Abstract
Obstacles to the expansion of cells with proliferative potential include the induction of cell death, telomere-based senescence, and the pRb and p53 tumor suppressors. Not infrequently, the molecular pathways regulating oncogenesis recapitulate aberrations of processes governing embryogenesis. The genetic network, consisting of the dachshund (dac), eyes absent (eya), eyeless, and sine oculis (so) genes, regulates cell fate determination in metazoans, with dac serving as a cointegrator through a So DNA-binding factor. Here, DACH1 inhibited oncogene-mediated breast oncogenesis, blocking breast cancer epithelial cell DNA synthesis, colony formation, growth in Matrigel, and tumor growth in mice. Genetic deletion studies demonstrated a requirement for cyclin D1 in DACH1-mediated inhibition of DNA synthesis. DACH1 repressed cyclin D1 through a novel mechanism via a c-Jun DNA-binding partner, requiring the DACH1 α-helical DS domain which recruits corepressors to the local chromatin. Analysis of over 2,000 patients demonstrated increased nuclear DACH1 expression correlated inversely with cellular mitosis and predicted improved breast cancer patient survival. The cell fate determination factor, DACH1, arrests breast tumor proliferation and growth in vivo providing a new mechanistic and potential therapeutic insight into this common disease.
The Drosophila dachshund (dac) gene is the founding member of the DACH subfamily of nuclear proteins, which conduct an essential role in promoting differentiation of the Drosophila eye and limb (42). The dac gene forms part of a retinal determination (RD) signaling pathway in Drosophila. The RD pathway requires eyeless (eya/Pax6), sine oculis (So, Six), eyes absent (eya/Eya), and dachshund (dac/Dach), which function together as genetic components of this network (24, 25, 36). In Drosophila, So functions as a DNA-binding factor and dac/eya are transcription cofactors. Although So/Six binding sites have been identified in RD signaling target gene promoters, genome-wide analysis of DACH1-regulated genes identified a preponderance of AP-1-responsive genes. Dachshund is expressed prior to photoreceptor differentiation and is required for retinal morphogenesis. Dachshund itself is sufficient for inducing retinal fates since targeted misexpression results in ectopic eye formation from non-neuronal tissues (13, 54).
Although the RD gene network is best known for its role in eye specification, these genes, either individually or as a network, are expressed in postmitotic cells and contribute to diverse developmental processes in many cell types in all metazoans. The So/Six family governs proliferation of progenitor populations prior to cell type specification. Misregulated expression of Six proteins occurs in human cancer (15, 21). HSIX1 is the human homologue of the Drosophila Six gene that was originally isolated through its enrichment during the S phase of the cell cycle. The Six gene in Drosophila functions as a DNA-binding component of the transcription factor complex. Six1 is overexpressed in breast cancer, and forced Six1 expression attenuates a G2 cell cycle check point (15, 21). Six1 has been implicated as a dual-function regulator of metastasis, enhancing poorly metastatic rhabdomyosarcoma tumors (66). The molecular mechanism by which the RD pathway regulates human tumorigenesis is poorly understood.
Orderly cell cycle progression of nontransformed cells is orchestrated by coordinated induction of cyclin-dependent kinases that assemble in temporally and spatially defined complexes within the cell (43). Sequential phosphorylation of key substrates, including the retinoblastoma (pRb) protein, by cyclin D1 and cyclin E-cdk complexes promotes the timely induction of cellular DNA synthesis (27). Oncogenic disruption of the cell cycle machinery, through amplification or disruption of the cell cycle proteins themselves, is a common finding in human breast cancer. The cyclin D1 gene encodes the regulatory subunit of a holoenzyme that contributes to the phosphorylation and inactivation of the pRb protein and is frequently overexpressed in human breast cancer epithelial cells. Specific oncogenic signals disrupt the cell cycle in a reproducible manner. Transformation by Ras requires the inactivation of the pRb and p53 pathways. Myc has activities compatible with the bypass of p21CIP1 (10, 14, 16, 28, 29, 51, 58) and/or activation of Arf and p53 (10), which impose a selection to the escape of cellular apoptosis (67). Genetic studies in mice have confirmed the fidelity of these molecular interactions in vivo. Thus, genetic deficiency for cyclin D1 provides resistance to Ras or ErbB2 but not c-Myc-induced tumorigenesis, whereas a distinct subset of genes antagonize Myc function including transforming growth factor β (TGF-β) and p21CIP1 (55).
