Abstract
This review focuses on the enzymes and pathways of RNA processing and degradation in Bacillus subtilis, and compares them to those of its gram-negative counterpart, Escherichia coli. A comparison of the genomes from the two organisms reveals that B. subtilis has a very different selection of RNases available for RNA maturation. Of 17 characterized ribonuclease activities thus far identified in E. coli and B. subtilis, only 6 are shared, 3 exoribonucleases and 3 endoribonucleases. Some enzymes essential for cell viability in E. coli, such as RNase E and oligoribonuclease, do not have homologs in B. subtilis, and of those enzymes in common, some combinations are essential in one organism but not in the other. The degradation pathways and transcript half-lives have been examined to various degrees for a dozen or so B. subtilis mRNAs. The determinants of mRNA stability have been characterized for a number of these and point to a fundamentally different process in the initiation of mRNA decay. While RNase E binds to the 5′ end and catalyzes the rate-limiting cleavage of the majority of E. coli RNAs by looping to internal sites, the equivalent nuclease in B. subtilis, although not yet identified, is predicted to scan or track from the 5′ end. RNase E can also access cleavage sites directly, albeit less efficiently, while the enzyme responsible for initiating the decay of B. subtilis mRNAs appears incapable of direct entry. Thus, unlike E. coli, RNAs possessing stable secondary structures or sites for protein or ribosome binding near the 5′ end can have very long half-lives even if the RNA is not protected by translation.
INTRODUCTION
In recent years, it has become clear that the common assumption that Bacillus subtilis is essentially Escherichia coli with a cell wall and the capacity to sporulate could not be further from the truth. While both organisms respond similarly to environmental stimuli, as, indeed, do the vast majority of mesophilic bacteria, the mechanisms underlying these responses have proven to be very different in practically every case examined. While an understanding of the mechanisms of RNA processing and degradation in B. subtilis to the depth available for E. coli is perhaps several years away, a simple comparison of the genomes of the two organisms (Table 1) provides sufficient evidence that the same will hold true for this particular aspect of cellular metabolism. In E. coli, a key enzyme of RNA decay is RNase E (178). It is an essential enzyme, responsible for the initial rate-limiting cleavage in the decay of many mRNAs (reviewed in references 77 and 121), as well as playing an important role in 5S and 16S rRNA processing (70, 128). However, neither the gene for RNase E, rne, nor that for oligoribonuclease, orn, also essential for E. coli viability (6, 71, 178), is present in B. subtilis. Also missing are the genes encoding a battery of exonucleases involved in tRNA maturation, namely, RNase T, RNase BN, and RNase D (124, 126). In contrast, B. subtilis, and other low-G+C gram-positive bacteria, have at least three enzymes identified thus far that are not found in E. coli: RNase M5, RNase Bsn and YhaM. Thus, only 6 of a total of 17 characterized RNase activities are shared between B. subtilis and E. coli: 3 exoribonucleases, PNPase, RNase R, and RNase PH, and 3 endoribonucleases, RNase III, RNase P, and RNase H (44). It is thought that the evolutionary split between E. coli and B. subtilis occurred more than a billion years ago, even before the split between plants and animals (177, 183, 231). Thus, it is difficult to go much deeper than this in evolutionary terms to compare the enzymes and pathways of bacterial RNA degradation of two well-studied organisms. It seems likely that the RNases and pathways of degradation that have been identified, or remain to be identified, in these two organisms will account for the vast majority of degradation mechanisms to be found in the whole eubacterial kingdom. By the same token, the enzymes and pathways these organisms have in common will probably be generally conserved throughout the eubacteria (see arguments in reference 67).
TABLE 1.
Comparison of RNases present in E. coli and B. subtilis
| Exoribonucleases | Endoribonucleases
|
||||||
|---|---|---|---|---|---|---|---|
| Name | Gene in:
|
Homologya (%) | Name | Gene in:
|
Homology (%) | ||
| E. coli | B. subtilis | E. coli | B. subtilis | ||||
| PNPase | pnp | pnpA | 52/69 | RNase III | rnc | rncS | 36/60 |
| RNase R | rnr | rnr | 39/59 | RNase P | rnpAB | rnpAB | 36/63 |
| RNase PH | rph | rph | 56/73 | RNase HI | rnhA | —b | |
| YhaM | yhaM | RNase HII | rnhB | rnhB | 48/65 | ||
| RNase II | rnb | RNase HIII | rnhC | ||||
| RNase D | rnd | RNase M5 | rnmV | ||||
| RNase BN | rbn | RNase Bsn | yurI | ||||
| RNase T | rnt | RNase E | rne | ||||
| Oligo-RNase | orn | RNase G | cafA | ||||
| RNase I | rna | ||||||
Percent homology is given as percent identity/percent similarity.
—, Although a protein with 46% similarity to E. coli RNase HI, YpdQ, exists in B. subtilis, it has been shown not to possess RNase H activity.
The decay of most mRNAs in E. coli is thought to be initiated by endonucleolytic cleavage by RNase E (158), an enzyme with relatively loose substrate specificity (AU-rich single-stranded regions) (63, 130, 147). This first cleavage is rate limiting and is dependent on the phosphorylation state of the 5′ end of the molecule; RNAs with 5′-triphosphates (primary transcripts) are cleaved much less efficiently than RNAs with 5′-monophosphates (resulting from a prior cleavage event) (137, 138, 233). Thus, once the initial cleavage event has occurred, RNAs are very rapidly cut into smaller pieces. While this process is generally cited as proceeding with an overall 5′-to-3′ directionality, specific examples are not so abundant. Indeed, a recent paper has suggested that RNase E may have intrinsic 3′-to-5′ directionality on short RNA fragments (66). In a limited number of cases, the initial endonucleolytic cleavage is catalyzed by RNase III (11, 143, 188, 194, 195), an enzyme with far more restricted specificity (double-stranded RNA hairpins with specific side bulges) (34, 118, 200, 212, 242). The RNA fragments generated by endonucleolytic cleavage are attacked by the exoribonucleases, polynucleotide phosphorylase (PNPase) and RNase II (22, 57, 86), which progressively remove individual nucleotides from the 3′ end of the molecules either phosphorolytically (PNPase) (115, 222) or hydrolytically (RNase II) (171). Both of these enzymes are significantly slowed by secondary structures (37, 38, 81, 148) and are thought to be allowed multiple attempts at degrading such structures by successive rounds of addition and degradation of poly(A) tails, which serve as “on-ramps” for the exonucleases, PNPase in particular (19, 39, 40, 42, 140, 175, 233). The association of RNase E, PNPase, enolase, and an RNA helicase (RhlB) for unwinding structured RNAs in a complex known as the degradosome (31, 41, 152, 190, 191) is thought to facilitate the coordination of the RNA decay process (133). The C-terminal half of RNase E provides the scaffolding for the assembly of this complex (223). There is also some evidence that poly(A) polymerase, the enzyme responsible for the addition of poly(A) tails, is associated with RNase E at substoichiometric levels (192), oiling the mechanism even further.
The absence of an obvious RNase E homolog in B. subtilis (119) suggests that the pathway of mRNA degradation in this organism is fundamentally different, if only from the standpoint of the degradosome. Although it is possible that some other protein acts as the scaffold for the formation of a complex between PNPase, enolase, and helicase, we have failed to demonstrate an association between these proteins in B. subtilis (D. Brechemier-Baey, H. Putzer, and C. Condon, unpublished results). In this review, I describe what is known about the different RNases found in B. subtilis, their substrates, and the different stability determinants identified thus far.
THE ENZYMES
The Exoribonucleases
Polynucleotide phosphorylase.
PNPase removes nucleotides processively from RNAs in the 3′-to-5′ direction (115, 222). The reaction is phosphorolytic; i.e., organic phosphate is consumed and 5′-diphosphate nucleosides are released on cleavage of each phosphodiester bond. PNPase activity was first clearly demonstrated in B. subtilis by Deutscher and Reuven (54) and shown to be the predominant degradative activity in cell extracts with poly(A) or poly(U) RNA as a substrate. This was in contrast to E. coli, where, although similar absolute levels of phosphorolytic activity were present, the predominant exonucleolytic activity was the hydrolytic activity of RNase II. These data were in support of previous experiments, based on the relative incorporation of 18O atoms into nucleotides and RNA, that suggested that a maximum of 20% of degradative activity on total RNA was hydrolytic in B. subtilis whereas around 70% was hydrolytic in E. coli (32, 61). Similar experiments to those of Deutscher and Reuven were later performed by Wang and Bechhofer (226). Although essentially the same results were obtained with poly(A) RNA in B. subtilis (i.e., the primary degradative activity was phosphorolytic), the major degradative activity of cell extracts on total cellular RNA was Mn2+ dependent and hydrolytic. It is likely that a significant proportion of rRNA in the preparations of total cellular RNA account for the difference (D. Bechhofer, personal communication), since rRNA is degraded primarily by the hydrolytic activity of RNase R (see below).
The gene encoding B. subtilis PNPase was identified through its role in competence development. A screen for mini-Tn10 insertions that prevented competence development identified the comR gene (136). The gene was sequenced and identified as the gene encoding PNPase, by virtue of its homology to the E. coli enzyme. Its role in competence is thought to be, paradoxically, through the increase in expression of the 27-kb surfactin operon (srfA) at a posttranscriptional level, although it is not clear whether this role is direct or indirect.
PNPase is a cold shock protein in E. coli (18, 142), and inactivation of the PNPase gene in both E. coli and B. subtilis results in a cold-sensitive phenotype, with cessation of growth at around 16°C (226). B. subtilis pnpA deletion mutants have a slightly increased half-life of total mRNA, a phenomenon that is exacerbated as the temperature decreases. They also show pleiotropic effects, such as increased sensitivity to tetracycline and filamentous growth, which are thought to reflect defects in the degradation of specific RNAs (226).
E. coli strains in which both major exonucleases, PNPase and RNase II, are inactivated are not viable (57). Curiously, however, B. subtilis, which does not have an RNase II enzyme, tolerates inactivation of the pnpA gene. Clearly other hydrolytic exonucleases compensate for the lack of RNase II activity.
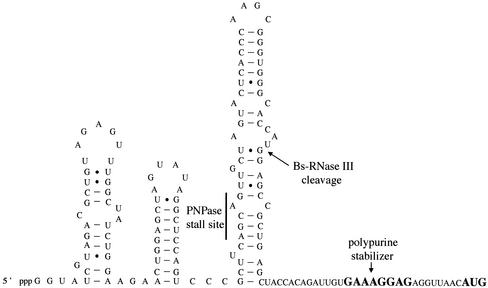
Although B. subtilis PNPase can functionally complement E. coli PNPase mutants (226), the B. subtilis and E. coli enzymes have different degradation properties on the same substrate. Mitra et al. showed that in time course reactions, a short RNA fragment from bacteriophage SP82 with a single-stranded 3′ region was degraded similarly by both enzymes (154). However, a longer fragment with a stable stem-loop at the 3′ end was a much poorer substrate for B. subtilis PNPase than for its E. coli counterpart, suggesting that B. subtilis PNPase might be more sensitive to structured RNA. An RNA structure than can actually halt B. subtilis PNPase progress both in vivo (15) and in vitro (154) has been found in the phage SP82 RNA (see Fig. 3).
FIG. 3.
Sequence and proposed secondary structure of the A-region at the 5′ end of phage SP82. The sites of RNase III cleavage and PNPase stalling (see the text) are indicated. Also shown is the polypurine sequence that functions as a 5′ stabilizer even in the absence of translation initiation at the AUG codon. Bs-RNase III, B. subtilis RNase III.
As in E. coli, the PNPase gene lies just downstream of the rpsO gene, encoding ribosomal protein S15. PNPase autoregulates its own expression in E. coli at the posttranscriptional level (199). Recently, it has been shown that this autoregulation occurs through degradation by PNPase of one strand of an RNA duplex, the product of cleavage of a stable secondary structure by RNase III in the intergenic region between the rpsO and pnp genes (100). Although a similar structure exists in the rpsO-pnpA intergenic region in B. subtilis, this structure is apparently not cleaved by B. subtilis RNase III (154). Thus, it remains to be seen whether and how B. subtilis PNPase might be autoregulated.
RNase R.
Although B. subtilis has no detectable RNase II activity, sequencing of the B. subtilis genome revealed the presence of a gene (yvaJ) whose product has a high degree of homology to both E. coli RNase II (28% identity and 48% similarity) and RNase R (39% identity and 59% similarity). Both RNase R and RNase II belong to the RNR family of exoribonucleases (244) and degrade RNA hydrolytically to small oligonucleotides. RNase R digests RNA processively (i.e., the enzyme does not release its substrate between hydrolytic cycles) to di- or trinucleotides (36). On the other hand, RNase II degradation is initially processive but becomes more distributive as the molecules become smaller, approaching an undigested core of 3 to 5 nucleotides (nt). RNase R has significantly more success in degrading highly structured RNAs such as rRNA, and this is thought to be its primary substrate in vivo. Oussenko and Bechhofer showed that inactivation of yvaJ essentially abolished Mg2+- dependent hydrolytic degradation of total cellular RNA by B. subtilis cell extracts and that while 23S rRNA was degraded by yvaJ+ extracts, it was left essentially intact when incubated with mutant extracts (179). This provided strong evidence that yvaJ encodes the B. subtilis RNase R protein, and the gene was thus renamed rnr. Although it shows a preference for Mg2+ as a divalent cation, RNase R can also function in the presence of Mn2+ ions and, as such, accounts for some 40% of the Mn2+-dependent exonuclease activity described by Wang and Bechhofer (226). B. subtilis strains with both pnpA and rnr genes inactivated are viable, although the double mutant grows about 25% slower than the parental pnpA strain does. This is in sharp contrast to E. coli, where pnp rnr double mutants are inviable (35).
RNase PH.
