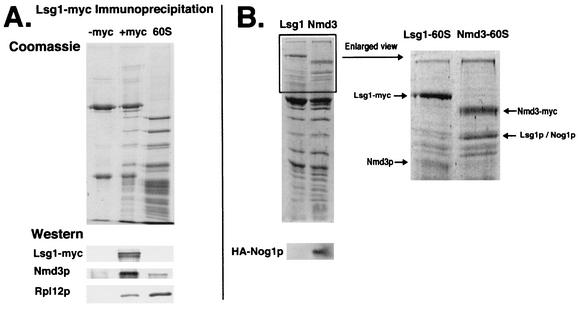
We previously demonstrated that free 60S subunits could be coimmunoprecipitated with 13xmyc-tagged Nmd3p (19). This coimmunoprecipitation is specific for tagged Nmd3p since the associated proteins were not seen in control purifications (19; see also below). Here, we have carried out an initial characterization of two of the proteins associated with the Nmd3p-bound 60S subunit. Several proteins specific to the coimmunoprecipitated complex were resolved from the large number of low-molecular-weight ribosomal proteins on polyacrylamide gels (Fig. 1A). Protein bands were excised, digested with trypsin, analyzed by MALDI-TOF mass spectrometry (W. M. Keck Facility), and identified by using ProFound (http://prowl.rockefeller.edu/cgi-bin/ProFound). Band 1 (116 kDa) contained translation elongation factor 3A (Ylr249wp). Band 2 contained two proteins that comigrated at 72 kDa: Nog1p (Ygl093wp), and Ygl099wp. We have named YGL099w LSG1 (for large-subunit GTPase). YGL099w has also been referred to as KRE35 (12, 31) based on the sensitivity of a heterozygous diploid to killer toxin (32). Although this mass spectrometry analysis was not quantitative; peptides representing greater coverage of Lsg1p were identified compared to Nog1p, suggesting that Lsg1p was the more abundant protein. Band 3 (66.5 kDa) contained the heat shock protein Ssb1p (Ydl229wp) and the putative ATP-dependent RNA helicase Ded1p (Yor204wp). Band 4 (65 kDa) contained the uncharacterized protein Ydr101cp related to methionyl aminopeptidases. Band 5 (35.6 kDa) was a faint band that contained GAPDH (glyceraldehyde-3-phosphate dehydrogenase) encoded by TDH3 (Ygr192cp). Considering that Ded1p and Tdh3p are abundant proteins, their presence was probably due to nonspecific contamination.
FIG. 1.
Immunoprecipitation of Nmd3p-60S complexes. (A) Extracts were prepared from AJY272 (Nmd3-myc) and immunoprecipitated as described previously (19). The proteins in the affinity purified complex were separated by SDS-PAGE (Fig. 1). 60S subunit proteins are indicated by asterisks. Protein not present in the purified 60S sample (labeled in Fig. 1) were excised, digested with trypsin, and analyzed by MALDI-TOF mass spectrometry. (B) RNA was extracted from the immunoprecipitated material prepared as in panel A and compared with total cellular RNA by Northern blotting as described in Materials and Methods. Relative amount indicates the ratio of signal for a given species in the immunoprecipitation (IP) compared to that seen in the total extract when normalized to 25S signals for large RNAs and 5.8S signals for small RNAs.
Nog1p (for nucleolar GTP-binding protein) is a recently characterized essential GTPase (33) that has been found in other preparations of affinity-purified pre-60S particles (1, 17, 35). Lsg1p has also been found in an Nmd3p-containing complex from a large-scale proteomic analysis of protein complexes in yeast (12). Lsg1p is a previously uncharacterized protein that belongs to the MMR/HSR1 family of GTP-binding proteins (pfam01926). In this family of proteins the order of the G motifs has been circularly permuted (26). Interestingly, two other members of this GTPase subfamily from yeast are Nug1p and Nog2p, both of which are found in nuclear pre-60S particles and are required for nuclear export of 60S subunits (1, 35).
We also examined the rRNA species present in the Nmd3-60S complex to determine the status of rRNA processing in the subunit bound by Nmd3p. rRNA was isolated from the immunoprecipitated complex and from total extract and relative levels of mature and intermediate rRNA species were compared by Northern blotting (Fig. 1B). rRNA signal strength was normalized to the 25S and 5.8S mature rRNA signals as internal controls to compensate for loading differences and to compare the level of enrichment present in the Nmd3p-60S immunoprecipitated rRNA. The Nmd3-myc-60S complex contained predominantly mature 25S and 5.8S rRNAs, a finding consistent with Nmd3p binding primarily to cytoplasmic 60S subunits (18). The slight enrichment of 27S and 7S precursors relative to 35S and 18S RNAs (Fig. 1B) may indicate that Nmd3p initially binds to a late nucleolar pre-60S particle.
Conditional mutants of NOG1 and LSG1.
