Abstract
The unfolded protein response (UPR) is an evolutionarily conserved mechanism by which all eukaryotic cells adapt to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Inositol-requiring kinase 1 (IRE1) and PKR-related ER kinase (PERK) are two type I transmembrane ER-localized protein kinase receptors that signal the UPR through a process that involves homodimerization and autophosphorylation. To elucidate the molecular basis of the ER transmembrane signaling event, we determined the x-ray crystal structure of the luminal domain of human IRE1α. The monomer of the luminal domain comprises a unique fold of a triangular assembly of β-sheet clusters. Structural analysis identified an extensive dimerization interface stabilized by hydrogen bonds and hydrophobic interactions. Dimerization creates an MHC-like groove at the interface. However, because this groove is too narrow for peptide binding and the purified luminal domain forms high-affinity dimers in vitro, peptide binding to this groove is not required for dimerization. Consistent with our structural observations, mutations that disrupt the dimerization interface produced IRE1α molecules that failed to either dimerize or activate the UPR upon ER stress. In addition, mutations in a structurally homologous region within PERK also prevented dimerization. Our structural, biochemical, and functional studies in vivo altogether demonstrate that IRE1 and PERK have conserved a common molecular interface necessary and sufficient for dimerization and UPR signaling.
Keywords: endoplasmic reticulum, protein structure, signal transduction, protein kinase, endoplasmic reticulum stress
The endoplasmic reticulum (ER) of eukaryotic cells is the cellular compartment where secretory and transmembrane proteins fold into their native conformations and undergo posttranslational modifications that are important for their structure and function. When protein folding in the ER is perturbed, a set of signal transduction pathways is activated to reduce the protein-folding load and increase folding capacity. These pathways are collectively termed the unfolded protein response (UPR) (1–4). To increase the folding capacity, synthesis of ER resident chaperones and folding catalysts is induced. To decrease the folding load in the ER, global mRNA translation is attenuated and clearance of misfolded proteins through ER-associated degradation is increased. UPR signaling is mediated by three ER resident transmembrane proteins: IRE1, PERK, and ATF6.
IRE1 is a type I transmembrane protein kinase receptor that also has a site-specific RNase activity that, upon activation, initiates a site-specific unconventional splicing reaction (5, 6). The substrate for IRE1 RNase in metazoans is Xbp1 mRNA, which encodes a basic leucine zipper transcription factor of the ATF/CREB family. XBP1 controls expression of genes containing an X-box element or a UPR element in their promoter regions (7–10). The IRE1-mediated splicing reaction introduces into XBP1 an alternative C terminus, thereby generating an XBP1 molecule that is a more potent transcriptional activator. Therefore, activation of IRE1 and its RNase increases the transcription of genes encoding ER chaperones and folding catalysts. In addition, the IRE1/XBP1 pathway is essential to activate genes encoding functions in ER-associated degradation (11). Two IRE1 genes exist in the mammalian genome, IRE1α and IRE1β (12, 13). IRE1α is expressed in all cells and tissues, whereas IRE1β expression is restricted to intestinal epithelial cells. IRE1α and IRE1β are structurally similar to TGF-β serine/threonine protein kinase receptors and mechanistically similar to receptor tyrosine kinases, where ligand binding-induced dimerization is a universal activation mechanism for the regulation of cytoplasmic activities. However, the UPR represents a novel intracellular ER transmembrane signaling pathway, which appears to employ a ligand-independent activation mechanism (14) in which the IRE1 N-terminal luminal domain (NLD) functions as an ER stress sensor. According to this model, under normal conditions IRE1 is maintained in a monomeric state through interaction of the NLD with the ER resident chaperone BiP (15–19). Upon ER stress, immunoglobin-binding protein (BiP)/glucose-regulated protein of 78 kDa (Grp78) (BiP) binds to unfolded proteins as they accumulate, permitting the released NLD to form homodimers. Dimerization of the NLD in turn leads to the activation of the protein kinase and RNase activities in the cytosolic domain of IRE1 (20, 21).
A complete understanding of the molecular mechanism for such an ER transmembrane signaling event requires a detailed knowledge of the high-resolution structure of the protein. In this study we seek to elucidate the structural basis for IRE1 receptor activation. Here we describe the crystal structure of the NLD of human IRE1α at 3.1 Å. The structure demonstrates that the NLD dimerizes through an extensive interface consisting largely of β-sheets, and this structural feature is conserved between IRE1 and PERK proteins. Mutation of critical residues within this interface prevented IRE1 dimerization and UPR signaling. Furthermore, mutations in a structurally homologous region of PERK also disrupted dimerization. In contrast to a recent study on the structure of the yeast Ire1p luminal domain (22), our results suggest that activation of the UPR by IRE1 and PERK is mediated by a common dimerization mechanism that does not require the direct binding of unfolded proteins.
