Abstract
The latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus (KSHV) is required for viral episome maintenance in host cells during latent infection. Two regions of the protein have been implicated in tethering LANA/viral episomes to the host mitotic chromosomes, and LANA chromosome-binding sites are subjects of high interest. Because previous studies had identified bromodomain protein Brd4 as the mitotic chromosome anchor for the bovine papillomavirus E2 protein, which tethers the viral episomes to host mitotic chromosomes (J. You, J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley, Cell 117:349-360, 2004, and J. You, M. R. Schweiger, and P. M. Howley, J. Virol. 79:14956-14961, 2005), we examined whether KSHV LANA interacts with Brd4. We found that LANA binds Brd4 in vivo and in vitro and that the binding is mediated by a direct protein-protein interaction between the ET (extraterminal) domain of Brd4 and a carboxyl-terminal region of LANA previously implicated in chromosome binding. Brd4 associates with mitotic chromosomes throughout mitosis and demonstrates a strong colocalization with LANA and the KSHV episomes on host mitotic chromosomes. Although another bromodomain protein, RING3/Brd2, binds to LANA in a similar fashion in vitro, it is largely excluded from the mitotic chromosomes in KSHV-uninfected cells and is partially recruited to the chromosomes in KSHV-infected cells. These data identify Brd4 as an interacting protein for the carboxyl terminus of LANA on mitotic chromosomes and suggest distinct functional roles for the two bromodomain proteins RING3/Brd2 and Brd4 in LANA binding. Additionally, because Brd4 has recently been shown to have a role in transcription, we examined whether Brd4 can regulate the CDK2 promoter, which can be transactivated by LANA.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human gamma-2 herpesvirus associated with the pathogenesis of Kaposi's sarcoma, primary effusion lymphomas, and multicentric Castleman's disease (8, 9, 37, 48). The virus persists in a latent state in tumor cells (18). During latent infection, the viral genomes are stably maintained as multicopy circular episomes in the nuclei of infected cells in which a small number of viral genes are expressed (6). The product of one of these latent genes, latency-associated nuclear antigen (LANA), encoded by open reading frame 73 (ORF 73), is also highly expressed in all forms of KSHV-related malignancies (15). LANA was initially identified as a hallmark of KSHV-infected cells and is characterized by its speckled, nuclear immunostaining (19, 26-28, 41).
LANA tethers KSHV viral genomes to host mitotic chromosomes to ensure maintenance of viral episomes in dividing cells (1, 2, 10, 49, 50, 57). This tethering mechanism involves concomitant interaction of LANA with both the viral episomes and the host metaphase chromosomes. The cis-acting viral DNA elements recognized by LANA on the viral genome have been well defined. LANA binds specifically to sequence motifs within the terminal repeat (TR) of the KSHV genome and tethers the viral episomes to host chromosomes (1, 2, 10, 20, 21, 50). The carboxyl-terminal domain of LANA is sufficient for this site-specific DNA binding (11, 21, 29, 34). Interestingly, both the amino and carboxyl termini of LANA have been implicated in LANA association with cellular chromatin. A chromosome-binding site has been mapped to amino acids (aa) 5 to 22, which mediate the specific interaction of LANA with chromatin during interphase and with chromosomes during mitosis (38). Deletion of this N-terminal chromosome-binding site abolishes the interaction of LANA with mitotic chromosomes (3, 38, 46). Mutational analysis further demonstrated that LANA aa 5 through 13 are sufficient for chromosome association of a green fluorescent fusion protein (3). N-terminal LANA directly binds histones H2A and H2B on the nucleosome surface to attach to chromosomes (4). However, additional evidence suggests that a second independent LANA chromosome-binding site maps to the C terminus (30) (M. Ballestas, T. Komatsu, and K. Kaye, 4th Int. Workshop KSHV and Related Agents, 2001). It has also been shown that deletion of aa 1129 to 1143 from LANA protein removes the tight association with nuclear heterochromatin (52). Furthermore, a naturally occurring C-terminal-truncated isoform of LANA, lacking the last 76 aa, fails to associate with either heterochromatin or full-length LANA in KSHV-infected cells (7).
Association of viral genomes with host mitotic chromosomes via a virus-encoded protein is a strategy employed by a number of different latent DNA viruses. Similar to KSHV LANA, the Epstein-Barr virus (EBV) EBNA1 and papillomavirus E2 proteins play a role in viral genome maintenance (5, 23, 24, 31, 47). Although unrelated in sequence, functional similarities exist among LANA, EBNA1, and E2 proteins. Each of these viral proteins binds its cognate binding sites on the viral DNA and tethers the viral genome to host mitotic chromosomes to ensure efficient segregation to progeny cells (1, 10, 31, 32). Each of these viral proteins also has roles in viral gene expression and viral DNA replication.
In previous studies, we have identified Brd4 (bromodomain-containing protein 4) as the major cellular receptor for bovine papillomavirus (BPV) E2 on mitotic chromosomes (58, 59). Brd4 is a member of the BET family of proteins that contain two bromodomains, involved in chromatin targeting, and an ET (extraterminal) domain of unknown function (14). Brd4 has been shown to bind to chromosomes with preference for acetylated histone H4 (13).
In this study, we have examined the possibility that LANA might also interact with Brd4 and in doing so possibly mediate some of its cellular functions. Our studies revealed that Brd4 interacts with LANA both in vivo and in vitro. The Brd4-binding sites map within the C-terminal domain of LANA; the ET domain of Brd4 interacts directly with the C-terminal domain of LANA. Brd4 associates with LANA and viral episomes in the punctate nuclear structures in cells that carry artificial viral episomes and in KSHV-infected cells. Our data thus provide an additional molecular target for LANA that may have a role in genome maintenance.
MATERIALS AND METHODS
Cell cultures and cell lines.
The primary PEL isolate BCLM was obtained from pleural fluid of a human immunodeficiency virus-positive patient. Cells were established in IMDM (GIBCO-BRL) media supplemented with 30% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B in 5% CO2 at 37°C for 1 week. The concentration of FBS in the growth medium was then reduced to 20% the following week. In the third week, FBS was reduced to 10% and fresh medium was introduced every 48 h. The BJAB and BJAB/F-LANA stable cells, BJAB/F-LANA carrying the artificial genome p8TR, and the BCLM cells were grown in RPMI (Invitrogen) containing 10% bovine growth serum (HyClone, Logan, Utah). BCLM was infected with KSHV and was not coinfected with EBV. C33A cells were maintained in monolayer culture in Dulbecco's modified Eagle's medium containing 10% fetal calf serum.
