Abstract
Cyclic nucleotide phosphodiesterases (PDEs) constitute a family of enzymes that degrade cAMP and cGMP. Intracellular cyclic nucleotide levels increase in response to extracellular stimulation by hormones, neurotransmitters, or growth factors and are down-regulated through hydrolysis catalyzed by PDEs, which are therefore candidate therapeutic targets. cAMP is a second messenger implicated in learning, memory, and mood, and cGMP modulates nervous system processes that are controlled by the nitric oxide (NO)/cGMP pathway. To investigate an association between genes encoding PDEs and susceptibility to major depressive disorder (MDD), we genotyped SNPs in 21 genes of this superfamily in 284 depressed Mexican Americans who participated in a prospective, double-blind, pharmacogenetic study of antidepressant response, and 331 matched controls. Polymorphisms in PDE9A and PDE11A were found to be associated with the diagnosis of MDD. Our data are also suggestive of the association between SNPs in other PDE genes and MDD. Remission on antidepressants was significantly associated with polymorphisms in PDE1A and PDE11A. Thus, we found significant associations with both the diagnosis of MDD and remission in response to antidepressants with SNPs in the PDE11A gene. We show here that PDE11A haplotype GAACC is significantly associated with MDD. We conclude that PDE11A has a role in the pathophysiology of MDD. This study identifies a potential CNS role for the PDE11 family. The hypothesis that drugs affecting PDE function, particularly cGMP-related PDEs, represent a treatment strategy for major depression should therefore be tested.
Keywords: gene association, pharmacogenetics, cGMP, SNP, Mexican American
Eleven different phosphodiesterase (PDE1–11; see Table 1) families have already been identified based on their substrate specificities, kinetic properties, allosteric regulators, inhibitor sensitivities, and amino acid sequences (1–10). Within each family, several genes and splice variants have been recognized (2, 11). Each family and members within a family exhibit distinct tissue and subcellular patterns (1, 3–5, 8, 9, 12). The hydrolysis of cAMP and cGMP are controlled by multiple PDEs, and they influence numerous pharmacological processes, including mediation of inflammation, ion channel function, muscle contraction, learning, differentiation, apoptosis, lipogenesis, glycogenolysis, and gluconeogenesis (13).
Table 1.
Distribution of genotyped SNPs by PDE families
| Family | Genes | Substrate | SNPs |
|---|---|---|---|
| PDE1 | 1A, 1B, 1C | cAMP/cGMP | 11 |
| PDE2 | 2A | cAMP/cGMP | 5 |
| PDE3 | 3A | cAMP/cGMP | 2 |
| PDE4 | 4A, 4B, 4C, 4D | cAMP | 21 |
| PDE5 | 5A | cGMP | 1 |
| PDE6 | 6A, 6C, 6D, 6G | cGMP | 8 |
| PDE7 | 7A, 7B | cAMP | 6 |
| PDE8 | 8A, 8B | cAMP | 5 |
| PDE9 | 9A | cGMP | 4 |
| PDE10 | 10A | cAMP/cGMP | 10 |
| PDE11 | 11A | cAMP/cGMP | 5 |
| Total | 21 | 78 |
As regulators of the ubiquitous second messengers cAMP and cGMP, PDEs modulate the transduction of various extracellular signals through the activation of cell-surface receptors. Intracellular concentrations of cyclic nucleotides increase and activate their target enzymes, which are PKA and PKG. These protein kinases are responsible for the phosphorylation of a number of substrates, such as ion channels, contractile proteins and transcription factors. In this manner, PDEs regulate key cellular functions and have fundamental and pharmacological interest: they have been acknowledged as important drug targets for the treatment of disparate diseases, such as congestive heart disease, depression, asthma, inflammation, and erectile dysfunction (14–17).
The PDE enzymes can be classified by their substrate (Table 1), whether cAMP-specific, cGMP-specific, or dual substrate (cAMP and cGMP) (14, 18, 19). The regulatory N terminus of these enzymes has considerable variation and includes regions that autoinhibit the catalytic domains and regions that control subcellular localization (20, 21). The N terminus may include a calmodulin-binding protein (PDE1), cGMP-binding sites (PDE2), phosphorylation sites for several protein kinases (PDE1–5), and a transducin-binding domain (PDE6).
The intense interest in PDE expression and activity during the last decade has advanced the understanding that in the brain, where regulation of second-messenger signaling is very complex, virtually all PDEs are expressed at high levels; their differential expression patterns and subcellular distributions are relevant to cell-to-cell communications and modulation of neuronal activity (22). Several lines of investigation suggest that PDE4, a cAMP-specific enzyme, should be considered a prime target for therapeutic intervention in a range of CNS disorders, including depression and impaired cognition (23–25). The selective inhibitor of PDE4 rolipram has been shown to produce antidepressant-like and memory-enhancing effects in animals (23, 25–27). Behavioral phenotype and pharmacological data of knockout mice support the concept that PDE4D might be involved in the mediation of depressive symptoms and antidepressant response (28).
Major depressive disorder (MDD) is one of the most common psychiatric disorders; both environmental and genetic factors contribute to its etiology. Heritability of MDD is estimated at 0.36–0.7 based on twin studies (29–35). The specific genetic substrates, precipitating environmental factors, and predictors of treatment response have not yet been elucidated. The multitude of cellular responses modulated by PDEs and the strong suggestion of a potential role of PDE4 in MDD support our main hypothesis that the PDE family is a strong candidate system for susceptibility to MDD. We genotyped SNPs in 21 genes encoding PDEs and tested their association in a study of MDD cases and matched controls. We also tested the secondary hypothesis that PDE genes are associated with antidepressant treatment response.