Results
Structure of the Human IRE1α NLD Monomer.
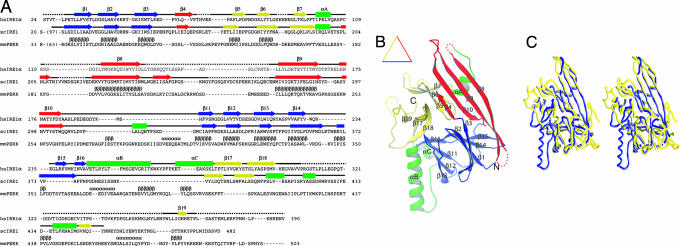
The x-ray structure of a major fragment of the human IRE1α NLD that comprises 367 amino acid residues (S24–V390) (Fig. 1A) was determined. The NLD monomer structure is organized into a triangular β-sheet cluster with three major β-structural motifs that occupy the three sides of a triangular plate (namely, the N, C, and M motifs) (Fig. 1B). Between these motifs are several inserted α-helices. The N motif on the bottom side contains the N terminus of the molecule (colored blue). The N motif has a β-barrel fold with a five-stranded antiparallel β-sheet stacking against a four-stranded mixed β-sheet. The motif on the top left side contains the C terminus of the molecule and is referred to as the C motif (colored yellow). The C motif contains a β-barrel fold, and it has two three-stranded antiparallel β-sheets stacking against each other. The middle motif on the top right side is referred to as the M motif, which is composed of an elongated four-stranded antiparallel β-sheet that runs nearly the entire length of the side (colored red). A pair of α-helices connects the N and C motifs at the bottom left corner of the triangle, and a short two-turn α-helix connects the M and C motifs (colored green). All three motifs pack tightly against each other with no apparent separation. Therefore, the entire monomer should be viewed as a single domain. Search for homologous protein folds using the DALI fold yielded no structures above noise level (23). Thus, the triangular β-cluster of the IRE1 NLD likely represents a new protein fold. Although the core triangular β-cluster fold is conserved between human IRE1α and yeast Ire1p NLDs (22), major structural differences were observed (Fig. 1C). In particular, whereas a long helix αB is inserted between β16 and αC in the human IRE1α NLD, this structure is absent in the yeast Ire1p NLD.
Fig. 1.
The crystal structure of human IRE1α NLD. (A) Sequence and secondary structure alignment of IRE1 and PERK. The NLD sequences of human IRE1α, S. cerevisiae Ire1p, and murine PERK were aligned by using the program T-Coffee (32). Secondary structural elements are indicated above the sequence: α-helices are drawn as rectangles, β-strands as arrows, other elements as solid lines, and structurally unobserved residues as dashed lines. These elements are colored based on their locations in the structure (see Results for details). Predicted α-helices and β-strands for PERK are indicated with Greek letters. (B) A ribbon drawing of the NLD monomer. The secondary structural elements are labeled and colored as in A: α-helices are lettered and drawn as coils, β-strands are numbered and drawn as arrows, and other elements are drawn as tubes. (C) A stereo diagram showing Cα trace superimposition of human IRE1α NLD (blue) and S. cerevisiae Ire1p NLD (yellow) structures. The programs Ribbons (33) and Grasp (34) were used to produce B and C, respectively.
Structure of the IRE1 NLD Dimer.
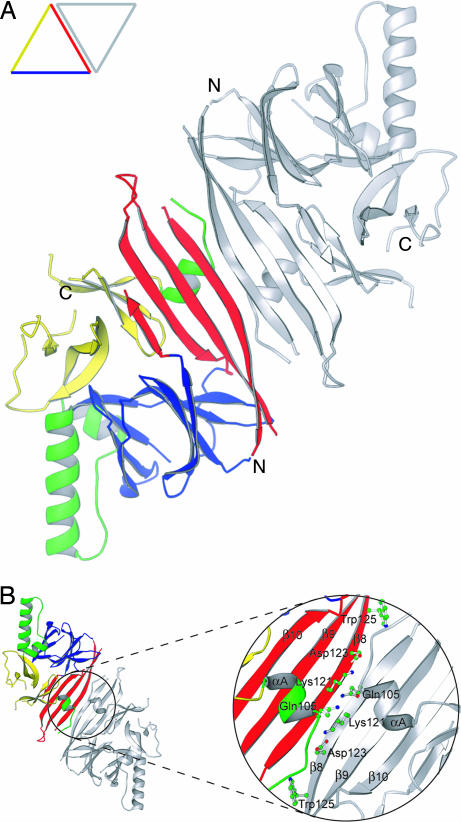
The NLD forms stable dimers in vitro with an apparent molecular mass of 96 kDa (24). Because the asymmetric unit of the crystal contains only one monomer, the dimer interface must span the crystallographic symmetry axis. By examining the crystal packing of one protomer against its neighbors, we identified a strong candidate for the dimer interface.
