Abstract
Although the protein-only hypothesis proposes that it is the conformation of abnormal prion protein (PrPSc) that determines strain diversity, the molecular basis of strains remains to be elucidated. In the present study, we generated a series of mutations in the normal prion protein (PrPC) in which a single glutamine residue was replaced with a basic amino acid and compared their abilities to convert to PrPSc in cultured neuronal N2a58 cells infected with either the Chandler or 22L mouse-adapted scrapie strain. In mice, these strains generate PrPSc of the same sequence but different conformations, as judged by infrared spectroscopy. Substitutions at codons 97, 167, 171, and 216 generated PrPC that resisted conversion and inhibited the conversion of coexpressed wild-type PrP in both Chandler-infected and 22L-infected cells. Interestingly, substitutions at codons 185 and 218 gave strain-dependent effects. The Q185R and Q185K PrP were efficiently converted to PrPSc in Chandler-infected but not 22L-infected cells. Conversely, Q218R and Q218H PrP were converted only in 22L-infected cells. Moreover, the Q218K PrP exerted a potent inhibitory effect on the conversion of coexpressed wild-type PrP in Chandler-infected cells but had little effect on 22L-infected cells. These results show that two strains with the same PrP sequence but different conformations have differing abilities to convert the same mutated PrPC.
Transmissible spongiform encephalopathies (TSE), or prion diseases, are lethal neurodegenerative diseases that include Creutzfeldt-Jakob disease in humans, scrapie in sheep, and bovine spongiform encephalopathy in cattle. The infectious agent, termed prion, is unique in that no agent-specific nucleic acid is detectable. The protein-only hypothesis proposes that this agent consists solely of an abnormal form of prion protein (PrPSc), which is produced by the conversion of the normal cellular prion protein (PrPC) and accumulates primarily in the lymphoreticular and central nervous system during the course of prion disease (41, 56). PrPC, a host-encoded glycoprotein anchored to the cell membrane by a glycosyl-phosphatidylinositol moiety, is expressed mainly in the central nervous system. PrPC is detergent soluble and proteinase K (PK) sensitive, whereas PrPSc is detergent insoluble and partially PK resistant (35). These different biophysical properties are thought to be due to different conformations of the two isoforms. PrPC is highly α-helical, but PrPSc has a large proportion of β-sheet structure (14, 38).
Various TSE strains with distinct biological characteristics have been identified in several mammalian species. These strains are characterized by different incubation periods and histopathological changes (9, 10). Generally, the phenotypic characteristics are maintained upon repeated passages in the same species with the same PrP amino acid sequence. In addition, previous reports showed that strain-specific biological characteristics remain unchanged after passages in cell cultures (2, 8). In contrast, changing to a species with a different PrP sequence often results in the emergence of a new strain (28, 29). The existence of multiple strains signifies that the infectious agent carries some form of strain-specific information that determines each strain's characteristics. One possibility is that this information stems from the distinct PrPSc conformation of each strain. Differences in the electrophoretic mobilities of PK-resistant PrPSc core fragments among strains are well documented (7, 16, 50). These different-sized PrPSc fragments are likely a consequence of differing conformations and thus different PK cleavage points. Conformational differences in β-sheet structures between strains have also been demonstrated by infrared (IR) spectroscopy (13, 52). Furthermore, Syrian hamster (SHa) PrPSc, when denatured, binds more anti-PrP antibody than when it is in its native form, and each strain can have distinct denatured versus nondenatured binding ratios (44). In addition, some Syrian hamster TSE strains are reported to differ in the extent of their PK resistance after partial denaturation with guanidine hydrochloride (39). These findings support the hypothesis that TSE strains have distinct PrPSc conformations. Moreover, cell-free conversion experiments have shown that different forms of PrPSc can induce strain-specific conformational changes in PrPC (6), and Jones and Surewicz recently reported that artificial amyloid fibrils of PrP23-144 from different species revealed strain-like behavior in vitro (25). Nevertheless, much remains to be learned about the mechanistic relationship between PrPSc conformational differences and the molecular basis of TSE strains.
Studies using transgenic mice and congenic mice have shown that several TSE strains differ in incubation periods in the same host (11, 23, 32). The molecular basis of this remains unresolved, but the conformation of PrPSc could influence incubation periods by affecting the efficiency and location of PrPSc formation. However, to date, there are little data on the influence of PrP mutations on PrPSc formation in vitro.
Because N2a58 cells overexpressing mouse PrP can be persistently infected with the Chandler or 22L prion strain (37), we chose to examine the strain-specific effect of PrP mutations on PrPSc formation in N2a58 cell cultures infected with the Chandler or 22L strain, designated Ch-N2a58 and 22L-N2a58, respectively. Although little is known about which amino acid residues of the PrP sequence correlate with the strain-specific formation of PrPSc, we noticed that mutations from glutamine to arginine or lysine in the C terminus of the PrP were related to the resistance of prion diseases (4, 47, 57) and inhibited the conversion of coexpressed wild-type PrP in Chandler-infected N2a cell cultures (26). In this study, we created a series of PrP mutations in which a single glutamine residue was replaced with an arginine residue and compared the effects of these mutations on PrPSc formation in Ch- and 22L-N2a58 cells. We demonstrated that specific amino acids residues in PrPC can allow or inhibit PrPSc formation in vitro for one strain but not another even though the amino acid sequence of PrPSc is the same in each strain. Our results suggest that each prion strain can interact with PrPC in a strain-specific manner, producing PrPSc with a strain-specific conformation and unique biological characteristics.
MATERIALS AND METHODS
Cell culture.
N2a58 cells overexpressing mouse PrP (PrP-a genotype, codons 108L and 189T) were prepared as described previously (37). To create N2a58 cells infected with either the Chandler/RML or 22L strain (Ch-N2a58 and 22L-N2a58), the cells were incubated with brain homogenates from ddY mice infected with each strain. After subcloning by limiting dilution, several PrPSc-positive clones were isolated. The cell clones producing the highest level of PrPSc were used for subsequent study. Both Ch-N2a58 and 22L-N2a58 cells stably expressed PrPSc for over 50 passages. Morphological appearances and growth characteristics of these prion-infected cells were indistinguishable from those of N2a58 cells (data not shown). All cells were cultured in Opti-MEM (Invitrogen) containing 10% fetal bovine serum and penicillin-streptomycin at 37°C in 5% CO2 and were split every 3 to 4 days at an 8- to 10-fold dilution.
Plasmid constructions.