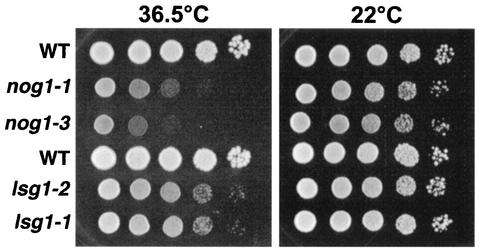
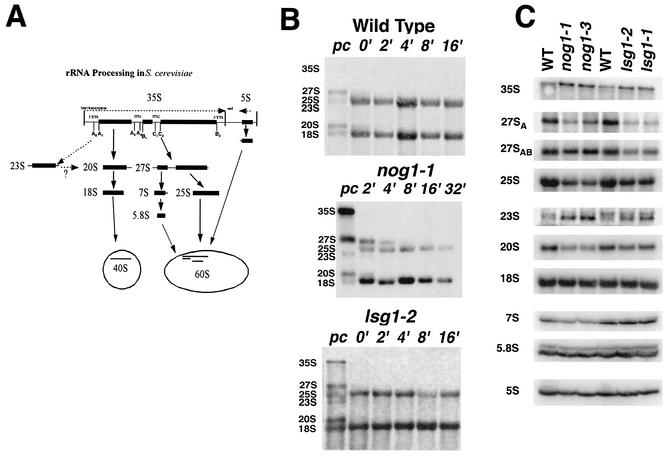
Disruption of NOG1 or LSG1 in two different genetic backgrounds was lethal and could be rescued by ectopic expression of plasmid-borne copies of NOG1 and LSG1, respectively (data not shown). This was consistent with previous reports that NOG1 and LSG1 are essential (13, 33). Consequently, we made conditional mutants of these genes. The glucose-repressible GAL1 promoter was integrated (27) into the genomic locus of NOG1 and LSG1. The resulting strains were unable to grow in the presence of glucose (data not shown). In addition, we created temperature-sensitive mutants of NOG1 and LSG1 by PCR mutagenesis. All mutants were severely impaired for growth at a restrictive temperature; however, the lsg1 mutants grew slightly better than the nog1 mutants, indicating less complete loss of function of the lsg1 alleles (Fig. 2).
FIG. 2.
Growth of temperature-sensitive mutants on plates. Tenfold serial dilutions of saturated cultures were spotted onto YPD plates and incubated for 3 days at the indicated temperatures. The strains tested were as follows: AJY1124 (GAL1::NOG1) containing plasmid pAJ290 (WT, first row), pAJ633 (nog1-1), or pAJ637 (nog1-3) and AJY1167 (LSG1::KanMX) containing pAJ289 (WT, fourth row), pAJ740 (lsg1-1), or pAJ741 (lsg1-2).
Nog1p and Lsg1p cosediment with free 60S subunits.
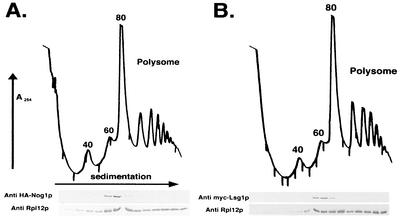
Nog1p and Lsg1p were identified by their association with the Nmd3p-60S complex. To examine their association with ribosomal subunits in more detail, we sought to determine whether Nog1p and Lsg1p cosedimented with 60S subunits or polyribosomes on sucrose gradients. Nog1p and Lsg1p were amino-terminally tagged with the hemagglutinin (HA) and the c-myc epitopes, respectively. Both HA-Nog1p and c-myc-Lsg1p fusion proteins were functional since they fully complemented deletion mutants (data not shown). Both proteins cosedimented in the position of free 60S subunits on sucrose gradients and were largely absent from polysomes (Fig. 3), a situation similar to Nmd3p sedimentation (18). The lack of accumulation of HA-Nog1p or c-myc-Lsg1p in the soluble fraction at the top of the gradient suggests that there is not a significant free pool of HA-Nog1p or c-myc-Lsg1p in the extracts. Sedimentation at the position of free 60S is typical of ribosome biogenesis factors associated with the 66S particle in the nucleolus or with the free 60S particle during nuclear or cytoplasmic maturation (17, 18, 31, 34, 47). The resolution of the sucrose gradients in Fig. 3 was not sufficient to distinguish 60S from 66S sedimentation. However, considering that Nog1p is nucleolar (33) and Lsg1p is cytoplasmic (see below), their sedimentation positions most likely reflect association with nucleolar 66S and cytoplasmic 60S species, respectively.
FIG. 3.
Nog1p and Lsg1p cosediment with free 60S subunits. Lysates were prepared in the presence of cycloheximide from strain AJY1124 (GAL1::NOG1) containing pAJ290 (HA-NOG1) (A) and strain AJY1126 (GAL1::LSG1) containing pAJ289 (c-myc-LSG1) (B) and then fractionated on 7 to 47% sucrose gradients by ultracentrifugation. The 40S, 60S, 80S, and polysome peaks are labeled in panel A. Fractions were collected while the absorbance at 254 nm was continuously monitored. Proteins were precipitated with trichloroacetic acid, separated on SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and immunoblotted for ribosomal protein Rpl12p and HA or c-myc as indicated.
Polysome analysis of nog1 and lsg1 mutants.
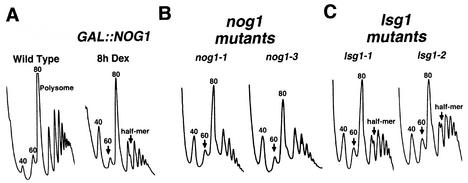
The cosedimentation of HA-Nog1p with 60S subunits and its localization to the nucleolus (33) strongly suggested a role for Nog1p in 60S subunit biogenesis. To examine this, we monitored free 60S levels after repressing NOG1 transcription. Repression of NOG1 resulted in a marked reduction of free 60S subunits that was evident after 8 h of repression (Fig. 4A). Temperature-sensitive nog1 mutants showed a similar reduction in 60S levels when incubated at the nonpermissive temperature (Fig. 4B). Some of the mutants showed half-mer polysomes, which appear as a shouldering of polysomal peaks. Half-mers reflect a defect in 60S subunit joining during translation initiation and can result from a deficiency in 60S subunit levels or from a specific joining defect. Other nog1 conditional mutants tested showed similar polysome defects (data not shown).
FIG. 4.
Polysome analysis. Extracts were prepared from mid-log-phase cultures and sedimented through 7 to 47% sucrose gradients. Gradients were analyzed by monitoring the absorbance at 254 nm. Arrows indicate reduced free 60S peaks. (A) Wild-type (CH1305, WT) and the glucose-repressible (AJY1118, GAL::NOG1) strains were cultured in galactose-containing medium. Glucose was added to 2%, and the cells were harvested after an additional 8 h. (B) AJY1124 (GAL1::NOG1) containing pAJ633 (nog1-1) or pAJ637 (nog1-3) were grown to log phase at 30°C, followed by incubation at a restrictive temperature (37°C) for 4 h. (C) AJY1167 (LSG1::KanMX) containing pAJ740 (lsg1-1) or pAJ741 (lsg1-2) were cultured as described for the nog1 mutants in panel B.