In the crystal lattice, two protomers of the NLD pack symmetrically across the top right side of the monomeric triangle, formed by the outside β-strand (β8) within the M motif and the preceding α-helix (αA) (Fig. 2A). Importantly, a similar molecular dimer structure is observed in the asymmetric unit of the yeast Ire1p NLD crystal (22) even though the rest of the crystal packing for the two structures is different, suggesting that dimerization via the β-strands in the M motif is conserved among the IRE1 family of proteins. Dimerization creates a diamond-shaped plate that is thin in the middle. The plate is slightly arched with an eight-stranded β-sheet formed by two neighboring M motifs on the convex side, a pair of interacting helices αA on the concave side, and helices αB and αC at the feet of the arch (Fig. 6, which is published as supporting information on the PNAS web site). Based on the location and projection of both C termini, we predict that the concave side of the NLD dimer is ER membrane-proximal. The dimer interface contains both polar and hydrophobic interactions (Fig. 2B). Four pairs of backbone hydrogen bonds are observed between the two antiparallel β8 strands. Lys-121 forms two backbone hydrogen bonds with its twofold symmetry-related partner. In addition, the amino group of the Lys-121 side chain appears to form two hydrogen bonds with backbone carbonyl groups of neighboring Val-104 and Ser-107, both of which are located in helix αA. The amide group of Asp-123 interacts with the carbonyl group of neighboring Gly-119. Gln-105 (located on helix αA) also interacts with its symmetry-related partner through hydrogen bond interaction of the side chains. Hydrophobic interactions also contribute to the stability of the dimer. Most notable is the insertion of the side chain of Trp-125 into a hydrophobic pocket formed by the side chains of Pro-108, Leu-116, and Met-118. The relatively low resolution of the structure precluded a more detailed description of the dimer interface.
Fig. 2.
The molecular dimer structure of human IRE1α NLD. (A) Ribbon drawing of the NLD dimer looking straight down the twofold axis of symmetry. One subunit is colored as in Fig. 1B, and the other is gray. In the displayed orientation both C termini of the dimer would project into the page. (B) An enlarged view of the dimer interface after rotating the dimer in A 180° around the horizontal axis. Side chains of residues examined in our mutagenesis studies are shown as ball-and-stick models. The program Ribbons (33) was used to produce both drawings.
In addition to the aforementioned antiparallel β-sheet interface, there are two additional areas of crystal contacts where the NLD protomers are related by the twofold crystallographic symmetry axis. Both contacts are significantly less extensive than the antiparallel β-sheet interface and involve secondary structural elements (predominantly helix αB) that are not conserved in the yeast Ire1p structure (22). Analysis of the molecular packing within the crystal lattice also did not support the existence of high-order NLD oligomers as proposed for the yeast protein (22). Any given pair of NLD dimers in the crystal lattice have their C termini projected in orthogonal or opposite directions, making oligomeric interactions on the ER membrane either sterically difficult or in need of membrane curvature (Fig. 6).
The Antiparallel β-Sheet Interface Mediates IRE1α NLD Dimerization.
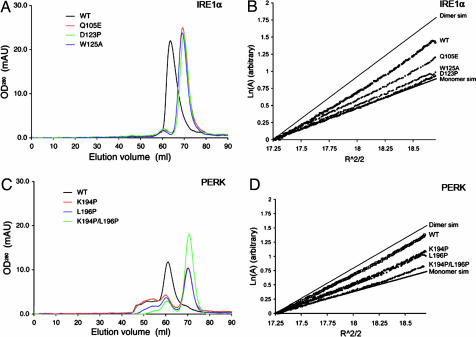
To test the importance of the antiparallel β-sheet interface in dimer formation, we generated mutations at specific residues located at the dimer interface and analyzed the oligomeric states of the mutants by gel-filtration chromatography and sedimentation equilibrium analytical ultracentrifugation. Specifically, K121P and D123P were designed to disrupt the β8 backbone hydrogen bond interactions between the monomers but preserve the β-sheet hydrogen bonding patterns within the M motif. Whereas W125A was designed to disrupt complementary hydrophobic interactions between the monomers, Q105E was constructed to introduce repulsive negative charges (Fig. 2B).
