Abstract
We have developed a method for isolation of random peptides that inhibit intracellular processes in bacteria. A library of random peptides expressed as fusions to Escherichia coli thioredoxin (aptamers) were expressed under the tight control of the arabinose-inducible PBAD promoter. A selection was applied to the library to isolate aptamers that interfered with the activity of thymidylate synthase (ThyA) in vivo. Expression of an aptamer isolated by this method resulted in a ThyA− phenotype that was suppressed by simultaneous overexpression of ThyA. Two-hybrid analysis showed that this aptamer is likely to interact with ThyA in vivo. The library also was screened for aptamers that inhibited growth of bacteria expressing them, and five such aptamers were characterized. Four aptamers were bacteriostatic when expressed, whereas one showed a bactericidal effect. Introduction of translational stop codons into various aptamers blocked their activity, suggesting that their biological effects were likely to be due to protein aptamer rather than RNA. Combinatorial aptamers provide a new genetic and biochemical tool for identifying targets for antibacterial drug development.
The emergence and spread of bacterial resistance to antibiotics has created a crisis in medicine. Many valuable antibiotics are losing effectiveness against common pathogens. The clinically useful antibiotics act on only a small number of cellular processes in bacteria; most affect cell wall, protein, or RNA synthesis, folate metabolism, or DNA gyrase. A number of natural peptide antibiotics synthesized by microbial or eukaryotic cells target bacterial cell membranes (1). However, the majority of essential genes are not targets of any known antibiotic. For Haemophilus influenzae, it has been estimated that about 20% of its genes are essential (2); clearly, most of these gene products are not affected by currently available antibiotics.
Most classes of antimicrobials have been discovered by screening natural products, their derivatives, or synthetic chemicals for their ability to inhibit bacterial growth. This approach has been highly productive, but may have limited the breadth of compounds discovered. Only a small number of drug classes have been described that are able to penetrate bacterial cells and then remain inside the cells long enough to inhibit an essential target molecule. New methods that circumvent the permeability barrier are needed to identify novel targets for antimicrobials and candidate compounds that inhibit those targets. One solution to this problem is to express inhibitory compounds directly inside target cells. For example, a library of random peptides fused to a carrier protein and expressed within yeast cells has been used to screen for protein “aptamers” that bind to cyclin-dependent kinase 2 (3) or the retinoblastoma protein (4) or that inhibit signaling through several pathways (5, 6). However, application of this approach to the identification of inhibitory peptides or aptamers within bacterial cells has not been reported.
We have developed a method we call “aptamer-based bacterial inhibition systems” (ABBIS), which allows the conditional expression within bacterial cells of a library of random peptides fused to a carrier protein. In this way, otherwise toxic aptamers can be identified among a library of random peptides, as those that display inhibitory activity only in the presence of inducer. We have shown that ABBIS-based screens can focus either on a specific molecular target (e.g., thymidylate synthase) or on a broader phenotype (growth inhibition) that depends on the function of multiple components. Our results indicate that it is possible to isolate aptamers that interfere with thymidylate synthase activity, inhibit cell growth, or cause cell death. The latter two phenotypes provide a starting point for the identification of potentially novel molecular targets for antimicrobial drug design.
Materials and Methods
Strains.
SM10 is a recA derivative of C600 that contains a chromosomal copy of plasmid RP4 that allows conjugal transfer of plasmids bearing oriT from plasmid RK2 (7). JB112 was constructed by P1kc transduction of the pir116 gene from BW25126 (8) and the recA1 gene from BW9301 (9) into C600 (thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21).
Construction of pBMT.
pBADmob was constructed by inserting oriT from plasmid pBSL237, a derivative of RK2 (10), into plasmid pBAD18 (11) at the ClaI site. The trxA gene of Escherichia coli XL-1 Blue was amplified by PCR and inserted between the EcoRI and XbaI sites of pBADmob to create pBMT.
Construction of the Library.
An oligonucleotide encoding 16 random amino acids was synthesized. The sequence is identical to that described previously (3), except that there are only 16 repeats of the nucleotide sequence NNK, where K is G or T. The use of K in the third position of the codon reduces the frequency of stop codons while preserving the diversity of amino acids. It also ensures that all stop codons are amber (UAG) codons, which can be suppressed in many strains, including SM10. A second strand was synthesized as described (3), and the double-stranded DNA was digested with AvaII. This material was ligated to RsrII-digested pBMT, and the mixture was used to transform DH10B (Life Technologies, Gaithersburg, MD) by electroporation. Pooled plasmid from the transformants was used to transform SM10 by electroporation.
Selection and Testing for ThyA−.