The polysome profiles of lsg1 mutants differed from those seen with nog1 mutants, showing a less severe 60S deficit and more pronounced half-mer peaks, findings that were particularly evident in the lsg1-2 mutant (Fig. 4C). A half-mer defect without a corresponding dramatic decrease in free 60S subunits has previously been interpreted as a subunit joining defect (7). Thus, Lsg1p may be required during or immediately prior to subunit joining, possibly releasing factors or mediating a conformational change to facilitate joining. However, because the lsg1 mutants were not as fully inhibited for growth compared to the nog1 mutants, the differences in the polysome profiles could reflect the different degrees to which 60S biogenesis was inhibited.
rRNA processing in nog1 and lsg1 mutants.
To determine what step in 60S biogenesis was affected in the nog1 mutants, we monitored rRNA processing kinetics in the mutants. rRNA is highly methylated immediately after transcription, allowing for pulse-labeling rRNA with l-[methyl-3H]methionine. A diagram of rRNA processing is shown in Fig. 5A. In wild-type cells, 35S pre-rRNA is rapidly processed through two large intermediates, 27S and 20S, into mature 25S, 5.8S, and 18S rRNAs (reviewed in reference 44). Kinetic analysis of wild-type rRNA processing at 37°C revealed 27S and 20S precursor rRNAs and mature 25S and 18S species prior to the addition of unlabeled methionine (Fig. 5B, wild type, lane pc). No 35S precursor was detected, demonstrating the rapid kinetics of 35S processing in wild-type cells (Fig. 5B). The ratio of 3H label in 25S rRNA compared to 18S rRNA 16 min after the addition of unlabeled methionine was 1.5.
FIG. 5.
rRNA Analysis. (A) Simplified schematic view of rRNA processing in S. cerevisiae. (B) [methyl-3H]methionine pulse-labeling of rRNA was carried out as previously described (18). Wild-type (CH1305), nog1-1 (AJY1124 containing pAJ633), and lsg1-2 (AJY1167 containing pAJ741 (lsg1-2) strains were grown to mid-log phase in selective medium lacking methionine at 30°C. Cells were shifted to 37°C for 2 h and labeled with l-[methyl-3H]methionine for 4 min and then chased with excess unlabeled methionine. Aliquots were collected and frozen 2 min before the addition of chase (lanes pc), immediately after the addition of chase (lanes 0′), and at the indicated times after the addition of chase. Total RNA was prepared and separated on formaldehyde-agarose gels. RNAs were transferred to nylon membranes, sprayed with En3Hance (Dupont) and exposed to X-ray film. (C) Northern blotting of pre- and mature rRNAs. The strains tested were as follows: left three lanes, AJY1124 containing pAJ290 (WT), pAJ633 (nog1-1), or pAJ637 (nog1-3); right three lanes, AJY1167 containing pAJ289 (WT), pAJ741 (lsg1-2), or pAJ740 (lsg1-1). Cultures were grown to early log phase at 26°C, followed by 3 h of incubation at 37°C. RNA was prepared and analyzed by Northern blotting as described in Materials and Methods.
The nog1-1 mutant showed an accumulation of 35S precursor and a delay in processing of 27S to 25S rRNA at nonpermissive temperature (Fig. 5B). Additionally, the mature 25S rRNA did not accumulate to the level of 18S rRNA (the ratio of 3H label in 25S compared to 18S rRNA reached 0.3). Also present transiently and in low abundance was a 23S species. This species is an aberrant 20S precursor indicative of altered cleavage of 35S (Fig. 5A).
The nog1 mutants were also tested for the accumulation of aberrant or immature rRNAs. Northern blotting was done with probes that were specific for known rRNA-processing intermediates. The nog1 mutants exhibited a slight accumulation of 35S and 23S species (Fig. 5C) consistent with the results of pulse-chase labeling. In contrast, 27SA, 25S, 20S and 7S were under-represented (Fig. 5C).
We also tested lsg1 mutants for defects in rRNA processing by using rRNA pulse radiolabeling and Northern blotting. At the prechase time point we observed a slight accumulation of 35S and 23S species (Fig. 5B, lsg1-2, lane pc), and 25S did not accumulate to wild-type levels. (The ratio of 3H label in 25S rRNA compared to 18S rRNA reached 0.5.) The higher ratio of 25S to 18S in lsg1 mutants compared to nog1 mutants may reflect the incomplete loss of function in the lsg1 mutants at a restrictive temperature. Northern analysis of pre-rRNA and mature rRNA species in lsg1 mutants showed a pattern of defects similar to that seen in nog1 mutants, although the decrease in 27S pre-rRNA was more pronounced in the lsg1 mutants, whereas the decrease in 7S was more pronounced in the nog1 mutants.
These results indicate that both Nog1p and Lsg1p are required for normal processing and accumulation of rRNA; however, the lack of significant accumulation of intermediates suggests that Nog1p and Lsg1p are not directly involved in rRNA processing. In particular, the cytoplasmic localization of Lsg1p (see below) clearly indicates an indirect role in nucleolar rRNA processing. Similar results have been found with dominant mutations in the export adapter Nmd3p (4, 18). It has previously been suggested that such indirect effects could arise from a general feedback mechanism, perhaps due to the failure in recycling nucleolar processing factors (44).
Nog1p and Lsg1p do not simultaneously bind 60S subunits in vivo.