Compared with the WT dimeric NLD protein, the monomer–dimer equilibria of D123P, W125A, and Q105E mutants were shifted toward the monomer species (Fig. 3A). Because the K121P mutant appeared as large protein aggregates on a gel-filtration column (data not shown), it is likely that protein folding is disrupted in this mutant. The sedimentation profiles of D123P, W125A, and Q105E in the centrifugation experiments were also consistent with the gel-filtration experiments (Fig. 3B). The dimer dissociation constants for the three mutants were 10- to 1,000-fold greater than that observed for the WT protein (Table 1). CD spectra for the three mutants were similar to that of the WT protein, indicating that the mutations did not cause major secondary structural changes within the protein (Fig. 7, which is published as supporting information on the PNAS web site). Interestingly, the hydrodynamic properties of alanine mutants at residues located in β8 (K121A and D123A) were similar to that of the WT protein (C.Y.L. and R.J.K., unpublished observation), indicating that side chain interactions of either K121 or D123 do not significantly contribute to dimer stability. The biochemical studies strongly suggest that the antiparallel β-sheet interface is responsible for IRE1α luminal domain dimerization.
Fig. 3.
The luminal domains of IRE1 and PERK share a similar dimerization mechanism. (A and C) Gel-filtration analysis. WT human IRE1α NLD eluted as a 158-kDa protein upon gel filtration (black line), and mutants Q105E (red line), D123P (green line), and W125A (blue line) eluted 6 ml later (A). WT murine PERK NLD eluted as a 181-kDa protein (black line), and mutants K194P (red line), L196P (blue line), and K194P/L196P (green line) eluted 10 ml later (C). (B and D) Analytical ultracentrifugation sedimentation equilibrium analysis. Point mutations in human IRE1α NLD (B) and murine PERK NLD (D) shift the dimer/monomer equilibrium toward the monomeric species. The IRE1α NLD mutant D123P and the PERK NLD double mutant K194P/L196P were exclusively monomeric.
Table 1.
Analytical ultracentrifugation sedimentation equilibrium data
| Protein | Oligomerization status | Kd for monomer/dimer equilibrium, μM |
|---|---|---|
| IRE1α NLD | ||
| WT | Dimer | 2.3 |
| Q105E | Monomer/dimer | 56 |
| D123P | Monomer | 3,000 |
| W125A | Monomer | 371 |
| PERK NLD | ||
| WT | Dimer | 0.53 |
| K194P | Monomer/dimer | 65 |
| L196P | Monomer/dimer | 94 |
| K194P/L196P | Monomer | 137 |
The Mechanism of Dimerization Is Conserved Between IRE1 and PERK.
Although the luminal domains of IRE1 and PERK share limited sequence homology (6% identity/similarity with <12% homology overall), secondary structure prediction (25) suggests that they likely have similar folds (Fig. 1A). Based on sequence alignment, we predicted that residues 189–201 of murine PERK represent the structural equivalent of the β-strands at the human IRE1α dimer interface. Therefore, Lys-194 and Leu-196 of PERK, corresponding to Lys-121 and Asp-123 of IRE1α NLD, were mutated to proline to test their roles for dimerization. Compared with the WT PERK NLD (residues Ala-32–Gly-322), both mutant proteins eluted at a later stage on a gel-filtration column, consistent with shifts in the monomer–dimer equilibria toward the monomeric species (Fig. 3C). Similarly, sedimentation equilibrium analysis of the two mutant proteins also revealed a pronounced shift in their monomer–dimer equilibria, with a 100-fold increase in dissociation constants (Fig. 3D and Table 1). The CD spectra for these mutants were not significantly different from those of the WT PERK NLD, indicating that it is unlikely that a significant structural change could account for the reduced dimer formation (Fig. 8, which is published as supporting information on the PNAS web site). Taken together, our results suggest that backbone hydrogen bonding of Lys-194 and Leu-196 within the PERK luminal domain are important determinants for dimerization and that a structurally similar dimer interface as observed in the IRE1α NLD is used in PERK. Interestingly, the double mutation (K194P/L196P) compromised but did not eliminate dimer formation of PERK, suggesting that other structural elements also contribute to dimerization of the PERK NLD.
Antiparallel β-Sheet Interactions Are Required for IRE1 Dimerization in Vivo.