Results
Cleaning and Filtering Steps.
Quality control.
The distribution of SNPs and genes across the chromosome after quality control can be seen in Table 1. In summary, 159 SNPs (80%) passed 7/7 plates; 15 (7%) passed 6/7 plates; 2 (1%) passed 5/7 plates; and 24 (12%) passed <5/7 plates. Most (92.6%) of the quality control duplicates matched. We eliminated 2.3% of observations, because they had missing data. Of our original dataset, 87.5% remained after this step.
Hardy–Weinberg Equilibrium (HWE).
We examined HWE in control and depressed groups separately. We excluded 18 SNPs that were not in HWE in the control group; we also excluded 5 SNPs that were monoallelic in at least one of the groups. After quality control and HWE steps, 153 SNPs remained for linkage disequilibrium (LD) analyses.
LD.
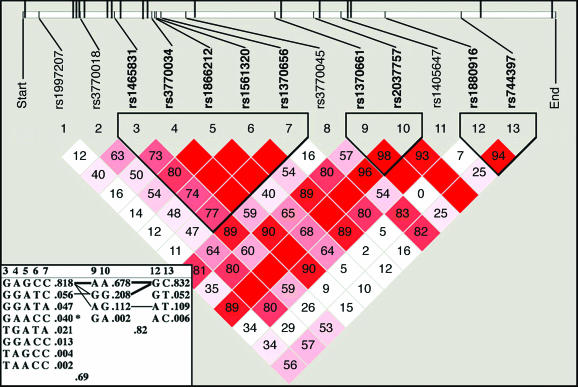
We assessed LD in each gene for control and depressed groups separately. We removed 75 of the 153 SNPs from further analysis, because they were in LD with an r2 of ≥80% with other SNPs within a specific gene. Fig. 1 illustrates the typical LD and haplotype block structure that were obtained.
Fig. 1.
LD pattern in gene PDE11A in the depressed group. Standard color scheme: white, D′ <1 and logarithm of odds (lod) <2; blue, D′ = 1 and lod <2; shades of pink/red, D′<1 and lod ≥2; bright red, D′ = 1 and lod ≥2. D′ values represent percentages and appeared inside each diamond; values of 100% are not labeled. At the top, the PDE11A gene structure is illustrated schematically by a long horizontal white rectangle. Short vertical lines indicate genotyped SNPs, which correspond to numbers 1–13 above the triangular image. Upward long vertical lines indicate exons (20 total some hidden because of scale). Three haplotype blocks were defined in our analyses: block 1 (38 kb, SNPs 3–7), block 2 (32 kb, SNPs 9–10), and block 3 (63 kb, SNPs 12–13). htSNPs are shown in bold. (Inset) Haplotypes are shown in blocks with frequency and connections from one block to the next; only htSNPs are displayed. Blocks are connected with thin lines if frequency is >5% and thick lines if >10%. Between the blocks, a value of multiallelic D′ is shown. D′ is a measure of the recombination between the two blocks. ∗ in haplotype GAACC denotes significant association to a diagnosis of major depression (P < 0.0001).
Data on 78 SNPs (of the initial 200) were used for further data analyses after quality control steps. The density of SNPs per PDE family and class was as follows: 10.7 SNPs per family of cAMP-specific PDEs, 6.6 SNPs per family of dual substrate (cAMP and cGMP) PDEs, and 4.3 SNPs per family of cGMP-specific PDE.
Haplotype Analyses.
The Four Gamete Rule formed three haplotype blocks in MDD and control groups for the PDE11A gene. In the control group, the first block was defined by four haplotype-tagging SNPs (htSNPs; rs1465831, rs3770034, rs1561320, and rs1370656), whereas in the MDD group, one additional htSNP was necessary to define it (rs1866212; Fig. 1). Haplotype blocks two and three were defined each by two htSNPs (rs1370661, rs2037757 and rs1880916, rs744397, respectively).
SNP Association with MDD.
Two SNPs (rs729861 in PDE9A and rs3770018 in PDE11A) were significant at the Bonferroni corrected significance level of <0.0006 for the test between control and depressed groups (Table 2).
Table 2.