B. subtilis RNase PH is encoded by the rph gene and shows 56% identity and 73% similarity to its E. coli counterpart. E. coli RNase PH degrades RNA phosphorolytically in the 3′-to-5′ direction (49, 53), like PNPase, to which it shows significant homology (44%). However, its activity seems to be restricted to the removal of the last few nucleotides from the 3′ end of tRNAs. Since this is a redundant function in E. coli (RNase T, RNase BN and RNase D, RNase II, and PNPase can fulfill essentially the same role [52, 126, 198]), rph mutants do not have a major phenotype. Mutations in RNase PH become inviable in E. coli only when combined with four other exonuclease mutations, i.e., rnt, rbn, rnd, and rnb (107, 108). Given that such enzyme redundancy does not appear to exist in B. subtilis, it is interesting that rph is not essential in this organism (49). E. coli PNPase RNase PH double mutants are much more cold sensitive than are PNPase mutants alone (243). After a shift to the lower temperature, ribosome synthesis is severely perturbed and 23S rRNA is rapidly degraded. This suggests some essential role for these two phosphorolytic exonucleases in rRNA metabolism in E. coli. It remains to be seen whether this is also true in B. subtilis.
YhaM.
The YhaM protein has recently been identified as an Mn2+-dependent 3′-to-5′ exonuclease, active on both RNA and single-stranded DNA (180). It was purified from a B. subtilis strain lacking PNPase and RNase R, and its amino acid sequence, and hence its gene, was identified by mass spectroscopy analysis. YhaM has a molecular mass of 35.5 kDa. It is also active in the presence of Co2+ but inactive in the presence of Mg2+ ions. Inactivation of the yhaM gene has little effect on exponential-phase bacterial growth either by itself or in combination with a pnpA mutation or a pnpA rnr double mutation. The triple mutant does, however, show longer lag times and increased cold sensitivity compared to its parent. A YhaM homolog from S. aureus, CBF1, was also shown to have 3′-to-5′ exoribonuclease activity. Interestingly, CBF1 binds cmp, the replication enhancer of plasmid pT181 (69), suggesting that it also plays a role in DNA metabolism.
YhaM is predicted to contain an OB-fold, found in a particular class of nucleic acid binding proteins (160), and a HD domain, the signature of metal-dependent phosphohydrolases (7). Indeed, YhaM was one of four B. subtilis proteins predicted to be RNases by Aravind and Koonin (8), based on the presence of these domains (see below). The OB-fold and HD domains are from residues 17 to 90 and from 160 to 279, respectively. Based on the conservation and relative position of these domains, 11 other orthologs of YhaM have been identified, and these are found only in gram-positive bacteria (180).
The size of YhaM would suggest that it is not the same as an Mn2+-dependent nuclease identified by Kerjan and Szulmajster (see below), which had a molecular mass of 72 kDa on denaturing gels (109, 110). Furthermore, YhaM was purified from late-log-phase cells whereas the Kerjan enzyme was isolated from sporulating cells. Whole-cell extracts of the triple pnpA rnr yhaM mutant still have Mn2+-dependent degradative activity (I. Oussenko and D. Bechhofer, personal communication), consistent with the idea that yet another Mn2+-dependent enzyme remains to be discovered. An Mn2+-dependent hydrolytic exonuclease which eluted from a gel filtration column at around 100 kDa but whose gene was never identified was also identified in B. subtilis by Deutscher and Reuven (54). The size of YhaM under nondenaturing conditions is not yet known.
The Endoribonucleases
RNase III.
RNase III activity was first identified in B. subtilis by Panganiban and Whiteley (186) based primarily on its size and substrate specificity (cleavage of double-stranded RNAs). The enzyme was purified from B. subtilis cell extracts, with the key step being chromatography on a poly(I)-poly(C) agarose column. The enzyme was initially reported to behave as a monomer of 27 kDa and to cleave double-stranded RNAs in the apical loop at the 5′ side of adenosine residues. A high degree of species specificity was also reported when the in vitro activity of B. subtilis RNase III was compared to its E. coli counterpart; B. subtilis RNase III could cleave only B. subtilis SP82 phage RNA and B. subtilis rRNA whereas E. coli RNase III could cleave only T7 phage RNA and E. coli rRNA under the assay conditions used. Using similar substrates, Mitra and Bechhofer obtained very different results (153). While in their hands E. coli RNase III was indeed unable to cleave SP82 RNA at the so-called A site, B. subtilis RNase III cleaved T7 RNA with specificity identical to that of the E. coli enzyme. Moreover, the so-called A, B, and C cleavage sites of B. subtilis RNase III in SP82 RNA were mapped to side bulges in the double-stranded RNAs rather than in the apical loops, with no preference for adenosine residues, more in line with what was expected from studies of the E. coli enzyme. This group also subsequently showed that B. subtilis RNase III could complement the defect in rRNA processing in E. coli rnc mutants in vivo (227). There is no obvious explanation for the difference between the studies of these two groups. The enzyme used was purified 2,000-fold in the Mitra and Bechhofer study (153), compared to 150-fold in the Panganiban and Whiteley study (186). It is possible that some contaminating factor affected substrate specificity in the earlier study.
The B. subtilis rnc gene was identified by its homology (60%) to E. coli RNase III (174). Plasmids containing this gene were shown to permit cleavage of the lambda N leader RNA in vivo, another known substrate of RNase III in E. coli (227). RNase III is an essential enzyme in B. subtilis, in contrast to E. coli, where rnc mutants are viable. All attempts to inactivate the B. subtilis rnc gene completely were unsuccessful, except in some rare cases where extragenic suppressor mutants were isolated (91, 227). The mapping of these suppressor mutations should provide some clues to why RNase III is essential in B. subtilis.
The E. coli rnc gene is autoregulated at the posttranscriptional level. Cleavage of a stable hairpin by RNase III, just upstream of the rnc coding sequence, destabilizes the RNA (11, 143, 144). Although a similar hairpin can be found about 100 nt upstream of the B. subtilis rnc gene, there is no evidence of cleavage of the rnc mRNA in vitro within 500 nt of the coding sequence (227). This is reminiscent of the situation that occurs with the B. subtilis gene encoding PNPase (above); i.e., the potential for autoregulation exists, but for some reason this particular mechanism is not exploited by B. subtilis. Presumably, an understanding of the different nature of the E. coli and B. subtilis RNA degradation pathways will shed light on this intrigue.
The construction of a strain conditionally expressing B. subtilis RNase III permitted an analysis of the role of RNase III in rRNA processing in B. subtilis (91). While inactivation of the rnc gene in E. coli allows one to detect an accumulation of a 30S rRNA precursor in ethidium bromide-stained agarose gels (227), one has to resort to Northern blot analysis to detect the small quantities of 30S rRNA that accumulate after rnc inactivation in B. subtilis (91). While this shows clearly that B. subtilis RNase III is involved in rRNA processing, it suggests that an alternative pathway for production of 16S and 23S rRNA exists in B. subtilis that is even more effective than that of E. coli. It also implies that the essential function of RNase III in B. subtilis is not rRNA processing.
The rnc gene of B. subtilis is located in an operon upstream of the smc (for “structural maintenance of chromosomes”) and srb (homolog of the signal recognition particle [SRP] α-subunit) genes. Interestingly, B. subtilis RNase III cleaves small cytoplasmic RNA (scRNA) precursor, a member of the SRP RNA family and a component of the SRP receptor, at 5′ and 3′ sites both in vitro (173) and in vivo (91). Depletion of scRNA leads to cell death in B. subtilis (163); thus, it was possible that maturation of this RNA was the essential function provided by B. subtilis RNase III. However, strains lacking the rnc gene and surviving due to an extragenic suppressor mutation (above) also fail to process scRNA, eliminating this possibility (91).
RNase M5.
Sogin and Pace identified an activity in B. subtilis cell extracts responsible for maturation of 5S rRNA, which they called RNase M5 (215). This enzyme cleaves the 5S rRNA precursor endonucleolytically on both sides of a double-stranded region to yield mature 5S rRNA in one step. Ribosomal protein L18, which binds 5S rRNA, is a cofactor in the reaction (218). Since the role of L18 can be replaced by 25% dimethyl sulfoxide, it is thought that L18 acts an RNA chaperone, putting the 5S precursor molecule in the correct conformation for cleavage (182). High concentrations of ribosomal protein L5, which also binds 5S rRNA, inhibit the cleavage reaction (218). RNase M5 has been estimated to be present at about 100 molecules per B. subtilis cell (43, 184). The enzyme was purified to homogeneity almost 20 years ago and estimated to be about 24 kDa in size (182). However, its gene was only recently identified. RNase M5 was repurified from B. subtilis, and after considerable difficulty in separating it from ribosomal protein L6, sufficient material was obtained to sequence its N terminus and identify its gene (43). The RNase M5 gene was one of previously unknown function, yabF, and was renamed rnmV. It is highly conserved throughout the low-G+C gram-positive kingdom and in the spirochete Borrelia burgdorferi. Interestingly, there is very little overlap between the organisms that have an RNase M5 enzyme and those that possess an RNase E-type enzyme to process their 5S rRNA. Only members Clostridium and Listeria species and Bacillus halodurans have both types of 5S rRNA maturases (44). Both enzymes are capable of processing Clostridium difficile 5S rRNA in vitro (43).
The N-terminal half of RNase M5 shows significant homology to a domain known as the Toprim domain, found at the active site of the archaeal reverse gyrases, members of the topoisomerase family, and the DNA primases (9). Topoisomerases cleave double-stranded DNA to allow strand passage while relaxing supercoils in the DNA and have even been shown to be capable of cleaving RNA under certain circumstances (55, 213). It thus seems likely that the cleavage mechanism of RNase M5 will prove to be very similar to that of the topoisomerases.
B. subtilis strains lacking a functional rnmV gene are viable and show only small growth rate defects (of the order of 25% in rich medium). These strains have long lag times that increase with increasing time spent in stationary phase (H. Putzer and C. Condon, unpublished results). No mature 5S rRNA whatsoever can be detected in ΔrnmV strains, showing that no other enzyme can take the place of RNase M5 (43). Precursor 5S rRNA molecules accumulate in both ribosomes and polysomes, suggesting that maturation of 5S rRNA is dispensable for ribosome function in B. subtilis at least.
Using gene array technology, it was recently shown that RNase M5 probably has no substrates in B. subtilis apart from the 5S rRNA precursor. Although the expression of two citric acid cycles enzymes, 2-oxoglutarate dehydrogenase and succinyl coenzyme A synthase, was significantly increased in rnmV mutants in late log phase, this effect appears to be indirect, via a mechanism as yet poorly understood (47). The apparent absence of other substrates for RNase M5 means that there is some feature of the 5S rRNA precursor induced by the binding of ribosomal protein L18 that is unique among B. subtilis RNAs.
Pace and coworkers showed that RNase M5 recognizes the double-stranded nature of the processing site rather than having particular sequence requirements (150, 217). Mutations which disrupted Watson-Crick base pairing at the cleavage site had a significant negative effect on the processing reaction, whereas compensatory mutations in the opposite strand restored cleavage (217). Of the mature 5S sequence, only the domain that binds ribosomal protein L18 and the helix where cleavage actually occurs are necessary for cleavage. The so- called prokaryotic loop (68), where L25 binds in E. coli, can be deleted without resulting in a significant effect on the maturation reaction (151).
Cleavage of 5S precursor by RNase M5 yields three fragments, mature 5S rRNA, a 21- nt 5′-fragment, and a 42-nt 3′ fragment. Schroeder et al. showed that the 5′ and 3′ cleavage products are very unstable in B. subtilis extracts and that their degradation occurs via a hydrolytic, rather than phosphorolytic, mechanism (211). This is of interest in light of the question whether exonucleolytic B. subtilis RNA degradation is largely phosphorolytic or hydrolytic in nature (above).
RNase P.
B. subtilis RNase P is a heterotetramer consisting of two protein subunits (encoded by rnpA) and two RNA subunits (encoded by rnpB) (65). The RNA subunit contains the catalytic site and, as such, constitutes a ribozyme (82), while the protein subunit facilitates substrate recognition (48, 120, 196). The primary role of RNase P is recognition and cleavage of tRNA precursors to create the mature tRNA 5′ ends. RNase P is essential in E. coli (202, 207, 229) and in B. subtilis (C. Condon and H. Putzer, unpublished results). The three-dimensional X-ray structure of B. subtilis RNase P protein (219) and a partial solution structure of the RNA moiety (122) are known, and detailed mechanistic studies of the recognition and cleavage process have been performed by several groups. The protein component shares an ancient fold with RNase PH (5, 8). The RNase P RNAs from E. coli and B. subtilis have significantly different secondary structure (99) and are the paradigms of type A and type B RNAs, respectively (25, 26, 83). This difference in structure apparently has an effect on the catalytic properties of the enzyme such as its monovalent- and divalent-cation requirements (82, 196, 228). It is interesting in this regard that B. subtilis RNase P RNA can complement a deletion of the E. coli equivalent (229).
RNase P from E. coli is known to have at least two substrates other than tRNA precursors. It is also involved in 4.5S RNA (an analog of scRNA) processing in E. coli (82), a task performed by RNase III in B. subtilis (above), and in processing of the Salmonella serovar Typhimurium his operon mRNA (3). Interestingly, while only E. coli RNase P RNA (M1 RNA) can cleave 4.5S RNA, this can be accomplished with either E. coli or B. subtilis RNase P protein as cofactor (82). No RNase P substrates other than tRNA precursors are known in B. subtilis.
RNase H.
RNase H cleaves RNA in RNA-DNA hybrid molecules endonucleolytically (106). Its primary function in E. coli seems to be to prevent aberrant DNA replication by degrading potential RNA primers of DNA synthesis at sites other than oriC (117, 172). E. coli has two genes encoding RNase H enzymes, rnhA and rnhB, encoding RNase HI and RNase HII, respectively (30, 97). Ninety percent of the cellular RNase H activity is encoded by rnhA (50). Inactivation of both enzymes leads to a temperature-sensitive phenotype in E. coli, although this is dependent on the genetic background. A genetic screen for B. subtilis genes, capable of complementing a temperature-sensitive E. coli rnhA rnhB mutant, yielded two positive clones (98). One was an obvious homolog of RNase HII, and the other was termed RNase HIII (rnhC), thus identifying a new family of RNase H genes in bacteria. A homolog of RNase HI (ypdQ), which was not picked up in the screen, can also be found in the B. subtilis chromosome. A plasmid expressing this gene does not complement the temperature-sensitive phenotype of the E. coli rnhA rnhB mutant, however (98), and this protein does not possess RNase H activity in vitro (176). Therefore, its function remains unknown. While inactivation of rnhA and rnhB together is possible in E. coli, giving at worst a temperature-sensitive phenotype, rnhB rnhC double mutants of B. subtilis are inviable, suggesting that the requirement of B. subtilis for RNase H in vivo is more stringent than that of E. coli (98).