E. coli SM10 bearing the aptamer library was plated on M9 glycerol medium containing ampicillin (50 μg/ml), arabinose (0.2%), thymine (0.01%), and trimethoprim (10 μg/ml). Pooled plasmid from surviving cells was transferred into fresh E. coli SM10, and these transformants then were subjected to a second round of selection. This process reduces the number of clones containing chromosomal mutations (in thyA or folA) that lead to trimethoprim resistance. Plasmid linkage of the phenotype was verified by a second transfer of the plasmid into SM10 or JB112.
Mutagenesis of pBMT101 and pBMT101Q.
Overlapping extension PCR was used to change the amber codons in plasmids pBMT101 and pBMT117 to GCG (alanine), CAG (glutamine), TCG (serine), or TAATAA (two ochre stop codons). Results were verified by sequence determination. Deletions of DNA encoding residues 1–7 (SYLCVTP), 4–10 (CVTPASQ), 7–13 (PASQLTE), and 10–16 (QLTEFWG) of the 16-aa insert in pBMT101Q were constructed similarly.
Construction of the α-ThyA and λcI-TrxA101Q Fusion Vectors.
Plasmids pACλcI-TrxA101Q, pACλcI-TrxA119, and pACλcI-TrxA express the respective λcI-TrxA fusion proteins under the control of the lacUV5 promoter. Plasmid pACλcI-TrxA101Q encodes λcI residues 1–236 fused to three alanine residues, which, in turn, are fused to residues 2–127 of TrxA101Q. pACλcI-TrxA119 encodes λcI residues 1–236 fused to three alanine residues, which, in turn, are fused to residues 2–127 of TrxA119. pACλcI-TrxA encodes λcI residues 1–236 fused to three alanine residues, which, in turn, are fused to residues 2–109 of wild-type TrxA. λcI-TrxA fusion vectors were constructed by cloning the appropriate NotI–BamHI-cut PCR products into NotI–BstYI-cut pACλcI32 (12). pACΔcI and pBRα have been described previously (13).
Plasmid pBRα-ThyA encodes residues 1–248 of the α subunit of E. coli RNA polymerase followed by three alanine residues fused to residues 1–264 of ThyA. Expression of the hybrid α-thyA gene in pBRα-ThyA is under the control of tandem lpp and lacUV5 promoters. The hybrid α-thyA gene was created by cloning the appropriate NotI–BamHI-cut PCR product into NotI–BamHI-cut pBRαLN (12).
Two-Hybrid Analysis.
To detect interactions between the α and λcI fusion proteins we used a λPRM-lacZ fusion that was introduced into E. coli strain MC1000 F′lacIq on the low-copy-number plasmid pPRMΔ-50 (13). Growth conditions and β-galactosidase assays were as described previously (13).
Construction of pYW4thyACm and pYW4ΔthyACm.
Plasmid pYW4 (Y. Wang and B. L. Wanner, unpublished results) contains the rhaB promoter, which is highly dependent on rhamnose for activity (14). The thyA gene was amplified by PCR from SM10 and then inserted between the NdeI and BamHI sites of pYW4. The NotI-ClaI fragment conferring gentamicin resistance then was replaced with a cassette conferring chloramphenicol resistance from pIADL46 (15). Plasmid pYW4ΔthyACm was constructed by deleting two NdeI fragments encoding the N-terminal 212 aa of ThyA from pYW4thyACm.
Screening for Toxic Aptamers.
Two methods were used. In the first, the library was plated on LB agar containing ampicillin, then replica-plated onto the same medium with or without 0.4% arabinose. In the second, the library was plated on LB containing ampicillin and 0.005% arabinose. Small colonies were picked and tested individually for growth in the presence or absence of arabinose.
Disc Diffusion.
Filter paper (Whatman no. 1) was soaked in 10% arabinose, dried, and cut into 6-mm discs. Overnight cultures were diluted 125-fold, and 0.2–0.3 ml was spread onto LB ampicillin agar. The filter paper disc was placed on the plate, which was incubated overnight at 37°C.
Growth Curves.
Overnight cultures of E. coli SM10 bearing various plasmids were diluted 104-fold into LB with ampicillin and shaken at 37°C. Arabinose was added after 2 hr. Growth was assessed by plating of serially diluted cultures in the absence of arabinose.
Insertion of Stop Codon Upstream of the Inserts.
To construct plasmid pBMTO, a single A-to-T nucleotide change was introduced into pBMT at codon 19 of the thioredoxin gene to produce a stop codon. This plasmid was digested with RsrII and BglI to remove the fragment encoding the C-terminal 74 aa of thioredoxin. The corresponding fragments from aptamer-encoding plasmids, including the inserted amino acids, were ligated to this vector to construct plasmids identical to the original aptamer-encoding plasmids except for the stop codon at amino acid 19.
Results
Construction of the ABBIS Library.