Recombinant plasmid construction.
pSG5 F-LANA has been described previously (1). Constructs of glutathione S-transferase (GST) fused to the N terminus of LANA aa 1 to 23 and GST-LANA aa 982 to 1162 were described previously (4, 29). To generate LANA aa 932 to 1162 fused to maltose-binding protein (MBP), sequence corresponding to LANA aa 932 to 1162 was amplified from pSGFLANA using PCR (1) to introduce EcoRI and XbaI sites on the 5′ and 3′ end and was subcloned into pMAL-c2X vector. The resulting construct was pMAL-c2X LANA aa 932-1162. To generate MBP-LANA aa 982-1162, the NruI/KpnI fragment of pSG5LANA (1) was first inserted into the EcoRV/KpnI sites of pBluescript (Stratagene). The EcoRI/BamHI fragment of the resulting construct was then subcloned into the EcoRI/BamHI sites of pEGFP-NLS (23) to generate the construct pGFP LANA 982-1162. The BglII/BamHI fragment of pGFP LANA 982-1162 was subcloned into the BamHI site of pMAL-c2X to generate pMAL-c2X LANA aa 982-1162. Constructs that encode subfragments of LANA were described previously (29). Additionally, pSGF LANA 778-980 was created by inserting an XbaI linker (CTAGTCTAGACTAG) to introduce a stop codon downstream of the NruI site in the pSGF LANA 778-1047 construct. For Brd4 expression, fragments of human Brd4 cDNA were amplified by PCR to introduce BamHI and NotI sites on the 5′ and 3′ ends and subcloned into the pcDNA4C vector using BamHI and NotI digestion. These plasmids were used for in vitro transcription and translation. pGEX-E2TA and pGEX-E2TR were from our laboratory plasmid bank. To generate the GST-Brd4 aa 471-594 and GST-Brd4 aa 595-730 constructs, the Brd4 cDNA fragments were amplified by PCR to incorporate BamHI and NotI sites on the 5′ and 3′ ends and were subcloned into pGEX-6P-1 vector using BamHI and NotI. GST-Brd2 aa 592-754 was described previously (39). All plasmid constructs were verified by DNA sequencing.
Immunoprecipitation (IP) and Western blot analysis.
For transient protein expression in C33A cells, 40% to 80% confluent cells growing in 10-cm dishes were transfected with 16 μg of plasmid DNA using FuGENE 6 Transfection Reagent (Roche). Cells were harvested 48 to approximately 72 h after transfection. Cytoplasmic and nuclear extracts were prepared as described previously (43).
For anti-FLAG IP, soluble extract proteins were mixed with 10 μl of anti-FLAG M2 agarose (Sigma) and rotated at 4°C for 7 h. For LANA IP, soluble extract proteins were incubated with 10 μl anti-LANA rat monoclonal antibody LN53 (Advanced Biotechnologies) at 4°C for 2 h prior to mixing with 10 μl protein A Sepharose for 7 h. The beads were washed three times with 1 ml ice-cold phosphate-buffered saline (PBS) containing 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and bound proteins were eluted with 30 μl of sodium dodecyl sulfate (SDS) sample buffer. Aliquots (15 μl) were resolved on an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel. Proteins were transferred to Immobilon-P (Millipore) and blotted with specific antibody to detect the protein of interest (enhanced chemiluminescence detection). Antibodies employed in the Western blot analysis were the rabbit polyclonal antibody against Brd4, C-MCAP, which has been described previously (14), and the anti-FLAG M2 monoclonal antibody was obtained from Sigma.
For anti-LANA coimmunoprecipitation of the Xpress-tagged Brd4 aa 471-730 fragment, 1 × 107 exponentially growing BJAB cells expressing full-length LANA (1) were transfected with 60 μg of either pcDNA4C or pcDNA4C encoding Brd4 aa 471-730. Two days posttransfection, cells were lysed in a standard lysis buffer (29) containing 400 mM NaCl. After incubation with 1 μg of LN53 at 4°C for 4 h, 10 μl of protein G agarose beads was added. After an additional 1 h of incubation at 4°C, the beads were washed thrice with the standard lysis buffer (29) and samples were analyzed on a standard SDS-8% PAGE gel. The proteins were transferred to a nitrocellulose membrane and probed with the anti-Xpress mouse monoclonal antibody from Invitrogen.
In vitro binding of Brd4 to GST-LANA.
[35S]Met-labeled Brd4 full-length protein or fragments were produced in vitro by a T7 polymerase transcription-coupled reticulocyte lysate translation system (TNT; Promega) per the manufacturer's instructions, using plasmid pcDNA4C, which carries the human Brd4 full-length or partial cDNA. GST fusion proteins were produced in Escherichia coli using the respective pGEX plasmids. A 10-μl aliquot of each translation mixture was mixed with 50 μl of immobilized GST fusion protein (∼1 μg of protein/μl beads) in 0.18 ml PBS prior to incubation at 4°C for 4 h. The beads were washed three times with 0.5 ml of 0.1 M KCl base buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 5 mM MgCl2, 0.1% Tween 20, 0.1 M KCl, 0.2 mM PMSF, and 0.5 mM dithiothreitol) and eluted with 80 μl of 1× SDS-PAGE sample buffer. An aliquot of each eluate (40 μl) was analyzed by SDS-PAGE and autoradiography.
Mapping the Brd4-binding sites on LANA protein.
Subfragments of LANA were in vitro translated and [35S]Met radiolabeled using a TNT coupled reticulocyte lysate system (Promega). GST-full-length Brd4 and GST protein alone were expressed in BL21(DE3) (Stratagene) and immobilized on the glutathione resin. Fifty microliters of GST-Brd4 or GST beads was incubated with 20 μl of in vitro-translated LANA proteins in 0.5 ml of lysis buffer (29). After incubation at 4°C for 4 h, the beads were washed five times in 0.5 ml of lysis buffer (29). The proteins bound to the beads were then analyzed by SDS-PAGE and autoradiography.
Direct binding of LANA C terminus to Brd4.
MBP-LANA aa 932-1162 and MBP-LANA aa 982-1162 fusions as well as MBP protein alone were expressed in E. coli and purified using the pMAL protein purification system (New England Biolabs) according to the manufacturer's manual. GST-Brd4 fusion proteins were produced in E. coli using the respective pGEX plasmids. Twenty micrograms of each MBP protein was mixed with 5 μl of immobilized GST fusion protein (∼1 μg of protein/μl beads) in 0.18 ml PBS prior to incubation at 4°C for 4 h. The beads were washed three times with 0.5 ml of 0.1 M KCl base buffer and eluted with 30 μl of SDS-PAGE sample buffer. An aliquot of each eluate (10 μl) was analyzed by SDS-PAGE and Coomassie blue staining.
Immunofluorescent staining.