RNase Bsn.
B. subtilis colonies growing on nutrient agar plates containing RNA produce a clear halo after addition of HCl, as a result of digestion of the RNA by one or more extracellular RNases. The gene coding for one of these RNases, RNase Bsn, was identified by shotgun cloning of chromosomal DNA fragments in B. subtilis and screening for colonies producing larger haloes than those transformed with the vector alone (162). The initial screen was performed with chromosomal DNA from B. subtilis strain IFO3034, and the gene encoding RNase Bsn was sequenced in its entirety. RNase Bsn shows 78% amino acid identity to the protein encoded by the yurI gene of B. subtilis strain W168 used in the sequencing project. We have recently shown that yurI mutants of B. subtilis W168 no longer have extracellular RNase activity (N. Vasnier, H. Putzer, and C. Condon, unpublished results), suggesting that the yurI gene encodes RNase Bsn. RNase Bsn has no apparent sequence specificity and can hydrolyze RNA endonucleolytically to yield 5′-phosphorylated oligonucleotides. A 51- to 53-amino-acid N-terminal peptide is removed on secretion of the enzyme. It is not known, however, whether the unprocessed protein is active within the cell.
RNase Bsn is also found in other Bacillus species. It has been discovered in Bacillus intermedius, where it has been termed binase II (84) to distinguish it from the better known guanyl-specific extracellular RNase, binase I. Binase I is a close homolog of barnase from Bacillus amyloliquifaciens, RNase Bp from Bacillus pumilus, RNase Bth from Bacillus thuringiensis, and RNase Bci from Bacillus circulans, for which structural studies abound (reviewed in reference 89). Although no homolog of barnase is found in B. subtilis, a gene potentially encoding its inhibitor, barstar, has been identified just upstream of the gene encoding the Lrp-like protein, AzlB (17).
Potential RNases
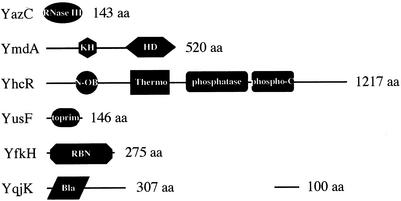
In addition to the yurI gene (above), six other B. subtilis genes of unknown function potentially encode RNases (Table 2). Three of these were identified by Aravind and Koonin, based on the presence of particular protein folds found in other known RNases (8). The product of the yazC gene is predicted to contain the all-helical fold of the RNase III family, for example. The YhcR protein (incorrectly labeled as YchR in reference 8) has an all-beta domain (the OB-fold) typically found in the thermonuclease family, and the ymdA gene product, like that of yhaM (above), is predicted to possess the all-helical HD (His Asp) domain found in the phosphohydrolase family of proteins. A fourth gene that can be added to this list is the yusF gene, which contains the Toprim domain found in RNase M5 (9). It is also possible, however, that this gene product is more functionally related to the topoisomerases or DNA primases. The yfkH gene product is classified in the RBN superfamily of exoribonucleases by Zuo and Deutscher (244). This superfamily includes E. coli RNase BN. Lastly, the yqjK gene encodes a strong homolog of RNase Z, an endoribonuclease responsible for the maturation of the 3′ end of tRNA molecules in plants and in the Archaea (208-210), and preliminary evidence suggests that it plays the same role in both B. subtilis and E. coli (C. Condon, H. Putzer, and A. Marchfelder, unpublished results). The domain structures of these potential RNases are shown in Fig. 1.
TABLE 2.
Potential B. subtilis RNases
FIG. 1.
Domain structure of potential B. subtilis RNases. aa, amino acids. Domain abbreviations: KH, ribonucleoprotein K homology domain; HD, His-Asp-containing domain of phosphoesterases; N-OB, oligonucleotide binding fold; Thermo, thermonuclease domain; Phospho-C, phosphatase C domain; RBN, RNase BN; Bla, β-lactamase domain.
RNases for Which Genes Are Not Yet Identified
A number of RNase activities, whose genes have not yet been identified, have been purified to various degrees from B. subtilis cell extracts. It is possible that some of the genes identified in the previous section will encode these functions. A broad-specificity enzyme called RNase C, because it preferentially degraded poly(C) RNA, was identified by Kennell and colleagues (141). This enzyme is characterized as an intracellular pyrimidine- specific endonuclease with a molecular mass of 15 kDa and a pH optimum of 6.2.
An Mn2+-stimulated exonuclease was purified from sporulating B. subtilis cells (109, 110). This RNase, which is sixfold more active in stationary phase than in log phase, has an estimated size of 150 kDa on glycerol gradients and 72 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Its 3′-to-5′ exonucleolytic activity is strongly stimulated by Mn2+ and inhibited by Ca2+, spermidine, and cGMP. The products of the hydrolytic cleavage reaction are nucleoside 5′-monophosphates. This enzyme appears to be distinct from the other characterized Mn2+-stimulated exonuclease of B. subtilis, YhaM, which has a molecular mass of 35.5 kDa on denaturing gels (above).
There have been a number of attempts to purify extracellular RNases from B. subtilis growth medium. A Ca2+-activated DNase/RNase was identified in cultures of B. subtilis SB19 by Kerr et al. (111, 112). This nuclease has a pH optimum of 9.5 for exonucleolytic degradation of RNA to nucleoside 3′-monophosphates. Apparently the same nuclease (Bs-II) and two others (Bs-IA and Bs-IB) were partially purified later by Kanamori et al. (103, 104). All three were Ca2+ dependent and had both RNase and DNase activity, leading to the production of nucleoside 3′-monophosphates. It is not clear whether these are distinct enzymes or different forms of the same enzyme. Based on the catalytic properties, they are apparently distinct from RNase Bsn, which produces 5′-phosphorylated oligonucleotides (see above). Two extracellular RNases were identified by Nakai et al. in the growth medium of B. subtilis Marburg (161), the sequenced strain, also known as W168. While one of these was Ca2+ activated, its pH optimum was only 5.0, it had no DNase activity, and the main product of the cleavage reaction was cyclic mononucleotides. The second had both RNase and DNase activity and a pH optimum of 9.3 and released 3′ mononucleotides, but unlike the enzyme identified by Kerr et al. (111, 112), it was apparently not stimulated by Ca2+ ions.
In the late 1950s to early 1970s, a number of other extracellular (169, 170, 201) and intracellular (168) RNases were described in B. subtilis H and K strains (235-238). The intracellular activity was strongly inhibited by ATP (or dATP), and the reaction products were cyclic 2′,3′-monophosphates. It was shown to have a molecular mass of around 25 kDa and a pH optimum of 5.7. While it turned out that the H and K strains were really Bacillus amyloliquifaciens species, this intracellular activity was also shown to exist in other B. subtilis strains, including B. subtilis natto (238), a close relative of B. subtilis W168, and in Bacillus cereus (221). Yamasaki et al. (238) also suggested that that the activity identified by Nakai et al. (161) (see above) corresponds to an excreted form of the ATP- inhibited intracellular RNase. It will be very interesting to see whether genes can be assigned to any of these functions in the future.
Polyadenylation
RNAs from both E. coli and B. subtilis are polyadenylated at their 3′ end (74, 75, 164, 216). Although the lengths of the tails are similar (10 to 20 A residues), B. subtilis contains intrinsically higher levels of poly(A) RNA than E. coli does (reviewed in reference 206). The first bacterial cDNA library, primed with oligo(dT), was made from B. subtilis mRNA (76, 105), but cDNA for only one specific mRNA was isolated, that of the hag gene, encoding flagellin (29). Interestingly, the site of poly(A) tail addition to hag mRNA was just upstream of the factor-independent transcription terminator, suggesting that, like E. coli, B. subtilis RNAs can be polyadenylated at sites of posttranscriptional processing of the primary transcript.
The addition of poly(A) tails destabilizes RNAs in E. coli (85, 90, 129, 155, 175, 234), and although it is assumed they have a similar effect in B. subtilis, this has not yet been directly demonstrated, mainly because the B. subtilis poly(A) polymerase gene has not yet been identified. The major poly(A) polymerase activity of E. coli, PAPI, is encoded by the pcnB gene (28). A residual poly(A) polymerase activity detectable in E. coli pcnB mutants, called PAPII, was reported to be encoded by the f310 gene (27), but this was subsequently convincingly shown not to be the case (156), and the activity has since been attributed to PNPase (157). Thus, PNPase appears to function as both a 3′-to-5′ exonuclease and, in the reverse reaction, a poly(A) polymerase. It is not clear what provokes the switch from degradation mode to polymerization mode in vivo; in vitro this can be accomplished by high nucleoside diphosphate concentrations. The tails added by PNPase are heterogeneous, with C and U residues being incorporated at a relatively high frequency.
B. subtilis does not have a true PAPI homolog. An apparent homolog, when more closely investigated, turned out to be nucleotidyltransferase, the enzyme that adds the CCA moiety to tRNA (193). It has been suggested that PNPase is the primary enzyme responsible for poly(A) tail synthesis in chloroplasts (241), which also lack a clear PAPI homolog. It will be interesting to see whether PNPase plays the same role in B. subtilis. Although two further poly(A) polymerase activities have been reported for a PNPase-minus B. subtilis strain (205), these enzymes were identified using the same filter binding assay that led to the erroneous attribution of the f310 gene in E. coli; the results therefore await further confirmation.
SUBSTRATES AND STABILITY DETERMINANTS
The degradation of only a dozen or so mRNAs has been studied in any detail in B. subtilis, and many of these are not endogenous to this organism. In some cases, similarities to degradation pathways of E. coli are evident. In others, however, it is quite clear that the mechanisms of the B. subtilis maturation and degradation process do not obey E. coli “rules.” In this section, I describe what is known about the processing of stable RNAs and the degradation of specific mRNAs, with a particular focus on the various stability determinants that have been identified.
Maturation of Stable RNAs
rRNA.
16S, 23S, and 5S rRNAs are cotranscribed from 10 rrn operons in B. subtilis, although, as in E. coli, full-length primary transcripts (30S) are not observed in wild-type cells. 30S precursor transcripts do, however, accumulate somewhat in RNase III mutant B. subtilis cells, although to significantly lower levels than those observed in E. coli rnc mutants (91), suggesting that cleavage of rRNA by RNase III in B. subtilis is less important. In both organisms, the RNase III cleavage can thus be bypassed by other enzymes. In E. coli, the endonucleases RNase E and RNase G combine to produce the mature 5′ end of 16S rRNA (128), which presumably accounts for the presence of significant quantities of mature 16S in cells completely lacking RNase III. 23S rRNA is not completely matured in E. coli rnc cells, however, but is nonetheless functional (113, 114). It is not yet known what enzymes are responsible for the production of mature 16S or 23S rRNA ends in B. subtilis.
B. subtilis 5S rRNA is cleaved by RNase M5 (see above) from a precursor molecule extending from the RNase III cleavage site at the 3′ side of 23S rRNA to the end of the operon (43, 215). No other enzyme appears to be capable of performing this cleavage reaction, since all 5S rRNA occurs in its precursor form in rnmV mutants in vivo. In E. coli, 5S rRNA is produced by cleavage of a 9S precursor first by RNase E, 3 nt on either side of the mature sequence (70), then by removal of the 3′ extension by RNase T (125). The enzyme responsible for removal of the 5′ extension is as yet unidentified.
tRNA.
tRNAs are processed at their 5′ ends by RNase P in both E. coli and B. subtilis. However, the enzymes producing the mature 3′ end differ somewhat. In E. coli, this task can be accomplished by any one of at least five exoribonucleases, RNase T, RNase BN, RNase D, RNase PH, and RNase II (126), following an initial endonucleolytic cleavage a few nucleotides downstream by RNase E, this being the function of RNase E that is essential for cell viability (127, 181). RNase PH is the only one of these enzymes to exist in B. subtilis, and its role in tRNA processing, if any, is unknown. A second, previously unsuspected pathway for generating mature 3′ ends of tRNAs, employing the endonuclease RNase Z, may also exist in both organisms and awaits confirmation (see above).
scRNA.
The stable B. subtilis RNA known as scRNA is structurally related to E. coli 4.5S RNA and eukaryotic 7SL RNA, both of which are components of the signal recognition particle in their respective organisms (135, 225). In B. subtilis, this RNA is synthesized as a precursor molecule with a ca. 40-nt extension at both ends (220). Whereas 4.5S RNA is processed by RNase P in E. coli (20), scRNA is processed by RNase III in B. subtilis (173). Cleavage of the scRNA precursor by RNase III yields an scRNA whose 5′ end is identical to that of the mature scRNA, whereas it contains four extra nucleotides at the 3′ end. It is not yet known which enzyme is responsible for the removal of these four nucleotides.
Stability Determinants of mRNAs
As will become evident in the following paragraphs, the 5′ end of transcripts is a major determinant of mRNA stability in B. subtilis, just as in E. coli. However, while access to the body of E. coli mRNAs through the 5′ end seems to be governed largely by the secondary structure, the phosphorylation state (mono- versus triphosphate), and active translation of the transcript, access to B. subtilis mRNAs can be inhibited by bound untranslating ribosomes and bound proteins, in addition to secondary structure. This suggests that while RNase E binds to the 5′ end and can loop to internal portions of the transcript to initiate degradation or can directly access cleavage sites more slowly in cases where a 5′ end is unavailable, the rate-limiting step of B. subtilis mRNA decay must be catalyzed by an enzyme which scans or tracks from the 5′ end uniquely. Specific examples of each of the different classes of 5′ mRNA stability determinants in B. subtilis are discussed in detail below. The role of the phosphorylation state of the 5′ end in B. subtilis has not yet been addressed.
mRNAs protected by ribosomes bound or stalled near the 5′ end.