The open reading frames of Syrian hamster PrP (SHa), human PrP (Hu), and mouse PrP containing the epitope for the 3F4 antibody (Mo3F4) were amplified by PCR with mouse DNA, MHM2/Mo3F4 PrP transgenic mouse DNA, SHa PrP transgenic mouse (3) DNA, and human DNA, respectively. Amplified fragments were inserted into the pcDNA3.1(+) vector (Invitrogen) between the BamHI and XbaI sites. Mouse PrP (PrP-a genotype) containing the epitope for the L42 antibody (MoL42) was introduced into mouse PrP by PCR-direct mutagenesis.
Mo3F4 PrP differs from the mouse PrP-a genotype by two amino acids, L108M and V111M, which are included in the epitope recognized by the 3F4 anti-PrP monoclonal antibody (Dako). MoL42 PrP has one amino acid substitution, W144Y, which is recognized by the L42 anti-PrP antibody (R-biopharm) (54). Since neither antibody reacts with mouse PrP, transfected recombinant PrP is distinguishable from endogenous mouse PrP.
Mo3F4 sequences with specific amino acid changes (Q90R, Q97R, Q159R, Q167R, Q171R, Q185R, Q185K, Q185H, Q185E, Q185L, Q211R, Q216R, Q218R, Q218K, Q218H, Q218E, Q218L, and Q222R) were generated by PCR mutagenesis. The resulting PCR fragments were subcloned into the pcDNA3.1(+) vector. To create MoL42 mutations with specific amino acid changes (Q185R, Q218R, Q218K, and Q218H), BamHI-BstPI fragments of the corresponding Mo3F4 mutants in the pcDNA3.1(+) vector were replaced by those of MoL42 PrP. The PrP sequences of all plasmids used in this study were confirmed by using the ABI PRISM 3100 genetic analyzer (Applied Biosystems), and no unexpected mutations were found.
Transfection and Western blotting.
N2a58, Ch-N2a58, and 22L-N2a58 cells were transiently transfected with various plasmid constructs (1 or 2 μg DNA per 6-cm dish) using the Effectene transfection reagent (QIAGEN) according to the manufacturer's instructions. To evaluate dominant-negative inhibition of PrPSc formation, cotransfections of two different PrP constructs were performed with a DNA ratio of 1:1 or 1:2. Indirect immunofluorescence of PrP and fluorescence imaging of pEGFP-C1 (Clontech) revealed that transfection efficiencies were around 40 to 60% and that the rate of the overlapping expression of two plasmids cotransfected was more than 90%.
After 72 h of transfection, cells from a 6-cm dish were lysed in 0.5 ml of lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, 0.5% sodium deoxycholate, and 1 mM EDTA). After cell debris and nuclei were removed by low-speed centrifugation, the protein concentration of the supernatant was measured by the BCA protein assay (Pierce). To detect PrPSc, the protein concentration of the supernatant was adjusted with lysis buffer to 1 mg/ml. Samples of equal protein concentrations and volumes were digested with 20 μg/ml of proteinase K at 37°C for 45 min, and the digestion was stopped by adding phenylmethylsulfonyl fluoride (2 mM). After 60 min of centrifugation at 20,400 × g, the pellet was dissolved with sample buffer (4 M urea, 4% sodium dodecyl sulfate, 100 mM dithiothreitol, 10% glycerol, 0.02% bromophenol blue, and 50 mM Tris-HCl [pH 6.8]), boiled, and then loaded onto a 15% polyacrylamide gel. Proteins were transferred onto a membrane (Immobilon P; Millipore). 3F4-positive PrPs were detected with 3F4 antibody, L42-positive PrPs were detected with L42 antibody, and total PrP was detected with mouse polyclonal anti-PrP antibody (designated SS). Immunoreactive bands were visualized using the ECL detection system (Amersham Biosciences). The expression level of transfected PrP in cell lysates (30 μg of total protein per lane) was also estimated by immunoblotting. Densitometric analysis of the film was performed using NIH Image software. The conversion efficiency of Mo3F4 was assigned as 100%, and the level of PrPSc formation in each 3F4-positive mutant was calculated relative to this value. In some experiments, the cell lysates with proteinase K treatment were digested with PNGase F (New England Biolabs).
IR spectroscopy of PrPSc.
PrPSc was isolated from the brains of mice affected by either 22L, Chandler, or 87V scrapie and treated with PK as described previously (13). For IR analysis, ∼3 μl of pelleted slurries containing at least 10 mg/ml PrPSc in a solution containing 20 mM sodium phosphate, 130 mM NaCl (pH 7.5), and 0.5% sulfobetaine was applied to a Golden Gate Single Reflection Diamond Attenuated Total Reflectance unit purged with dry air and covered to prevent sample evaporation. Data collection was performed using a System 2000 IR instrument (Perkin-Elmer). Test conditions were as follows: 20°C, 4.00-cm−1 resolution, 2-cm/s optical path difference velocity, 1,000 scans, and 0.5-cm−1 data interval. The detector was an nb1 MCT detector cooled by liquid nitrogen. Primary spectra were obtained by subtracting the spectra of the corresponding buffer or buffer with additives and water vapor and by adjusting the baseline and normalizing for comparable absorbance of different concentrations of PrP. Second-derivative spectra were calculated from the primary spectra using 13 data points. The software used for spectral analyses was Spectrum v2.00 (Perkin-Elmer).
RESULTS
Mo3F4 PrP converts to PrPSc with similar efficiency in Ch-N2a58 and 22L-N2a58 cells.
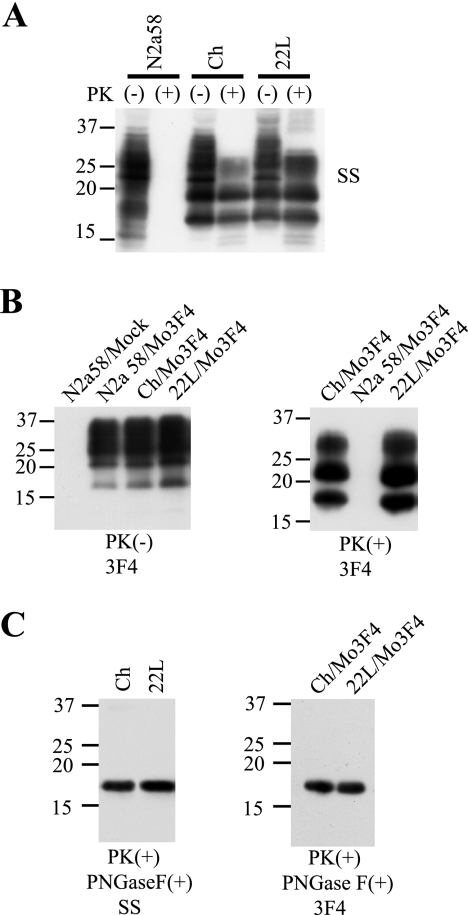
Prior to creating PrP mutants, we first confirmed that our starting Mo3F4 vector could convert to PrPSc in cells persistently infected with the Chandler or 22L mouse-adapted scrapie strain (Ch-N2a58 and 22L-N2a58 cells, respectively). The presence of endogenous mouse PrPSc in the Ch-N2a58 and 22L-N2a58 cells was confirmed by immunoblotting with the mouse polyclonal anti-PrP SS antibody. Similar amounts of endogenous mouse PrPSc accumulated in Ch-N2a58 and 22L-N2a58 cells, while none was detected in uninfected N2a58 cells (Fig. 1A). In the transfected cells, PK-resistant PrPSc derived from Mo3F4 PrP was distinguished from endogenous mouse PrPSc by immunoblotting with the monoclonal anti-PrP 3F4 antibody (Fig. 1B). PK-resistant PrPSc core fragments from Ch-N2a58 and 22L-N2a58 cells were treated with PNGase F to remove asparagine-linked glycosylation and immunoblotted with either SS or 3F4 antibody. No differences in gel migration patterns were seen (Fig. 1C).