Cells cultured in suspension were streaked onto coverslips and air dried in a 37°C incubator. After fixation with 3% paraformaldehyde in PBS, cells were incubated in blocking/permeabilization buffer (0.5% Triton X-100 and 3% bovine serum albumin in PBS) for 10 min at room temperature and were stained with primary antibodies at room temperature for 60 min. For Brd4/LANA double staining, anti-Brd4 rabbit polyclonal antibody, N-MCAP (1/500 dilution) (14), and anti-LANA rat monoclonal antibody LN53 (1/200 dilution; Advanced Biotechnologies) were used. For Brd2/LANA double staining, an anti-Brd2 peptide rabbit polyclonal antibody (1/500 dilution) was used with LN53. The rabbit polyclonal antibody was raised against Brd2 synthetic peptide acetylated-CVSNPKKPERVTNQLQYLHK-amide (Biosource Inc., Hopkinton, MA) and then purified from serum by antipeptide immunoaffinity chromatography. After incubation, cells were washed three times with blocking/permeabilization buffer and incubated with Alexa Fluor 594 goat anti-rabbit immunoglobulin G (IgG) (1/1,000 dilution; Molecular Probes) and a goat anti-rat IgG fluorescein isothiocyanate (FITC) conjugate (1/200 dilution; Southern Biotech) for an additional 60 min. Cells were counterstained with 0.3 μM 4′,6′-diamidino-2-phenylindole (DAPI) and examined using a Zeiss LSM 510 Meta UV upright confocal microscope and associated Zeiss LSM 510 software.
Reporter assays.
Ten million BJAB B lymphoma cells were suspended in 400 μl of RPMI containing 10% bovine growth serum (BGS) and no antibiotics. After a 10-min incubation at room temperature, 10 μg of −683CDK2/LUC luciferase (55) and 5 μg of pEGFP (Clontech Laboratories, CA) were added to the cells; 15 μg of pSG5 F-LANA (1) and/or 5 μg, 10 μg, 15 μg, 20 μg, or 25 μg of pcDNA4CBrd4 was also added where indicated. Cells were electroporated with a Gene Pulser (Bio-Rad, CA) at 220 V and 960 μF. After electroporation, cells were resuspended in 10 ml of RPMI containing 10% BGS. Thirty-six hours posttransfection, cells were washed once in phosphate-buffered saline and lysed in 400 μl of complete cell lysis buffer (Promega, WI). Luciferase assays were performed as recommended by the manufacturer (Promega) using an Optocomp 1 luminometer (MGM Instruments, CT). Transfection efficiencies were normalized by percentage of cells expressing green fluorescent protein as determined by a fluorescent-activated cell sorter.
RESULTS
LANA interacts with Brd4.
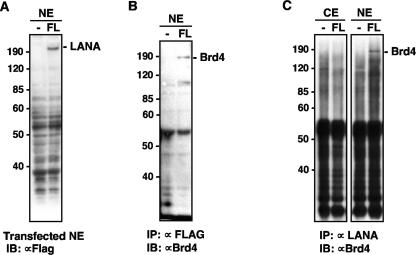
The papillomavirus E2 protein, EBNA1 of EBV, and LANA of KSHV have critical roles in viral genome maintenance in dividing cells through similar noncovalent protein-protein interactions with mitotic chromosomes (1, 10, 32). Since Brd4 binds the papillomavirus E2 proteins and mediates plasmid maintenance for BPV, we examined whether LANA might also bind Brd4. FLAG-tagged LANA proteins were transiently expressed in transfected C33A cells and tested for the ability to bind Brd4 by coimmunoprecipitation. Although the expression level of LANA was low (Fig. 1A), FLAG-LANA was able to interact with endogenous Brd4, as demonstrated by coimmunoprecipitation of Brd4 with anti-FLAG beads. No Brd4 was observed in cells transfected with the empty vector (Fig. 1B), demonstrating that the interaction was specific for LANA. Brd4 was also coimmunoprecipitated with FLAG-LANA by the LANA-specific LN53 antibody (Fig. 1C). Although the anti-LANA immunoprecipitation pulled down many nonspecific bands in both the negative control and the FLAG-LANA-transfected cells, a high-molecular-weight band corresponding to the size of Brd4 was detected only in the LANA-transfected sample using an anti-Brd4 antibody (Fig. 1C).
FIG. 1.
KSHV LANA protein binding to Brd4. (A) Expression of FLAG-tagged LANA in C33A cells. Nuclear extracts (NE) from C33A transfected with a FLAG-LANA construct (FL) or empty vector (−) were analyzed on an SDS-PAGE gel and immunoblotted with anti-FLAG M2 antibody. Molecular masses of the markers are indicated on the left. (B) Anti-Brd4 immunoblot analysis of anti-FLAG immunoprecipitation. NE of C33A cells transfected with FLAG-LANA plasmid (FL) or empty vector (−) were immunoprecipitated with anti-FLAG M2 beads. Western blot with an anti-Brd4 rabbit polyclonal antibody, C-MCAP, demonstrated that human Brd4 protein is present in the sample coimmunoprecipitated with LANA. (C) Anti-Brd4 immunoblot analysis of cellular proteins coimmunoprecipitated with LANA. C33A cells were transfected with FLAG-LANA plasmid (FL) or empty vector (−). Cytoplasmic extracts (CE) and nuclear extracts (NE) immunoprecipitated with anti-LANA antibody LN53 were immunoblotted with the Brd4 antibody C-MCAP. IB, immunoblot.
Brd4 aa 471-730 interacts with the C terminus of LANA.
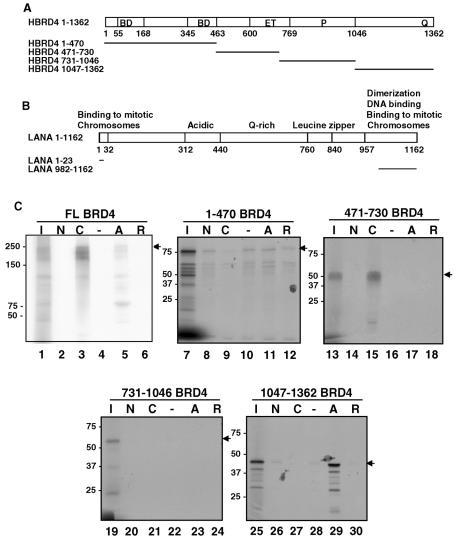
Both the LANA N terminus and C terminus exert important functions in KSHV episome maintenance. We therefore assayed whether LANA bound to Brd4 through each of these regions. We used constructs wherein GST was fused to the LANA N terminus (aa 1 to 23) or C terminus (aa 982 to 1162) (Fig. 2B). Because Brd4 binds the full-length bovine papillomavirus E2 protein E2TA but not to the truncated E2TR protein (58), GST-E2TA and GST-E2TR served as positive and negative controls, respectively (Fig. 2C, lanes 5 and 6). Under the same conditions, Brd4 bound to the GST-LANA C terminus but not to the GST-LANA N terminus or to GST alone (Fig. 2C, lanes 2 to 4). To map the domain of Brd4 engaged by the LANA C terminus, subfragments of Brd4 were tested for binding to the GST-LANA fusions (Fig. 2A and C). Among the four fragments of Brd4 tested, Brd4 aa 471-730 bound specifically to the GST-LANA C terminus but not to the other GST fusions tested (Fig. 2C, lanes 13 to 18). As a control, Brd4 aa 1047-1362 bound specifically to GST-E2TA, confirming the previously established Brd4-binding specificity for E2TA (Fig. 2C, lanes 25 to 30). None of the other Brd4 fragments showed any specific binding to the GST-LANA fusion proteins tested (Fig. 2C). These data established that Brd4 binds to the C terminus of LANA through the region of aa 471 to 730.