A number of RNAs have been proposed to be stabilized by the binding or stalling of ribosomes near the 5′ end in B. subtilis. The ermA, ermC, and phage SP82 mRNAs have been characterized in the greatest detail. The cryIIIA mRNA also provides a convincing example of this phenomenon. The stability of the gsiB mRNA has also been proposed to be increased by ribosome binding near the 5′ end (102), but the data in this case are much more preliminary and are not discussed here. The aprE mRNA is stabilized by both ribosome binding and secondary structure near its 5′ end and is discussed in a subsequent section.
(i) ermA and ermC mRNAs.
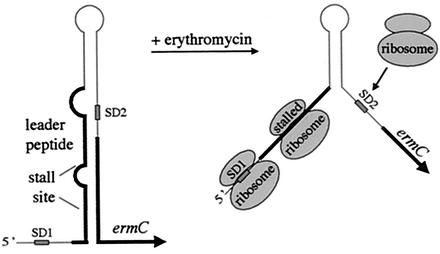
Regulation of the Staphylococcus aureus ermA and ermC genes has been studied in some detail in B. subtilis to take advantage of the genetics offered by this system. These genes encode structurally similar rRNA methylases that confer resistance to macrolide, lincosamide, and streptogramin B antibiotics. Expression of ermC is subject to three different levels of posttranscriptional control: translational attenuation, mRNA stabilization, and translational autoregulation (reviewed in references 12 and 13), whereas ermA expression has so far been shown to be governed only by the first two. Subinhibitory concentrations of the inducer, erythromycin, cause pausing of ribosomes at a particular site on a leader peptide (146). This opens up a stem-loop structure that normally sequesters the ribosome binding site of the erm gene, thus allowing translation of the methylase (80, 145), a mechanism known as translational attenuation (Fig. 2). Whereas only one leader peptide exists in the ermC leader, it is thought that in ermA, ribosome stalling on one leader peptide permits the translation of a second peptide, stalling on which, in turn, permits translation of the methylase gene (159). The ermC gene is also subject to translational autoregulation: the methylase is thought to bind to an RNA structure in the leader that resembles its 23S rRNA substrate to cause inhibition of translation (51), although subsequent work suggested that the methylase binding site extends beyond this zone (24). Lastly, the stalling of the ribosome on the leader peptide causes a 7- to 20-fold stabilization of erm RNA (14, 214) or, indeed, any heterologous RNA which is fused to the stalling sequence (14, 56, 203). Remarkably, stabilization of the erm mRNA is independent of translation of the methylase gene (14).
FIG. 2.
Model of ermC regulation by translational attenuation and mRNA stabilization. The proposed structure of the ermC leader and the position of the leader peptide and ribosome- stalling site are shown to the left. SD1 and SD2 refer to the SD sequences of the leader peptide and ermC methylase, respectively. In the presence of erythromycin, ribosomes stalled on the leader peptide open up the RNA structure and allow ribosome access to SD2. At the same time, the stalled ribosomes protect the transcript from decay.
The question of whether the proximity of the stalling sequence (codons 5 to 9) to the 5′ end of the erm RNA (<10 nt upstream of the ribosome binding site) is important for the stabilizing effect was addressed by cloning heterologous extensions between the promoter and the leader peptide for both ermA (204) and ermC (16). In both cases, a full-length unstable RNA and a shorter stable species were detected in the presence of erythromycin. The downstream stabilized portion had the same 5′ end regardless of the sequence context or the point of transcription initiation. For ermC, the 5′ end of the stable fragment mapped to 57 or 58 nt upstream of the ribosome binding site of the leader peptide ermC (16), while for ermA, the 5′ ends of the stabilized species were mapped to the stalling sites of the two leader peptides, with the major signal corresponding to the stalling site of the first leader peptide (204). In recent experiments, with a deletion derivative of the ermC mRNA, Drider et al. also mapped a major 5′ end to the stall site of the leader peptide, although this 5′ end could not be detected in the wild-type context (60). In no case could the mRNA fragment upstream of the stall site be detected, even after overexpression of the erm RNA.
The simplest explanation for these observations would be that the downstream portion of the erm mRNA is protected from degradation by a 5′-to-3′ exonuclease by the stalled ribosome (12, 16, 204), although no such activity has been detected in bacteria. Alternatively, an endonucleolytic cleavage required to start the decay process in a distal portion of the mRNA may somehow be inhibited by the presence of a stalled ribosome at the 5′ end (60).
To address the issue of whether the 5′ end of the RNA fragment stabilized in the presence of erythromycin was generated by endonucleolytic cleavage or by processive nuclease activity degrading from the 5′ end, a second stalling site was cloned upstream of both ermA and ermC derivatives with 5′ extensions. In both cases, the extended transcript was stabilized in the presence of erythromycin, and this, it was argued, ruled out endonuclease cleavage at the internal stall site (16, 204). It is still possible, however, that unless the 5′ end is accessible and bound by such an endonuclease, cleavage is inhibited at the internal stall site, analogous to the mechanism of RNase E cleavage. Drider et al. have suggested that cleavage in the stall sequence of the deletion derivative of the ermC transcript is catalyzed by a ribosome-associated endonuclease (60), similar to the activity described by Loomis, Moseley, and coworkers to be responsible for cleavage of the daa transcript in E. coli (131, 132).
The cat mRNA from S. aureus, encoding a chloramphenicol-inducible chloramphenicol acetyltransferase, is regulated very similarly to the erm genes and has also been studied in B. subtilis. As in the erm system, expression of cat depends on antibiotic- induced stalling of ribosomes during synthesis of a short leader peptide upstream of the cat gene. The stalling of ribosomes causes a 15- to 20-fold increase in the cat mRNA half-life (<0.5 to 8 mins), even in the absence of cat translation (58). By further analogy to the erm system, stalling of the ribosome is associated with processing of the cat RNA 14 nt upstream of the stall site in the leader peptide. It has not been shown, however, whether the cat system can function as a general 5′ stabilizer by fusion with heterologous RNAs.
(ii) Phage SP82 mRNA.
One of the best-studied RNAs in B. subtilis is the 5′-proximal portion of the phage SP82 RNA. It was initially chosen for study because it contained several cleavage sites for B. subtilis RNase III (153, 185). Cloning of one of the SP82 RNase III sites (the so-called A-region [Fig. 3]) in the ermC mRNA resulted in a destabilization (half-life <5 min) of the portion of the mRNA upstream of the cleavage site normally stabilized by ribosome stalling, while the portion downstream remained stable (half-life, >20 min) (56). When the A-region was moved nearer to the 5′ end of the ermC transcript, the half-life of the ermC mRNA increased further (half-life, >40 min) (95). This region can confer stability on other heterologous mRNAs such as lacZ or penA, provided that it is cloned at or near the 5′ end. Thus, the SP82 A-region functions as a 5′ RNA stabilizer. mRNA stabilization is not a general property of RNase III sites in B. subtilis, however, since portions of the SP82 mRNA containing two other RNase III cleavage sites (the so-called B- and C-regions) do not stabilize ermC mRNA when inserted in the same fashion as the A-region. Furthermore, cleavage of the A-region by B. subtilis RNase III per se is not necessary for the stabilizing effect, since an RNA beginning at the first nucleotide downstream of the A-region cleavage site also has a half-life of 40 min. These experiments eliminated the possibility that RNase III binding to the 5′ end of the newly cleaved mRNA inhibited its decay. Deletion analysis of the A-region showed that the stability determinant was a short polypurine sequence which resembles a ribosome binding site. This sequence shows perfect complementarity to an 8-nt stretch corresponding to the anti-Shine-Dalgarno (SD) sequence at the 3′ end of 16S rRNA. Although an initiation codon lies downstream of the polypurine sequence in its native context, a construct with no possibility of translation initiation was also stabilized by the A-region. Although it has yet to be shown definitively that the polypurine sequence functions as a ribosome binding site, e.g., by toeprint analysis, it appears that the SP82 5′ RNA stabilizer and the erm stalling sequence (above) play very similar roles in preventing RNA decay. It is also interesting that the SP82 polypurine element does not function as an RNA stabilizer in E. coli (cited in reference 95).
(iii) cryIIIA mRNA.
A 5′ RNA stabilizer very similar to the polypurine sequence of SP82 RNA (see above) was subsequently identified in the cryIIIA mRNA. The cry RNAs encode different Bacillus thuringiensis crystal proteins that are pathogenic to insect larvae, and their degradation in B. subtilis has been studied. Transcription of these genes is activated at the onset of sporulation (123). The 5′ end of the cryIIIA mRNA was mapped to 129 nt upstream of the translation initiation codon. This was initially thought to be the start point of transcription but was subsequently shown to be a strong site of RNA processing (1). The leader region just downstream of T-129 was shown to act as a 5′ mRNA stabilizer by deletion analysis of lacZ fusions (2). This region conferred a >sixfold increase in the half-life of the cryIIIA mRNA and an eightfold increase in half-life of lacZ mRNA. The key element is a perfect SD sequence (called Stab-SD for “stabilizing SD”) 5 nt from the 5′ end of the processed mRNA. It is thought that the binding of a ribosome to the Stab-SD stabilizes the downstream sequence, even in the absence of translation, rather like the role of the stalled ribosome in the erm system. Stab-SD sequences and function were predicted to exist in several other cryIII genes and in the cwp and inlAB genes of Bacillus brevis and Listeria monocytogenes, respectively. Since the site of RNA cleavage is covered by the ribosome, it seems likely that the RNase that catalyzes this particular cleavage is the same ribosome-associated nuclease that cleaves the ermC and ermA RNAs at their respective stall sites.
mRNAs protected by proteins bound near the 5′ end.
Given that the binding or stalling of ribosomes near the 5′ end of mRNAs significantly increases their stability, it is not surprising that the binding of proteins or the presence of strong secondary structures in this region would have a similar effect. The glpD mRNA, discussed here, is thought to provide an example of protein-mediated mRNA stabilization.
(i) glpD mRNA.
The glpD gene encodes glycerol-3-phosphate (G3P) dehydrogenase and is controlled by transcription antitermination (92, 93). GlpP protein is thought to bind the glpD leader in the presence of G3P and promote terminator read through by stabilizing a mutually exclusive antiterminator structure (73) that resembles the RAT family of B. subtilis antiterminator structures (10). The glpD mRNA is also stabilized by the presence of GlpP and G3P (72). The combination of these two effects leads to a 10- to 50-fold increase in glpD expression after induction by G3P. Mutations were isolated in the terminator structure that allowed constitutive expression of glpD and hence growth on glycerol in the absence of GlpP. The increase in glpD expression is temperature sensitive and is related to a temperature-dependent degradation of the RNA. The half-life of wild-type glpD RNA at 32°C is twice that at 45°C (4.5 and 2.3 min respectively). In two particular terminator mutants isolated, the half-life of the glpD mRNA was decreased three- to four-fold at 32°C (to 1.1 to 1.4 min), and was almost unmeasurable at 45°C (<0.3 min), thus explaining the temperature-sensitive growth on glycerol. Interestingly, the stability of the mutant RNAs could be completely restored by overproduction of GlpP, consistent with the idea that the binding of GlpP to the glpD leader somehow protects it from nuclease attack, although this remains to be shown directly.
The glpD mRNA is also interesting in that it is one of only a few RNAs whose degradation has been compared directly in B. subtilis and E. coli. Wild-type and mutant (temperature sensitive) translational fusions of the glpD leader to lacZ were integrated into the chromosomes of both organisms. In B. subtilis, the mutant fusion transcript showed the same temperature dependency as the native glpD mRNA in the absence of GlpP, showing that the glpD leader is the major stability determinant for both transcripts (187). In E. coli, however, both the wild- type and mutant glpD-lacZ mRNAs were relatively stable (half-lives, 3 to 4 min) at 42°C. Furthermore, overproduction of GlpP did not lead to an increase in the stability of the mutant transcript as it did in B. subtilis. Lastly, the authors showed that the terminator structure of the glpD leader is a target for RNase III cleavage in E. coli but that no significant cleavage occurs at this site in B. subtilis. A difference in the specificities of E. coli and B. subtilis RNase III has been noted previously (153). This RNA thus provides an excellent example of an RNA with very different fates in E. coli and B. subtilis.
mRNAs protected by secondary structure at the 5′ end.
Some B. subtilis RNAs are thought to be stabilized by secondary structure at or near the 5′ end. The best-characterized and most convincing example is the aprE gene mRNA. Although the stability of two other mRNAs, the thrS and gapA mRNAs, is also thought to be governed by secondary structure at the 5′ end, the evidence in these cases is more preliminary and awaits confirmation.
(i) aprE mRNA.
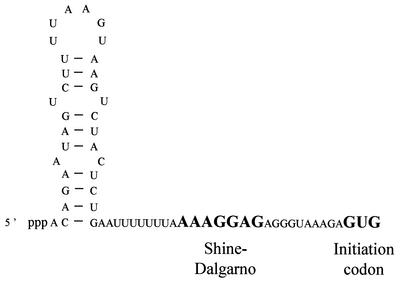
The aprE gene encodes subtilisin, an extracellular proteolytic enzyme produced in early stationary phase. The aprE mRNA is particularly stable, with a half-life of the order of 25 minutes (88). Fusions of the aprE leader to lacZ have a similar half-life; thus, the determinant for this extreme RNA stability is contained within the leader region. Mutations which deleted or altered a predicted stem-loop at the extreme 5′ end of the aprE leader (Fig. 4) reduced the half-life fivefold. While the stability of the aprE mRNA was initially thought to be growth phase dependent (197), according for its rapid disappearance after dilution of stationary-phase cultures into fresh medium, this was subsequently shown not to be the case; aprE has the same half-life in both log-phase and stationary-phase cultures (88). Striking similarities were noted between the polypurine-rich stabilizer sequence of SP82 and the sequence around the aprE ribosome binding site. Indeed, as with SP82, mutations predicted to diminish the free energy of interaction between 16S rRNA and the aprE ribosome binding site resulted in a decrease in half-life (fourfold) whether or not the RNA was translated (87). Thus, both a 5′ RNA structure and bound untranslating ribosomes contribute to the high levels of aprE mRNA stability.