To express random peptides within bacterial cells, we constructed a library of peptides fused to a carrier protein, which provides stability, conformational constraint, and targeting to a specific subcellular location. In accordance with previous publications using analogous constructs in yeast (3, 4), we have adopted the term “aptamer” to describe the resulting fusion proteins. To test aptamers for activity in various assays and cell types, we have constructed a vector that places aptamer expression under the tight control of the E. coli araBAD promoter (11). The PBAD promoter is positively regulated by AraC. Hence, it is in the “off state” when the plasmid first enters cells, eliminating issues of zygotic induction, which might otherwise be lethal for cells carrying certain aptamers. The vector is derived from pBAD18 (11), with the addition of an origin of transfer (oriT of plasmid RK2) to allow efficient transfer by conjugation. The vector also contains the E. coli thioredoxin gene (trxA). Thioredoxin was chosen as a carrier protein for random peptides because of its stability, ease of purification, and the presence of a convenient restriction site in the region encoding the active site of the enzyme. Insertions into this site are tolerated (16), and tethering of both ends of the inserted peptide provides a degree of conformational constraint to the peptide that is not present in fusions to the N or C terminus of a protein. This may be important, given the lack of conformational stability of short peptides that are not tethered (17). Conformational stability may increase the affinity of a peptide for its target and may eventually make structural studies more practical by reducing the number of possible conformations of the peptide.
An oligonucleotide with restriction sites at the ends and a central segment encoding 16 random amino acids was then cloned into a restriction site that corresponds to the active site of thioredoxin (the disulfide loop). Similar constructions have been used to make thioredoxin fusions that have been selected for several biological activities (3, 16, 18). A 16-aa random peptide has approximately 6.6 × 1020 possible sequences. Based on the number of bacterial transformants, our construction resulted in a library of approximately 107 different peptides expressed as internal fusion proteins in thioredoxin. This represents a tiny fraction of the possible sequence diversity; thus, there should be virtually no duplication of sequences within the 107 clones.
Selection for Thymidylate Synthase (ThyA) Inhibitors.
We used the library to isolate aptamers that inhibit the function of ThyA of E. coli. Cells that lack ThyA activity are resistant to trimethoprim (TMP) and require exogenous thymine for growth (19). To select for cells that were phenotypically ThyA− in the presence of arabinose, the library was plated on M9 glycerol containing arabinose, thymine, and trimethoprim. Two clones were isolated that showed both arabinose-dependent thymine auxotrophy and arabinose-dependent trimethoprim resistance. These were designated pBMT101 and pBMT117. Because pBMT117 conferred a weaker phenotype than pBMT101, we characterized the latter plasmid further. Growth test results for SM10 carrying pBMT101 on various media are shown in Table 1. Cells containing pBMT101 require thymine and are trimethoprim-resistant only in the presence of arabinose. Neither a control aptamer (encoded by pBMT119) nor thioredoxin without an insertion produced these phenotypes, even though these were expressed at levels similar to the active aptamer (data not shown). Because expression of the active aptamer causes the cell to have both phenotypes, it is likely that the aptamer inhibits thymidylate synthase activity.
Table 1.
Effects of aptamers on growth in selective media
| Additions to medium | pBMT119 | pBMT101 |
|---|---|---|
| None | + | + |
| ara | + | − |
| ara, thymine | + | + |
| Thymine, TMP | − | − |
| ara, thymine, TMP | − | + |
E. coli SM10 bearing the indicated plasmids were streaked onto minimal medium and incubated at 37° for 2–3 days. Plasmid pBMT119 encodes a control aptamer, and pBMT101 encodes the inhibitor of ThyA. +, growth; −, no growth; ara, arabinose; TMP, trimethoprim.
The amino acid sequences of the inserts were: SYLCVTPAS*LTEFWG (pBMT101) and SGNPYACL*SGPNLCV (pBMT117), where * is an amber stop codon. The translational stops in these two aptamers were predicted to be partially suppressed by insertion of glutamine in strain SM10 because this strain expresses a supE suppressor tRNA. We therefore used site-directed mutagenesis to change the amber codon in pBMT101 to glutamine (pBMT101Q), serine (pBMT101S), alanine (pBMT101A), or two nonsuppressible ochre stop codons (pBMT101Oc2). Results of growth tests with these plasmids are shown in Table 2. These data show that the aptamer encoded by pBMT101 can inhibit ThyA with glutamine, alanine, or serine present at position 10 in the 16-aa insert. Interruption of the aptamer with a chain-terminating ochre codon abolishes ThyA inhibitory activity.
Table 2.