Allele frequency for SNPs significantly (*) associated and likely [(∼), P < 0.05] to be associated with depression (MDD) when compared to control (CT)
| Gene | SNP | P value | SNP class | Allele | Minor allele frequency | |
|---|---|---|---|---|---|---|
| MDD | CT | |||||
| PDE11A | rs3770018(*) | 0.0005 | Intron | A → C | 0.058 | 0.11 |
| PDE9A | rs729861(*) | 0.0006 | Intron | T → C | 0.39 | 0.29 |
| PDE5A | rs3775845(∼) | 0.007 | Intron | A → G | 0.33 | 0.26 |
| PDE10A | rs717602(∼) | 0.009 | Intron | A → G | 0.46 | 0.38 |
| PDE2A | rs370013(∼) | 0.01 | Intron | A → G | 0.50 | 0.43 |
| PDE6C | rs650058(∼) | 0.01 | Intron | C → T | 0.41 | 0.48 |
| PDE10A | rs220818(∼) | 0.01 | Intron | T → C | 0.29 | 0.23 |
| PDE10A | rs676389(∼) | 0.03 | Intron | T → C | 0.24 | 0.23 |
| PDE6C | rs701865(∼) | 0.03 | Nonsynon | T → A | 0.46 | 0.40 |
Seven other SNPs had a P value ≤0.05. Those SNPs were located in four genes: PDE2A (rs376724), PDE5A (rs3775845), PDE6C (rs650058, rs701865), and PDE10A (rs220818, rs676389, and rs717602). The presence of multiple independent signals in PDE6C and PDE10A further strengthens the likelihood of an association with MDD. Table 2 shows genotype frequencies for significant SNPs in the depressed and control groups. The odds ratio for being depressed was 2.1 [95% confidence interval (C.I.) 1.3–3.3] for individuals homozygous (AA) for the major allele for rs3770018 in the PDE11A gene and 0.6 (95% C.I. 0.4–0.8) for individuals homozygous (TT) for the major allele for rs729861 in the PDE9A gene. An odds ratio of 1.4 indicates that a person with the minor allele is 40% more likely to be in the depressed group than not. Likewise, an odds ratio of 0.5 indicates that a person is half as likely to be depressed as not.
SNP Association with Antidepressant Response.
Two SNPs in the PDE family had a P value <0.05 when tested for association with attaining remitter and nonremitter status within the entire depressed group treated with either desipramine or fluoxetine (Table 3). They were located in the PDE1A (rs1549870) and PDE11A (rs1880916) genes. The odds ratio for attaining remitter status was 4.6 (95% C.I. 1.6–13.6) for individuals homozygous (G/G) for the major allele for rs1880916 in the PDE1A gene and 3.2 (95% C.I. 1.2722–8.0092) for individuals heterozygous (A/G) for rs1880916 in the PDE11A gene.
Table 3.
Allele frequency table between remitter (R) and nonremitter (NR) groups for SNPs significantly associated to drug response at P < 0.05
| Treatment | Gene | SNP | P value | Allele | Minor allele frequency | |
|---|---|---|---|---|---|---|
| Fluoxetine or desipramine | PDE1A | rs1549870 | 0.005 | G → A | 0.03* | 0.12† |
| PDE11A | rs1880916 | 0.04 | G → A | 0.16* | 0.074† | |
| Fluoxetine alone | PDE1A | rs1549870 | 0.007 | G → A | 0.022‡ | 0.14§ |
| PDE8B | rs884162 | 0.02 | C → T | 0.09‡ | 0.0§ | |
| PDE6A | rs2544934 | 0.03 | A → T | 0.17‡ | 0.054§ | |
| PDE11A | rs1880916 | 0.03 | G → A | 0.16‡ | 0.036§ | |
| PDE11A | rs3770018 | 0.04 | A → T | 0.076‡ | 0.0§ | |
| Desipramine alone | PDE1C | rs992185 | 0.006 | A → C | 0.47¶ | 0.24‖ |
| PDE1C | rs30585 | 0.02 | T → G | 0.47¶ | 0.26‖ |
*R (n = 82).
†NR (n = 61).
‡R (n = 46).
§NR (n = 28).
¶R (n = 36).
‖NR (n = 33).
Although each group was small, we also analyzed antidepressant response by drug and found that different SNPs and genes were associated with attaining remitter status in fluoxetine and desipramine treatment groups.
Fluoxetine Treatment.
Five SNPs located in four genes were associated with remission during fluoxetine treatment (Table 3). SNPs in PDE1A (rs1549870), PDE6A (rs2544934), PDE8B (rs884162), and PDE11A (rs1880916 and rs3770018) had a difference in allele frequency with a P value ≤0.05 for remitters and nonremitters within the subjects treated with fluoxetine. Both SNPs associated with remission in the entire depression group were also associated with remission in the fluoxetine-treated subjects. The odds ratio for remission in the fluoxetine treatment for rs1549870 was 8.8 (1.7118–45.2382) for the major genotype; for rs1880916, 5.12 (95% C.I. 1.0602–24.738) for the heterozygous genotype; and for rs2544934, 4.4 (95% C.I. 1.1608–17.0161) for the heterozygous genotype. These confidence intervals are wide, and these results await confirmation in larger samples.
Desipramine Treatment.
Two SNPs were associated with remission during desipramine treatment (Table 3). These SNPs (rs30585 and rs992185) were located in the PDE1C gene. The odds ratio for remission with desipramine treatment for rs30585 was 5.16 (95% C.I. 1.0258–26.0228) for the minor genotype and for rs992185, 4.6 (95% C.I. 1.66–12.7) for the heterozygous genotype.
Haplotype Association with MDD.
In the PDE11A gene, haplotype GAACC in block 1 (Fig. 1) was found to be significantly associated with a diagnosis of depression (P < 0.0001). The frequency of haplotype GAACC in the depressed group was 4.1%, and it was not present in the control group. No haplotype was found to be significantly associated with response to antidepressants.