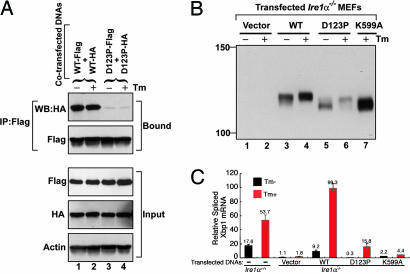
To further test the requirement for the antiparallel β-sheet in dimer formation, WT and D123P mutant full-length human IRE1α tagged with Flag and HA epitopes were coexpressed in mammalian cells, and their interaction was monitored by immunoprecipitation and immunoblot analysis (Fig. 4A). WT and D123P mutant IRE1α proteins were expressed at comparable levels and immunoprecipitated equally efficiently. Immunoprecipitation with anti-Flag antibody and Western blot with anti-HA antibody demonstrated a significant interaction between coexpressed HA- and Flag-tagged WT proteins (Fig. 4A). Treatment of the cells with tunicamycin (Tm), an inhibitor of N-linked glycosylation that activates the UPR, did not further increase the interaction of WT IRE1α molecules, likely a consequence of IRE1α overexpression that spontaneously activates the UPR (12). By contrast, the amount of HA-tagged D123P mutant IRE1α that coimmunoprecipitated with Flag-tagged mutant was dramatically reduced, and Tm treatment also did not further increase coimmunoprecipitation. These results show that the D123P mutant expressed in mammalian cells is defective in dimerization and indicate that the NLD dimer interface is required for dimerization of intact IRE1 in vivo.
Fig. 4.
Human IRE1α mutant D123P is defective in dimerization, autophosphorylation, and RNase activity. (A) COS-1 cells were cotransfected with plasmids expressing human IRE1α (WT or D123P) tagged with Flag and HA epitopes. Lysates were prepared from cells treated with or without Tm for 4 h and subjected to immunoprecipitation using anti-Flag monoclonal antibody and then analyzed by Western blot using anti-HA antibody. (B) Ire1α−/− MEF cell lines stably expressing human IRE1α WT or mutant proteins were treated with or without Tm for 5 h. Cell lysates were prepared and subjected to immunoprecipitation by using anti-IRE1α antibody and then analyzed by Western blot with antibodies against IRE1α. (C) Quantitative real-time RT-PCR analysis of spliced Xbp1 transcripts. WT and Ire1α−/− MEF cell lines stably expressing WT or mutant human IRE1α proteins were treated with Tm (10 μg/ml) for 5 h, and RNA was isolated for quantitative real-time RT-PCR by using a primer set flanking the intron in the Xbp1 mRNA. The columns and bars represent the means and standard deviations of three independent experiments.
Dimerization of IRE1 Stimulates Autophosphorylation and UPR Signaling.
Because we were able to disrupt dimerization of full-length IRE1α in vivo, we further analyzed the functional requirement of dimerization for protein kinase activation. Autophosphorylation of IRE1α was measured by analysis of the upward mobility shift observed upon SDS/PAGE and Western blot analysis (12). To circumvent ambiguity in interpretation due to the presence of low levels of endogenous WT IRE1α in the cells, we studied Ire1α−/− mouse embryonic fibroblast (MEF) cell lines that stably express human WT, dimerization-defective D123P, or the kinase-defective K599A IRE1α mutant proteins. The K599A mutant IRE1α lacks kinase activity and is not phosphorylated in the absence or presence of Tm (12). Compared with the K599A mutant, the mobility of WT IRE1α was slightly reduced and was further reduced upon Tm treatment (Fig. 4B). Therefore, overexpressed WT IRE1α was partially phosphorylated and, after Tm treatment, became hyperphosphorylated. The D123P mutant IRE1α comigrated with K599A kinase-defective IRE1α, indicative of the absence of phosphorylation. Upon Tm treatment, D123P IRE1α did become phosphorylated (Fig. 4B, lane 5 vs. lane 6), although not to the same extent as WT IRE1α. These findings indicate that dimerization stimulates autophosphorylation, which is required for hyperphosphorylation of IRE1α.
To elucidate the role of IRE1α dimerization in the activation of its RNase activity, we analyzed IRE1α-mediated UPR signaling in both yeast and mammalian cells. Because the human IRE1α NLD functions as a replacement for the yeast Ire1p NLD to signal the UPR in Saccharomyces cerevisiae (14), we analyzed yeast Ire1p harboring the NLD of either WT human IRE1α or the D123P mutant. Compared with the WT chimeric Ire1p, UPR signaling from the chimera harboring the D123P mutation was reduced ≈10-fold (Fig. 9, which is published as supporting information on the PNAS web site), suggesting a requirement for dimerization in signaling from the human–yeast chimeric protein in yeast.