Effects of various aptamers on the ThyA phenotype
| Plasmid | Additions to growth medium
|
Interpretation | ||
|---|---|---|---|---|
| Ara | Ara+Thy | Ara+Thy+TMP | ||
| pBMT | + | + | − | No effect |
| pBMT101 | − | + | + | ThyA inhibitor |
| pBMT101Q | − | + | + | ThyA inhibitor |
| pBMT101A | − | + | + | ThyA inhibitor |
| pBMT101S | − | + | + | ThyA inhibitor |
| pBMT101Oc2 | + | + | − | No effect |
E. coli SM10 bearing the indicated plasmids were streaked onto minimal medium and incubated at 37° for 2–3 days. +, growth; −, no growth; ara, arabinose; Thy, thymine; TMP, trimethoprim. Cells bearing any of these plasmids grew in minimal medium without additions.
To determine whether only a portion of the 16-mer insert is required for inhibition, a series of overlapping deletions in pBMT101Q, each of which removed 7 of the 16 inserted amino acids, was constructed and analyzed. When residues 1–7, 4–10, 7–13, or 10–16 of the inserted amino acids were deleted, no inhibition of thymidylate synthase was observed in vivo (data not shown). Thus, most, if not all, of the 16-aa insert is required for activity.
Excess ThyA Expression Suppresses Effects of the ThyA Inhibitor.
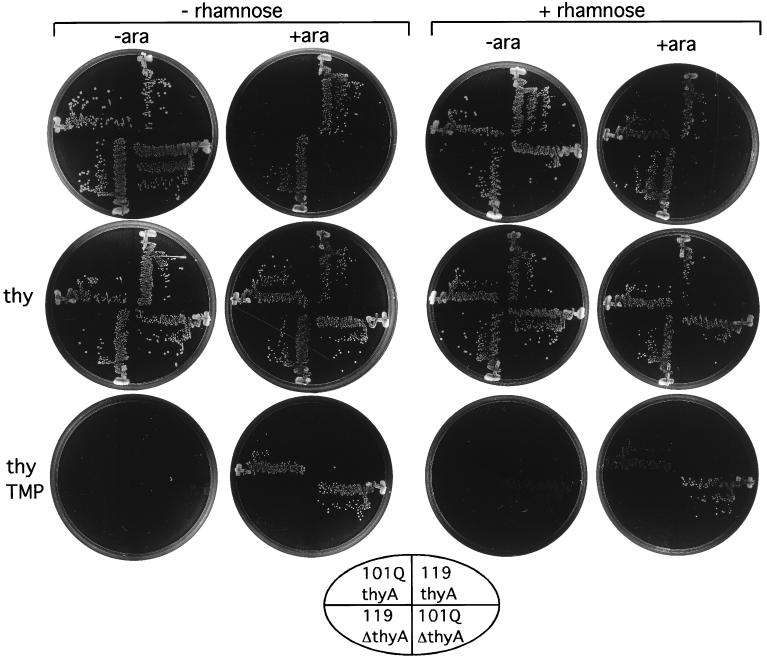
Rapid genetic methods for identifying the target of an inhibitory aptamer would provide additional utility for the ABBIS method. For example, overexpression of an aptamer's target might be predicted to reverse the phenotype attributed to the aptamer (20). To demonstrate the feasibility of this approach, and to verify that ThyA is the target of the aptamer encoded by pBMT101Q, we set up a system for inducing expression of both ThyA and the aptamer in the same cells under independent control. The thyA gene of E. coli SM10 was inserted into a vector (pYW4thyACm) that allowed its expression under the control of exogenous rhamnose. Expression of thymidylate synthase blocked thymine auxotrophy and partially blocked trimethoprim resistance induced by the aptamer, as would be expected if the aptamer exerts its effects through thymidylate synthase (Fig. 1). The control plasmid pYW4ΔthyACm, from which most of the coding sequence of thyA had been deleted, did not confer this effect.
Figure 1.
Increased expression of ThyA blocks the effect of the ThyA inhibitory aptamer. E. coli JB112 bearing the plasmids indicated below were plated on M9 glycerol containing carbenicillin (50 μg/ml) and chloramphenicol (20 μg/ml). Additions to the medium were rhamnose (0.32%), ara (arabinose, 0.02%), thy (thymine, 0.01%), or TMP (trimethoprim, 2 μg/ml). Plasmids pBMT101Q and pBMT119 express aptamers in response to arabinose, and plasmid pYW4thyACm expresses thymidylate synthase in response to rhamnose. Plasmid pYW4ΔthyACm is a derivative of pYW4thyACm, in which most of the thyA coding sequence has been deleted. Grid: 101Q, anti-ThyA aptamer plasmid pBMT101Q; 119, control aptamer plasmid pBMT119; thyA, plasmid pYW4thyACm expressing ThyA; ΔthyA, control plasmid pYW4ΔthyACm.