Discussion
We found that SNPs in PDE genes are associated with MDD and antidepressant treatment response. PDEs constitute a complex family of enzymes that are essential regulators of intracellular cyclic nucleotide signaling, which have a central role in neuronal signal transduction. Through a series of rigorous processes of data cleaning, filtering steps, and analyses, we have identified two SNPs (in PDE9A and -11A genes) associated with a diagnosis of MDD and two other SNPs (in PDE1A and -11A genes) associated with treatment response. Interestingly, the PDE11A gene was associated both with drug response and depression, but different SNPs were associated with diagnosis and drug response. Almost all of the PDEs that we identified as relevant for disease or drug response catalyze cGMP; only one gene (PDE8B) identified in our study is a cAMP-specific PDE gene.
Association with MDD.
Two SNPs in the PDE gene family have significantly different allele frequencies between control and depressed groups. Those SNPs were located in PDE9A and -11A genes (Table 2). PDE9A belongs to the class of cGMP-specific enzymes, and PDE11A catalyzes both cAMP and cGMP. Our data also indicate that two other members of the cGMP-specific enzymes (PDE5A and -6C) and two other members of the dual substrate (cAMP and cGMP) class of PDEs (PDE2A and -10A; refs. 14, 18, and 19) are likely to be associated with MDD. Intriguingly, five of six of these PDEs (PDE2A, -5A, -6C, -10A, and -11A) are classified as GAF-PDEs (GAF, cGMP-binding and stimulated phosphodiesterase, Anabaena adenylate cyclases, and Escherichia coli Fh1A; ref. 36). High amino acid sequence similarity (42–51%) is found in the catalytic region of GAF-PDEs, and catalytic domain phylogenic tree analysis of human PDEs demonstrates evolutionary relatedness among the GAF-PDE family and suggests that these genes have a common ancestor gene. Our findings are further supported by haplotype analyses of PDE11A, which showed that haplotype GAACC in block 1 is significantly associated with a diagnosis of MDD (P < 0.0001; Fig. 1).
Association with Drug Response.
Two SNPs (rs30585 in PDE1A and rs992185 in PDE11A) have significantly different allele frequencies between remitters and nonremitters within the entire depressed group. PDE1A and -11A hydrolyze cAMP and cGMP (14, 18, 19). Individuals who have the G/G genotype for rs30585 or the A/G genotype for rs992185 are, respectively, 4.6 and 3.2 times more likely to attain remission in our sample. These two SNPs also have significantly different allele frequencies between remitters and nonremitters within the fluoxetine group but not within the desipramine group (Table 3). Different SNPs and genes were significantly associated with remitters and nonremitters in fluoxetine- and desipramine-treated patients. Three additional SNPs (rs2544934 in PDE6A, rs884162 in PDE8B, and rs3770018 in PDE11A) were also significantly associated with drug response in the fluoxetine group. Genes associated with response to fluoxetine are located in two chromosomal regions, 2q31–32 and 5q14–31. Two SNPs (rs30585 and rs992185) in the PDE1C gene were associated with treatment response in the desipramine group.
Potential CNS Role of Significant Genes.
Many PDEs are expressed in high concentrations in the brain; their differential expression and subcellular compartmentalization are very suggestive that they are important in fine-tuning neuronal activity and controlling distinct physiological processes and signaling pathways.
The CNS roles for many PDEs remain elusive. Of all significant genes identified in our study, only PDE9A has a known potential role in CNS; it is relevant to cognition and neurodegeneration (22). Interestingly, all of the PDE genes that we have identified as likely to be associated with MDD have potential roles in the CNS (22): cognition and neurodegeneration (PDE2A), cognition and depression (PDE5A), retinal degeneration (PDE6C), and Huntington's disease (PDE10A; refs. 37 and 38).
Conclusions
Our data indicate that PDE genes that modulate intracellular levels of cGMP are the predominant class of PDE associated with the diagnosis and treatment outcome of major depression. All but one PDE gene (PDE8B is cAMP-specific) we identified were either cGMP-specific or dual-substrate enzymes. The cAMP-specific PDE8B, which is high-affinity and rolipram-insensitive, was associated with treatment response in our fluoxetine-treated group, but surprisingly none of the SNPs we examined in cAMP-specific PDE genes were significantly associated with diagnosis, even though in our study, SNP density was higher for that class of PDE genes.
Unexpectedly, we found that polymorphisms in the PDE11A gene are significantly associated with the diagnosis of MDD and treatment response, which strongly suggests the involvement of this enzyme in the biology of depression. PDE11, the newest member of the mammalian PDE family, was characterized 6 years ago (9). This family has a single gene (PDE11A) that has four splicing variants (PDE11A1–A4). The expression and function of this gene are not well understood, but it appears to have a role in spermatogenesis (39); however, no potential CNS role had been previously contemplated for PDE11 (22). PDE11A is phylogenetically related to GAF-containing PDEs: PDE2, -5, -6, and -10; it closely resembles PDE5 by sequence (50% identity and 70% similarity in the catalytic domain) and is located in chromosome 2q31–32 (for a recent review, refer to ref. 40). Thus, our data support the involvement of chr 2q31–32 in the susceptibility for MDD and in antidepressant response.
Pharmacological and genetic studies have indicated that cGMP could be the central mediator of the effects NO/cGMP in several brain regions (41–43). cGMP has several target proteins, including cGMP-regulated cation channels and cGMP-dependent PKs. Two cGMP-PK genes (types 1 and 2) that have been described in mammals are widely distributed in the brain (42, 44). cGMP has been implicated in neuronal maturation (45–47), directional guidance of growth cones (48–50), and learning and memory tasks (51–53). Recently, Horvath et al. (54) described inactivating mutations of the PDE11A gene in a condition predisposing to the development of adrenocortical hyperplasia leading to Cushing syndrome.