To determine whether Nog1p and Lsg1p bind to 60S at the same time or sequentially in the Nmd3p-mediated export pathway, we used coimmunoprecipitation and Western blotting. Lsg1-myc efficiently coimmunoprecipitated 60S subunits (Fig. 6A, +myc), a finding consistent with the cosedimentation of Lsg1p with free 60S subunits in sucrose gradients. The immunoprecipitated subunits were free of detectable 18S rRNA (data not shown), indicating that Lsg1p binds specifically to the free 60S subunit, a result similar to the binding previously reported for Nmd3p (19). Comparison of the immunoprecipitated proteins with purified 60S subunits revealed similar protein profiles. Western blotting confirmed the presence of Nmd3p and ribosomal protein Rpl12p. We were unable to immunoprecipitate HA-Nog1p. However, when Nmd3-myc and Lsg1-myc immunoprecipitations were performed from cells expressing HA-Nog1p, epitope-tagged Nog1p was detected only in the Nmd3-myc precipitates and not in the Lsg1-myc precipitates (Fig. 6B). Thus, Nog1p and Lsg1p do not bind to the 60S subunit simultaneously. This conclusion is consistent with the localization of Nog1p and Lsg1p to the nucleus and cytoplasm, respectively (see below).
FIG. 6.
Coimmunoprecipitation of free 60S by Lsg1p. (A) Extracts from strain CH1305 (wild-type) with pRS416 (−myc) and pAJ901 (+myc) were incubated with monoclonal antibody 9e10 (anti-myc) antibody (Covance) and protein A-beads. Precipitated proteins were eluted from the beads by heating in 1× Laemmli sample buffer and separated by SDS-PAGE. Purified 60S subunit proteins were used for comparison. Proteins were stained with Coomassie blue or transferred to nitrocellulose. Western blotting was performed against Lsg1-myc, Nmd3p, and Rpl12p to assay their presence on the Lsg1p-bound 60S subunit. (B) Extracts from strain AJY272 (NMD3-myc) containing pAJ625 (HA-NOG1) and strain CH1305 containing pAJ901 (LSG1-myc) and pAJ625 were subjected to immunoprecipitation as described in panel A. Precipitated proteins were separated by SDS-PAGE and stained with Coomassie blue or transferred to nitrocellulose for Western blotting for HA-Nog1p.
Lsg1p is a cytoplasmic protein that does not shuttle in a Crm1p-dependent manner.
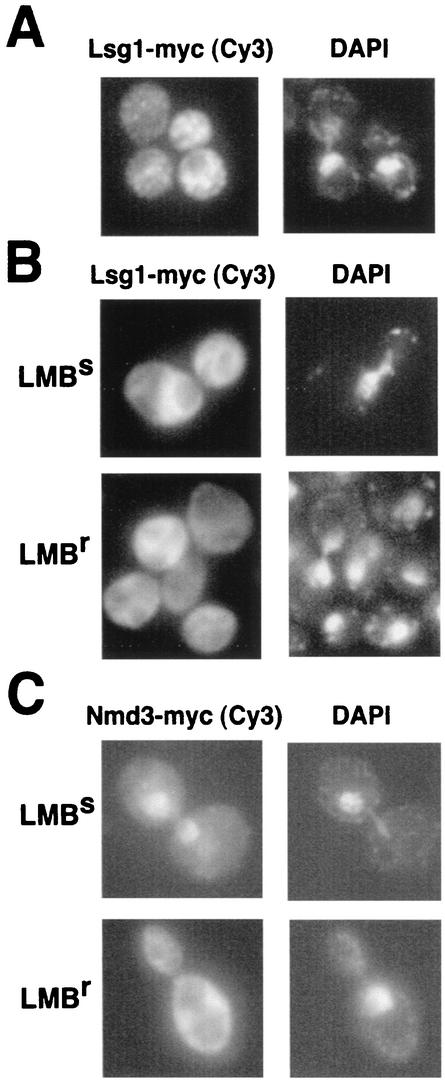
Nog1p is a nucleolar protein (33) (data not shown), but the intracellular localization of Lsg1p had not been characterized. By using indirect immunofluorescence we found that Lsg1-myc was cytoplasmic and not evident in the nucleus (Fig. 7A). Inspection of the amino acid sequence of Lsg1p revealed a putative bipartite nuclear localization signal in the amino terminus of the protein (amino acids 8 to 24). However, this sequence is not conserved among related proteins, and deletion of amino acids 2 to 24 had no apparent effect on cell growth (unpublished data), a finding consistent with our conclusion that Lsg1p is cytoplasmic.
FIG. 7.
Lsg1p is cytoplasmic and does not shuttle in a Crm1-dependent fashion. (A) Strain CH1305 (wild-type) containing pAJ901 (LSG1-myc) was grown to mid-log phase, and immunofluorescence analysis was performed as described in Materials and Methods. (B) Strains MNY7 (LMB resistant [LMBr]) and MNY8 (LMB sensitive [LMBs]) were transformed with pAJ901. (C) As a control, the LMB-dependent nuclear accumulation of Nmd3-myc was monitored in MNY7 and MNY8 containing pAJ538 (NMD3-myc) (20). Strains were cultured and treated with LMB as described in Materials and Methods.
Nevertheless, it was possible that Lsg1p, like Nmd3p, shuttles in and out of the nucleus. Nuclear export of the 60S subunit depends on the NES of Nmd3p (11, 20) and the receptor Crm1p (11, 20, 38). Upon treatment of LMB-sensitive yeast cells with LMB, both 60S and Nmd3p accumulate in the nucleus (11, 20). Similarly, we would expect Lsg1p to accumulate in the nucleus if it were a shuttling protein that was exported from the nucleus bound to the nascent 60S subunit. Lsg1-myc was expressed in both LMB-sensitive and LMB-resistant strains. After treatment with LMB, Lsg1-myc remained cytoplasmic in both strains (Fig. 7B). In a control experiment, Nmd3-myc was readily trapped in the nucleus after the addition of LMB (Fig. 7C), as previously demonstrated (11, 20). These results indicate that Lsg1p does not shuttle in a Crm1p-dependent manner and is restricted to the cytoplasm. Similarly, expression of a dominant-negative Ran mutant (GSP1-G21V) leads to nuclear accumulation of Nmd3p but not Lsg1p (M. West and A. W. Johnson, unpublished data). We conclude that Lsg1p binds to the 60S subunit after it is exported to the cytoplasm and that Lsg1p is one of the last proteins acting in the 60S subunit biogenesis pathway.