FIG. 2.
Mapping of the LANA-binding domain on Brd4 protein. (A) Diagram of human Brd4 (HBRD4) full-length protein and the fragments used to map the LANA binding domain. The full-length Brd4 and each indicated fragment was translated and labeled by [35S]Met using in vitro transcription and translation (TNT). BD, bromodomain. (B) GST-tagged LANA constructs for testing Brd4 binding. Shown are the domain structures of LANA protein and the fragments of LANA fused to GST (LANA aa 1-23, LANA-N; LANA aa 982-1162, LANA-C). (C) In vitro binding of Brd4 to LANA protein. Each Brd4 TNT product was tested for LANA binding using GST-LANA-N (N) or GST-LANA-C (C) immobilized on glutathione resin. GST-E2TA (A) and GST-E2TR (R) beads were used as positive and negative controls, respectively. GST alone (−) was used as a negative control. Aliquots (40 μl) of sample eluted from GST fusion beads were resolved by SDS-PAGE along with 25% of the input sample (lanes I), and they were detected by autoradiography. Arrows indicate the size of full-length translation products. FL, full length.
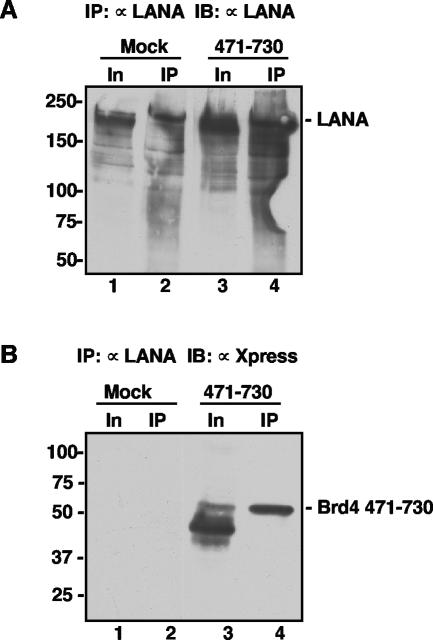
To further examine the LANA-Brd4 interaction, a pcDNA4C construct that encoded the Xpress-tagged Brd4 aa 471-730 fragment was transfected into BJAB cells stably expressing FLAG-LANA. Immunoprecipitation of LANA pulled down a distinct band corresponding to the Brd4 aa 471-730 fragment in the Brd4 aa 471-730-transfected sample but not in the mock transfection (Fig. 3B). A Western blot using anti-LANA antibody detected a similar amount of immunoprecipitated LANA in both samples (Fig. 3A). Notably, although the Brd4 aa 471-730 fragment was expressed as a series of bands migrating closely with the 50-kDa molecular mass marker (Fig. 3B, lane 3), only the slowest migrating species coimmunoprecipitated with LANA. Since the Western blot antibody recognizes the Xpress tag at the N terminus of Brd4 aa 471-730, LANA binding might depend upon determinants located close to the C-terminal end of this fragment. Indeed, further analysis showed that LANA binds to the ET domain located within the C-terminal half of the Brd4 aa 471-730 fragment (see below).
FIG. 3.
Brd4 aa 471-730 interacts with LANA in vivo. (A) Anti-LANA immunoblot detection of the input and immunoprecipitated LANA protein. Exponentially growing BJAB cells (1 × 107) expressing full-length LANA (1) were transiently transfected with either a pcDNA4C plasmid expressing His-Xpress-Brd4 aa 471-730 (471-730) or empty vector (Mock). Forty-eight hours after transfection, cells were lysed as described previously (29) and were immunoprecipitated with 1 μg of anti-LANA antibody LN53. The immunoprecipitated samples (IP) were analyzed with the input sample (In) on an SDS-8% PAGE gel and blotted with an anti-LANA polyclonal human serum to show that anti-FLAG immunoprecipitation of LANA was not affected by Brd4 aa 471-730 expression. (B) Anti-Xpress immunoblot detection of the interaction between LANA and Brd4 aa 471-730. Anti-LANA-immunoprecipitated samples were blotted with an anti-Xpress antibody. The Brd4 aa 471-730 fragment was specifically coimmunoprecipitated with LANA in the transfected cells but not in the “mock” transfection. Similar to the full-length Brd4, the Brd4 aa 471-730 fragment migrated as a protein larger than the deduced molecular mass of 26 kDa. IB, immunoblot.
Brd4-binding sites map to LANA aa 475 to 777 and aa 982 to 1162.
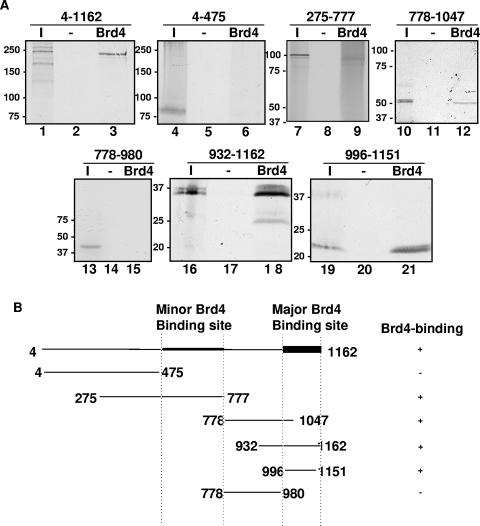
Although the C terminus of LANA was sufficient to bind Brd4, it was not clear if other regions of LANA might also be involved in binding Brd4. We therefore tested a series of subfragments spanning the complete sequence of LANA for Brd4 binding (Fig. 4). GST full-length Brd4 purified from E. coli was immobilized on glutathione beads before incubation with the in vitro-translated fragments of LANA (Fig. 4A). As anticipated, GST-Brd4 bound full-length LANA as well as the LANA C-terminal aa 932 to 1162 and aa 996 to 1151 fragments. In addition, LANA aa 275-777 and LANA aa 778-1047 showed a low level of binding to GST-Brd4. LANA aa 4-475 and LANA aa 778-980 did not bind GST-Brd4. None of the fragments bound GST as a negative control. As summarized in Fig. 4B, the Brd4-binding sites mapped to two different regions of LANA, located at aa 475 to 777 and aa 982 to 1162. Quantitation of the binding indicated that ∼50% of the LANA C terminus bound to Brd4, whereas only 20% of LANA aa 275-777 and an even smaller fraction (7.5%) of LANA aa 778-1047 bound to Brd4 under similar conditions. We therefore concluded that LANA aa 982-1162 represents the major Brd4-binding site and that LANA aa 475-777 constitutes a minor Brd4-binding site.