Further studies are necessary to establish whether polymorphisms in PDE2A, -5A, -6C, and -10A genes contribute to susceptibility to MDD. Our studies have not exhaustively examined the involvement of PDE polymorphisms in MDD or antidepressant treatment response; therefore, we cannot reject the role of any PDE gene in the genetic or pharmacogenetic of MDD. The contributions of other SNPs in the PDE family of genes should also be further scrutinized, especially in the PDE4 gene family, because PDE4D-regulated cAMP signaling may play a role in the pathophysiology and pharmacotherapy of depression (24, 25). Although we examined 17 SNPs in this gene of the PDE4 family, we have not found an association with diagnosis of MDD or drug response. Regretfully, the limited size of our sample does not permit us to comprehensively explore and detect the likely gene–gene interactions within the PDE family. Such interactions are present if an allele or SNP in one gene influences the effect of a SNP in a second gene. In a larger sample, such explorations could be conducted statistically by using stepwise logistic regression models that include effects for the SNPs within individual genes along with their interactions. In addition to detecting interactions among the SNPs already identified as significant, these analyses might also reveal SNPs within genes that play only interactive roles and thus have not yet been detected as significant.
This study identifies a potential CNS function for the PDE11 family, specifically the susceptibility for major depression and antidepressant drug response. Our results support the need for large-scale comprehensive studies focused on the role of PDE genes on the susceptibility to major depression and antidepressant treatment response. These findings suggest that drugs targeted to affect PDE function, particularly cGMP-related PDEs, could represent a new treatment strategy for major depression and should therefore be tested.
Methods
Study Population.
The study population consisted of 284 depressed subjects enrolled in a pharmacogenetic study of antidepressant treatment response to desipramine or fluoxetine. We also studied 331 age- and sex-matched control subjects recruited from the same Mexican-American community in Los Angeles and studied by the same bilingual clinical research team at the Center for Pharmacogenomics and Clinical Pharmacology at the University of California, Los Angeles (55). Controls were in general good health but were not screened for medical or psychiatric illness. All patients were Mexican-American men and women aged 21–68 years, with a current episode of major depression as diagnosed by DSM-IV (56). In this study, all Mexican-American subjects had at least three grandparents born in Mexico (57). We used diagnostic and ratings instruments that have been fully validated in English and Spanish, and all assessments were conducted in the subjects' primary language.
Inclusion criteria included DSM-IV diagnosis of a current unipolar major depressive episode, with a 21-item Hamilton Depression Rating Scale (58) score of ≥18 with item number 1 (depressed mood) rated ≥2. There was no anxiety threshold for inclusion. Subjects with any primary axis I disorder other than MDD (e.g., dementia, psychotic illness, bipolar disorder, and adjustment disorder), electroconvulsive therapy in the last 6 mo, or previous lack of response to desipramine or fluoxetine were excluded. Because anxiety can be a manifestation of depression, patients who met criteria for depression and also anxiety disorders were not excluded. Exclusion criteria included active medical illnesses that could be etiologically related to the ongoing depressive episode (e.g., untreated hypothyroidism, cardiovascular incident within the past 6 mo, uncontrolled hypertension, or diabetes), current active suicidal ideation with a plan and strong intent, pregnancy, lactation, current use of medications with significant CNS activity that interferes with electroencephalogram activity (e.g., benzodiazepines) or any other antidepressant treatment within the 2 wk before enrollment, illicit drug use and/or alcohol abuse in the last 3 mo, or current enrollment in psychotherapy.
All patients had an initial comprehensive psychiatric and medical assessment and, if enrolled, had 9 wk of structured followup assessments. The study consists of two phases: a 1-wk single-blind placebo lead-in phase to minimize the impact of placebo responders followed, if subjects continue to meet the inclusion criteria after phase 1, by random assignment to one of the two treatment groups: fluoxetine 10–40 mg/day or desipramine 50–200 mg/day, administered in a double-blind manner for 8 wk, with blind dose escalation based on clinical outcomes. In the depressed group, 230 subjects received treatment in our double-blind clinical trial. Of those, 122 patients were treated with desipramine [83 female (F), 39 male (M)], and 108 were treated with fluoxetine (71F, 37M). Sixty-nine patients treated with desipramine (45F, 24M), and 72 treated with fluoxetine (52F, 20M) completed our 8-wk treatment with weekly data collection.
Genomic DNA Collection.
At the initial visit, blood samples were collected into EDTA (K2EDTA) BD Vacutainer EDTA tubes (Becton Dickinson, Franklin Lakes, NJ), and genomic DNA was isolated by using Gentra Puregene DNA purification kits (Gentra Systems, Indianapolis, IN).
Antidepressant Treatment Response.
Our primary clinical outcome measure within the depressed group receiving antidepressant treatment was the Hamilton Depression Rating Scale (HAM-D21). Treatment response was classified into two categories, remission and nonremission status, based on the final (week 8) HAM-D21 score. Remission was defined as having a final HAM-D21 score of <8.
SNP Genotyping Methods.