Lsg1p binds to recycling cytoplasmic 60S subunits.
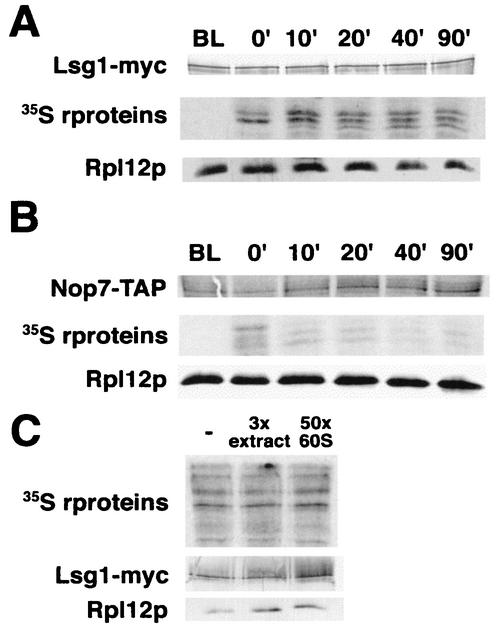
To examine the kinetics of Lsg1p binding to the free 60S pool, we used a combination of pulse-chase labeling and immunoprecipitation of subunits bound to Lsg1p. If Lsg1p bound only to nascent 60S, the label in the r-proteins bound by Lsg1p should chase out of the Lsg1-60S complex quickly, resulting in a large drop in specific activity. On the other hand, if Lsg1p bound recycling 60S subunits, the specific activity seen at later time points should not be dramatically reduced. As shown in Fig. 8A, the specific activity of label in r-proteins bound by Lsg1p peaked 10 min after chase and dropped only twofold after an additional 80 min. Since the cells were growing throughout the time course of this experiment, the twofold drop in specific activity could be accounted for by dilution with newly made unlabeled subunits. In a similar experiment with Nmd3p, we previously observed a significantly greater drop in specific activity of Nmd3p-bound subunits during the course of the experiment (19), a finding consistent with the conclusion that Nmd3p initially binds to subunits earlier than does Lsg1p.
FIG. 8.
Time course of labeled subunit binding to Lsg1p in vivo. Cultures of CH1305 containing pAJ901 (LSG1-myc) (A) and NOP7-TAP (B) were pulse-labeled with l-[35S]methionine for 5 min as described in Materials and Methods. At the indicated time points aliquots were removed, extracts prepared, and tagged proteins were affinity purified by using anti-myc antibodies for Lsg1-myc or as described previously for NOP7-TAP (17). Precipitated complexes were subjected to SDS-PAGE and dried for autoradiography or transferred to nitrocellulose for Western blotting. (C) Extracts were prepared from 35S-labeled CH1305 containing pAJ901. Extracts were incubated with a 3-fold excess of unlabeled wild-type extract or with a 50-fold excess of purified unlabeled 60S. Lsg1-myc and associated 60S proteins were immunoprecipitated and analyzed as described in panel A.
As a control demonstrating the kinetics of an early binding 60S biogenesis factor, we utilized a Nop7p-TAP fusion, which binds to a 66S nucleolar pre-60S subunit (17). The specific activity of Nop7-TAP purified 60S subunits was maximal at the earliest time point.
In order to rule out the possibility that the relatively constant level of 35S label in the Lsg1p immunoprecipitations was due to exchange of subunits on Lsg1p during the immunoprecipitation we performed a competition experiment. l-[35S]methionine-labeled extract containing Lsg1-myc was mixed with unlabeled yeast extract prepared from cells expressing wild-type Lsg1p or with an excess of purified 60S subunits. 60S subunits were then coimmunoprecipitated with Lsg1-myc. If Lsg1p were readily exchangeable, we would expect to see the amount of radiolabel in the immunoprecipitated subunits drop as the labeled subunits exchanged with the pool of excess cold subunits. There was no significant drop in 35S signal when either a threefold excess of unlabeled extract (Fig. 8C, lane 2) or a 50-fold excess of purified 60S subunits (Fig. 8C, lane 3) was added. We estimate that fewer than 10% of the 60S subunits are exchanging on Lsg1p during immunoprecipitation. Thus, the twofold drop in specific activity in Fig. 8A is not due to rapid exchange of subunits and must reflect the relatively constant specific activity of the 60S pool bound by Lsg1p in cells.
Ribosome export.
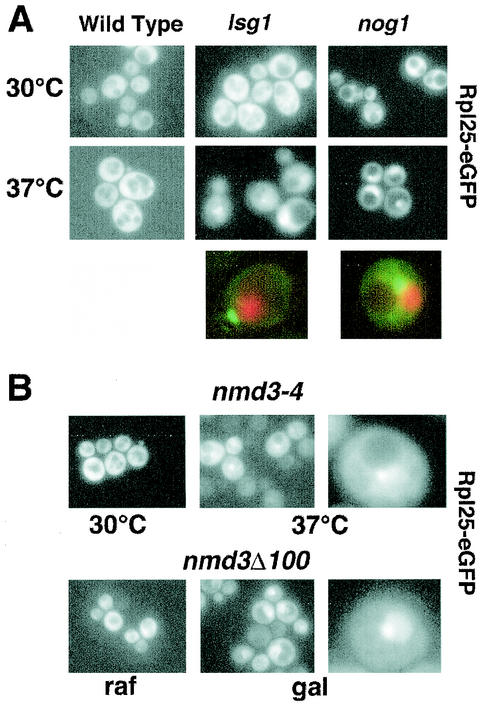
Nascent ribosomal subunits assembled in the nucleolus are transported through the nucleoplasm to the cytoplasm. This intranuclear movement requires a number of additional proteins, including the ATPase and GTPases Rix7p, Nug1p, and Nog2p (1, 10, 35). Conditional mutants in these factors result in retention of the ribosomal reporter Rpl25-eGFP in the nucleolus or nucleoplasm. Using this fluorescent reporter protein, which is functional and is incorporated into 60S subunits (11), we observed a strong nucleolar accumulation in nog1-3 and nog1-1 mutants (Fig. 9A), suggesting that Nog1p is also required for release of subunits from the nucleolus.