FIG. 4.
Mapping of the Brd4-binding domain on the LANA protein. (A) In vitro binding of LANA fragments to Brd4 protein. LANA and each indicated LANA subfragment were translated in vitro and labeled using [35S]Met. Each TNT reaction was tested for binding using bacterially expressed GST-Brd4 protein immobilized on glutathione resin. GST alone (−) was used as a negative control. Aliquots of sample eluted from GST fusion beads were resolved by SDS-PAGE along with 20% of the input sample (lanes I) and were detected by autoradiography. (B) Summary of the interactions of various regions of LANA with GST-Brd4.
Brd4 interacts directly with the LANA C terminus.
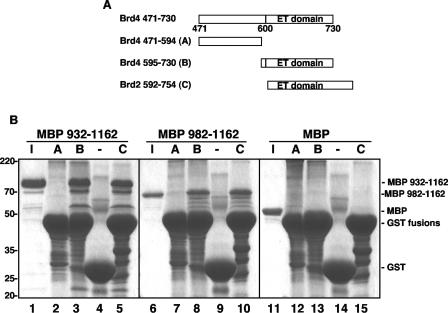
Several LANA-interacting proteins have been identified previously, and most were shown to interact with LANA in cellular or reticulocyte lysates (10, 30, 33, 36, 39, 42). To test whether LANA and Brd4 interacted directly, we examined the binding of recombinant Brd4 and LANA fragments expressed and purified from E. coli. The LANA C-terminal fragments aa 932 to 1162 and aa 982 to 1162 were expressed as MBP fusion proteins and purified to near homogeneity from E. coli (Fig. 5B, lanes 1 and 6). In addition, to further map the LANA binding site within the Brd4 aa 471-730 region, the fragment was divided into aa 471 to 594 (fragment A), which contains no previously defined domain, and aa 595 to 730 (fragment B), which contains the ET domain (Fig. 5A). Each fragment was expressed and purified as a GST fusion protein from E. coli (Fig. 5B). The purified MBP-LANA fragments were incubated with equal amounts of the GST-Brd4 fragments or GST protein alone and immobilized on glutathione beads. After extensive washing, the bound proteins were eluted from the beads with SDS sample buffer and analyzed by SDS-PAGE. As shown in Fig. 5B, both MBP-LANA aa 932-1162 and MBP-LANA aa 982-1162 showed specific binding to GST-Brd4 aa 595-730 but not to GST-Brd4 aa 471-594 or to GST alone. In addition, MBP alone did not bind any of the GST fusion proteins tested, indicating that the binding was mediated by the LANA C terminus. These data demonstrated that Brd4 interacts directly with the C terminus of LANA and that this binding is mediated by the ET domain. LANA has previously been shown to interact with the ET domain of RING3/Brd2 in insect cell lysate (39). To examine whether the interaction between the C terminus of LANA and the RING3/Brd2 ET domain was direct, we also examined the binding of the MBP-LANA C-terminal fragments with the ET domain of RING3/Brd2 fused to GST protein (Fig. 5A, fragment C). The data showed that both of the LANA C-terminal fragments could bind to the RING3/Brd2 ET domain directly (Fig. 5B, lane 5 and 10). Therefore, the ET domains of RING3/Brd2 and Brd4 are both able to bind the C terminus of LANA directly.
FIG. 5.
Direct interaction of Brd4 with the LANA C terminus. (A) GST-tagged Brd4 and Brd2 constructs for testing LANA binding. Shown are the domain structures of Brd4 aa 471-730 and the Brd2 ET domain region examined in this experiment. (B) In vitro direct binding of Brd4 to the LANA C terminus. Each MBP-LANA product was tested for binding on GST-Brd4 aa 471-594 (A), GST-Brd4 aa 595-730 (B), GST-Brd2 aa 592-754 (C), or GST protein alone (−) immobilized on glutathione resin. Aliquots (10 μl) of sample eluted from beads were resolved by SDS-PAGE along with 30% of the input sample (I) and were detected by Coomassie blue staining.
LANA and Brd4 colocalize on host mitotic chromosomes in cells stably expressing LANA that carry artificial KSHV episomes.
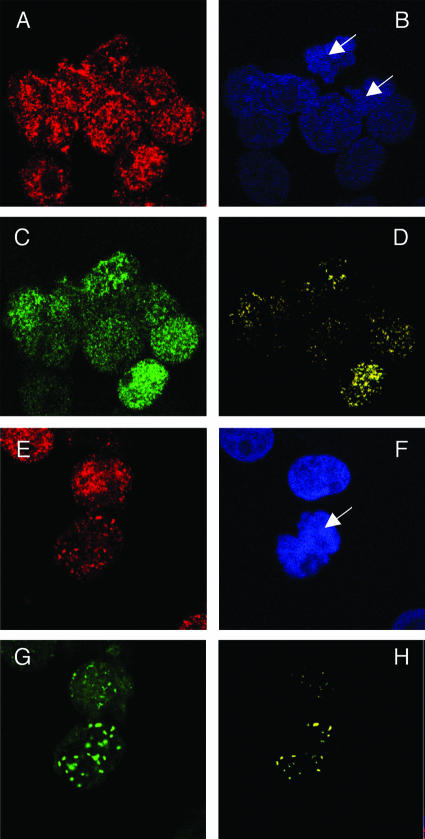
In the absence of KSHV DNA, LANA diffusely stains the nucleus as well as mitotic chromosomes. In contrast, in the presence of the KSHV DNA, LANA is colocalized with the viral genomes in punctate dots both in interphase nuclei and on mitotic chromosomes (1, 10, 50). Since Brd4 is a mitotic chromosome-associated protein (14), we examined the localization of LANA and Brd4 in BJAB cells that stably express FLAG-tagged LANA (BJAB/F-LANA). Brd4 showed punctate nuclear staining on both interphase nuclei and mitotic chromosomes (Fig. 6A). LANA expression was detected in ∼50% of the stable cells. In the LANA-positive cells, LANA staining was rather diffuse but the signal also appeared to accumulate in discrete speckles around the Brd4 dots (Fig. 6C), displaying a partial colocalization with Brd4 on both interphase nuclei and mitotic chromosomes (Fig. 6D).