SNPs were selected from 21 of the 25 genes in the PDE family, located across 14 chromosomes. We selected an average of 10 intragenic SNPs per gene from dbSNP (build 121). SNP assays were designed and typed with the Golden Gate assay as part of a 1,536 multiplex reaction (59). DNAs with poor results (50% GC score <0.65) were removed as well as loci with a low clustering score (<0.3). The threshold for retaining individual genotype calls was set to a Genecall score of 0.25.
Cleaning and Filtering Steps.
SNP quality control.
Our data analysis plan included a series of data cleaning steps followed by a series of filtering steps to identify a list of significantly associated SNPs. Only data generated by SNP assays that were successfully genotyped on at least 80% of samples were included. Data quality was assessed by duplicate DNAs (n = 26) across all plates. Genotypes from nonmatching duplicates were dropped; they were also dropped if they had one missing data point.
HWE.
We used the HWE equation (p2 + 2pq + q2 = 1; p is the frequency of the dominant allele, and q is the frequency of the recessive allele for a trait controlled by a pair of alleles) to determine the probable genotype frequencies in our study populations. Deviation from HWE was tested separately for the control and depressed groups by using the ALLELE procedure in SAS/Genetics 9.1.3 (SAS Institute, Cary, NC). PROC ALLELE uses the notation and methods described by Weir (60). SNPs that were not in HWE in the control group (P < 0.05) and SNPs that were monoallelic in both groups were excluded.
LD among SNPs.
Pairwise LD was calculated within each gene for all SNPs that passed quality control measures by using the r2 measure. An r2 cutoff of ≥80% was used to remove redundant SNPs from the analysis (Fig. 1). The Four Gamete Rule was used to identify haplotype blocks. This method of haplotype block definition assumes no recombination within a block but does allow for recombination between blocks (61). LD measures were assessed by using Haploview, Version 3.2 (ref. 62; Broad Institute, Cambridge, MA).
Haplotype Analyses.
Haplotype block analyses and haplotype population frequency estimation were performed by using Haploview, Version 3.2 (Broad Institute) and by applying the Four Gamete Rule (61). Blocks are formed by consecutive markers where only three gametes are observed. Analyses were initially performed for depressed and control groups separately. Further haplotype analyses were conducted with the depressed and control groups combined to test whether a certain haplotype was associated with a diagnosis of depression. htSNPs were defined in Haploview by using aggressive tagging (two- and three-marker haplotypes). This method selects a minimal set of markers where all other alleles to be captured are correlated (r2 ≥ 0.8) with a marker in that set. Then, the use of multimarker tests expands the set and includes additional markers that capture alleles not otherwise captured in the initial pairwise tagging. All haplotypes >0% were examined, and nontagging SNPs within haplotype blocks were omitted from the final analyses and figures (Fig. 1 Inset).
Statistical Analyses.
SNP Analyses.
Allele, genotype and allelic trend association tests were performed by using PROC CASE CONTROL in SAS/Genetics 9.1.3 (SAS Institute). PROC CASE CONTROL is designed to test for differences in frequency of marker data when random samples are available from populations affected and unaffected by disease and is based on case-control tests for biallelic markers described by P. D. Sasieni (63). The following criteria were used to identify a list of SNPs statistically associated with a diagnoses of depression: (i) SNPs were in HWE in the control group; (ii) the minor allele frequency in the control group was ≥5%; and (iii) multiple testing was corrected by using Bonferroni correction, which set the significance level at P value ≤0.0006 for tests between control and depressed groups. We tested our secondary hypothesis using similar criteria to identify a list of SNPs associated with treatment response: (i) SNPs were in HWE in the control group; (ii) the minor allele frequency in the control group was ≥5%; (iii) P value ≤0.05 for allele test between remitter and nonremitter groups was used. Because of the small sample size, this part of the analyses is preliminary.
Odds ratios.
We compared the odds of having depression given the homozygous major, homozygous minor, or heterozygous genotype for SNPs associated with diagnoses of depression. Similarly, we compared the odds of attaining remission given the homozygous major, homozygous minor, or heterozygous genotype for SNPs associated with treatment response. Odds ratios were calculated by using PROC FREQ in SAS/Genetics 9.1.3 (SAS Institute.
Acknowledgments
We are grateful for the contributions of Israel Alvarado, Rita Jepson, Lorraine Garcia-Teague, Patricia Reyes, Isabel Rodriguez, and Gabriela Marquez for their clinical work and Kristopher Irizarry for helping with the bioinformatics component. This work was supported by National Institutes of Health Grants GM61394, RR017365, MH062777, RR000865, K30HL04526, K12RR17611, RR16996, HG002500, and DK063240 and by an award from the Dana Foundation. P.D., P.W., and M.D. are supported by the Wellcome Trust.
Abbreviations
- PDE
phosphodiesterase
- MDD
major depressive disorder
- HWE
Hardy–Weinberg Equilibrium
- LD
linkage disequilibrium
- htSNP
haplotype-tagging SNP
- C.I.
confidence interval
- GAF
cGMP-binding PDE, Anabaena adenylyl cyclase, and E. coli Fh1A domain.
Footnotes
Author contributions: M.-L.W. and J.L. designed research; M.-L.W., F.W., P.D., P.W., M.D., and J.L. performed research; F.W., P.D., P.W., M.D., and R.M.C. contributed new reagents/analytic tools; M.-L.W., F.W., R.M.C., and J.L. analyzed data; and M.-L.W., S.M.M., and J.L. wrote the paper.