FIG. 9.
Rpl25-eGFP localization. (A) Rpl25-eGFP was expressed in strain CH1305 (wild-type), strain AJY1124 (GAL1::NOG1) carrying pAJ637 (nog1-3), and strain AJY1167 (LSG1::KanMX) carrying pAJ740 (lsg1-1). Fresh overnight cultures were diluted twofold into fresh medium. After 30 min at 30°C, each culture was divided, and one half of each shifted to 37°C for 3 h. Enlarged examples compare the localization of the Rpl25-eGFP with that of DNA stained with Hoechst 34442. For the colocalization experiments, the strains were AJY1124 (GAL1::NOG1) carrying pAJ633 (nog1-1) and AJY1167 (LSG1::KanMX) carrying pAJ740 (lsg1-1). GFP fluorescence (green) and Hoechst fluorescence (red) were visualized in separate channels, artificially colored, and merged. (B) Rpl25-eGFP was expressed in AJY734 (nmd3-4) at a permissive (30°C) or a nonpermissive (37°C) temperature for 3 h or coexpressed in CH1305 containing pAJ368 (GAL1::NMD3Δ100) (20) in the presence of the inducer galactose (gal) or in the presence of the noninducing sugar raffinose (raf). Enlarged examples of cells are shown.
For comparison, we examined the localization of Rpl25-eGFP in nmd3 mutants. We previously showed that Rpl25-GFP accumulates in the nucleus when it is coexpressed under the control of the strong GAL1 promoter with a dominant-negative nmd3 mutant that lacked the C-terminal 100 amino acids (nmd3Δ100) (20). However, we were unable to assay the localization of Rpl25-GFP in temperature-sensitive nmd3 mutants due to low signal strength. If the more sensitive Rpl25-eGFP reporter is used, nucleolar accumulation with a weaker signal in the nucleoplasm can be seen in both the temperature-sensitive nmd3-4 mutant and the dominant-negative nmd3Δ100 (Fig. 9B).
Since Lsg1p is cytoplasmic and does not shuttle in a Crm1-dependent manner, it is unlikely to be directly involved in 60S export. Nevertheless, we examined the localization of Rpl25-eGFP in lsg1 mutants. As with the nog1 mutants, Rpl25-eGFP strongly accumulated in the nucleolus in lsg1-1 and lsg1-2 mutant cells when incubated at a restrictive temperature (Fig. 9A). To examine the specificity of the Rpl25-eGFP reporter for export defects, we monitored its localization in a temperature-sensitive mutant of translation initiation factor 3, prt1-1. After a shift to a restrictive temperature Rpl25-eGFP did not accumulate in the nucleolus or nucleus (data not shown). Thus, the nucleolar accumulation of Rpl25-eGFP in nog1 and lsg1 mutants appears to be due to a specific block in subunit export, a finding consistent with the reduced levels of 60S subunits but not of 40S subunits in these mutants. Interestingly, Nmd3p does not accumulate in the nucleus in nog1 and lsg1 mutants (data not shown), suggesting that the defect in these mutants is upstream of binding the export adapter.
The effect of Lsg1p on ribosomal subunit export is likely indirect and could be explained if Lsg1p is necessary for recycling an export factor(s) that accompanies the nascent subunit to the cytoplasm. The failure to recycle such proteins would lead to their depletion from the nucleus. For example, loss of Nmd3p from the nucleus should yield a phenotype similar to that seen in a nmd3 loss-of-function mutant. Similarly, a failure to recycle Tif6p exported from the nucleus on 60S subunits results in the accumulation of Rpl25-eGFP in the nucleus (37). We tested several candidate proteins for their possible accumulation in the cytoplasm upon shifting temperature-sensitive lsg1 mutants to a restrictive temperature. GFP fusions were made with Nog1p, Nog2p, Nug1p, and Tif6p. Of these proteins, only Tif6p is found associated with the Lsg1p-bound subunit (G. Kallstrom and A. Johnson, unpublished results) and is reported to shuttle (3, 37). None of these proteins relocated to the cytoplasm or was lost from the nucleus in lsg1 mutants (data not shown). The two additional proteins, Arx1p and Ybr267wp, that have been reported to be in the Lsg1p-60S complex (12) are unlikely to be responsible for the arrest of 60S export in lsg1 mutants since neither protein is essential (unpublished results). We also examined the localization of Nmd3p in lsg1 mutants. Nmd3p is predominantly cytoplasmic in wild-type cells; however, the addition of GFP to the C terminus of Nmd3p causes a modest nuclear localization without significantly affecting function (unpublished observation). No change in the localization of Nmd3-GFP ectopically expressed from a plasmid was observed in lsg1 temperature-sensitive mutants (data not shown).
DISCUSSION
Pre-60S subunits.
Mass spectrometry has recently been used to identify proteins associated with pre-60S species (1, 9, 17, 35). From such work a dynamic pathway of association and dissociation of processing factors on the pre-60S species can be developed (9). Here, we have focused on two GTP-binding proteins, Lsg1p and Nog1p, that we identified by their copurification with the Nmd3p-bound 60S subunit. Because Nmd3p shuttles, the 60S subunits bound by Nmd3p are a mixture of nascent subunits from the nucleus and cytoplasm and mature recycling cytoplasmic subunits (18, 19).