FIG. 1.
Formation of Mo3F4-derived PrPSc in Ch-N2a58 and 22L-N2a58 cells. (A) Western blot using polyclonal anti-PrP antibody SS in N2a58, Ch-N2a58 (Ch), and 22L-N2a58 (22L) cells without (−) or with (+) PK treatment. (B) Expression levels of Mo3F4 PrP (left panel) and detection of Mo3F4-derived PrPSc (right panel) were measured by Western blot using monoclonal antibody 3F4. Mock, untransfected cells. (C) After consecutive treatments of PK and PNGase F, untransfected (left panel) and Mo3F4 PrP-transfected cells (right panel) were analyzed by Western blotting using SS and 3F4 antibodies, respectively. Molecular mass markers are indicated in kilodaltons on the left side of each panel.
Q218K PrP does not convert and inhibits PrPSc formation from coexpressed Mo3F4 PrP in Ch-N2a58 but not in 22L-N2a58 cells.
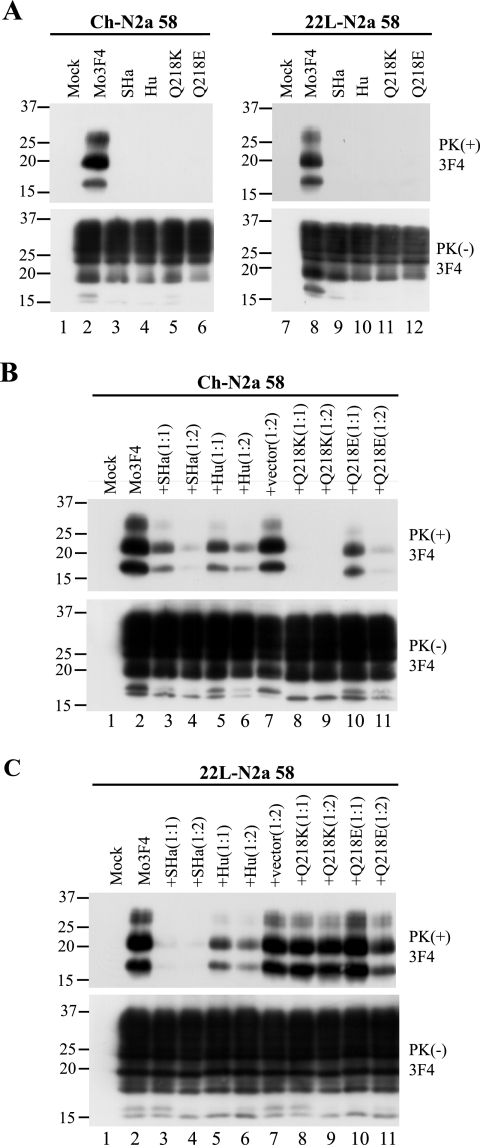
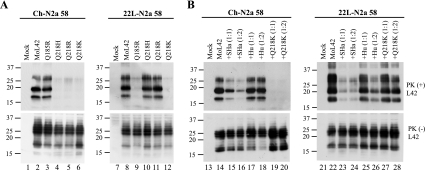
In order to compare the consequences of changes in the PrP primary structure between Ch-N2a58 and 22L-N2a58 cells, we first examined the PrPSc formation of two heterologous PrPs, Syrian hamster (SHa) and human (Hu) PrPs, and two Mo3F4 mutated PrPs with a single amino acid substitution at codon 218, Q218K and Q218E, in the infected cells transfected with the corresponding expression vectors. The 3F4 antibody detected SHa, Hu, and Mo3F4 mutated PrPs expressed in Ch-N2a58 and 22L-N2a58 cells at a level similar to that of wild-type Mo3F4 PrP (Fig. 2A, lower panels). SHa, Hu, Q218K, and Q218E PrP did not convert to PrPSc in Ch-N2a58 (Fig. 2A, upper panel, lanes 3 to 6) and 22L-N2a58 (Fig. 2A, upper panel, lanes 9 to 12) cells.
FIG. 2.
Strain-dependent inhibitory effect of Q218K mutation on PrPSc formation of wild-type Mo3F4. (A) Conversion to 3F4-positive PrPSc (upper panels) and expression of Mo3F4, SHa, Hu, Q218K, and Q218E (lower panels) were measured by Western blot using 3F4 antibody. The 3F4 epitope was present in all these constructs. (B and C) The inhibitory effect of the constructs was determined by cotransfection with Mo3F4 in the DNA ratio of 1:1 or 1:2. The blots were probed with 3F4 antibody. Mock, untransfected cells; +vector(1:2), cotransfection of Mo3F4 and pcDNA3.1(+) at a 1:2 ratio.
To evaluate the inhibitory effect of these heterologous and mutated PrPs, each expression vector was cotransfected with that of Mo3F4 PrP at a DNA ratio of 1:1 or 1:2. As seen previously (26), in Ch-N2a58 cells, Q218K PrP completely inhibited the accumulation of PrPSc derived from Mo3F4 PrP even at a DNA ratio of 1:1 (Fig. 2B, upper panel, lanes 8 and 9). SHa, Hu, and Q218E PrP also revealed a dose-dependent inhibitory effect but to a lesser extent (Fig. 2B, upper panel, lanes 3 to 6, 10, and 11). In remarkable contrast, in 22L-N2a58 cells, Q218K PrP had little effect on Mo3F4 PrPSc accumulation (Fig. 2C, upper panel, lanes 8 and 9). The inhibitory effect of Q218E PrP in 22L-N2a58 cells was also very weak (Fig. 2C, upper panel, lanes 10 and 11). Conversely, SHa PrP inhibited Mo3F4-derived PrPSc formation to a greater extent in 22L-N2a58 cells than in Ch-N2a58 cells (Fig. 2C, upper panel, lanes 3 and 4). These results were reproduced in three independent experiments (Table 1). These data suggest that the inhibition of PrPSc formation by some of these PrP molecules, especially Q218K, is strain specific.