To analyze the requirement for IRE1 dimerization in activation of its RNase, we used quantitative RT-PCR with primers specific to the spliced Xbp1 mRNA and monitored Xbp1 mRNA splicing in Ire1α−/− MEFs that stably express WT or mutant IRE1α. Tm treatment of WT MEFs produced a 3-fold increase in spliced Xbp1 mRNA (Fig. 4C). The spliced Xbp1 mRNA in mock-transfected Ire1α−/− MEFs was reduced by at least 10-fold, indicating the specificity of the RT-PCR assay. Basal Xbp1 mRNA splicing was restored by expression of WT IRE1α in Ire1α−/− MEFs, and Tm treatment further increased Xbp1 mRNA splicing by 8-fold. By contrast, expression of the D123P mutant IRE1α in Ire1α−/− MEFs did not restore basal Xbp1 mRNA splicing, although Tm treatment did significantly increase the level of spliced Xbp1 transcripts. Finally, expression of the K599A kinase-defective IRE1α in Ire1α−/− MEFs also did not restore Xbp1 mRNA splicing, confirming the requirement for protein kinase activity in activation of the RNase activity. It is interesting to note that the degree of IRE1α phosphorylation (Fig. 4B) correlated with the extent of Xbp1 mRNA splicing in these transfected Ire1α−/− MEFs, indicating that autophosphorylation promotes RNase activity. Finally, Xbp1 mRNA splicing measured by luciferase reporter assay also independently confirmed that the D123P mutation significantly reduced the efficiency of Xbp1 mRNA splicing (Fig. 10, which is published as supporting information on the PNAS web site). Based on these findings, we conclude that dimerization leads to both the autophosphorylation and the RNase activities of IRE1α.
Discussion
We have identified a conserved dimerization interface within the luminal domains of IRE1 and PERK that is stabilized by hydrogen bonds and hydrophobic interactions. Our results demonstrate that dimerization of the luminal domain via this novel interface is required by IRE1 and PERK for dimerization and autophosphorylation to signal the UPR in mammalian cells. In a recent structural and mutagenesis analysis of the S. cerevisiae Ire1p (22), it was proposed that unfolded protein-induced oligomerization mediates receptor activation. In this model, unfolded proteins act as a scaffold by binding to the MHC-like grooves located on the concave side of IRE1 monomers to promote oligomer assembly. However, our findings provide compelling evidence to suggest that peptide binding is neither probable nor necessary for homodimer or oligomer formation. First, the purified NLD can form dimers in vitro in the absence of other proteins. Because individual mutations within the M motif alone were sufficient to disrupt dimerization in vitro and in vivo, it is unlikely that a second high-affinity intermolecular interaction site exists that would be required for oligomer assembly. Second, the MHC-like groove present in the human NLD crystal structure is too narrow to permit peptide binding. In fact, the αA helices that form the walls of the MHC-like groove are spatially much closer together in the human NLD than those observed in the yeast NLD. The symmetry-related Gln-105 residues in the αA helices of the human NLD form a hydrogen bond that contributes significantly to dimer stability and blocks access to the proposed peptide-binding groove at its midway point (Fig. 11, which is published as supporting information on the PNAS web site). In addition, mutagenesis studies of the yeast Ire1p NLD suggested that three residues (Met-229, Phe-285, and Tyr-301) in the MHC-like groove could potentially contribute to peptide binding (22). However, two of the corresponding residues in the human IRE1α NLD structure (Tyr-161 and Tyr-179) are buried, and the third one is a nonconserved methionine-to-lysine replacement (Lys-121). Therefore, it is unlikely that these three residues in human IRE1α would be involved in peptide binding. Finally, the NLD dimer is oriented in the ER membrane in a manner that would make it difficult for peptide-induced oligomers to form. Projection of the molecule's C termini in both the yeast and human structures suggests that the proposed groove would directly face the ER membrane instead of the lumen (Fig. 6). Although this configuration does not eliminate the possibility of peptide binding, it does make it more difficult and less likely to occur in vivo. In addition, crystal-packing analysis of the human IRE1α structure also suggests that it would be sterically difficult to assemble a linear array of IRE1 monomers on the ER membrane. Given the high degree of conservation between human IRE1 and yeast Ire1p primary amino acid sequences and three-dimensional structures, and the observation that the human NLD as well as an isolated basic leucine zipper dimerization domain can substitute for the yeast NLD to signal the UPR in yeast (14), we believe that in both species IRE1 activation does not require peptide-induced formation of higher-order oligomeric structures.