FIG. 4.
Sequence and proposed secondary structure of the aprE 5′ stabilizer. The SD sequence and initiation codon are in bold capital letters.
(ii) thrS mRNA.
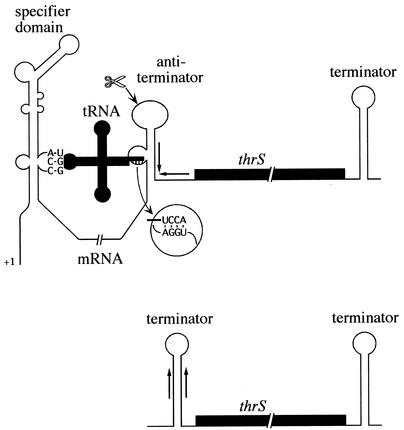
The threonyl-tRNA synthetase gene, thrS, is a member of the T-box family, a family of around 250 genes from gram-positive bacteria (79) that are regulated by a tRNA-dependent antitermination mechanism in response to starvation for a specific amino acid. In this type of control mechanism, uncharged tRNA is thought to stabilize an antiterminator structure in the RNA in preference to a mutually exclusive transcription terminator upstream of the structural gene (78, 189). The effect of antitermination on expression of the thrS gene is amplified by endonucleolytic cleavage within the antiterminator structure (Fig. 5) and a subsequent fivefold stabilization of the downstream coding portion of the mRNA (45). Cleavage at or near this site is conserved in at least five other members of this family, suggesting that it plays a generally important role in this type of control. When the B. subtilis thrS gene is expressed in E. coli, cleavage at this site is dependent on RNase E, suggesting that B. subtilis has a functional analog of this enzyme (46) despite the lack of an obvious homolog on the chromosome. (A parallel can perhaps be drawn with the DNA recombination and repair systems of E. coli and B. subtilis, where the role of the C and D subunits of the E. coli RecBCD complex is played by the nonhomologous B subunit of the B. subtilis AddAB complex [33].) The RNase E-like cleavage site has been mapped to a site in the apical loop of the antiterminator structure, 9 nt upstream of the transcription terminator stem, and thus the stabilized portion of the transcript is flanked on both its 5′ and 3′ sides by transcription terminators. Although it is likely that this explains the increased stability, this has not yet been directly demonstrated. Cleavage of the thrS leader in B. subtilis is much more efficient under inducing conditions, suggesting that the mRNA is preferentially cut when in the antiterminator conformation (45). Paradoxically, however, the cleavage can no longer be detected in a mutant RNA lacking the downstream half of the terminator but maintaining the integrity of the antiterminator. It is likely that the lack of the stabilizing hairpin at the 5′ end prevents detection of the cleavage product.
FIG. 5.
Model for regulation of thrS gene expression. Uncharged tRNAThr interacts with the thrS leader to pull the leader into the antitermination conformation. Specificity for tRNAThr is afforded by the ACC codon bulged out of the specifier domain. The antiterminator is stabilized by Watson-Crick base pairing between the CCA end of the tRNA and the complementary sequence bulged out of the antiterminator. Cleavage (scissors) in the loop of the antiterminator structure results in a shorter RNA flanked by factor-independent transcription terminators, which is much more stable than the full-length transcript.
(iii) gapA operon.
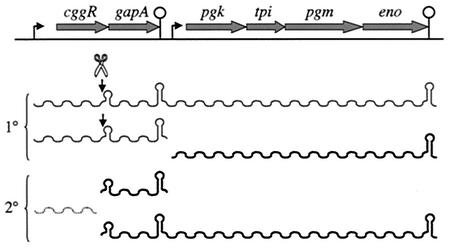
The gapA operon contains six genes involved in the interconversion of triose phosphates of the glycolytic pathway, from glyceraldehyde-3-phosphate and dihydroxyacetone phosphate to phosphoenolpyruvate (Fig. 6). Three primary transcripts are specified by this operon: a bicistronic cggR gapA transcript, a hexacistronic cggR gapA pgk tpi pgm eno transcript (both of which are repressed by the product of the first cistron, cggR) and a tetracistronic pgk tpi pgm eno transcript, expressed constitutively at low levels (134). The most abundant transcript, however, is a monocistronic gapA transcript, extending from an endonucleolytic cleavage site mapped to the 3′ end of cggR to a transcription terminator at the 3′ end of gapA. The cggR portion of the primary transcript is rapidly degraded (half-life, <0.3 min), while the gapA portion has a significantly longer half-life (ca. 3.5 min). This makes physiological sense, with much greater quantities of GapA than of CggR repressor being required by the cell. This is one of the few clear cases of differential stability of coding segments of an RNA in B. subtilis. Another example is found in the dnaK operon (94), while examples abound in E. coli and other bacteria (3, 116, 165, 167). The processing site was mapped to an AU-rich sequence, 1 to 2 nt upstream of a weak hairpin (ΔG = −9.1 kcal/mol). Thus, this RNA is predicted to have a hairpin structure at both the 5′ and 3′ ends, similar to the processed thrS mRNA, which may account for its increased stability. Although the gapA- processing site has some similarity to that of E. coli RNase E, it is not cleaved by RNase E in vitro (C. Condon, H. Putzer, and G. Stulke, unpublished data).
FIG. 6.
Structure and transcripts of the gapA operon. The primary (1°) and secondary (2°) transcripts (wavy lines) are indicated. Cleavage (scissors) of either the bicistronic or hexacistronic transcript just upstream of the secondary structure indicated results in shorter, stable 3′ fragments and an unstable cggR transcript. The relative abundance of the different transcripts is reflected in the relative thickness of the wavy lines.
The degradation of only four other B. subtilis RNAs has been studied, to various degrees, and these RNAs do not fall into any of the above categories. The pur leader transcript and cryIa mRNAs may provide examples of mRNAs stabilized by structures at their 3′ ends (62, 224), while the stability determinants of the sdh and xynA mRNAs are still unknown (4, 149).
GENERAL PRINCIPLES
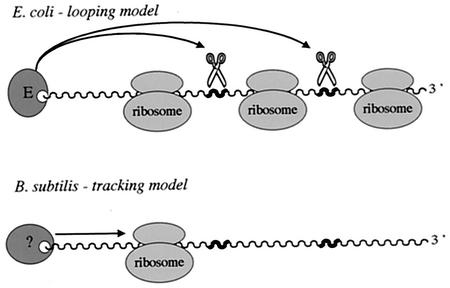
It is clear from the specific examples described above that the 5′ end of transcripts is the major determinant of mRNA stability in B. subtilis, where access to the body of the mRNA can be inhibited by bound untranslating ribosomes, bound proteins, and secondary structure. In E. coli, ribosomes bound but not translating the 5′ end of mRNAs do not have a stabilizing effect on downstream sequences. An untranslated lacZ mRNA with 11 nt complementary to the 3′ end of 16S rRNA, which would be predicted to be highly stabilized in B. subtilis, is relatively unstable in E. coli, except for the 80 nt to which the ribosome binds (101). As mentioned above, the SP82 polypurine element also loses its 5′-stabilizing capacity in E. coli. Thus, while the principle of the 5′ end governing overall stability is shared between E. coli and B. subtilis, the mechanism by which this is achieved is clearly not the same, highlighting a fundamental difference between the pathways of RNA decay in these two organisms. In E. coli, active translation is required for an effect of ribosomes on mRNA stability (239, 240; for recent review, see reference 59). In this case, decay is thought to be inhibited by the passage of ribosomes through the coding sequence (23, 96, 139, 166), which masks cleavage sites of nucleases that can access the mRNA directly or which bind to the 5′ end and can loop to internal sites in the mRNA (Fig. 7). Although the enzyme which initiates RNA decay in B. subtilis has not yet been identified, a good deal of accumulated evidence allows us to predict that on binding to, or activation by, a free 5′ end, it is prevented from scanning or tracking to sites downstream of road blocks, even if the RNA is otherwise completely denuded of ribosomes. While E. coli RNase E prefers an accessible 5′ end, it can also cleave RNAs with no available end, i.e., circular RNAs (138). The binding of ribosomes and subsequent stabilization of mRNAs in B. subtilis in the absence of translation would suggest that a broad-specificity direct-access endonuclease does not exist in this organism or is at least not very active. A similar conclusion was recently reached by Dreyfus and Joyce (59). Although there is evidence for a functional analog of RNase E in B. subtilis in terms of its specificity of substrate cleavage (46), nothing is known about its pathway of substrate binding or how it might scan for sites. The fact that the introduction of an RNase III cleavage site into the body of the B. subtilis ermC mRNA results in its destabilization (56) also suggests that the ability of at least this type of 5′ stabilizer to function as such depends on the accessibility of the mRNA to alternative pathways of degradation.
FIG. 7.
Comparison of the modes of action of E. coli RNase E and the hypothetical 5′-end-dependent RNase of B. subtilis. RNase E can bind to the 5′ end of transcripts (wavy lines) and loop to internal sites (thick portions) or can access cleavage sites directly. Accumulated evidence suggests that the B. subtilis enzyme is strictly 5′ end dependent, i.e., cannot access sites directly, and tracks along the RNA in the 5′-to-3′ direction to find cleavage sites. Thus, while active translation is required to inhibit cleavage by RNase E, static ribosomes can inhibit cleavage by the B. subtilis equivalent.
The effect of mutations that increase the free energy of interaction between the ribosome and the SD sequence can be mimicked by the addition of very low levels of tetracycline (0.25 μg/ml) to B. subtilis cell cultures (230). This results in the stabilization of a number of cellular mRNAs including polC, sigA, veg, and tet(L) genes, the last of which encodes a membrane-bound ion transport protein that affords resistance to tetracycline itself. When the mechanism was probed further, the results suggested that the effect of tetracycline is to somehow increase the affinity of the ribosome to the SD sequence. Thus, the increase in stability is probably due to a general inhibition of nuclease access to cellular mRNAs.
Both E. coli and B. subtilis use secondary structure to protect the 5′ end of mRNAs. The best example in B. subtilis is the aprE gene, where removal of a 5′ hairpin had a very deleterious effect on stability. Interestingly, aprE also seems to use bound ribosomes to stabilize the 5′ end. Two other examples where secondary structure at the 5′ end seems to be the most likely explanation for increased mRNA stability are the thrS and gapA mRNAs. In thrS, nine unpaired nucleotides remain at the 5′ end; in both aprE and gapA, there are only 1 to 2 nt. It is worth noting that 9-nt extension upstream of a 5′ structure would destabilize such an mRNA in E. coli; single-stranded extensions of more than 4 nt neutralized the stabilizing effect of a hairpin upstream of the ompA mRNA by allowing the binding of RNase E (21, 64). Thus, the functional analogy between E. coli RNase E and the hypothetical 5′-end-recognizing nuclease of B. subtilis does not extend to the number of nucleotides required for access to transcripts.
The phenomenon of 5′-end protection of mRNA provides an “in-hindsight” justification for the use of reporter genes to study the expression of genes that prove to be regulated at the posttranscriptional level by RNA processing and degradation: mRNAs from fusions to reporter genes very often have same or similar half-lives to the native mRNAs. Otherwise, this might have intuitively seemed an unwise choice of experimental tool.
The ermC, ermA, cat, and cryIIIA mRNAs, all of which are stabilized by stalled or stationary ribosomes, also have characterized RNA-processing sites that fall within the region normally protected from RNase access by the ribosome. This suggests that the nuclease that cleaves at this site is actually associated with the ribosome (59, 60). It should be remembered, however, that none of these RNAs are endogenous to B. subtilis; thus, its more general role may be to inactivate RNAs that are no longer translatable, rather than stabilizing them. The sites of RNA cleavage of these four mRNAs bear no resemblance to one another, suggesting that the ribosome-associated nuclease has no sequence specificity but is simply activated by the stalled ribosome to cut at that particular location. While it is formally possible that the 5′-end-dependent tracking nuclease and the ribosome-associated nuclease are one and the same enzyme, the fact that some B. subtilis RNAs can be stabilised by secondary structure or bound proteins in their untranslated leader regions suggests they are more likely to be different enzymes. Cleavage by the activity associated with the stalled ribosome would thus reinforce the protection from the 5′-end-dependent nuclease by removing its attachment site.
Protection of the 3′ end has been less well studied for B. subtilis than for E. coli. E. coli transcription terminators and other stable secondary structures inhibit the progress of both PNPase and RNase II. The cryIAa transcription terminator serves as a stabilizer of heterologous RNAs in B. subtilis (232), suggesting that this phenomenon may be shared by both organisms even though PNPase is the only major exonuclease they have in common. It remains to be seen, however, how general this phenomenon will be.
CONCLUDING REMARKS
These are exciting times for the field of RNA decay in gram-positive organisms, B. subtilis in particular. The advent of genome sequencing has helped confirm what some have suspected all along, i.e., that B. subtilis goes about dealing with the same problems in a very different way from E. coli. The identification of the analog for RNase E, i.e., the broad-specificity nuclease which carries out the rate-limiting cleavage of B. subtilis mRNAs, perhaps by tracking from the 5′ end, will be the key to solving the mystery of how B. subtilis approaches the task of RNA processing and degradation. It can probably be safely assumed that the ability to degrade RNA completely to mononucleotides is an essential process in all forms of life. In E. coli, this task is performed by a combination of endonucleolytic cleavages followed by exonucleolytic degradation to small oligoribonucleotides, which are then digested into mononucleotides by oligoribonuclease. Inactivation of both major exonucleases RNase II and PNPase together or of oligoribonuclease is lethal in E. coli. A triple pnpA rnr yhaM mutant of B. subtilis is viable, however, suggesting that at least one other major exonuclease remains to be discovered in this organism. Lastly, there are data pointing to the existence of a ribosome-associated nuclease in B. subtilis, whose identity remains unknown. Perhaps these are some of the enzymes identified in the section on potential RNases or perhaps they contain new folds and their identification will thus require a more biochemical approach. Either way, it is reassuring to know that the identification and characterization of these enzymes will open up significant new fields of research, different from those for E. coli.