Two-Hybrid Detection of Interaction Between ThyA and Aptamer.
Aptamer targets could also be identified by using powerful genetic methods for detecting protein–protein interactions, termed two-hybrid screens (5, 12, 21, 22). To test this concept, we measured the protein–protein interaction between ThyA and the TrxA101Q aptamer, using an E. coli-based, two-hybrid system (13, 23). This system is based on the finding that a sufficiently strong interaction between two proteins can activate transcription of a reporter gene if one of the interaction partners is fused to a DNA-binding protein and the other partner is fused to one of the subunits of RNA polymerase (RNAP). Accordingly, we replaced the C-terminal domain of the RNAP α subunit with ThyA and fused the TrxA101Q aptamer to λcI (a DNA-binding protein). We compared the level of reporter gene expression in cells containing the λcI-TrxA101Q fusion protein in the absence or presence of the α-ThyA fusion protein. Table 3 shows that λcI-TrxA101Q stimulated transcription from the reporter (a PRM-lacZ fusion) six times more efficiently in the presence of the α-ThyA protein than in its absence. This effect was specific for λcI-TrxA101Q, because the presence of the α-ThyA fusion protein did not alter the ability of the λcI-TrxA fusion protein to activate transcription. Two fusion proteins (λcI-Gal4 and α-Gal11P) previously shown to interact efficiently (23) served as a positive control for this experiment. These data indicate that the aptamer probably binds directly to ThyA, although this interaction is weaker than the interaction between the positive control proteins.
Table 3.
Two-hybrid analysis of aptamer–ThyA interaction
| α Fusion partner | λcI fusion partner | β-Galactosidase |
|---|---|---|
| ThyA | TrxA101Q | 299 (±10) |
| None | TrxA101Q | 46 (±0.6) |
| ThyA | TrxA | 100 (±1) |
| None | TrxA | 118 (±0.2) |
| Gal11P | Gal4 | 739 (±5) |
Cells expressing RNA polymerase α-subunit and λ cI fused to the indicated proteins were induced with 20 μM isopropyl β-d-thiogalactoside, and β-galactosidase activity was determined. Data shown are the mean and range of duplicates.
Identification of Aptamers that Block Bacterial Growth.
Most clinically useful antibacterials inhibit growth or kill bacteria. Identification of molecules that are toxic to bacteria, or identification of the cellular targets on which the toxic molecules act, may be valuable in the development of new antibacterial compounds. We therefore screened a portion of the library for aptamers toxic to the cells. In contrast to the selection for inhibitors of thymidylate synthase, no knowledge of the aptamer's cellular target was needed for this screen. Two methods were used. In the first, colonies were replica-plated onto medium with or without 0.4% arabinose, and colonies inhibited by arabinose were chosen for further study. In the second, the library was plated on medium with 0.05% arabinose, which gives incomplete induction of aptamer expression, and small colonies were picked. We screened approximately 1.8 × 104 colonies and tested candidates by placing a filter paper disc impregnated with arabinose on a lawn of bacteria. The diameter of the zone of inhibition around the disc indicates the sensitivity to growth inhibition by arabinose (Fig. 2A). After eliminating those clones that contained frameshift mutations, we obtained five clones that were severely inhibited by arabinose. Three had a single 16-mer insert, and two had a pair of inserts in tandem (for a total of 34 aa, because the restriction sites at the ends of the inserts encode another 2 aa). The deduced amino acid sequences of the inserts are: FRSVCWALTLLGTTFL (JM7), IRMCLRWSCFCLLTLV (JB63), LWFTEVRGHGWRYKVG (KO1), AAYLPRVSMFSWSGCLGPCLLWLFLSLVLCAYLF (KO19), and VTLKASVRWYGGGGDTGPRVSWAWVVMLVWAVTL (KO52). Examination of these sequences did not reveal consistent trends in hydrophobicity, nor did they differ substantially from a random sample of sequences from the library.
Figure 2.
Effects of aptamers that block cell growth. (A) Arabinose-impregnated discs were placed on a lawn of E. coli SM10 containing plasmids expressing the indicated aptamers. (B) Arabinose (final concentration, 0.1%) was added at 2 hr to exponentially growing cultures of SM10 containing plasmids expressing the indicated aptamers. Samples were withdrawn and viable counts in colony-forming units (cfu) were determined. □, No arabinose; ⋄, arabinose added.
To quantify the effects of aptamer expression on cell growth, arabinose was added to exponentially growing cultures, and growth was followed (Fig. 2B). A control aptamer had no effect on growth. Some of the toxic aptamers caused arrest of growth, whereas one caused cell death. Microscopical examination of the cultures revealed that some of the toxic aptamers caused filamentation of the bacteria (data not shown).