TABLE 1.
Summary of results
| Mutations | Chandler
|
22L
|
||
|---|---|---|---|---|
| Conversiona | Inhibitionb | Conversiona | Inhibitionb | |
| SHa | − | + | − | 2+ |
| Hu | − | + | − | + |
| Q90R | + | NA | + | NA |
| Q97R | − | 2+ | − | 2+ |
| Q159R | + | NA | + | NA |
| Q167R | − | 2+ | − | + |
| Q171R | − | 2+ | − | 2+ |
| Q185R | + | NA | − | 2+ |
| Q185K | 2+ | NA | − | 2+ |
| Q185H | + | NA | +/− | + |
| Q185E | − | + | − | +/− |
| Q185L | +/− | + | +/− | + |
| Q211R | + | NA | + | NA |
| Q216R | − | + | − | 2+ |
| Q218R | − | 2+ | + | NA |
| Q218K | − | 2+ | − | − |
| Q218H | − | 2+ | + | NA |
| Q218E | − | + | +/− | − |
| Q218L | +/− | + | + | NA |
| Q222R | + | NA | + | NA |
PrPSc formation of each 3F4-positive construct was quantified by densitometric analysis. The percent conversion of Mo3F4 was assigned as 100%, and the relative scores compared with Mo3F4 are shown. 2+ indicates >200%; + indicates 50 to 200%; +/− indicates 10 to 50%; − indicates <10%. Each value represents the mean of two or three replicates.
Relative inhibitory effect on PrPSc formation of conversion-defective mutated PrPs was assessed. 2+ indicates >80% inhibition of Mo3F4 PrPSc formation (in the DNA ratio of 1:1); + indicates 50 to 80% inhibition (1:1); +/− indicates <50% inhibition (1:1) and >50% inhibition (1:2); − indicates <50% inhibition (1:1 and 1:2). Each value represents the mean of two or three replicates. NA, not applicable.
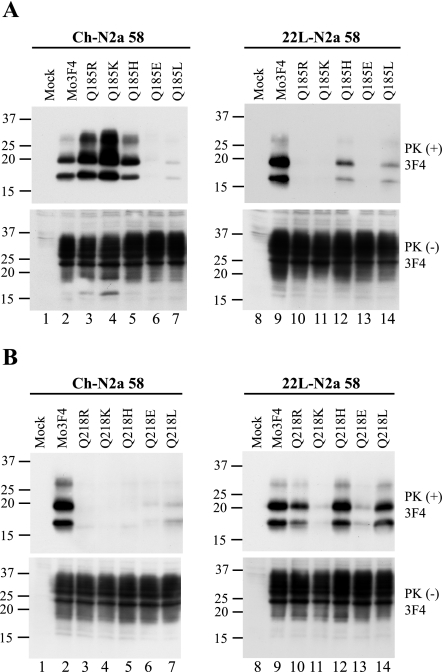
Q185R PrP converts to PrPSc in Ch-N2a58 but not 22L-N2a58 cells.
To further analyze the strain-specific effect of PrP mutations on PrPSc formation in Ch-N2a58 and 22L-N2a58 cells, we generated nine 3F4-positive mutated PrPs with a single arginine substitution for each glutamine residue in the C-terminal half and examined their conversion efficiencies in the infected cells (see Fig. 6). The Q90R, Q159R, Q211R, and Q222R PrPs readily converted to PrPSc in Ch-N2a58 and 22L-N2a58 cells (Fig. 3A, upper panels), whereas the Q97R, Q167R, Q171R, and Q216R PrPs failed to convert in both cell lines (Fig. 3A, upper panels). These conversion-defective mutated PrPs potently inhibited the accumulation of wild-type Mo3F4-derived PrPSc in both cell lines (Fig. 3B, upper panels). Interestingly, Q185R PrP efficiently converted in Ch-N2a58 cells but not in 22L-N2a58 cells (Fig. 3A), where it actually had an inhibitory effect (Fig. 3B, upper panel, and Table 1).
FIG. 6.
Amino acid sequence of Mo3F4 and the position of mutations. The amino acid residue number is based on Mo3F4 PrP. The secondary structures in mouse PrPC are indicated in white boxes at the top. Boxed residues indicate the representative mutations tested; + indicates that conversion occurred in the two cells; − indicates that conversion did not occur in the two cells; “C” indicates Chandler-specific conversion; “L” indicates 22L-specific conversion.
FIG. 3.
Strain-specific effects of Q185R mutation on PrPSc formation. (A) Conversion to 3F4-positive PrPSc (upper panels) and expression of Mo3F4, Q90R, Q97R, Q159R, Q167R, Q171R, Q185R, Q211R, Q216R, and Q222R (lower panels) were measured by Western blotting using 3F4 antibody. The 3F4 epitope was present in all these constructs. (B) Inhibitory effects of constructs that did not convert were determined by cotransfection with Mo3F4 at a 1:1 or 1:2 DNA ratio. The blots were probed with 3F4 antibody.
Substitutions of various amino acid species at codons 185 and 218 differentially affect PrPSc formation between Ch- and 22L-N2a58 cells.
To further examine the effect of amino acid substitutions at codons 185 and 218, where strain-specific effects were observed, we replaced each glutamine residue (Q) with various amino acid species, including basic amino acids (R, K, and H), an acidic amino acid (E), and a hydrophobic amino acid (L). As shown in Fig. 4A and Table 1, Q185K, Q185H, and Q185R PrP readily converted to PrPSc in Ch-N2a58 cells. Interestingly, the amount of Q185K-derived PrPSc accumulation in Ch-N2a58 cells was higher than that of Mo3F4-derived PrPSc, suggesting a more efficient conversion of these mutated PrPs in the cells. In contrast, in 22L-N2a58 cells, little PrPSc derived from Q185R, Q185K, and Q185H PrP accumulated. Q185E and Q185L PrP minimally converted to PrPSc in both Ch- and 22L-N2a58 cells.
FIG. 4.