Acknowledgments
I thank D. Bechhofer, A. J. Carpousis, M. Dreyfus, I. Iost, H. Putzer, P. Regnier, B. Rutberg, and M. Uzan for their comments and suggestions to improve the manuscript.
This work was supported by funds from the CNRS (UPR 9073), in association with Université Paris VII, and the Ministère de l'Education Nationale.
REFERENCES
- 1.Agaisse, H., and D. Lereclus. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13:97-107. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse, H., and D. Lereclus. 1996. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20:633-643. [DOI] [PubMed] [Google Scholar]
- 3.Alifano, P., F. Rivellini, C. Piscitelli, C. M. Arraiano, C. B. Bruni, and M. S. Carlomagno. 1994. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 8:3021-3031. [DOI] [PubMed] [Google Scholar]
- 4.Allmansberger, R. 1996. Degradation of the Bacillus subtilis xynA transcript is accelerated in response to stress. Mol. Gen. Genet. 251:108-112. [DOI] [PubMed] [Google Scholar]
- 5.Anantharaman, V., E. V. Koonin, and L. Aravind. 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30:1427-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apirion, D., and A. B. Lassar. 1978. A conditional lethal mutant of Escherichia coli which affects processing of ribosomal RNA. J. Biol. Chem. 253:1738-1742. [PubMed] [Google Scholar]
- 7.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 8.Aravind, L., and E. V. Koonin. 2001. A natural classification of ribonucleases. Methods Enzymol. 341:3-28. [DOI] [PubMed] [Google Scholar]
- 9.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aymerich, S., and M. Steinmetz. 1992. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl. Acad. Sci. USA 89:10410-10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardwell, J. C., P. Regnier, S. M. Chen, Y. Nakamura, M. Grunberg-Manago, and D. L. Court. 1989. Autoregulation of RNase III operon by mRNA processing. EMBO J. 8:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bechhofer, D. H. 1990. Triple post-transcriptional control. Mol. Microbiol. 4:1419-1423. [DOI] [PubMed] [Google Scholar]
- 13.Bechhofer, D. H. 1993. 5′ mRNA stabilisers, p. 31-52. In J. G. Belasco and G. Brawerman (ed.), Control of messenger RNA stability. Academic Press, Inc., San Diego, Calif.
- 14.Bechhofer, D. H., and D. Dubnau. 1987. Induced mRNA stability in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 84:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bechhofer, D. H., and W. Wang. 1998. Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J. Bacteriol. 180:5968-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belitsky, B. R., M. C. Gustafsson, A. L. Sonenshein, and C. Von Wachenfeldt. 1997. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol 179:5448-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beran, R. K., and R. W. Simons. 2001. Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 39:112-125. [DOI] [PubMed] [Google Scholar]
- 19.Blum, E., A. J. Carpousis, and C. F. Higgins. 1999. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 274:4009-4016. [DOI] [PubMed] [Google Scholar]
- 20.Bothwell, A. L. M., R. L. Garber, and S. Altman. 1976. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5S RNA. J. Biol. Chem. 251:7709-7716. [PubMed] [Google Scholar]
- 21.Bouvet, P., and J. G. Belasco. 1992. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360:488-491. [DOI] [PubMed] [Google Scholar]
- 22.Braun, F., E. Hajnsdorf, and P. Regnier. 1996. Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3′ hairpin. Mol. Microbiol. 19:997-1005. [DOI] [PubMed] [Google Scholar]
- 23.Braun, F., J. Le Derout, and P. Regnier. 1998. Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J. 17:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breidt, F., and D. Dubnau. 1990. Identification of cis-acting sequences required for translational autoregulation of the ermC methylase. J. Bacteriol. 172:3661-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown, J. W. 1999. The Ribonuclease P Database. Nucleic Acids Res. 27:314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown, J. W., and N. R. Pace. 1992. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 20:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao, G. J., J. Pogliano, and N. Sarkar. 1996. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:11580-11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao, G. J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 89:10380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao, G. J., and N. Sarkar. 1993. Poly(A) RNA in Bacillus subtilis: identification of the polyadenylylation site of flagellin mRNA. FEMS Microbiol. Lett. 108:281-285. [DOI] [PubMed] [Google Scholar]
- 30.Carl, P. L., L. Bloom, and R. J. Crouch. 1980. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J. Bacteriol. 144:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpousis, A. J., G. Van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNAse E and PNPase: evidence for a specific association between two enzymes important in mRNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 32.Chaney, S. G., and P. D. Boyer. 1972. Incorporation of water oxygens into intracellular nucleotides and RNA. II. Predominantly hydrolytic RNA turnover in Escherichia coli. J. Mol. Biol. 64:581-591. [DOI] [PubMed] [Google Scholar]
- 33.Chedin, F., S. D. Ehrlich, and S. C. Kowalczykowski. 2000. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J. Mol. Biol. 298:7-20. [DOI] [PubMed] [Google Scholar]
- 34.Chelladurai, B., H. Li, K. Zhang, and A. W. Nicholson. 1993. Mutational analysis of a ribonuclease III processing signal. Biochemistry 32:7549-7558. [DOI] [PubMed] [Google Scholar]
- 35.Cheng, Z.-F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273:14077-14080. [DOI] [PubMed] [Google Scholar]
- 36.Cheng, Z. F., and M. P. Deutscher. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 277:21624-21629. [DOI] [PubMed] [Google Scholar]
- 37.Coburn, G. A., and G. A. Mackie. 1996. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem. 271:15776-15781. [DOI] [PubMed] [Google Scholar]
- 38.Coburn, G. A., and G. A. Mackie. 1996. Overexpression, purification, and properties of Escherichia coli ribonuclease II. J. Biol. Chem. 271:1048-1053. [DOI] [PubMed] [Google Scholar]
- 39.Coburn, G. A., and G. A. Mackie. 1998. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 279:1061-1074. [DOI] [PubMed] [Google Scholar]
- 40.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 41.Coburn, G. A., X. Miao, D. J. Briant, and G. A. Mackie. 1999. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 13:2594-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen, S. N. 1995. Surprises at the 3′ end of prokaryotic RNA. Cell 80:829-832. [DOI] [PubMed] [Google Scholar]
- 43.Condon, C., D. Brechemier-Baey, B. Beltchev, M. Grunberg-Manago, and H. Putzer. 2001. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function. RNA 7:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Condon, C., and H. Putzer. 2002. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 30:5339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Condon, C., H. Putzer, and M. Grunberg-Manago. 1996. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in B. subtilis. Proc. Natl. Acad. Sci. USA 93:6992-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Condon, C., H. Putzer, D. Luo, and M. Grunberg-Manago. 1997. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol. 268:235-242. [DOI] [PubMed] [Google Scholar]
- 47.Condon, C., J. Rourera, D. Brechemier-Baey, and H. Putzer. 2002. Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis. J. Bacteriol. 184:2845-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crary, S. M., S. Nirajanakumari, and C. A. Fierke. 1998. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry 37:9409-9416. [DOI] [PubMed] [Google Scholar]
- 49.Craven, M. G., D. J. Henner, D. Alessi, A. T. Schauer, K. A. Ost, M. P. Deutscher, and D. I. Friedman. 1992. Identification of the rph (RNase PH) gene of Bacillus subtilis: evidence for suppression of cold-sensitive mutations in Escherichia coli. J. Bacteriol. 174:4727-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crouch, R. J. 1990. Ribonuclease H: from discovery to 3D structure. New Biol. 2:771-777. [PubMed] [Google Scholar]
- 51.Denoya, C. D., D. H. Bechhofer, and D. Dubnau. 1986. Translational autoregulation of ermC 23S rRNA methyltransferase expression in Bacillus subtilis. J. Bacteriol. 168:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deutscher, M. P. 1993. Promiscuous exoribonucleases of Escherichia coli. J. Bacteriol. 175:4577-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deutscher, M. P., G. T. Marshall, and H. Cudny. 1988. RNase PH: an Escherichia coli phosphate-dependent nuclease distinct from polynucleotide phosphorylase. Proc. Natl. Acad. Sci. USA 85:4710-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deutscher, M. P., and N. B. Reuven. 1991. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 88:3277-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiGate, R. J., and K. J. Marians. 1992. Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J. Biol. Chem. 267:20532-20535. [PubMed] [Google Scholar]
- 56.DiMari, J. F., and D. H. Bechhofer. 1993. Initiation of mRNA decay in Bacillus subtilis. Mol. Microbiol. 7:705-717. [DOI] [PubMed] [Google Scholar]
- 57.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dreher, J., and H. Matzura. 1991. Chloramphenicol-induced stabilization of cat messenger RNA in Bacillus subtilis. Mol. Microbiol. 5:3025-3034. [DOI] [PubMed] [Google Scholar]
- 59.Dreyfus, M., and S. Joyce. 2002. The interplay between translation and mRNA decay in procaryotes. In J. Lapointe and L. Brakier-Gingras (ed.), Translational mechanisms. [Online.] Landes Bioscience, Georgetown, Tex.http://www.eurekah.com/abstract.php?chapid=670&bookid=659&catid=654.
- 60.Drider, D., J. M. DiChiara, J. Wei, J. S. Sharp, and D. H. Bechhofer. 2002. Endonuclease cleavage of messenger RNA in Bacillus subtilis. Mol. Microbiol. 43:1319-1329. [DOI] [PubMed] [Google Scholar]
- 61.Duffy, J. J., S. G. Chaney, and P. D. Boyer. 1972. Incorporation of water oxygens into intracellular nucleotides and RNA. I. Predominantly non-hydrolytic RNA turnover in Bacillus subtilis. J. Mol. Biol. 64:565-579. [DOI] [PubMed] [Google Scholar]
- 62.Ebbole, D. J., and H. Zalkin. 1988. Detection of pur operon-attenuated mRNA and accumulated degradation intermediates in Bacillus subtilis. J. Biol. Chem. 263:10894-10902. [PubMed] [Google Scholar]
- 63.Ehretsmann, C., A. J. Carpousis, and H. M. Krisch. 1992. Specificity of Escherichia coli endoribonuclease RNAse E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 6:149-159. [DOI] [PubMed] [Google Scholar]
- 64.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 65.Fang, X. W., X. J. Yang, K. Littrell, S. Niranjanakumari, P. Thiyagarajan, C. A. Fierke, T. R. Sosnick, and T. Pan. 2001. The Bacillus subtilis RNase P holoenzyme contains two RNase P RNA and two RNase P protein subunits. RNA 7:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng, Y., T. A. Vickers, and S. N. Cohen. 2002. The catalytic domain of RNase E shows inherent 3′ to 5′ directionality in cleavage site selection. Proc. Natl. Acad. Sci. USA 99:14746-14751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forsburg, S. L. 1999. The best yeast? Trends Genet. 15:340-344. [DOI] [PubMed] [Google Scholar]
- 68.Fox, G. W., and C. R. Woese. 1975. 5S RNA secondary structure. Nature 256:505-507. [DOI] [PubMed] [Google Scholar]
- 69.Gennaro, M. L., and R. P. Novick. 1986. cmp, a cis-acting plasmid locus that increases interaction between replication origin and initiator protein. J. Bacteriol. 168:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghora, B. K., and D. Apirion. 1978. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell 15:1055-1066. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh, S., and M. P. Deutscher. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. USA 96:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glatz, E., R.-P. Nilsson, L. Rutberg, and B. Rutberg. 1996. A dual role for the Bacillus subtilis leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol. Microbiol. 19:319-328. [DOI] [PubMed] [Google Scholar]
- 73.Glatz, E., M. Persson, and B. Rutberg. 1998. Antiterminator protein GlpP of Bacillus subtilis binds to glpD leader mRNA. Microbiology 144:449-456. [DOI] [PubMed] [Google Scholar]
- 74.Gopalakrishna, Y., D. Langley, and N. Sarkar. 1981. Detection of high levels of polyadenylate-containing RNA in bacteria by the use of a single-step RNA isolation procedure. Nucleic Acids Res. 9:3545-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gopalakrishna, Y., and N. Sarkar. 1982. Characterization of polyadenylate- containing ribonucleic acid from Bacillus subtilis. Biochemistry 21:2724-2729. [DOI] [PubMed] [Google Scholar]
- 76.Gopalakrishna, Y., and N. Sarkar. 1982. The synthesis of DNA complementary to polyadenylate-containing RNA from Bacillus subtilis. J. Biol. Chem. 257:2747-2750. [PubMed] [Google Scholar]
- 77.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 78.Grundy, F. J., and T. M. Henkin. 1993. Transfer RNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74:475-482. [DOI] [PubMed] [Google Scholar]
- 79.Grundy, F. J., T. R. Moir, M. T. Haldeman, and T. M. Henkin. 2002. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 30:1646-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gryczan, T. J., G. Grandi, J. Hahn, R. Grandi, and D. Dubnau. 1980. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 8:6081-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guarneros, G., and C. Portier. 1990. Different specificities of ribonuclease II and polynucleotide phosphorylase in 3′ mRNA decay. Biochimie 72:771-777. [DOI] [PubMed] [Google Scholar]
- 82.Guerrier-Takada, C., K. Gardiner, T. Marsh, N. Pace, and S. Altman. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849-857. [DOI] [PubMed] [Google Scholar]
- 83.Haas, E. S., A. B. Banta, J. K. Harris, N. R. Pace, and J. W. Brown. 1996. Structure and evolution of ribonuclease P RNA in Gram-positive bacteria. Nucleic Acids Res. 24:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hahnen, E., L. Znamenskaya, D. Koczan, I. Leshchinskaya, and G. Hobom. 2000. A novel secreted ribonuclease from Bacillus intermedius: gene structure and regulatory control. Mol. Gen. Genet. 263:571-580. [DOI] [PubMed] [Google Scholar]
- 85.Hajnsdorf, E., F. Braun, J. Haugel-Nielsen, and P. Regnier. 1995. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hajnsdorf, E., and P. Regnier. 1999. E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol. 286:1033-1043. [DOI] [PubMed] [Google Scholar]
- 87.Hambraeus, G., K. Karhumaa, and B. Rutberg. 2002. A 5′ stem-loop and ribosome binding site but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology 148:1795-1803. [DOI] [PubMed] [Google Scholar]
- 88.Hambraeus, G., M. Persson, and B. Rutberg. 2000. The aprE leader is a determinant of extreme mRNA stability in Bacillus subtilis. Microbiology 146:3051-3059. [DOI] [PubMed] [Google Scholar]
- 89.Hartley, R. W. 1997. Barnase and barstar, p. 51-100. In J. D'Alessio and J. F. Riordan (ed.), Ribonuclease: structures and functions. Academic Press, Inc., San Diego, Calif.