Translational Termination Prevents Aptamer Activity.
Selection of libraries of random RNAs, as well as proteins, has yielded biologically active molecules. Our library was constructed to identify proteins with biological activities, but expression of the aptamer requires expression of a messenger RNA that also has the potential for activity. To confirm that the effects of aptamer expression were due to the protein rather than the mRNA, we constructed plasmid pBMTO, which contains an A-to-T change at codon 19 of trxA, creating a UAA stop codon at amino acid 19. DNA encoding the random peptide insertions and the C-terminal portion of the thioredoxin gene from three of the toxic aptamer genes and from plasmids pBMT101Q (the ThyA inhibitor) and pBMT119 (the control aptamer) were introduced into this vector. In each case, induction with arabinose did not reproduce the activity associated with the aptamer before the introduction of the stop codon (data not shown). In addition, a pair of ochre codons inserted in a different location (within the synthetic insert) in pBMT101 also blocked its function (Table 2). Together with our two-hybrid results, these data suggest that the biological effects of the aptamers are mediated by aptamer protein rather than RNA. However, we cannot rule out the possibility that translational termination prevents aptamer activity by reducing mRNA stability.
Discussion
The method described here (ABBIS) provides a simple approach to the isolation of combinatorial peptides (aptamers) that display biologically interesting activities when expressed inside bacterial cells. Although similar methods have been developed successfully for use in yeast (3–6), these methods lacked features that facilitate their use in bacterial systems, such as positively controlled, inducible expression of aptamer production. This property allowed inhibitory and blatantly toxic aptamers to be identified with comparative ease. Furthermore, by expressing aptamers within the cells, we have bypassed the permeability barrier imposed by the cell wall and membrane. This may lead to the identification of targets that have not come out of screens in which the drugs are applied to the outside of cells. We have also used ABBIS to isolate an aptamer that activates the Pho regulon in E. coli (J.H.B., J.J.M., B. Wanner, and M. Perez, unpublished data). Thus, aptamers can be used either to inhibit or to activate selected bacterial cellular processes.
In the case of the aptamer that inhibits thymidylate synthase, a selection was available that was highly specific for a particular molecular target, enabling us to isolate a molecule that inhibited that enzyme. Inhibitors of thymidylate synthase are of interest for several reasons. First, this system serves as a model to validate the feasibility of aptamer-based intracellular enzyme inhibition. Second, thymidylate synthase serves an important physiologic role in bacterial cells. In addition to being required for growth on minimal medium, it is required for virulence in several species, including the intracellular pathogens Shigella flexneri (24) and Legionella pneumophila (25). The requirement for thymidylate synthase function in vivo probably accounts for the finding that most trimethoprim-resistant clinical isolates have mutations in folA (dihydrofolate reductase) rather than thyA (26). Thus, thymidylate synthase is a potential target for antibiotics. Finally, thymidylate synthase is also a target for anticancer drugs such as 5-fluorouracil. Because a human thymidylate synthase gene can complement an E. coli thyA mutant (27), ABBIS could be used to identify peptide inhibitors of human thymidylate synthase as well. Similarly, thymidylate synthase genes from such diverse sources as Trypanosoma brucei (28), Cryptococcus neoformans (29), and varicella zoster virus (30) can complement an E. coli thyA mutant, raising the possibility of using ABBIS to develop inhibitors of thymidylate synthase from these important pathogens.
The aptamer encoded by plasmid pBMT101Q clearly inhibits ThyA in vivo. This is shown by two features of cells expressing this aptamer. First, these cells require exogenous thymine, which reflects the failure of the cells to make dTMP from dUMP. Second, they are resistant to trimethoprim. These two characteristics reflect different aspects of the pathway. Because expression of the aptamer causes the cell to have both phenotypes (trimethoprim resistance and thymine requirement), it is very likely that the aptamer acts on thymidylate synthase. An alternative possibility, that the aptamer inhibits production of dUMP, the substrate of thymidylate synthase, is unlikely because there are two pathways for the production of dUMP (31).
Two-hybrid analysis suggests that the aptamer that inhibits thymidylate synthase may act by binding directly to the enzyme. It is not clear whether the effect on enzymatic activity is mediated by competitive or noncompetitive inhibition or whether binding of the aptamer might simply accelerate degradation of the enzyme. In addition, it is possible that the interaction detected by this method is mediated by a third, intermediary protein. Confirmation of binding of the aptamer to the enzyme will require direct biochemical analysis of the interaction.