Strain-specific PrPSc formation of Q185R, Q185K, Q218R, and Q218H mutated PrP. (A) Conversion to 3F4-positive PrPSc (upper panels) and expression of Mo3F4, Q185R, Q185K, Q185H, Q185E, and Q185L (lower panels) were measured by Western blot using 3F4 antibody. The 3F4 epitope was present in all these constructs. (B) Western blotting of Mo3F4, Q218R, Q218K, Q218H, Q218E, and Q218L was done as in A.
The introduction of substitutions at codon 218, including basic amino acids (R, K, and H), resulted in the loss of conversion in Ch-N2a58 cells (Fig. 4B). Conversely, in 22L-N2a58 cells, Q218R, Q218H, and Q218L efficiently converted to PrPSc. Q218K PrP did not convert in either cell line (Fig. 4B) and failed to inhibit wild-type Mo3F4 PrPSc formation (Fig. 2C).
To determine whether different cellular localizations of the mutated PrPs might be the cause of the different conversion effects, we examined the mutated PrPs with indirect immunofluorescence using the 3F4 antibody. Mo3F4, Q185R, Q218R, and Q218K all localized to the cell surface of Ch-N2a58 and 22L-N2a58 cells (data not shown). In addition, phosphatidylinositol-specific phospholipase C treatment removed the mutated PrPs from the cell surface (data not shown). These results demonstrate that the localization of the mutated PrPs cannot account for their strain-specific properties.
Strain-specific properties of the PrP mutations are independent of antibody epitopes.
To assess whether the 3F4 epitope can influence the strain-specific properties of the mutated PrPs, we replaced the 3F4 epitope with the L42 epitope (W144Y), because others have previously shown that MoL42 PrP, like Mo3F4 PrP, readily converted to PrPSc in ScN2a cells (55). Expression of the L42-positive PrPs, MoL42, Q185R, Q218H, Q218R, and Q218K PrPs, was confirmed by Western blotting using the L42 antibody (Fig. 5A, lower panels). The conversion efficiencies of these L42-positive mutated PrPs were similar to those of 3F4-positive mutated PrPs (Fig. 5A, upper panels). Moreover, as shown in Fig. 5B, Q218K PrP again showed strain-dependent effects on MoL42-derived PrPSc. The data shown in Fig. 5 indicate that changing from a 3F4 epitope to an L42 epitope in the mutant PrPs does not significantly affect their strain-specific effects on PrPSc formation.
FIG. 5.
Strain-specific effects of L42-positive mutated PrPs on PrPSc formation. (A) Conversion to L42-positive PrPSc (upper panels) and expression of MoL42, Q185R, Q218H, Q218R, and Q218K (lower panels) were measured by Western blot using L42 antibody. The L42 epitope was present in all these constructs. (B) Inhibitory effects of SHa, Hu, and Q218K were determined by cotransfection with MoL42 at a 1:1 or 1:2 DNA ratio. The blots were probed with the L42 antibody. Molecular mass markers are indicated in kilodaltons on the left side of each panel.
22L and Chandler PrPSc have different conformations by IR spectra.
To assess whether there are any detectable differences in structure between 22L and Chandler PrPSc, we performed IR spectroscopy. The amide I region (1,600 to 1,700 cm−1) of protein IR spectra is sensitive to differences in protein secondary structure. Although it is difficult to make complete and unequivocal assignments of IR amide I bands, predominantly α-helical and β-sheet proteins have absorption maxima of 1,653 to 1,657 cm−1 and 1,615 to 1,640 cm−1, respectively, in water-based (as opposed to D2O-based) media (see Fig. 7). Unfolded or random-coil polypeptides tend to have absorbance maxima near 1,645 to 1,650 cm−1, and turn structures tend to absorb between 1,660 and 1,680 cm−1. Absorbance maxima are represented by negative deflections in the second-derivative spectra shown in Fig. 7. Previous studies have shown that the infrared spectrum of PrPSc of different hamster TSE strains can vary markedly despite being composed of PrP molecules of the same amino acid sequence (13, 52). Consistent with that theme, PK-treated PrPSc isolated from the brains of mice with 22L and Chandler scrapie differed in the IR spectral region commonly ascribed to the β-sheet region (see Fig. 7). For comparison, PrPSc from another mouse scrapie strain, 87V, is also shown to have a distinct infrared spectrum in the β-sheet region. In contrast, hemoglobin, a highly α-helical protein, has very little absorbance in the β-sheet region. These results provide direct spectroscopic evidence for differences in conformation between 22L, Chandler, and 87V PrPSc.
FIG. 7.
Comparison of 22L, Chandler, and 87V PrPSc by infrared spectroscopy. Second-derivative Fourier transform IR spectra are shown for PK-treated PrPSc samples isolated from the brains of mice affected by either 22L, Chandler, or 87V scrapie. Spectral differences, especially in the β-sheet region of the spectrum, provide evidence that PrPSc proteins associated with these murine-adapted scrapie strains have distinct conformations. For comparison, a highly α-helical protein, hemoglobin, gives strong absorbance (represented by a negative deflection) at ∼1,657 cm−1, with only minor absorbance in the β-sheet region. Similar results were obtained from at least two independent preparations of each strain of PrPSc.
DISCUSSION
In this study, we found evidence that TSE strain characteristics depend on their conformation. We showed that substitutions at codons 185 and 218 resulted in strain-specific PrPSc formation in cultured neuronal cells infected with two mouse-adapted scrapie prion strains, Chandler and 22L. While others previously demonstrated conformational differences between strains (13, 39, 44, 52), and some strain-specific differences in conformation have been observed in cell-free conversions (6), synthetic amyloid fibrils (25), and purified recombinant Saccharomyces cerevisiae Sup35 (31, 49), our results are the first to be obtained from a cell culture comparison of strain effects on the conversion of a panel of mutant PrPc molecules.
The amino acid sequence of PrP can certainly influence the efficiency of transmission of the infectious agent to a new host species (45), but this “species barrier” cannot be explained on the basis of sequence heterogeneity alone. Our results demonstrate that TSE strains with the same sequence have various abilities to convert the PrPc mutants at codons 185 and 218, implying a sequence-independent cause of strain specificity. Although the most efficient conversions are expected to occur between PrPc and PrPSc with identical sequences, our Q185K mutation promoted PrPSc formation in Ch-N2a58 cells at a rate higher than that of the homologous wild-type PrPc (Table 1), indicating that sequence homology between PrPc and PrPSc does not necessarily guarantee the most efficient PrPSc formation.