FIG. 6.
Colocalization of Brd4 and LANA on mitotic chromosomes in BJAB cells stably expressing LANA. BJAB/F-LANA cells stably expressing LANA (A to D) and BJAB/F-LANA stable cells carrying the p8TR artificial KSHV episomes (E to H) were double stained with an anti-Brd4 rabbit polyclonal antibody, N-MCAP, and the anti-LANA Rat monoclonal antibody, LN53. The staining was detected by incubation with an Alexa Fluor 594 goat anti-rabbit IgG (A and E) and a goat anti-rat IgG FITC conjugate (C and G), respectively. Cells were also counter stained with DAPI to identify nuclei and mitotic chromosomes (B and F) (the arrows indicate mitotic chromosomes). Cells were examined under a Zeiss LSM 510 UV upright confocal microscope, and the colocalized Brd4 and LANA staining was visualized using Zeiss LSM 510 software (D and H). Colocalization of Brd4 and LANA protein was distinctly observed on mitotic chromosomes as bright yellow dots (H).
We further examined the BJAB/F-LANA cells that carry an artificial KSHV episome, p8TR, which contains eight copies of the KSHV TR element (3). The cells that carry the viral episomes could easily be distinguished from the neighboring cells with no viral DNA, because LANA concentrated at sites of KSHV episome localization along mitotic chromosomes and stained as bright punctate dots (Fig. 6G). Most significantly, the merge of the Brd4 and LANA signals demonstrated a specific colocalization of these two proteins in the punctate dots on mitotic chromosomes in cells harboring viral episomes (Fig. 6H).
LANA and Brd4 colocalize on host mitotic chromosomes in KSHV-infected cells.
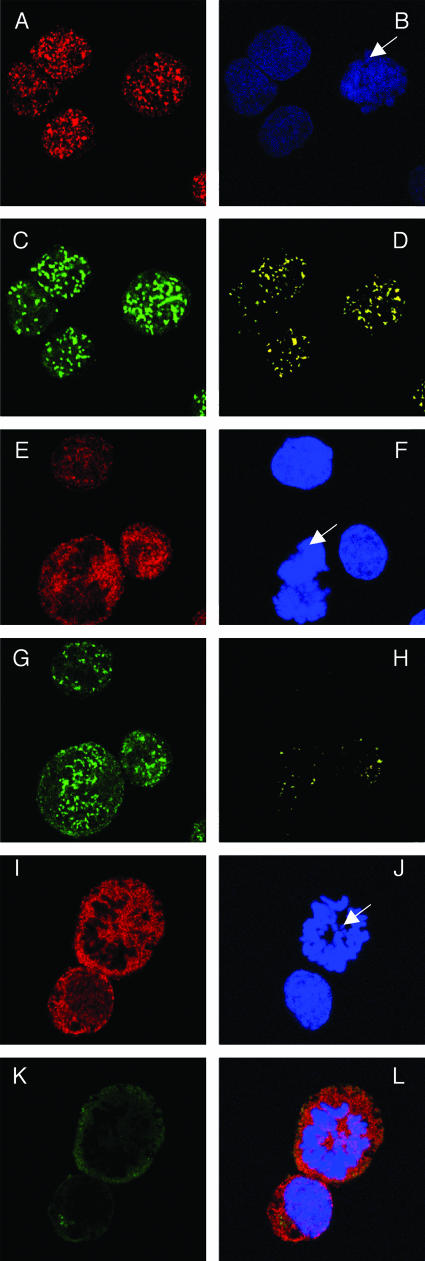
To confirm the LANA and Brd4 colocalization, we further examined the localization of LANA and Brd4 in KSHV-infected BCLM cells. Consistent with the previous observation (1), LANA concentrated at sites of KSHV DNA both in interphase nuclei and along mitotic chromosomes in metaphase, producing a characteristic stippled pattern (Fig. 7C). As in BJAB cells, Brd4 stained as punctate nuclear dots on both mitotic chromosomes and interphase nuclei (Fig. 7A). Merging of LANA and Brd4 signals demonstrated a clear colocalization of the two proteins in a similar dotted pattern (Fig. 7D).
FIG. 7.
Colocalization of Brd4 and LANA on mitotic chromosomes in KSHV-positive BCLM cells. (A to D) BCLM cells were double stained with N-MCAP (A) and LN53 (C) as described in the legend to Fig. 6. Cells were also counterstained with DAPI (B) (the arrow indicates a mitotic chromosome). Colocalization of Brd4 and LANA was distinctly observed on mitotic chromosomes as bright yellow dots (D). (E to H) BCLM cells were double stained with an anti-Brd2 peptide rabbit polyclonal antibody and LN53. The staining was detected by incubation with a Alexa Fluor 594 goat anti-rabbit IgG (E) and a goat anti-rat IgG FITC conjugate (G), respectively. The staining of DAPI is shown in panel F. The overlay signal of panels E and G is shown in panel H. In contrast to the LANA staining, a majority of Brd2 staining is excluded from the mitotic chromosomes (E and H). (I to L) KSHV-negative BJAB cells were double stained for Brd2 and LANA as for panels E to H. The overlay signal is shown in panel L. In contrast to the status in KSHV-positive cells, the Brd2 staining is almost completely excluded from mitotic chromosomes.
Since RING3/Brd2 also directly interacts with LANA (Fig. 5), we examined the localization of RING3/Brd2 protein in the BCLM cells. Surprisingly, RING3/Brd2 stained in a diffuse manner in interphase nuclei and was mostly excluded from the mitotic chromosomes (Fig. 7E). Merging the RING3/Brd2 and LANA staining showed very little colocalization of the two proteins on both interphase nuclei and mitotic chromosomes (Fig. 7H). In the KSHV-negative BJAB cells, RING3/Brd2 was even more diffusely localized on interphase nuclei and was almost completely excluded from mitotic chromosomes (Fig. 7I to L). Similar results were obtained using a second anti-Brd2 polyclonal antibody (data not shown). RING3/Brd2 has been previously shown to be distributed evenly throughout the interphase nucleus (12). Compared with the RING3/Brd2 staining in the absence of LANA, the limited Brd2/LANA colocalization in the KSHV-positive cells (Fig. 7H) suggested a LANA-mediated recruitment of RING3/Brd2 protein to mitotic chromosomes, as previously suggested (36).
Brd4 downregulates LANA activation of the CDK2 promoter.