In contrast, screening for aptamers that inhibit colony formation led to the isolation of multiple aptamers with unknown targets. We have not yet identified any of the targets of the toxic aptamers, but this problem should be amenable to genetic or biochemical analysis. The demonstration that a phenotype (ThyA−) induced by an aptamer can be suppressed by overexpression of the target (ThyA) suggests that multicopy suppression may be a useful approach to this problem. The demonstration of an interaction between ThyA and its inhibitory aptamer in a two-hybrid system also raises the possibility of using this method to identify proteins that interact with aptamers with unknown targets. Such an approach has been used successfully to identify the target of aptamers that affect signal transduction pathways in yeast (5).
A number of limitations apply to the ABBIS method. First, it is not intended to produce useful antibacterial drugs directly. Aptamers are proteins, which would have poor entry into cells, poor oral bioavailability, potential immunogenicity, and high cost. Although it is possible that synthetic cyclic peptides that correspond to the combinatorial peptide in an aptamer might have the same biochemical activity as the protein-based aptamer, these, too, might not penetrate bacterial cells. Rather, ABBIS provides inhibitory aptamers that are intended to be used to identify potential targets for antimicrobials. These targets then can be used in more conventional screens for novel drugs that specifically bind to or inhibit the target. Alternatively, once a target is identified, structural information on the target (obtained through x-ray crystallography) may allow the application of molecular drug design techniques to identify small molecules that bind to and inhibit the target. In this application, the inhibitory aptamer can be thought of as a “lead compound” in the context of structural studies. Thus, by studying the structure of an aptamer–target complex, one might identify a likely binding site for a drug and, possibly, drug side chains and stereochemistry that might be appropriate to explore. Converting a protein lead compound to a “drug-like” chemical with good activity and acceptable cell penetration is a complex task, but considerable progress has been made in this area (32). First, small molecules have been discovered that are able to mimic the effects of large polypeptides (33) or interfere with protein–protein interactions (34). Second, efforts to develop small-molecule drugs by starting with a peptide and converting it to a “peptidomimetic” have succeeded in a number of cases, most notably HIV protease inhibitors (35–37) and angiotensin II receptor antagonists (38).
It also is worth noting that we do not yet know what contribution thioredoxin makes to the activity of the aptamer. This protein was chosen partly because it should hold the aptamer in a constrained conformation, so it is possible that the combinatorial peptide's activity will be lost if it is produced alone or fused to a different protein. In addition, thioredoxin might contribute to an aptamer's activity by participating directly in interactions with a target molecule. Even if this is the case, it should not interfere with the applications of aptamers that we have described.
Because the ABBIS approach involves screening or selecting for arabinose-dependent phenotypes, the affinities of the aptamers for their targets must be defined by the quantities of the aptamer present in the cells in their induced (with arabinose) and uninduced (without arabinose) states. Although this promoter is tightly regulated (11), the residual low level of expression in the absence of arabinose may limit the potency of compounds that come out of our screens. There is a positive side to this problem, in that the relatively high level of expression of the aptamers in the induced state means that we should be able to detect aptamers even if they have weak activity (low affinity for their targets), which presumably are more abundant in the library than those with strong activity. Such weak aptamers then can be optimized by mutagenesis to obtain more active ones.
In conclusion, ABBIS provides a powerful, new genetic tool for the design of custom inhibitors of bacterial cellular processes, including those required for growth, viability, virulence, or other nonessential phenotypes of interest. We anticipate the use of biologically active aptamers to define targets for the development of new classes of antimicrobial agents.
Acknowledgments
We thank Barry Wanner for helpful advice, strains, and plasmids, Heather Losey for constructing pACλcI-TrxA, and Kelly O'Brien for technical assistance. This work was supported by Grant AI26289 from the National Institutes of Health (J.J.M.), Grant MCB-9728765 from the National Science Foundation (A.H.), and an established investigatorship from the American Heart Association (A.H.). J.H.B. was supported by a Howard Hughes Medical Institute Postdoctoral Fellowship. S.L.D. was supported by a Charles A. King Trust postdoctoral fellowship.