The locations of residues 185 and 218 within the secondary structure of PrP may explain why mutations at these sites revealed strain-specific differences in conversion. The nuclear magnetic resonance structure of mouse PrP contains three α-helices comprised of residues 144 to 154, 175 to 193, and 200 to 219; two β-strands containing residues 128 to 131 and 161 to 164; and a disulfide bridge between C178 and C213, linking helices 2 and 3 (42). Amino acid 185 is in helix 2, and residue 218 is in the C-terminal portion of helix 3 (Fig. 6). Helices 2 and 3 and their disulfide bridge are crucial for PrPSc formation (22, 36), and many point mutations associated with familial human prion diseases are located within or adjacent to these two helices (41). One such mutation, D178N, is seen in two clinicopathologically distinct diseases, fatal familial insomnia and Creutzfeldt-Jakob disease, the phenotype being determined by the methionine-valine polymorphism at codon 129 of the same mutant allele (21). The striking influence of codon 129 on the D178N mutation phenotype suggests that there may be a modifiable electrostatic interaction or hydrogen bonding between residues 129 and 178 in human PrP (1, 43). Of note, anti-PrP antibody binding studies have revealed that the main conformational differences between PrPc and PrPSc actually lie toward the N-terminal region in residues 90 to 120, while the C-terminal regions, including helices 2 and 3, remain accessible to antibody in both forms of PrP, implying that their conformation is not significantly altered during conversion (40). This is also consistent with the maintenance of significant α-helical secondary structure content in PrPSc (13, 14, 38). In addition, a conformation-dependent immunoassay has localized the primary structural differences among PrPSc strains to their N termini (44). Such observations suggest that helices 2 and 3 may be involved in intra- or intermolecular interactions with the N-terminal domain during PrPSc formation and may influence the ultimate conformational change of the N terminus, perhaps through an altered β-sheet structure. In keeping with this, our IR spectra detected a difference in β-sheet structures between 22L-PrPSc and Chandler-PrPSc (Fig. 7). If these distinct N-terminal domains had differing interactions with helices 2 and 3, particularly around residues 185 and 218, then our mutations may have created structural changes that were compatible with only one of the strains. For example, the introduction of Q185R into helix 2 of mouse PrPc may have interfered with the conformational change of its N-terminal domain into 22L-PrPSc via steric hindrance and/or electrostatic incompatibility while still allowing its conversion into Chandler-PrPSc. These strain-specific interactions between the N-terminal domain and helices 2 and 3 are likely quite localized, as mutations at other sites did not reveal any strain differences.
In addition to the location of the mutant residues, the nature of their amino acid change may have contributed to our observations. Similar mutations have been studied to a great extent in yeast, where the translation termination factor Sup35 can aggregate and self-propagate in a prion-like manner. The introduction of point mutations into Sup35 often prevents its aggregation and can block the phenotype of cells that contain aggregated Sup35 (the yeast prion state or [PSI]) in a dominant inhibitory fashion. Interestingly, random mutagenesis of Sup35 has revealed that most of these mutants have a glutamine or serine replaced with an arginine (18). Likewise, our PrP mutants, which contained an arginine instead of a glutamine, frequently did not convert and also inhibited the conversion of wild-type PrP. The large, charged arginine side chain most likely has a disruptive effect on the protein-protein interactions that are necessary for aggregation and/or conversion.
Another interesting relationship between our study and those of Sup35 is that in both settings, select mutants have shown strain-specific conversion or aggregation behaviors. When Sup35 aggregates, various levels of translation through premature stop codons can occur, which produces different [PSI] phenotypes (53, 58). [PSI] strains are heritable and have distinct biological properties that can be propagated in the same yeast genetic background (20). A few Sup35 mutants that displayed different levels of dominant inhibition of [PSI], depending on the variant to which they were exposed, have been discovered (19, 30), just as our codon 185 and 218 mutants showed different conversion rates depending on the PrPSc strain to which they were exposed. The analogous results suggest that various prion types may share similar strain determinants.
A second possibility that could account for the strain-specific properties of our PrP mutations is that the alterations affected interactions between PrP and a strain-specific agent or a host factor. The heterodimer model of the protein-only hypothesis suggests that an as-yet-unidentified host factor, protein X, is responsible for the behavior of a number of dominant inhibitory forms of PrPc (26, 51). Interestingly, codon 218 is one of the proposed binding sites for protein X; therefore, a mutation at this site should result in similar conversion rates in cells from the same line, which would have the same protein X. However, in our study, there was a dramatic difference in Q218R and Q218H mutant PrPSc formation in the same cell line infected with either the Chandler or the 22L strain. Moreover, we and others (55) have shown that PrP mutations with inhibitory effects on conversion are not restricted to the proposed protein X binding sites. There are several previous studies that demonstrated the importance of sulfated glycosaminoglycans (5, 12, 46, 48, 59) and the laminin receptor in PrPSc formation (33), and more recently, in vitro PrPSc formation experiments using brain homogenates revealed that host-encoded RNA molecules facilitated PrPSc formation (17). However, to fully explain how the same mutant PrPc can convert differently when exposed to two PrPSc strains, any invoked factor must be associated with the strain itself. For example, the virus or virino hypothesis (15, 27, 34) proposes that agent-encoded nucleic acids are the true determinants of strain diversity. Unfortunately, evidence for such nucleic acids is lacking.
It should be noted that the strain-specific effects were not related to cloning artifacts or the influence of introduced epitopes. Our results were reproduced in other clones and mass cultures prior to cloning (data not shown). In addition, changing the epitope tag from 3F4 to L42 in the mutated PrPs did not affect the strain-specific effects on PrPSc formation (Fig. 5). This indicated that the properties observed were due only to the codon substitutions.
The best explanation for our data lies with the seeding model hypothesis, which proposes that mutated PrPc, which is unable to convert, forms a heteropolymer with wild-type PrPc and PrPSc, which prevents the conversion of both wild-type and mutated PrPc. Cell-free conversion studies with purified mouse and hamster PrP isoforms have revealed that heterologous PrPc, which itself cannot convert, can directly interfere with the conversion of homologous PrPc into PrPSc. Furthermore, mouse PrPc can form heteropolymers with hamster PrPc and PrPSc and vice versa (24).
In conclusion, we have shown that mutations at codons 185 and 218 in mouse PrPc reveal strain-specific effects on PrPSc formation in cell culture. The conversion differences and IR data suggest that distinct conformations underlie the characteristics of these strains, although the presence of an unidentified strain-specific cofactor cannot be excluded. Further study of these mutants may lead to a better understanding of the structure of PrPSc and the process by which it is formed. This, in turn, will help advance the knowledge of the molecular basis of TSE strains.
Acknowledgments
We thank Nobuhiko Okimura for technical assistance, Tsuyoshi Mori for support of indirect immunofluorescence of PrP, and Gregory Raymond for providing brain-derived PrPSc.