- 90.He, L., F. Soderbom, E. G. Wagner, U. Binnie, N. Binns, and M. Masters. 1993. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol. 9:1131-1142. [DOI] [PubMed] [Google Scholar]
- 91.Herskowitz, M. A., and D. H. Bechhofer. 2000. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol. 38:1027-1033. [DOI] [PubMed] [Google Scholar]
- 92.Holmberg, C., and B. Rutberg. 1991. Expression of the gene encoding glycerol-3- phosphate dehydrogenase (glpD) in Bacillus subtilis is controlled by antitermination. Mol. Microbiol. 5:2891-2900. [DOI] [PubMed] [Google Scholar]
- 93.Holmberg, C., and L. Rutberg. 1992. An inverted repeat preceding the Bacillus subtilis glpD gene is a conditional terminator of transcription. Mol. Microbiol. 6:2931-2938. [DOI] [PubMed] [Google Scholar]
- 94.Homuth, G., A. Mogk, and W. Schumann. 1999. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol. Microbiol. 32:1183-1197. [DOI] [PubMed] [Google Scholar]
- 95.Hue, K. K., S. D. Cohen, and D. H. Bechhofer. 1995. A polypurine sequence that acts as a 5′ mRNA stabilizer in Bacillus subtilis. J. Bacteriol. 177:3465-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iost, I., and M. Dreyfus. 1995. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 14:3252-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Itaya, M. 1990. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc. Natl. Acad. Sci. USA 87:8587-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Itaya, M., A. Omori, S. Kanaya, R. J. Crouch, T. Tanaka, and K. Kondo. 1999. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J. Bacteriol. 181:2118-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.James, B. D., G. J. Olsen, J. S. Liu, and N. R. Pace. 1988. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell 52:19-26. [DOI] [PubMed] [Google Scholar]
- 100.Jarrige, A. C., N. Mathy, and C. Portier. 2001. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 20:6845-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joyce, S. A., and M. Dreyfus. 1998. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Biol. 282:241-254. [DOI] [PubMed] [Google Scholar]
- 102.Jurgen, B., T. Schweder, and M. Hecker. 1998. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol. Gen. Genet. 258:538-545. [DOI] [PubMed] [Google Scholar]
- 103.Kanamori, N., N. Cozzarelli, and R. Okazaki. 1973. Extracellular nucleases of Bacillus subtilis. II. The nucleases as 5′-end-group reagents. Biochim. Biophys. Acta 335:173-184. [Google Scholar]
- 104.Kanamori, N., K. Sakabe, and R. Okazaki. 1973. Extracellular nucleases of Bacillus subtilis. I. Purification and properties. Biochim. Biophys. Acta 335:155-172. [Google Scholar]
- 105.Karnik, P., Y. Gopalakrishna, and N. Sarkar. 1986. Construction of a cDNA library from polyadenylated RNA of Bacillus subtilis and the determination of some 3′-terminal sequences. Gene 49:161-165. [DOI] [PubMed] [Google Scholar]
- 106.Keller, W., and R. Crouch. 1972. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc. Natl. Acad. Sci. USA 69:3360-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kelly, K. O., and M. P. Deutscher. 1992. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J. Bacteriol. 174:6682-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kelly, K. O., N. B. Reuven, Z. Li, and M. P. Deutscher. 1992. RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J. Biol. Chem. 267:16015-16018. [PubMed] [Google Scholar]
- 109.Kerjan, P., and J. Szulmajster. 1974. Intracellular ribonuclease activity in stationary phase cells of Bacillus subtilis. Biochem. Biophys. Res. Commun. 59:1079-1087. [DOI] [PubMed] [Google Scholar]
- 110.Kerjan, P., and J. Szulmajster. 1976. Isolation and properties of a cyclic guanosine-monophosphate sensitive intracellular ribonuclease from Bacillus subtilis. Biochimie 58:533-541. [DOI] [PubMed] [Google Scholar]
- 111.Kerr, I. M., J. R. Chien, and I. R. Lehman. 1967. Exonucleolytic degradation of high molecular weight deoxyribonucleic acid and ribonucleic acid to nucleoside 3′-phosphates by a nuclease from Bacillus subtilis. J. Biol. Chem. 242:2700-2708. [PubMed] [Google Scholar]
- 112.Kerr, I. M., E. A. Pratt, and I. R. Lehman. 1965. Exonucleolytic degradation of high-molecular-weight DNA and RNA to nucleoside 3′-phosphates by a nuclease from B. subtilis. Biochem. Biophys. Res. Commun. 20:154-162. [DOI] [PubMed] [Google Scholar]
- 113.King, T. C., and D. Schlessinger. 1983. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J. Biol. Chem. 258:12034-12042. [PubMed] [Google Scholar]
- 114.King, T. C., R. Sirdeshmukh, and D. Schlessinger. 1984. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc. Natl. Acad. Sci. USA 81:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Klee, C. B., and M. F. Singer. 1968. The processive degradation of individual polyribonucleotide chains. II. Micrococcus lysodeikticus polynucleotide phosphorylase. J. Biol. Chem. 243:923-927. [PubMed] [Google Scholar]
- 116.Klug, G., C. W. Adams, J. Belasco, B. Doerge, and S. N. Cohen. 1987. Biological consequences of segmental alterations in mRNA stability: effects of deletion of the intercistronic hairpin loop region of the Rhodobacter capsulatus puf operon. EMBO J. 6:3515-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kogoma, T., N. L. Subia, and K. von Meyenburg. 1985. Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol. Gen. Genet. 200:103-109. [DOI] [PubMed] [Google Scholar]
- 118.Krinke, L., and D. L. Wulff. 1990. The cleavage specificity of RNase III. Nucleic Acids Res. 18:4809-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 120.Kurz, J. C., S. Niranjanakumari, and C. A. Fierke. 1998. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry 37:2393-2400. [DOI] [PubMed] [Google Scholar]
- 121.Kushner, S. R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184:4658-4665; discussion, 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Leeper, T. C., M. B. Martin, H. Kim, S. Cox, V. Semenchenko, F. J. Schmidt, and S. R. Van Doren. 2002. Structure of the UGAGAU hexaloop that braces Bacillus RNase P for action. Nat. Struct. Biol. 9:397-403. [DOI] [PubMed] [Google Scholar]
- 123.Lereclus, D., A. Delécluse, and M.-M. Lecadet. 1993. Diversity of Bacillus thuringiensis toxins and genes, p. 37-69. In P. Entwistle, J. S. Cory, M. J. Bailey, and S. Higg (ed.), Bacillus thuringiensis, an environmental biopesticide: theory and practice. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 124.Li, Z., and M. P. Deutscher. 1994. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J. Biol. Chem. 269:6064-6071. [PubMed] [Google Scholar]
- 125.Li, Z., and M. P. Deutscher. 1995. The tRNA processing enzyme RNase T is essential for maturation of 5S rRNA. Proc. Natl. Acad. Sci. USA 92:6883-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li, Z., and M. P. Deutscher. 1996. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86:503-512. [DOI] [PubMed] [Google Scholar]
- 127.Li, Z., and M. P. Deutscher. 2002. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 8:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li, Z., S. Reimers, S. Pandit, and M. P. Deutscher. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J. 21:1132-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin-Chao, S., T. T. Wong, K. J. McDowall, and S. N. Cohen. 1994. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 269:10797-10803. [PubMed] [Google Scholar]
- 131.Loomis, W. P., J. T. Koo, T. P. Cheung, and S. L. Moseley. 2001. A tripeptide sequence within the nascent DaaP protein is required for mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 39:693-707. [DOI] [PubMed] [Google Scholar]
- 132.Loomis, W. P., and S. L. Moseley. 1998. Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Mol. Microbiol. 30:843-853. [DOI] [PubMed] [Google Scholar]
- 133.Lopez, P. J., I. Marchand, S. A. Joyce, and M. Dreyfus. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33:188-199. [DOI] [PubMed] [Google Scholar]
- 134.Ludwig, H., G. Homuth, M. Schmalisch, F. M. Dyka, M. Hecker, and J. Stulke. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41:409-422. [DOI] [PubMed] [Google Scholar]
- 135.Luirink, J., S. High, H. Wood, A. Giner, D. Tollervey, and B. Dobberstein. 1992. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature 359:741-743. [DOI] [PubMed] [Google Scholar]
- 136.Luttinger, A., J. Hahn, and D. Dubnau. 1996. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19:343-356. [DOI] [PubMed] [Google Scholar]
- 137.Mackie, G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720-723. [DOI] [PubMed] [Google Scholar]
- 138.Mackie, G. A. 2000. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 275:25069-25072. [DOI] [PubMed] [Google Scholar]
- 139.Makarova, O. V., E. M. Makarov, R. Sousa, and M. Dreyfus. 1995. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl. Acad. Sci. USA 92:12250-12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Marujo, P. E., E. Hajnsdorf, J. Le Derout, R. Andrade, C. M. Arraiano, and P. Regnier. 2000. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA 6:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mathur, S., V. J. Cannistraro, and D. Kennell. 1993. Identification of an intracellular pyrimidine-specific endoribonuclease from Bacillus subtilis. J. Bacteriol. 175:6717-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mathy, N., A. C. Jarrige, M. Robert-Le Meur, and C. Portier. 2001. Increased expression of Escherichia coli polynucleotide phosphorylase at low temperatures is linked to a decrease in the efficiency of autocontrol. J. Bacteriol. 183:3848-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Matsunaga, J., M. Dyer, E. L. Simons, and R. W. Simons. 1996. Expression and regulation of the rnc and pdxJ operons of Escherichia coli. Mol. Microbiol. 22:977-989. [DOI] [PubMed] [Google Scholar]
- 144.Matsunaga, J., E. L. Simons, and R. W. Simons. 1996. RNase III autoregulation: structure and function of rncO, the posttranscriptional “operator”. RNA 2:1228-1240. [PMC free article] [PubMed] [Google Scholar]
- 145.Mayford, M., and B. Weisblum. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J. 8:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mayford, M., and B. Weisblum. 1989. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J. Mol. Biol. 206:69-79. [DOI] [PubMed] [Google Scholar]
- 147.McDowall, K. J., S. Lin-Chao, and S. N. Cohen. 1994. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269:10790-10796. [PubMed] [Google Scholar]
- 148.McLaren, R. S., S. F. Newbury, G. S. Dance, H. C. Causton, and C. F. Higgins. 1991. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol. 221:81-95. [PubMed] [Google Scholar]
- 149.Melin, L., H. Friden, E. Dehlin, L. Rutberg, and A. von Gabain. 1990. The importance of the 5′-region in regulating the stability of sdh mRNA in Bacillus subtilis. Mol. Microbiol. 4:1881-1889. [DOI] [PubMed] [Google Scholar]
- 150.Meyhack, B., B. Pace, and N. R. Pace. 1977. Involvement of precursor-specific segments in the in vitro maturation of Bacillus subtilis precursor 5S ribosomal RNA. Biochemistry 16:5009-5015. [DOI] [PubMed] [Google Scholar]
- 151.Meyhack, B., and N. R. Pace. 1978. Involvement of the mature domain in the in vitro maturation of Bacillus subtilis precursor 5S ribosomal RNA. Biochemistry 17:5804-5809. [DOI] [PubMed] [Google Scholar]
- 152.Miczak, A., V. R. Kaberdin, C.-L. Wei, and S. Lin-Chao. 1996. Proteins associated with RNAse E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. USA 93:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mitra, S., and D. Bechhofer. 1994. Substrate specificity of an RNAse III-like activity from Bacillus subtilis. J. Biol. Chem. 269:31450-31456. [PubMed] [Google Scholar]
- 154.Mitra, S., K. Hue, and D. H. Bechhofer. 1996. In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Mol. Microbiol. 19:329-342. [DOI] [PubMed] [Google Scholar]
- 155.Mohanty, B. K., and S. R. Kushner. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34:1094-1108. [DOI] [PubMed] [Google Scholar]
- 156.Mohanty, B. K., and S. R. Kushner. 1999. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol. Microbiol. 34:1109-1119. [DOI] [PubMed] [Google Scholar]
- 157.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′→5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mudd, E. A., H. M. Krisch, and C. F. Higgins. 1990. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol. Microbiol. 4:2127-2135. [DOI] [PubMed] [Google Scholar]
- 159.Murphy, E. 1985. Nucleotide sequence of ermA, a macrolide-lincosamide- streptogramin B determinant in Staphylococcus aureus. J. Bacteriol. 162:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Murzin, A. G. 1993. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakai, M., Z. Minami, T. Yamazaki, and A. Tsugita. 1965. Studies on the nucleases of a strain of Bacillus subtilis. J. Biochem. 57:96-99. [DOI] [PubMed] [Google Scholar]
- 162.Nakamura, A., Y. Koide, H. Miyazaki, A. Kitamura, H. Masaki, T. Beppu, and T. Uozumi. 1992. Gene cloning and characterization of a novel extracellular ribonuclease of Bacillus subtilis. Eur. J. Biochem. 209:121-127. [DOI] [PubMed] [Google Scholar]
- 163.Nakamura, K., Y. Imai, A. Nakamura, and K. Yamane. 1992. Small cytoplasmic RNA of Bacillus subtilis: functional relationship with human signal recognition particle 7S RNA and Escherichia coli 4.5S RNA. J. Bacteriol. 174:2185-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nakazato, H., S. Venkatesan, and M. Edmonds. 1975. Polyadenylic acid sequences in E. coli messenger RNA. Nature 256:144-146. [DOI] [PubMed] [Google Scholar]
- 165.Newbury, S. F., N. H. Smith, and C. F. Higgins. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131-1143. [DOI] [PubMed] [Google Scholar]
- 166.Nilsson, G., J. G. Belasco, S. N. Cohen, and A. von Gabain. 1987. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc. Natl. Acad. Sci. USA 84:4890-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nilsson, P., and B. E. Uhlin. 1991. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNaseE-dependent endonucleolytic processing. Mol. Microbiol. 5:1791-1799. [DOI] [PubMed] [Google Scholar]