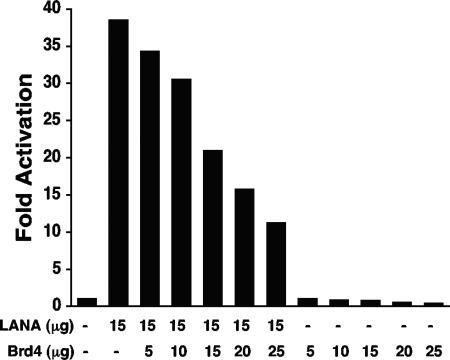
Since both LANA and Brd4 are implicated in cell proliferation (16, 17, 25, 40, 53, 56), we assayed whether these proteins might both be involved in the regulation of the CDK2 promoter. As previously reported (54), LANA activated the CDK2 promoter approximately 38-fold compared to the reporter alone (Fig. 8). However, coexpression of Brd4 diminished the LANA transactivation. Increasing amounts of Brd4 caused successively larger degrees of repression of LANA activation. While transfection of 5 μg of Brd4 only mildly reduced LANA activation to ∼34-fold over that of the reporter, 25 μg of Brd4 resulted in reduction of LANA activation to ∼11-fold. Since these results were consistent with Brd4 repressing CDK2 promoter activity, we assayed Brd4's effects in the absence of LANA (Fig. 8). Transfection of 5 μg, 10 μg, 15 μg, 20 μg, and 25 μg of Brd4 into cells resulted in ∼1-, ∼0.8-, ∼0.76-, ∼0.5-, and ∼0.4-fold CDK promoter activity, respectively, compared to that of reporter alone. Therefore, expression of Brd4 inhibits CDK2 promoter activity and similarly the LANA activation of the CDK2 promoter.
FIG. 8.
Brd4 downregulates the CDK2 promoter and LANA transactivation of the CDK2 promoter. BJAB B-lymphoma cells were transfected with the indicated plasmids. Thirty-six hours posttransfection, luciferase activity was determined and compared to the activity after transfection with reporter alone to determine the fold activation or repression. The experiment shown is representative of more than 10 experiments. The reduction in activation was not due to diminished LANA expression, as determined by Western blotting.
DISCUSSION
Noncovalent association of specific virally encoded DNA-binding proteins with host cell mitotic chromosomes appears to be the principal strategy employed by a number of episomal DNA viruses to ensure that their genomes are enclosed within reformed nuclear envelopes and thus maintained in progeny cells during latent infection (1, 23, 24, 47). Understanding the mechanisms involved in the binding of these proteins to mitotic chromosomes is therefore crucial. In KSHV, LANA plays a critical role in tethering viral genomes to the host mitotic chromosomes (1, 10). The molecular mechanism underlying this virus-host protein-protein interaction is, however, complex.
In a previous study, we identified Brd4 as the host mitotic chromosome receptor for the bovine papillomavirus E2 protein (58, 59). In this present study, we show that this mitotic chromosome-associated protein also interacts with KSHV LANA. Interestingly, in preliminary experiments, we were not able to observe an interaction between Brd4 and the EBNA1 protein of EBV (data not shown), even though EBNA1, like BPV-1 E2 and KSHV LANA, is required for episome maintenance. The C terminus of LANA contributes the major binding site for Brd4, whereas the highly acidic central repeats of LANA constitute a minor binding site for Brd4. We demonstrate that binding of Brd4 with the C terminus of LANA is mediated by a direct protein-protein interaction that involves the ET domain of Brd4. The significance of this interaction was demonstrated by the colocalization of LANA and Brd4 in both BJAB cells stably expressing LANA and in the KSHV-infected cells (Fig. 6 and 7). The LANA-Brd4 colocalization was most prominently observed on the host mitotic chromosomes in cells containing KSHV DNA or artificial viral episomes.
In addition to Brd4, the bromodomain protein RING3/Brd2 has been shown to interact with LANA (39). Similar to Brd4, RING3/Brd2 contains two bromodomains and an ET domain. The C terminus of LANA has been previously shown to interact with the ET domain of RING3/Brd2 in an insect cell lysate (39). Further studies showed that although RING3/Brd2 was primarily localized to the euchromatin and dissociated from the chromosomes during mitosis in several KSHV-negative cell lines, it is relocated almost completely to mitotic chromosomes in KSHV-infected body cavity lymphoma cells (36). Consistent with some of these results, we showed that LANA interacts directly with the ET domain of RING3/Brd2 in vitro. In addition, in KSHV-uninfected cells, RING3/Brd2 localizes to the interphase nucleus but primarily does not localize to chromosomes in mitosis (Fig. 7I). However, in KSHV-infected human B lymphocytes, we found very little colocalization between LANA and RING3/Brd2 on host mitotic chromosomes (Fig. 7E to H). This observation was confirmed by using our anti-Brd2 polyclonal antibody and an antibody raised against a Brd2 peptide to analyze the RING3/Brd2 localization. Further analysis revealed that the Brd2 antibody used in the previous study (36) cross-reacts with Brd4 protein (data not shown and T. F. Schulz, personal communication), indicating that the initially reported immunofluorescent staining actually detected both RING3/Brd2 and Brd4 signals (36). The fact that the RING3/Brd2 protein is nearly completely excluded from the mitotic chromosome in KSHV-negative cell lines and merely became weakly engaged with mitotic chromosomes in KSHV-infected cells suggests that LANA recruits some of the RING3/Brd2 protein to host mitotic chromosomes, as previously proposed (36). Dey et al. observed an interaction of RING3/Brd2 with mitotic chromosomes (13). It is possible that the association requires unknown ancillary factors that may be missing in KSHV-infected B lymphocytes. In contrast to RING3/Brd2, Brd4 remains associated with condensed chromosomes throughout mitosis (14) (Fig. 6 and 7). Although similar in sequence, the distinct localization patterns of RING3/Brd2 and Brd4 pointed to unique functional roles for each protein.
For the full-length LANA protein, the N terminus is considered to be the dominant chromosome-binding region (3, 30, 38, 46). However, the C terminus appears to constitute a second independent chromosome-binding site (30). The LANA C terminus localizes to dots on mitotic chromosomes in the absence of episomes (K. M. Kaye, unpublished data). It has also been shown that the LANA C-terminal domain is necessary and sufficient for LANA to localize to the discrete nuclear speckles characteristic of the native protein in interphase in the absence of episomes (7, 38, 44, 50, 52). These findings indicate that the C terminus and its binding partners specify LANA's localization to nuclear speckles, although interaction with RING3/Brd2 is not sufficient for the speckle formation (44). We found that the LANA C terminus directly interacts with mitotic chromosome-associated Brd4 (Fig. 5), and the bulk of both proteins colocalize in the punctate nuclear dots on interphase nuclei as well as mitotic chromosomes. Further studies will be needed, however, to test the hypothesis that Brd4 is the actual binding target for the LANA C terminus on mitotic chromosomes.