This work was supported by the 21st Century COE Program of Nagasaki University and grants from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and the Ministry of Health, Labor, and Welfare, Japan. V.L.S. acknowledges support from the Alberta Heritage Foundation for Medical Research through a clinical fellowship award.
REFERENCES
- 1.Alonso, D. O., S. J. DeArmond, F. E. Cohen, and V. Daggett. 2001. Mapping the early steps in the pH-induced conformational conversion of the prion protein. Proc. Natl. Acad. Sci. USA 98:2985-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arima, K., N. Nishida, S. Sakaguchi, K. Shigematsu, R. Atarashi, N. Yamaguchi, D. Yoshikawa, J. Yoon, K. Watanabe, N. Kobayashi, S. Mouillet-Richard, S. Lehmann, and S. Katamine. 2005. Biological and biochemical characteristics of prion strains conserved in persistently infected cell cultures. J. Virol. 79:7104-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi, R., N. Nishida, K. Shigematsu, S. Goto, T. Kondo, S. Sakaguchi, and S. Katamine. 2003. Deletion of N-terminal residues 23-88 from prion protein (PrP) abrogates the potential to rescue PrP-deficient mice from PrP-like protein/Doppel-induced neurodegeneration. J. Biol. Chem. 278:28944-28949. [DOI] [PubMed] [Google Scholar]
- 4.Belt, P. B., I. H. Muileman, B. E. Schreuder, R. Bos-de Ruijter, A. L. Gielkens, and M. A. Smits. 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 76:509-517. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Zaken, O., S. Tzaban, Y. Tal, L. Horonchik, J. D. Esko, I. Vlodavsky, and A. Taraboulos. 2003. Cellular heparan sulfate participates in the metabolism of prions. J. Biol. Chem. 278:40041-40049. [DOI] [PubMed] [Google Scholar]
- 6.Bessen, R. A., D. A. Kocisko, G. J. Raymond, S. Nandan, P. T. Lansbury, and B. Caughey. 1995. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 375:698-700. [DOI] [PubMed] [Google Scholar]
- 7.Bessen, R. A., and R. F. Marsh. 1992. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 66:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkett, C. R., R. M. Hennion, D. A. Bembridge, M. C. Clarke, A. Chree, M. E. Bruce, and C. J. Bostock. 2001. Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J. 20:3351-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruce, M. E. 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49:822-838. [DOI] [PubMed] [Google Scholar]
- 10.Bruce, M. E. 2003. TSE strain variation. Br. Med. Bull. 66:99-108. [DOI] [PubMed] [Google Scholar]
- 11.Bruce, M. E., I. McConnell, H. Fraser, and A. G. Dickinson. 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72:595-603. [DOI] [PubMed] [Google Scholar]
- 12.Caughey, B., and G. J. Raymond. 1993. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 67:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caughey, B., G. J. Raymond, and R. A. Bessen. 1998. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J. Biol. Chem. 273:32230-32235. [DOI] [PubMed] [Google Scholar]
- 14.Caughey, B. W., A. Dong, K. S. Bhat, D. Ernst, S. F. Hayes, and W. S. Caughey. 1991. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry 30:7672-7680. [DOI] [PubMed] [Google Scholar]
- 15.Chesebro, B. 2003. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull. 66:1-20. [DOI] [PubMed] [Google Scholar]
- 16.Collinge, J., K. C. Sidle, J. Meads, J. Ironside, and A. F. Hill. 1996. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383:685-690. [DOI] [PubMed] [Google Scholar]
- 17.Deleault, N. R., R. W. Lucassen, and S. Supattapone. 2003. RNA molecules stimulate prion protein conversion. Nature 425:717-720. [DOI] [PubMed] [Google Scholar]
- 18.DePace, A. H., A. Santoso, P. Hillner, and J. S. Weissman. 1998. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93:1241-1252. [DOI] [PubMed] [Google Scholar]
- 19.Derkatch, I. L., M. E. Bradley, P. Zhou, and S. W. Liebman. 1999. The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI+] prion in yeast. Curr. Genet. 35:59-67. [DOI] [PubMed] [Google Scholar]
- 20.Derkatch, I. L., Y. O. Chernoff, V. V. Kushnirov, S. G. Inge-Vechtomov, and S. W. Liebman. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldfarb, L. G., R. B. Petersen, M. Tabaton, P. Brown, A. C. LeBlanc, P. Montagna, P. Cortelli, J. Julien, C. Vital, W. W. Pendelbury, M. Haltia, P. R. Wills, J. J. Hauw, P. E. McKeever, L. Monari, B. Schrank, G. D. Swergold, L. Autilio-Gambetti, D. C. Gajdusek, E. Lugaresi, and P. Gambetti. 1992. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: disease phenotype determined by a DNA polymorphism. Science 258:806-808. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann, L. M., and B. Caughey. 1998. The importance of the disulfide bond in prion protein conversion. Neuroreport 9:2457-2461. [DOI] [PubMed] [Google Scholar]
- 23.Hill, A. F., M. Desbruslais, S. Joiner, K. C. Sidle, I. Gowland, J. Collinge, L. J. Doey, and P. Lantos. 1997. The same prion strain causes vCJD and BSE. Nature 389:448-450, 526. [DOI] [PubMed] [Google Scholar]
- 24.Horiuchi, M., S. A. Priola, J. Chabry, and B. Caughey. 2000. Interactions between heterologous forms of prion protein: binding, inhibition of conversion, and species barriers. Proc. Natl. Acad. Sci. USA 97:5836-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, E. M., and W. K. Surewicz. 2005. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell 121:63-72. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko, K., L. Zulianello, M. Scott, C. M. Cooper, A. C. Wallace, T. L. James, F. E. Cohen, and S. B. Prusiner. 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. USA 94:10069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimberlin, R. H. 1982. Scrapie agent: prions or virions? Nature 297:107-108. [DOI] [PubMed] [Google Scholar]
- 28.Kimberlin, R. H., S. Cole, and C. A. Walker. 1987. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J. Gen. Virol. 68:1875-1881. [DOI] [PubMed] [Google Scholar]
- 29.Kimberlin, R. H., C. A. Walker, and H. Fraser. 1989. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J. Gen. Virol. 70:2017-2025. [DOI] [PubMed] [Google Scholar]
- 30.King, C. Y. 2001. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol. 307:1247-1260. [DOI] [PubMed] [Google Scholar]