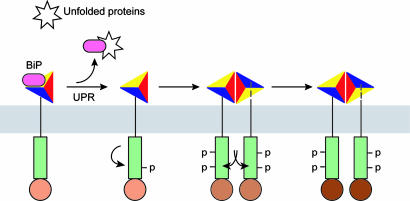
Therefore, our findings instead favor a negative regulatory dimerization model that does not require the direct binding of unfolded protein (14). In this model, inactive IRE1α is maintained in a monomeric state by interaction with BiP within the ER lumen. As unfolded proteins accumulate, they sequester BiP from IRE1α. BiP release would permit IRE1 dimer formation through hydrogen bonding and extensive hydrophobic interactions at the dimer interface. Based on results from our functional studies (Fig. 4), BiP release would initiate the first phase of dimerization-independent intramolecular autophosphorylation. Subsequently, dimerization-induced intermolecular autophosphorylation would lead to complete activation of the RNase activity. Thus, our findings indicate that the activation of IRE1α occurs in stages and that each stage correlates with increasing degrees of autophosphorylation and RNase function (Fig. 5).
Fig. 5.
Model for staged activation of IRE1. The model depicts that several steps mediate IRE1 activation including BiP release, initial intramolecular autophosphorylation, dimerization, and dimerization-induced trans-autophosphorylation (see Discussion for details). Increasing degrees of IRE1 autophosphorylation cause higher levels of RNase activity indicated as darker shades of brown.
Previous studies support the notion that IRE1 and PERK are regulated by a common activation mechanism. For example, the PERK NLD can substitute for the yeast Ire1p NLD to signal the UPR in yeast, even though yeast genome does not have a PERK homologue (14). In addition, BiP interacts with both IRE1 and PERK and is a negative regulator of UPR activation (15, 26). Although our structural studies did not identify a BiP interaction site, deletion studies in yeast and human IRE1, as well as in human PERK (17, 18, 27), suggest that a conserved BiP interaction site exists in the region C-terminal to residue Val-307 in human IRE1α. It is interesting to note that the potential BiP interaction site in the crystal structure of human IRE1α is spatially conserved with that in the structure of yeast Ire1p (22). However, in both structures these regions were not resolved, suggesting that they are structurally flexible. Kimata et al. (17) demonstrated that Ire1p with a deletion in this BiP binding region displayed ER stress-induced activation in yeast, suggesting that BiP release from yeast Ire1p cannot lead to full activation of the UPR. Thus, we propose that BiP release may prime IRE1 for subsequent dimerization that may be further influenced by a secondary effect induced upon ER stress. Finally, our studies suggest that, although PERK has significantly diverged from IRE1 in evolution, it has conserved essential structural motifs in the NLD required for dimerization. Therefore, the unique dimerization interface formed between the antiparallel β-sheets from each monomer is likely a general characteristic used for activation of the ER stress-sensing protein kinase receptors. Further elucidation of the common features and differences between the IRE1 and PERK dimerization interfaces should lead to identification of compounds that selectively inhibit or activate only one sensor. As evidence continues to accumulate regarding the significance of UPR signaling in health and disease (28, 29), the ability to independently modulate IRE1 and PERK signaling will likely lead to new modes of therapeutic intervention in disease states.
Materials and Methods
Plasmid Construction and Protein Preparation.
The human IRE1α NLD core domain (residues 24–390) was constructed as described previously (18). Full-length human IRE1α NLD (residues 24–446) and murine PERK NLD (residues 32–322) were cloned into a pET-43.1b (+) derivative vector (Novagen, Darmstadt, Germany). Because the full-length PERK NLD did not fold well when expressed in Escherichia coli, we used limited protease digestion to identify a stable fragment of PERK NLD (residues 32–322) that could efficiently express in E. coli. All mutations were generated by PCR using the QuikChange method (Stratagene, La Jolla, CA) and confirmed by DNA sequencing.
All proteins were expressed in E. coli strain BL21 (DE3) as N-terminal NusA-tagged proteins containing a poly-His tag and the tobacco etch virus (TEV) recognition site. Selenomethionyl human IRE1 NLD (residues 24–390) was expressed in E. coli B834 (DE3) in M9 minimal medium supplemented with 75 mg/ml selenomethionine. Proteins were purified by using two Ni2+-NTA columns with TEV protease removal of the NusA tag in between, followed by one anion-exchange column (Resource Q, Amersham Biosciences, Piscataway, NJ), and one gel-filtration column (Superdex 200, Amersham Biosciences).
Crystallization and Data Collection.