- 168.Nishimura, S., and B. Maruo. 1960. Intracellular ribonuclease from Bacillus subtilis. Biochim. Biophys. Acta 40:355-357. [DOI] [PubMed] [Google Scholar]
- 169.Nishimura, S., and M. Nomura. 1958. Ribonuclease of Bacillus subtilis. Biochim. Biophys. Acta 30:430-431. [DOI] [PubMed] [Google Scholar]
- 170.Nishimura, S., and M. Nomura. 1959. Ribonuclease of Bacillus subtilis. J. Biochem. 46:161-167. [Google Scholar]
- 171.Nossal, N. G., and M. F. Singer. 1968. The processive degradation of individual polyribonucleotide chains. I. Escherichia coli ribonuclease II. J. Biol. Chem. 243:913-922. [PubMed] [Google Scholar]
- 172.Ogawa, T., G. G. Pickett, T. Kogoma, and A. Kornberg. 1984. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc. Natl. Acad. Sci. USA 81:1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Oguro, A., H. Kakeshita, K. Nakamura, K. Yamane, W. Wang, and D. H. Bechhofer. 1998. Bacillus subtilis RNase III cleaves both 5′- and 3′-sites of the small cytoplasmic RNA precursor. J. Biol. Chem. 273:19542-19547. [DOI] [PubMed] [Google Scholar]
- 174.Oguro, A., H. Kakeshita, H. Takamatsu, K. Nakamura, and K. Yamane. 1996. The effect of Srb, a homologue of the mammalian SRP receptor alpha-subunit, on Bacillus subtilis growth and protein translocation. Gene 172:17-24. [DOI] [PubMed] [Google Scholar]
- 175.O'Hara, E. B., J. A. Chekanova, C. A. Ingle, Z. R. Kushner, E. Peters, and S. R. Kushner. 1995. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:1807-1811. [DOI] [PMC free article] [PubMed]
- 176.Ohtani, N., M. Haruki, M. Morikawa, R. J. Crouch, M. Itaya, and S. Kanaya. 1999. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry 38:605-618. [DOI] [PubMed] [Google Scholar]
- 177.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ono, M., and M. Kuwano. 1979. A conditional lethal mutation in an E. coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129:343-357. [DOI] [PubMed] [Google Scholar]
- 179.Oussenko, I. A., and D. H. Bechhofer. 2000. The yvaJ gene of Bacillus subtilis encodes a 3′-to-5′ exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J. Bacteriol. 182:2639-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Oussenko, I. A., R. Sanchez, and D. H. Bechhofer. 2002. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J. Bacteriol. 184:6250-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ow, M. C., and S. R. Kushner. 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 16:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pace, B., D. A. Stahl, and N. R. Pace. 1984. The catalytic element of a ribosomal RNA-processing complex. J. Biol. Chem. 259:11454-11458. [PubMed] [Google Scholar]
- 183.Pace, N. R., G. J. Olsen, and C. R. Woese. 1986. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell 45:325-326. [DOI] [PubMed] [Google Scholar]
- 184.Pace, N. R., and B. Pace. 1990. Ribosomal RNA terminal maturase: ribonuclease M5 from Bacillus subtilis. Methods Enzymol. 181:366-374. [DOI] [PubMed] [Google Scholar]
- 185.Panganiban, A. T., and H. R. Whiteley. 1983. Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA. Cell 33:907-913. [DOI] [PubMed] [Google Scholar]
- 186.Panganiban, A. T., and H. R. Whiteley. 1983. Purification and properties of a new Bacillus subtilis RNA processing enzyme. Cleavage of phage SP82 mRNA and Bacillus subtilis precursor rRNA. J. Biol. Chem. 258:12487-12493. [PubMed] [Google Scholar]
- 187.Persson, M., E. Glatz, and B. Rutberg. 2000. Different processing of an mRNA species in Bacillus subtilis and Escherichia coli. J. Bacteriol. 182:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Portier, C., L. Dondon, M. Grunberg-Manago, and P. Regnier. 1987. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J. 6:2165-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Putzer, H., S. Laalami, A. A. Brakhage, C. Condon, and M. Grunberg-Manago. 1995. Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis: induction, repression and growth-rate regulation. Mol. Microbiol. 16:709-718. [DOI] [PubMed] [Google Scholar]
- 190.Py, B., H. Causton, E. A. Mudd, and C. F. Higgins. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14:717-729. [DOI] [PubMed] [Google Scholar]
- 191.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 192.Raynal, L. C., and A. J. Carpousis. 1999. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 32:765-775. [DOI] [PubMed] [Google Scholar]
- 193.Raynal, L. C., H. M. Krisch, and A. J. Carpousis. 1998. The Bacillus subtilis nucleotidyltransferase is a tRNA CCA-adding enzyme. J. Bacteriol. 180:6276-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Regnier, P., and M. Grunberg-Manago. 1989. Cleavage by RNase III in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol. 210:293-302. [DOI] [PubMed] [Google Scholar]
- 195.Regnier, P., and M. Grunberg-Manago. 1990. RNase III cleavages in non-coding leaders of Escherichia coli transcripts control mRNA stability and genetic expression. Biochimie 72:825-834. [DOI] [PubMed] [Google Scholar]
- 196.Reich, C., G. J. Olsen, B. Pace, and N. R. Pace. 1988. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science 239:178-181. [DOI] [PubMed] [Google Scholar]
- 197.Resnekov, O., L. Rutberg, and A. von Gabain. 1990. Changes in the stability of specific mRNA species in response to growth stage in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:8355-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Reuven, N. B., and M. P. Deutscher. 1993. Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. FASEB J. 7:143-148. [DOI] [PubMed] [Google Scholar]
- 199.Robert-Le Meur, M., and C. Portier. 1992. E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 11:2633-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Robertson, H. D. 1982. Escherichia coli ribonuclease III cleavage sites. Cell 30:669-672. [DOI] [PubMed] [Google Scholar]
- 201.Rushizky, G. W., A. E. Greco, R. W. Hartley Jr., and H. A. Sober. 1963. Studies on B. subtilis ribonuclease. I. Characterization of enzymatic specificity. J. Biol. Chem. 2:787-793. [DOI] [PubMed] [Google Scholar]
- 202.Sakano, H., S. Yamada, T. Ikemura, Y. Shimura, and H. Ozeki. 1974. Temperature sensitive mutants of Escherichia coli for tRNA synthesis. Nucleic Acids Res. 1:355-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sandler, P., and B. Weisblum. 1988. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J. Mol. Biol. 203:905-915. [DOI] [PubMed] [Google Scholar]
- 204.Sandler, P., and B. Weisblum. 1989. Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5′-to-3′ nucleolytic cleavage of the ermA transcript. J. Bacteriol. 171:6680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sarkar, B., G. J. Cao, and N. Sarkar. 1997. Identification of two poly(A) polymerases in Bacillus subtilis. Biochem. Mol. Biol. Int. 41:1045-1050. [DOI] [PubMed] [Google Scholar]
- 206.Sarkar, N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66:173-197. [DOI] [PubMed] [Google Scholar]
- 207.Schedl, P., and P. Primakoff. 1973. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl. Acad. Sci. USA 70:2091-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schierling, K., S. Rosch, R. Rupprecht, S. Schiffer, and A. Marchfelder. 2002. tRNA 3′ end maturation in archaea has eukaryotic features: the RNase Z from Haloferax volcanii. J. Mol. Biol. 316:895-902. [DOI] [PubMed] [Google Scholar]
- 209.Schiffer, S., M. Helm, A. Theobald-Dietrich, R. Giege, and A. Marchfelder. 2001. The plant tRNA 3′ processing enzyme has a broad substrate spectrum. Biochemistry 40:8264-8272. [DOI] [PubMed] [Google Scholar]
- 210.Schiffer, S., S. Rosch, and A. Marchfelder. 2002. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 21:2769-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schroeder, E., J. McKibbin, M. L. Sogin, and N. R. Pace. 1977. Mode of degradation of precursor-specific ribonucleic acid fragments by Bacillus subtilis. J. Bacteriol. 130:1000-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Schweisguth, D. C., B. S. Chelladurai, A. W. Nicholson, and P. B. Moore. 1994. Structural characterization of a ribonuclease III processing signal. Nucleic Acids Res. 22:604-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sekiguchi, J., and S. Shuman. 1997. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell 1:89-97. [DOI] [PubMed] [Google Scholar]
- 214.Shivakumar, A. G., J. Hahn, G. Grandi, Y. Kozlov, and D. Dubnau. 1980. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc. Natl. Acad. Sci. USA 77:3903-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sogin, M. L., and N. R. Pace. 1974. In vitro maturation of precursors of 5S ribosomal RNA from Bacillus subtilis. Nature 252:598-600. [DOI] [PubMed] [Google Scholar]
- 216.Srinivasan, P. R., M. Ramanarayanan, and E. Rabbani. 1975. Presence of polyriboadenylate sequences in pulse-labeled RNA of Escherichia coli. Proc. Natl. Acad. Sci. USA 72:2910-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stahl, D. A., B. Meyhack, and N. R. Pace. 1980. Recognition of local nucleotide conformation in contrast to sequence by a rRNA processing endonuclease. Proc. Natl. Acad. Sci. USA 77:5644-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stahl, D. A., B. Pace, T. Marsh, and N. R. Pace. 1984. The ribonucleoprotein substrate for a ribosomal RNA-processing nuclease. J. Biol. Chem. 259:11448-11453. [PubMed] [Google Scholar]
- 219.Stams, T., S. Niranjanakumari, C. A. Fierke, and D. W. Christianson. 1998. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science 280:752-755. [DOI] [PubMed] [Google Scholar]
- 220.Struck, J. C., R. K. Hartmann, H. Y. Toschka, and V. A. Erdmann. 1989. Transcription and processing of Bacillus subtilis small cytoplasmic RNA. Mol. Gen. Genet. 215:478-482. [DOI] [PubMed] [Google Scholar]
- 221.Sugi, M., M. Yamasaki, T. Mizukami, and G. Tamura. 1980. ATP-sensitive ribonuclease of Bacillus cereus. J. Biol. Chem. 255:943-948. [PubMed] [Google Scholar]
- 222.Thang, M. N., W. Guschlbauer, H. G. Zachau, and M. Grunberg-Manago. 1967. Degradation of transfer ribonucleic acid by polynucleotide phosphorylase. I. Mechanism of phosphorolysis and structure of RNA. J. Mol. Biol. 26:403-421. [Google Scholar]
- 223.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Vazquez-Cruz, C., and G. Olmedo-Alvarez. 1997. Mechanism of decay of the crylAa mRNA in Bacillus subtilis. J. Bacteriol. 179:6341-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Walter, P., R. Gilmore, and G. Blobel. 1984. Protein translocation across the endoplasmic reticulum. Cell 38:5-8. [DOI] [PubMed] [Google Scholar]
- 226.Wang, W., and D. H. Bechhofer. 1996. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 178:2375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang, W., and D. H. Bechhofer. 1997. Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci. J. Bacteriol. 179:7379-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Warnecke, J. M., R. Held, S. Busch, and R. K. Hartmann. 1999. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 290:433-445. [DOI] [PubMed] [Google Scholar]
- 229.Waugh, D. S., and N. R. Pace. 1990. Complementation of an RNase P RNA (rnpB) gene deletion in Escherichia coli by homologous genes from distantly related eubacteria. J. Bacteriol. 172:6316-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wei, Y., and D. H. Bechhofer. 2002. Tetracycline induces stabilization of mRNA in Bacillus subtilis. J. Bacteriol. 184:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Woese, C. R. 1994. There must be a prokaryote somewhere: microbiology's search for itself. Microbiol. Rev. 58:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wong, H. C., and S. Chang. 1986. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc. Natl. Acad. Sci. USA 83:3233-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Xu, F., and S. N. Cohen. 1995. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374:180-183. [DOI] [PubMed] [Google Scholar]
- 234.Xu, F., S. Lin-Chao, and S. N. Cohen. 1993. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc. Natl. Acad. Sci. USA 90:6756-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yamasaki, M., and K. Arima. 1967. Regulation of intracellular ribonuclease of Bacillus subtilis by ATP and ADP. Biochim. Biophys. Acta 139:202-204. [DOI] [PubMed] [Google Scholar]
- 236.Yamasaki, M., and K. Arima. 1969. Intracellular ribonuclease of Bacillus subtilis: specific inhibition by ATP and dATP. Biochem. Biophys. Res. Commun. 37:430-436. [DOI] [PubMed] [Google Scholar]
- 237.Yamasaki, M., and K. Arima. 1970. ATP-inhibited ribonuclease of Bacillus subtilis. II. Effect of adenosine triphosphate and other compounds. Biochim. Biophys. Acta 209:475-483. [DOI] [PubMed] [Google Scholar]
- 238.Yamasaki, M., K. Yoshida, and K. Arima. 1970. ATP-inhibited ribonuclease of Bacillus subtilis. I. Purification and general properties. Biochim. Biophys. Acta 209:463-474. [DOI] [PubMed] [Google Scholar]
- 239.Yarchuk, O., I. Iost, and M. Dreyfus. 1991. The relation between translation and mRNA degradation in the lacZ gene. Biochimie 73:1533-1541. [DOI] [PubMed] [Google Scholar]
- 240.Yarchuk, O., N. Jacques, J. Guillerez, and M. Dreyfus. 1992. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J. Mol. Biol. 226:581-596. [DOI] [PubMed] [Google Scholar]
- 241.Yehudai-Resheff, S., M. Hirsh, and G. Schuster. 2001. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21:5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhang, K., and A. W. Nicholson. 1997. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. USA 94:13437-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhou, Z., and M. P. Deutscher. 1997. An essential function for the phosphate-dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J. Bacteriol. 179:4391-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]