Conflict of interest statement: M.-L.W. and J.L. have filed a patent based on the work reported in this paper.
References
- 1.Beavo JA. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 2.Beavo JA, Conti M, Heaslip RJ. Mol Pharmacol. 1994;46:399–405. [PubMed] [Google Scholar]
- 3.Soderling SH, Bayuga SJ, Beavo JA. Proc Natl Acad Sci USA. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Biochem Biophys Res Commun. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi M, Matsushima K, Ohashi H, Tsunoda H, Murase S, Kawarada Y, Tanaka T. Biochem Biophys Res Commun. 1998;250:751–756. doi: 10.1006/bbrc.1998.9379. [DOI] [PubMed] [Google Scholar]
- 6.Soderling SH, Bayuga SJ, Beavo JA. J Biol Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 7.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. J Biol Chem. 1998;273:15559–15564. doi: 10.1074/jbc.273.25.15559. [DOI] [PubMed] [Google Scholar]
- 8.Soderling SH, Bayuga SJ, Beavo JA. Proc Natl Acad Sci USA. 1999;96:7071–7076. doi: 10.1073/pnas.96.12.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, Hetman J, Beavo JA, Phillips SC. Proc Natl Acad Sci USA. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon YH, Heo YS, Kim CM, Hyun YL, Lee TG, Ro S, Cho JM. Cell Mol Life Sci. 2005;62:1198–1220. doi: 10.1007/s00018-005-4533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolger GB. Cell Signal. 1994;6:851–859. doi: 10.1016/0898-6568(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 12.Houslay MD. Semin Cell Dev Biol. 1998;9:161–167. doi: 10.1006/scdb.1997.0221. [DOI] [PubMed] [Google Scholar]
- 13.Perry MJ, Higgs GA. Curr Opin Chem Biol. 1998;2:472–481. doi: 10.1016/s1367-5931(98)80123-3. [DOI] [PubMed] [Google Scholar]
- 14.Mehats C, Andersen CB, Filopanti M, Jin SL, Conti M. Trends Endocrinol Metab. 2002;13:29–35. doi: 10.1016/s1043-2760(01)00523-9. [DOI] [PubMed] [Google Scholar]
- 15.Rotella DP. Nat Rev Drug Discov. 2002;1:674–682. doi: 10.1038/nrd893. [DOI] [PubMed] [Google Scholar]
- 16.Conti M, Nemoz G, Sette C, Vicini E. Endocr Rev. 1995;16:370–389. doi: 10.1210/edrv-16-3-370. [DOI] [PubMed] [Google Scholar]
- 17.Torphy TJ. Am J Respir Crit Care Med. 1998;157:351–370. doi: 10.1164/ajrccm.157.2.9708012. [DOI] [PubMed] [Google Scholar]
- 18.Beavo JA, Brunton LL. Nat Rev Mol Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 19.Conti M. Mol Endocrinol. 2000;14:1317–1327. doi: 10.1210/mend.14.9.0534. [DOI] [PubMed] [Google Scholar]
- 20.Houslay MD, Adams DR. Biochem J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnenburg WK, Seger D, Kwak KS, Huang J, Charbonneau H, Beavo JA. J Biol Chem. 1995;270:30989–31000. doi: 10.1074/jbc.270.52.30989. [DOI] [PubMed] [Google Scholar]
- 22.Menniti FS, Faraci WS, Schmidt CJ. Nat Rev Drug Discov. 2006;5:660–670. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- 23.Houslay MD. Prog Nucleic Acid Res Mol Biol. 2001;69:249–315. doi: 10.1016/s0079-6603(01)69049-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HT, O'Donnell JM. Psychopharmacology (Berl) 2000;150:311–316. doi: 10.1007/s002130000414. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HT, Crissman AM, Dorairaj NR, Chandler LJ, O'Donnell JM. Neuropsychopharmacology. 2000;23:198–204. doi: 10.1016/S0893-133X(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 26.Perez J, Tardito D, Racagni G, Smeraldi E, Zanardi R. Mol Psychiatry. 2001;6:44–49. doi: 10.1038/sj.mp.4000795. [DOI] [PubMed] [Google Scholar]
- 27.O'Donnell JM, Frith S, Wilkins J. J Pharmacol Exp Ther. 1994;271:246–254. [PubMed] [Google Scholar]
- 28.Zhang HT, Huang Y, Jin SL, Frith SA, Suvarna N, Conti M, O'Donnell JM. Neuropsychopharmacology. 2002;27:587–595. doi: 10.1016/S0893-133X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 29.Torgersen S. Arch Gen Psychiatry. 1986;43:222–226. doi: 10.1001/archpsyc.1986.01800030032003. [DOI] [PubMed] [Google Scholar]
- 30.McGuffin P, Katz R, Rutherford J. Psychol Med. 1991;21:329–335. doi: 10.1017/s0033291700020432. [DOI] [PubMed] [Google Scholar]
- 31.McGuffin P, Katz R, Watkins S, Rutherford J. Arch Gen Psychiatry. 1996;53:129–136. doi: 10.1001/archpsyc.1996.01830020047006. [DOI] [PubMed] [Google Scholar]
- 32.Kendler KS, Gardner CO, Neale MC, Prescott CA. Psychol Med. 2001;31:605–616. doi: 10.1017/s0033291701003907. [DOI] [PubMed] [Google Scholar]