- 31.King, C. Y., and R. Diaz-Avalos. 2004. Protein-only transmission of three yeast prion strains. Nature 428:319-323. [DOI] [PubMed] [Google Scholar]
- 32.Korth, C., K. Kaneko, D. Groth, N. Heye, G. Telling, J. Mastrianni, P. Parchi, P. Gambetti, R. Will, J. Ironside, C. Heinrich, P. Tremblay, S. J. DeArmond, and S. B. Prusiner. 2003. Abbreviated incubation times for human prions in mice expressing a chimeric mouse-human prion protein transgene. Proc. Natl. Acad. Sci. USA 100:4784-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leucht, C., S. Simoneau, C. Rey, K. Vana, R. Rieger, C. I. Lasmezas, and S. Weiss. 2003. The 37 kDa/67 kDa laminin receptor is required for PrP(Sc) propagation in scrapie-infected neuronal cells. EMBO Rep. 4:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manuelidis, L. 2003. Transmissible encephalopathies: speculations and realities. Viral Immunol. 16:123-139. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, R. K., M. P. McKinley, K. A. Bowman, M. B. Braunfeld, R. A. Barry, and S. B. Prusiner. 1986. Separation and properties of cellular and scrapie prion proteins. Proc. Natl. Acad. Sci. USA 83:2310-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muramoto, T., M. Scott, F. E. Cohen, and S. B. Prusiner. 1996. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc. Natl. Acad. Sci. USA 93:15457-15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishida, N., D. A. Harris, D. Vilette, H. Laude, Y. Frobert, J. Grassi, D. Casanova, O. Milhavet, and S. Lehmann. 2000. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 74:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan, K. M., M. Baldwin, J. Nguyen, M. Gasset, A. Serban, D. Groth, I. Mehlhorn, Z. Huang, R. J. Fletterick, F. E. Cohen, and S. B. Prusiner. 1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 90:10962-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peretz, D., M. R. Scott, D. Groth, R. A. Williamson, D. R. Burton, F. E. Cohen, and S. B. Prusiner. 2001. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 10:854-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peretz, D., R. A. Williamson, Y. Matsunaga, H. Serban, C. Pinilla, R. B. Bastidas, R. Rozenshteyn, T. L. James, R. A. Houghten, F. E. Cohen, S. B. Prusiner, and D. R. Burton. 1997. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J. Mol. Biol. 273:614-622. [DOI] [PubMed] [Google Scholar]
- 41.Prusiner, S. B., M. R. Scott, S. J. DeArmond, and F. E. Cohen. 1998. Prion protein biology. Cell 93:337-348. [DOI] [PubMed] [Google Scholar]
- 42.Riek, R., S. Hornemann, G. Wider, M. Billeter, R. Glockshuber, and K. Wuthrich. 1996. NMR structure of the mouse prion protein domain PrP(121-321). Nature 382:180-182. [DOI] [PubMed] [Google Scholar]
- 43.Riek, R., G. Wider, M. Billeter, S. Hornemann, R. Glockshuber, and K. Wuthrich. 1998. Prion protein NMR structure and familial human spongiform encephalopathies. Proc. Natl. Acad. Sci. USA 95:11667-11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Safar, J., H. Wille, V. Itri, D. Groth, H. Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4:1157-1165. [DOI] [PubMed] [Google Scholar]
- 45.Scott, M., D. Foster, C. Mirenda, D. Serban, F. Coufal, M. Walchli, M. Torchia, D. Groth, G. Carlson, S. J. DeArmond, D. Westaway, and S. B. Prusiner. 1989. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59:847-857. [DOI] [PubMed] [Google Scholar]
- 46.Shaked, G. M., Z. Meiner, I. Avraham, A. Taraboulos, and R. Gabizon. 2001. Reconstitution of prion infectivity from solubilized protease-resistant PrP and nonprotein components of prion rods. J. Biol. Chem. 276:14324-14328. [DOI] [PubMed] [Google Scholar]
- 47.Shibuya, S., J. Higuchi, R. W. Shin, J. Tateishi, and T. Kitamoto. 1998. Codon 219 Lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 43:826-828. [DOI] [PubMed] [Google Scholar]
- 48.Snow, A. D., R. Kisilevsky, J. Willmer, S. B. Prusiner, and S. J. DeArmond. 1989. Sulfated glycosaminoglycans in amyloid plaques of prion diseases. Acta Neuropathol. (Berlin) 77:337-342. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka, M., P. Chien, N. Naber, R. Cooke, and J. S. Weissman. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428:323-328. [DOI] [PubMed] [Google Scholar]
- 50.Telling, G. C., P. Parchi, S. J. DeArmond, P. Cortelli, P. Montagna, R. Gabizon, J. Mastrianni, E. Lugaresi, P. Gambetti, and S. B. Prusiner. 1996. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274:2079-2082. [DOI] [PubMed] [Google Scholar]
- 51.Telling, G. C., M. Scott, J. Mastrianni, R. Gabizon, M. Torchia, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 1995. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 83:79-90. [DOI] [PubMed] [Google Scholar]
- 52.Thomzig, A., S. Spassov, M. Friedrich, D. Naumann, and M. Beekes. 2004. Discriminating scrapie and bovine spongiform encephalopathy isolates by infrared spectroscopy of pathological prion protein. J. Biol. Chem. 279:33847-33854. [DOI] [PubMed] [Google Scholar]
- 53.Uptain, S. M., and S. Lindquist. 2002. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56:703-741. [DOI] [PubMed] [Google Scholar]
- 54.Vorberg, I., A. Buschmann, S. Harmeyer, A. Saalmuller, E. Pfaff, and M. H. Groschup. 1999. A novel epitope for the specific detection of exogenous prion proteins in transgenic mice and transfected murine cell lines. Virology 255:26-31. [DOI] [PubMed] [Google Scholar]
- 55.Vorberg, I., M. H. Groschup, E. Pfaff, and S. A. Priola. 2003. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J. Virol. 77:2003-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissmann, C. 2004. The state of the prion. Nat. Rev. Microbiol. 2:861-871. [DOI] [PubMed] [Google Scholar]
- 57.Westaway, D., V. Zuliani, C. M. Cooper, M. Da Costa, S. Neuman, A. L. Jenny, L. Detwiler, and S. B. Prusiner. 1994. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie. Genes Dev. 8:959-969. [DOI] [PubMed] [Google Scholar]
- 58.Wickner, R. B., H. K. Edskes, B. T. Roberts, U. Baxa, M. M. Pierce, E. D. Ross, and A. Brachmann. 2004. Prions: proteins as genes and infectious entities. Genes Dev. 18:470-485. [DOI] [PubMed] [Google Scholar]
- 59.Wong, C., L. W. Xiong, M. Horiuchi, L. Raymond, K. Wehrly, B. Chesebro, and B. Caughey. 2001. Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein. EMBO J. 20:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]