LANA is a highly acidic protein composed of a basic N terminus, an internal acidic repeat region, and a basic C-terminal domain. The charge structure suggested that LANA might have strong internal attractions. Ballestas et al. have provided data indicating that subnuclear localization of LANA is altered in the presence of the viral genomes (1). Although the altered localization is likely due to a high concentration of LANA-binding sites on KSHV DNA, the conformation of LANA may also be altered upon DNA binding. In this study, the colocalization of LANA with Brd4 was most distinctly observed in KSHV-infected cells, in which both proteins appeared in high-density punctate dots on mitotic chromosomes (Fig. 7A to D). In the absence of viral episomes, LANA localizes rather diffusely on the chromosomes and only partially overlaps with Brd4 protein present in nuclear dots (Fig. 6A to D). Upon introduction of artificial viral genomes that contain multiple TR repeats, the diffuse localization of LANA on the chromosomes changes to punctate staining (Fig. 6E to H), and LANA becomes clearly colocalized with Brd4 in punctate nuclear dots. The clear colocalization of LANA with Brd4 in the presence of viral genomes (Fig. 6H and 7D) could be due to increased detection of concentrated Brd4 at sites of LANA bound to DNA. Alternatively, certain functional domains within the LANA C terminus might be exposed and/or activated following LANA binding to viral DNA. For instance, binding to viral genomes could alter the structure of LANA and enhance Brd4 binding. The in vitro system we established in this study should permit further dissection of the mechanisms involved in the regulation of LANA-Brd4 interactions.
As a mitotic chromosome-associated protein, Brd4 provides a natural docking site for episomal viral genomes. Brd4 tethers BPV-1 E2 and the viral genomes to mitotic chromosomes through its C-terminal domain (58, 59). Experiments in which the binding of E2 to Brd4 was disrupted established that Brd4 is essential for BPV-1 E2/viral DNA binding to mitotic chromosomes (58, 59). However, the mechanism of LANA-mediated chromosomal tethering appears to be more complex than the mechanism for tethering BPV-1 E2, and it seems to require more complex protein-protein interactions. DEK1 has been proposed to tether C-terminal LANA to chromosomes (30). The data presented here provide an additional cellular factor through which LANA and KSHV genomes might access host chromosomes.
The finding that LANA and Brd4 are both involved in regulating the CDK2 promoter is highly intriguing. Brd4 plays an important role in cellular growth control and cell cycle progression (22, 35). Recently, Brd4 has been implicated in the positive regulation of RNA polymerase II-dependent transcription (25, 56), and we have found that Brd4 is required for papillomavirus E2-mediated transcriptional activation (45). LANA is also a multifunctional protein involved in modulation of both viral and cellular gene expression as well as the regulation of cellular proliferation and apoptosis (51). The findings here suggest that it is possible that Brd4 is involved in some of these functions of LANA and as such contribute to the pathogenesis of KSHV-associated disorders.
Acknowledgments
We thank the members of our laboratories for helpful discussions, Carrie Wu and Katherine Newcomb for technical assistance, and Keiko Ozato for the Brd4 antibodies.
This work has been supported by grants from the National Cancer Institute to P.M.H. (P01CA050661 and R01CA116720) and to K.M.K. (CA082036). J.Y. has been supported by a fellowship from the Charles A. King Trust, Bank of America, cotrustees.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856-861. [DOI] [PubMed] [Google Scholar]
- 5.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 7.Canham, M., and S. J. Talbot. 2004. A naturally occurring C-terminal truncated isoform of the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus does not associate with viral episomal DNA. J. Gen. Virol. 85:1363-1369. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, M. A., Jr., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, M. A., Jr., C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 12.Denis, G. V., and M. R. Green. 1996. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 10:261-271. [DOI] [PubMed] [Google Scholar]
- 13.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fakhari, F. D., J. H. Jeong, Y. Kanan, and D. P. Dittmer. 2006. The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J. Clin. Investig. 116:735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 18.Ganem, D. 1997. KSHV and Kaposi's sarcoma: the end of the beginning? Cell 91:157-160. [DOI] [PubMed] [Google Scholar]
- 19.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 20.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 21.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 26.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 28.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 29.Komatsu, T., M. E. Ballestas, A. J. Barbera, B. Kelley-Clarke, and K. M. Kaye. 2004. KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology 319:225-236. [DOI] [PubMed] [Google Scholar]
- 30.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 33.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 34.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maruyama, T., A. Farina, A. Dey, J. Cheong, V. P. Bermudez, T. Tamura, S. Sciortino, J. Shuman, J. Hurwitz, and K. Ozato. 2002. A mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 22:6509-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattsson, K., C. Kiss, G. M. Platt, G. R. Simpson, E. Kashuba, G. Klein, T. F. Schulz, and L. Szekely. 2002. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 83:179-188. [DOI] [PubMed] [Google Scholar]
- 37.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 38.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 41.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakakibara, S., K. Ueda, K. Nishimura, E. Do, E. Ohsaki, T. Okuno, and K. Yamanishi. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 78:7299-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweiger, M. R., J. You, and P. M. Howley. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 80:4276-4285. [DOI] [PMC free article] [PubMed]
- 46.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 49.Szekely, L., F. Chen, N. Teramoto, B. Ehlin-Henriksson, K. Pokrovskaja, A. Szeles, A. Manneborg-Sandlund, M. Lowbeer, E. T. Lennette, and G. Klein. 1998. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J. Gen. Virol. 79:1445-1452. [DOI] [PubMed] [Google Scholar]
- 50.Szekely, L., C. Kiss, K. Mattsson, E. Kashuba, K. Pokrovskaja, A. Juhasz, P. Holmvall, and G. Klein. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80:2889-2900. [DOI] [PubMed] [Google Scholar]
- 51.Verma, S. C., and E. S. Robertson. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155-163. [DOI] [PubMed] [Google Scholar]
- 52.Viejo-Borbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. A domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 77:7093-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe, T., M. Sugaya, A. M. Atkins, E. A. Aquilino, A. Yang, D. L. Borris, J. Brady, and A. Blauvelt. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J. Virol. 77:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong, L. Y., G. A. Matchett, and A. C. Wilson. 2004. Transcriptional activation by the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J. Virol. 78:10074-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie, R. L., S. Gupta, A. Miele, D. Shiffman, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 2003. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. J. Biol. Chem. 278:26589-26596. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 57.Ye, F. C., F. C. Zhou, S. M. Yoo, J. P. Xie, P. J. Browning, and S. J. Gao. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78:11121-11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]
- 59.You, J., M. R. Schweiger, and P. M. Howley. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 79:14956-14961. [DOI] [PMC free article] [PubMed] [Google Scholar]