Nog1p has been found associated with several pre-60S species (1, 9, 17, 35), indicating that it loads onto the nascent subunit quite early during 60S biogenesis in the nucleolus and remains associated with the subunit throughout the course of assembly. However, the rRNA processing defects of nog1 mutants are similar to the defects observed in mutants of some late-acting nuclear 60S biogenesis factors, including NUG1, NOG2, and TIF6 (1, 2, 35). It has been suggested that such rRNA processing defects are nonspecific and due to feedback inhibition of the processing pathway (44). For example, the failure to recycle limiting nucleolar proteins that accumulate on intermediate species could lead to defects earlier in the pathway. Consistent with this, we observed the accumulation of Rpl25-eGFP in the nucleolus in nog1 mutants, suggesting a defect in release of pre-60S from the nucleolus. Although Nog1p appears to load onto the subunit at an early stage of 60S maturation, it may not be required until later in the biogenesis pathway. Interestingly, Nmd3-GFP did not accumulate in the nucleus in nog1 mutant cells, suggesting that the subunits accumulated in the nucleolus of nog1 mutants are not competent for Nmd3p binding.
Because we found Nog1p on the Nmd3p-bound subunit, Nog1p can remain on the subunit at least until the export adapter is loaded. This is in apparent contradiction with analyses determining the protein composition of pre-60S particles affinity purified with other trans-acting factors, including Nog2p, Nsa3p, Nug1p, and Sda1p (1, 31, 35). Nmd3p was not detected in these complexes. Since Nug1p and Nog2p are present in the nucleoplasm, as well as in the nucleolus, and Sda1p is nucleoplasmic, these results have been interpreted to indicate that Nmd3p loads onto the subunit at a late step in the nucleoplasm, after release from the nucleolus. However, it is possible that Nmd3p is lost from these complexes during affinity purification due to weak initial binding to the 60S subunit and that a subsequent nucleoplasmic step stabilizes its interaction with the subunit.
The Nog1p-bound pre-60S particle contains other GTPases, including Nug1p and Nog2p (1, 35), that are also required for 60S biogenesis, indicating that multiple GTPases are required for multiple events in parallel during ribosome assembly. Further work is needed to determine where these GTPases bind on the subunit and whether the binding of any of the GTPases is mutually exclusive. Mutual exclusion or binding to the same sites would suggest sequential loading of GTPases acting at the same site. Although Lsg1p, Nug1p, and Nog2p are structurally related to one another and belong to a subfamily of GTPases in which the GTPase motifs are circularly permuted (26), Nug1 and Nog2p appear to be able to bind to the nascent subunit simultaneously (35), indicating that their binding is not mutually exclusive.
Since the steady-state distribution of Nmd3p in yeast is largely cytoplasmic (18), Nmd3p-bound 60S subunits are enriched in cytoplasmic subunits. We found that Lsg1p was an abundant protein in this complex, a finding consistent with recent mass spectrometric identification of Nmd3p associated with Lsg1p (12, 31). We did not detect Nog1p associated with the Lsg1p-bound 60S subunit, indicating that Nog1p is released by the time Lsg1p binds and that these two GTPases bind to the subunit sequentially.
Ribosome export.
Deletion of the NES of Nmd3p traps the 60S subunit reporter protein Rpl25-GFP in the nucleus (11, 20). Using the more sensitive reporter Rpl25-eGFP (11), we first examined the localization of 60S subunits in various nmd3 mutants. The conditional loss-of-function mutant nmd3-4 (18) and a C-terminal deletion of 100 amino acids (Nmd3Δ100) led to nucleolar retention of Rpl25-eGFP with some signal in the nucleoplasm. These results are consistent with studies of human Nmd3p (hNmd3); in HeLa cells GFP-hNmd3 accumulates in the nucleolus and in the nucleoplasm in the presence of LMB, whereas the truncation mutant comparable to yeast Nmd3Δ100 accumulates predominantly in the nucleolus (40a). These results suggest that Nmd3p may first bind to the nascent 60S subunit in the nucleolus. This interpretation is also consistent with the slight enrichment of 7S and 27S pre-rRNAs and the presence of Nog1p in the Nmd3p-bound subunit. Thus, the export adapter Nmd3p may be required for the release of a stable subunit from that compartment (see Fig. 10).
FIG. 10.
Simplified diagram of Nmd3p and Lsg1p binding to 60S subunits. Nmd3p loads onto the pre-60S subunit in the nucleus, possibly as the nascent subunit emerges from the nucleolus. Nmd3p remains associated with the subunit during export to the cytoplasm. Tif6p, Arx1p, Crm1(Xpo1p), and Ran(Gsp1p) also accompany the subunit during export but are not shown for clarity. In the cytoplasm, Lsg1p likely binds to the newly exported free 60S subunit, and we suggest that it is required for recycling an as-yet-unidentified factor to the nucleus. Both Nmd3p and Lsg1p are released prior to or upon subunit joining. When released, Nmd3p can reenter to the nucleus for another round of subunit export or bind to a recycling free 60S subunit after translation termination. Lsg1p, on the other hand, is restricted to the cytoplasm and, after it is released from a subunit, it can bind to a newly exported 60S subunit or to a recycling free 60S subunit as with Nmd3p.
In addition to Nmd3p, we have shown that Nog1p is required for release of nascent 60S subunits from the nucleolus. This finding is consistent with Nog1p acting late in the biogenesis pathway. Nog1p and Nmd3p can be added to a growing list of proteins, including Mak21p/Noc1p and Noc2p, that are required for the release of subunits from the nucleolus (28). How the function of these proteins is coordinated and whether they work together or sequentially to control release of the subunit is not known. Since Nmd3p provides the export signal for the large subunit, the addition of Nmd3p may provide a quality control check of subunit assembly, with additional factors contributing to such a functional check.