- 33.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. Arch Gen Psychiatry. 1993;50:863–870. doi: 10.1001/archpsyc.1993.01820230054003. [DOI] [PubMed] [Google Scholar]
- 34.Bierut LJ, Heath AC, Bucholz KK, Dinwiddie SH, Madden PA, Statham DJ, Dunne MP, Martin NG. Arch Gen Psychiatry. 1999;56:557–563. doi: 10.1001/archpsyc.56.6.557. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan PF, Neale MC, Kendler KS. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 36.Yuasa K, Kanoh Y, Okumura K, Omori K. Eur J Biochem. 2001;268:168–178. doi: 10.1046/j.1432-1327.2001.01866.x. [DOI] [PubMed] [Google Scholar]
- 37.Hebb AL, Robertson HA, Denovan-Wright EM. Neuroscience. 2004;123:967–981. doi: 10.1016/j.neuroscience.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Hu H, McCaw EA, Hebb AL, Gomez GT, Denovan-Wright EM. Eur J Neurosci. 2004;20:3351–3363. doi: 10.1111/j.1460-9568.2004.03796.x. [DOI] [PubMed] [Google Scholar]
- 39.Wayman C, Phillips S, Lunny C, Webb T, Fawcett L, Baxendale R, Burgess G. Int J Impot Res. 2005;17:216–223. doi: 10.1038/sj.ijir.3901307. [DOI] [PubMed] [Google Scholar]
- 40.Makhlouf A, Kshirsagar A, Niederberger C. Int J Impot Res. doi: 10.1038/sj.ijir.3901441. in press. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Robinson PJ. J Neurochem. 1997;68:443–456. doi: 10.1046/j.1471-4159.1997.68020443.x. [DOI] [PubMed] [Google Scholar]
- 42.Feil S, Zimmermann P, Knorn A, Brummer S, Schlossmann J, Hofmann F, Feil R. Neuroscience. 2005;135:863–868. doi: 10.1016/j.neuroscience.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann F. J Biol Chem. 2005;280:1–4. doi: 10.1074/jbc.R400035200. [DOI] [PubMed] [Google Scholar]
- 44.Feil R, Feil S, Hofmann F. Trends Mol Med. 2005;11:71–75. doi: 10.1016/j.molmed.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Scholz NL, Truman JW. Functional Neuroanatomy of the Nitric Oxide System. In: Steinbusch HWM, De Vente J, Vincent SR, editors. Amsterdam: Elsevier; 2000. pp. 417–411. [Google Scholar]
- 46.Bicker G. Arch Insect Biochem Physiol. 2001;48:100–110. doi: 10.1002/arch.1062. [DOI] [PubMed] [Google Scholar]
- 47.Scholz NL, de Vente J, Truman JW, Graubard K. J Neurosci. 2001;21:1610–1618. doi: 10.1523/JNEUROSCI.21-05-01610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song H, Ming G, He Z, Lehmann M, McKerracher L, Tessier-Lavigne M, Poo M. Science. 1998;281:1515–1518. doi: 10.1126/science.281.5382.1515. [DOI] [PubMed] [Google Scholar]
- 49.Song HJ, Poo MM. Curr Opin Neurobiol. 1999;9:355–363. doi: 10.1016/s0959-4388(99)80052-x. [DOI] [PubMed] [Google Scholar]
- 50.Polleux F, Morrow T, Ghosh A. Nature. 2000;404:567–573. doi: 10.1038/35007001. [DOI] [PubMed] [Google Scholar]
- 51.Kendrick KM, Guevara-Guzman R, Zorrilla J, Hinton MR, Broad KD, Mimmack M, Ohkura S. Nature. 1997;388:670–674. doi: 10.1038/41765. [DOI] [PubMed] [Google Scholar]
- 52.Bernabeu R, Cammarota M, Izquierdo I, Medina JH. Braz J Med Biol Res. 1997;30:961–965. doi: 10.1590/s0100-879x1997000800008. [DOI] [PubMed] [Google Scholar]
- 53.Izquierdo LA, Vianna M, Barros DM, Mello e Souza T, Ardenghi P, Sant'Anna MK, Rodrigues C, Medinam JH, Izquierdo I. Neurobiol Learn Mem. 2000;73:141–149. doi: 10.1006/nlme.1999.3925. [DOI] [PubMed] [Google Scholar]
- 54.Horvath A, Boikos S, Giatzakis C, Robinson-White A, Groussin L, Griffin KJ, Stein E, Levine E, Delimpasi G, Hsiao HP, et al. Nat Genet. 2006;38:794–800. doi: 10.1038/ng1809. [DOI] [PubMed] [Google Scholar]
- 55.Licinio J, O'Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, Lake S, Tantisira KG, Weiss ST, Wong ML. Mol Psychiatry. 2004;9:1075–1082. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 56.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 57.Hazuda HP, Comeaux PJ, Stern MP, Haffner SM, Eifler CW, Rosenthal M. Am J Epidemiol. 1986;123:96–112. doi: 10.1093/oxfordjournals.aje.a114228. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton M. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, et al. Cold Spring Harbor Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 60.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- 61.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Am J Hum Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 63.Sasieni PD. Biometrics. 1997;53:1253–1261. [PubMed] [Google Scholar]