Crystals of human IRE1α NLD core domain (residues 24–390) were grown at 20°C by using the hanging-drop vapor diffusion method by mixing the protein (10 mg/ml) with an equal volume of reservoir solution containing 3% 2-methyl-2,4-pentanediol, 3–3.5% polyethylene glycol 20000, and 100 mM Pipes (pH 6.5). Crystals belong to space group P6522, with unit cell dimensions a = b = 185.01 Å and c = 67.69 Å, and contain one NLD molecule in the asymmetric unit. Diffraction data were collected by using the DND-CAT ID-5 beam line at the Advanced Photon Source in Argonne National Laboratory (Argonne, IL). Multiwavelength anomalous diffraction data were collected at three wavelengths. All data were processed by using DENZO, and intensities were scaled by using SCALEPACK (30).
Structural Determination and Refinement.
Experimental phases were obtained by using the multiwavelength anomalous diffraction method with selenomethionine derivatives (31). Structure refinement was performed by using CNS with cross-validation. The yeast Ire1p NLD structure (kindly provided by Peter Walter, University of California, San Francisco, CA) was also helpful in resolving some of the ambiguity in model building. The current model consists of residues 29–65, 71–85, 91–110, 116–130, 153–307, and 358–368, with an R factor of 26.8% and a free R factor of 31.6%. Of all of the nonglycine residues, none are located in the disallowed regions on the Ramachandran plot. Crystallographic statistics are summarized in Table 2, which is published as supporting information on the PNAS web site.
For in vitro and in vivo characterization of human IRE1α, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Peter Walter and colleagues for providing yeast Ire1p structural coordinates before publication; X. Zhou and K. Yoshino-Koh for excellent technical assistance; D. T. Rutkowski (University of Michigan, Ann Arbor, MI) for reagents and critical reading of the manuscript; J. Mitchell for assistance in preparation of the manuscript; A. Ludlam for help with the analytical ultracentrifugation experiments; P. Graf and U. Jakob for access and help with the CD experiments; and Z. Wawrzak for access and help at the Advanced Photon Source DuPont–Northwestern–Dow Collaborative Access Team 5ID-D beam line. The DuPont–Northwestern–Dow Collaborative Access Team is supported by E. I. DuPont de Nemours, Dow Chemical Company, U.S. National Science Foundation Grant DMR-9304725, and the State of Illinois through the Department of Commerce and the Board of Higher Education (Grant IBHE HECA NWU 96). This work was supported in part by National Institutes of Health Grants DK042394 and HL052173 (to R.J.K.) and DK065980 (to Z.X.). Z.X. is a Pew Scholar in Biomedical Sciences. R.J.K. is an investigator at the Howard Hughes Medical Institute.
Abbreviations
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- NLD
N-terminal luminal domain
- Tm
tunicamycin
- MEF
mouse embryonic fibroblast.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates of the human IRE1α NLD have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2HZ6).
References
- 1.Schroder M, Kaufman RJ. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 2.Patil C, Walter P. Curr Opin Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- 3.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 4.Mori K. Traffic. 2003;4:519–528. doi: 10.1034/j.1600-0854.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 5.Cox JS, Shamu CE, Walter P. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Ma W, Gething MJ, Sambrook J. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 7.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 9.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 12.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CY, Schroder M, Kaufman RJ. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 15.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 16.Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Mol Biol Cell. 2003;14:2559–2569. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. J Cell Biol. 2004;167:445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu CY, Xu Z, Kaufman RJ. J Biol Chem. 2003;278:17680–17687. doi: 10.1074/jbc.M300418200. [DOI] [PubMed] [Google Scholar]
- 19.Oikawa D, Kimata Y, Takeuchi M, Kohno K. Biochem J. 2005;391:135–142. doi: 10.1042/BJ20050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Proc Natl Acad Sci USA. 1997;94:4289–4294. doi: 10.1073/pnas.94.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shamu CE, Walter P. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 22.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CY, Wong HN, Schauerte JA, Kaufman RJ. J Biol Chem. 2002;277:18346–18356. doi: 10.1074/jbc.M112454200. [DOI] [PubMed] [Google Scholar]
- 25.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 26.Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman RJ. J Biol Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- 27.Ma K, Vattem KM, Wek RC. J Biol Chem. 2002;277:18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman RJ. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiseman RL, Balch WE. Trends Mol Med. 2005;11:347–350. doi: 10.1016/j.molmed.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Hendrickson WA, Ogata CM. Methods Enzymol. 1997;276:494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- 32.Notredame C, Higgins DG, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 33.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 34.Nicholls A, Sharp KA, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.