Considering that Lsg1p is cytoplasmic, it is surprising that lsg1 mutants trap Rpl25-eGFP in the nucleolus. However, a similar phenotype was reported recently for a ria1/efl1 mutant in which Tif6p recycling to the nucleus is inhibited, resulting in a block in the 60S maturation pathway in the nucleus (37). We suggest that the effect of lsg1 mutants on nucleolar 60S biogenesis is due to a similar failure in the recycling of a biogenesis factor(s) that remains bound to the subunit during transport from the nucleus (Fig. 10). In an attempt to identify the putative factor, we made fusions of GFP to candidate proteins, but none showed relocalization to the cytoplasm. Although a failure to reimport Nmd3p would be expected to cause nucleolar accumulation of Rpl25-eGFP, as was observed in an nmd3-4 conditional mutant, we have not observed relocalization of Nmd3-GFP in lsg1 mutants. Consequently, the factor responsible for the arrest of 60S export observed in lsg1 mutants remains to be identified.
Does Lsg1p have a role in translation?
Mutants of LSG1 clearly have a defect in 60S biogenesis and export; however, we have also provided data that Lsg1p binds to cytoplasmic free 60S subunits that are recycling in translation, suggesting an additional role in subunit recycling. Current models for eukaryotic translation focus on assembly of the 40S subunit-containing preinitiation complex at the initiation codon, with 60S joining being regulated by release of factors from the 40S subunit that prevent 60S binding. It is possible that Lsg1p triggers the release of factors from the 60S subunit in response to interaction with the preinitiation complex. Such a function could couple translation initiation with 60S subunit biogenesis.
Acknowledgments
We thank C. Nguyen for making lsg1 mutants, M. Yoshida for providing LMB, E. Hurt for providing the Rpl25-eGFP vector, and J. Ballesta for anti-Rpl12p antibody. Mass spectrometry was performed by HHMI Biopolymer Laboratory and W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. We thank N.-J. Hung and M. West for preparation of the GFP fusions and for helpful comments on the manuscript.
This work was supported by NIH grant GM53655 to A. Johnson.
REFERENCES
- 1.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 2.Basu, U., K. Si, J. R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bécam, A. M., F. Nasr, W. J. Racki, M. Zagulski, and C. J. Herbert. 2001. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol. Gen. Genet. 266:454-462. [DOI] [PubMed] [Google Scholar]
- 4.Belk, J. P., F. He, and A. Jacobson. 1999. Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA 5:1055-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dragon, F., J. E. G. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar, D. A., F. Dragon, S. J. Lee, and S. J. Baserga. 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 97:13027-13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisinger, D. P., F. A. Dick, and B. L. Trumpower. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 17:5136-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faber, A. W., M. Van Dijk, H. A. Raue, and J. C. Vos. 2002. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA 8:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 10.Gadal, O., D. Strauss, J. Braspenning, D. Hoepfner, E. Petfalski, P. Philippsen, D. Tollervey, and E. Hurt. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin, A. C., M. Bösche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, and B. Huhse. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 13.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Véronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. André, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, and D. J. Garfinkel. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, D., A. St. John, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gleizes, P. E., J. Noaillac-Depeyre, I. Léger-Silvestre, F. Teulières, J. Y. Dauxois, D. Pommet, M. C. Azum-Gelade, and N. Gas. 2001. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 155:923-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 17.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 18.Ho, J., and A. W. Johnson. 1999. NMD3. encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2389-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, J. H.-N., G. Kallstrom, and A. W. Johnson. 2000. Nascent 60S subunits enter the free pool bound by Nmd3p. RNA 6:1625-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, J. H. N., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Karl, T., K. Önder, R. Kodzius, A. Pichová, H. Wimmer, A. Thür, H. Hundsberger, M. Löffler, T. Klade, A. Beyer, M. Breitenbach, and L. Koller. 1999. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr. Genet. 34:419-429. [DOI] [PubMed] [Google Scholar]
- 23.Kranz, J. E., and C. Holm. 1990. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA 87:6629-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of assembly of ribosomal subunits in yeast. Biochem. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 25.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 26.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. r. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 30.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page, N., M. Gerard-Vincent, P. Menard, M. Beaulieu, M. Azuma, G. J. P. Dijkgraaf, H. Li, J. Marcoux, T. Nguyen, T. Dowse, A. Sdicu, and H. Bussey. 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163:875-894. [DOI] [PMC free article] [PubMed]
- 33.Park, J. H., B. C. Jensen, C. T. Kifer, and M. Parsons. 2001. A novel nucleolar G-protein conserved in eukaryotes. J. Cell Sci. 114:173-185. [DOI] [PubMed] [Google Scholar]
- 34.Russell, D. W., and L. L. Spremulli. 1979. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J. Biol. Chem. 254:8796-8800. [PubMed] [Google Scholar]
- 35.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 38.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11:3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapman, J., and R. J. Planta. 1976. Maturation of ribosomes in yeast. I. Kinetic analysis by labeling of high molecular weight rRNA species. Biochim. Biophys. Acta 442:265-274. [DOI] [PubMed] [Google Scholar]
- 40.Trapman, J., J. Retel, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 40a.Trotla, C. R., E. Lund, L. Kahan, A. W. Johnson, and J. E. Dahlberg. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 41.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412-1416. [PubMed] [Google Scholar]
- 42.Udem, S. A., and J. R. Warner. 1972. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 65:227-242. [DOI] [PubMed] [Google Scholar]
- 43.van Hoof, A., P. Lennertz, and R. Parker. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 19:1357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 45.Warner, J. R. 1971. The assembly of ribosomes in yeast. J. Biol. Chem. 246:447-454. [PubMed] [Google Scholar]
- 46.Xue, Y., X. Bai, I. Lee, G. Kallstrom, J. Ho, J. Brown, A. Stevens, and A. W. Johnson. 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 20:4006-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zanchin, N., and D. S. Goldfarb. 1999. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]