Abbreviations
- ABBIS
aptamer-based bacterial inhibition systems
- ThyA
thymidylate synthase
- TMP
trimethoprim
- aa
amino acid
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040573397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040573397
References
- 1.Kelley K J. Nat Biotechnol. 1996;14:587–590. doi: 10.1038/nbt0596-587. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Wu Z, Fields S. Nucleic Acids Res. 1995;23:1152–1156. doi: 10.1093/nar/23.7.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman T C, Smith D L, Sorger P K, Drees B L, O'Rourke S M, Hughes T R, Roberts C J, Friend S H, Fields S, Murray A W. Science. 1999;285:591–595. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 6.Caponigro G, Abedi M R, Hurlburt A P, Maxfield A, Judd W, Kamb A. Proc Natl Acad Sci USA. 1998;95:7508–7513. doi: 10.1073/pnas.95.13.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon R, Priefer U, Puhler A. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 8.Metcalf W W, Jiang W, Daniels L L, Kim S K, Haldimann A, Wanner B L. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 9.Wanner B L. In: Methods in Molecular Genetics. Adolph K W, editor. Vol. 3. Orlando, FL: Academic; 1994. pp. 291–310. [Google Scholar]
- 10.Alexeyev M F, Shokolenko I N. BioTechniques. 1995;19:22–24. , 26. [PubMed] [Google Scholar]
- 11.Guzman L M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J C, Kornacker M G, Hochschild A. Methods. 2000;20:80–94. doi: 10.1006/meth.1999.0908. [DOI] [PubMed] [Google Scholar]
- 13.Dove S L, Joung J K, Hochschild A. Nature (London) 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 14.Haldimann A, Daniels L L, Wanner B L. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva J C, Haldimann A, Prahalad M K, Walsh C T, Wanner B L. Proc Natl Acad Sci USA. 1998;95:11951–11956. doi: 10.1073/pnas.95.20.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaVallie E R, DiBlasio E A, Kovacic S, Grant K L, Schendel P F, McCoy J M. Bio/Technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 17.Corey M J, Corey E. Proc Natl Acad Sci USA. 1996;93:11428–11434. doi: 10.1073/pnas.93.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Z, Murray K S, Van Cleave V, LaVallie E R, Stahl M L, McCoy J M. Bio/Technology. 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 19.Neuhard J, Kelln R A. In: Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Neidhardt F C, Curtiss R I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Shaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 580–599. [Google Scholar]
- 20.Kolonin M G, Finley R L., Jr Proc Natl Acad Sci USA. 1998;95:14266–14271. doi: 10.1073/pnas.95.24.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimova G, Pidoux J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelletier J N, Campbell-Valois F X, Michnick S W. Proc Natl Acad Sci USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dove S L, Hochschild A. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed Z U, Sarker M R, Sack D A. Vaccine. 1990;8:153–158. doi: 10.1016/0264-410x(90)90139-d. [DOI] [PubMed] [Google Scholar]
- 25.Mintz C S, Chen J X, Shuman H A. Infect Immunol. 1988;56:1449–1455. doi: 10.1128/iai.56.6.1449-1455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huovinen P, Sundstrom L, Swedberg G, Skold O. Antimicrob Agents Chemother. 1995;39:279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davisson V J, Sirawaraporn W, Santi D V. J Biol Chem. 1989;264:9145–9148. [PubMed] [Google Scholar]
- 28.Gamarro F, Yu P L, Zhao J, Edman U, Greene P J, Santi D. Mol Biochem Parasitol. 1995;72:11–22. doi: 10.1016/0166-6851(95)00059-a. [DOI] [PubMed] [Google Scholar]
- 29.Livi L L, Edman U, Schneider G P, Greene P J, Santi D V. Gene. 1994;150:221–226. doi: 10.1016/0378-1119(94)90430-8. [DOI] [PubMed] [Google Scholar]
- 30.Thompson R, Honess R W, Taylor L, Morran J, Davison A J. J Gen Virol. 1987;68:1449–1455. doi: 10.1099/0022-1317-68-5-1449. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad S I, Kirk S H, Eisenstark A. Annu Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 32.Fairlie D P, West M L, Wong A K. Curr Med Chem. 1998;5:29–62. [PubMed] [Google Scholar]
- 33.Tian S S, Lamb P, King A G, Miller S G, Kessler L, Luengo J I, Averill L, Johnson R K, Gleason J G, Pelus L M, et al. Science. 1998;281:257–259. doi: 10.1126/science.281.5374.257. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Gao J, Satoh T, Friedman T M, Edling A E, Koch U, Choksi S, Han X, Korngold R, Huang Z. Proc Natl Acad Sci USA. 1997;94:73–78. doi: 10.1073/pnas.94.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McQuade T J, Tomasselli A G, Liu L, Karacostas V, Moss B, Sawyer T K, Heinrikson R L, Tarpley W G. Science. 1990;247:454–456. doi: 10.1126/science.2405486. [DOI] [PubMed] [Google Scholar]
- 36.Meek T D, Lambert D M, Dreyer G B, Carr T J, Tomaszek T A, Jr, Moore M L, Strickler J E, Debouck C, Hyland L J, Matthews T J, et al. Nature (London) 1990;343:90–92. doi: 10.1038/343090a0. [DOI] [PubMed] [Google Scholar]
- 37.Roberts N, Martin J, Kinchington D, Broadhurst A, Craig J, Duncan I, Galpin S, Handa B, Kay J, Krohn A, et al. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 38.Greenlee W J, Siegl P K S. Annu Rep Med Chem. 1991;26:63–72. [Google Scholar]