Obstacles to the expansion of cells with proliferative potential include the induction of cell death, telomere-based senescence, and the pRb and p53 tumor suppressors. Not infrequently, the molecular pathways regulating oncogenesis, recapitulate aberrations of processes governing embryogenesis (1). The transcription factors encoded by the homeobox gene family play a vital role in growth and differentiation during normal development. This diverse group of proteins is divided into groups based on similarity among the homeodomain box and includes the MSX, Engrailed, PAX, and the SIX families. Deregulated homeobox gene expression is well known in cancer, with genes normally expressed in undifferentiated cells upregulated in cancer. Homeobox genes known to be deregulated in cancer include HOX, MSX, HSIX1, GBX2, the PAX genes, CDX2, NKX3.1, and BARX2. Recent microarray analysis studies demonstrated DACH1-repressed proliferative signaling in cultured cells (62). We examined the expression and function of DACH1 in normal and tumorous breast epithelium. We demonstrate DACH1 blocks cellular proliferation and contact-independent growth and identify a new mechanism by which DACH1 regulates cellular proliferation and growth in vivo.
MATERIALS AND METHODS
Plasmid construction.
The expression plasmids for DACH1, DACH1 DS-domain alone (DS), or DACH1 DS-domain deleted (ΔDS) (which includes an N-terminal Flag peptide), the reporter plasmid 3-TP lux, 3XCRE luc the ponasterone-regulated expression system, and the full-length wild-type and mutant cyclin D1 promoter reporter constructions were previously described (5, 6, 62). The Flag-tagged DACH1 cDNA was subcloned into the MSCV-internal ribosome entry site (IRES)-green fluorescent protein (GFP). The SKI cDNAs was subcloned into the 3x Flag-CMV7.1 vector (Sigma, St. Louis, MO).
Cell sorting and DNA synthesis analysis.
Magnetic activated cell sorting to enrich for transfected cells was conducted as previously described (7). Cell cycle parameters were determined by using laser scanning cytometry. Cells were processed by standard methods using propidium iodide staining of cell DNA. Each sample was analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson Biosciences, Mansfield, MA) using a 488-nm laser. Histograms were analyzed for cell cycle compartments using ModFit version 2.0 (Verity Software House, Topsham, ME). A minimum of 20,000 events was collected to maximize statistical validity of the compartmental analysis.
Western blot analysis, tissue microarrays, immunohistochemistry, and purification of DACH1 antibody.
Western blot analysis with antibodies to cyclin D1, cyclin E, cdk4, phosphorylated pRB (residue amino acid Ser708), Flag, p21CIP1/WAF1, p27KIP1, c-Jun, CREB, and the loading control guanine dissociation inhibitor (GDI) and immunohistochemistry for cyclin D1 and DACH1 were performed as previously described (35, 62). Affinity-purified polyclonal antibody to DACH1 was produced by Affinity BioReagents through immunizing rabbits with the peptide ERTIQDGRLYLKTTVMY. Human breast tissue microarrays were from Imgenex (IMH-304) and NCI CBCTR 2001 TMA#2. The human breast cancer prognostic tissue microarray was previously described (2). This array consists of 2,197 formalin-fixed, paraffin-embedded breast cancer tissues with clinical-pathological characteristics, clinical follow-up information, and 283 normal/precancerous tissues. The level of DACH1 protein expression in tumors and normal human breast cancer in the TMAs was categorized by a semiquantitative score of the immunostaining intensity by light microscopy evaluation, using a standard methodology that determined a range of staining intensities from negative to strong with intermediate grades. The intensity of immunoperoxidase staining was scored as 0 (negative), 1+ (a minimal to low level of positive staining), 2+ (moderate expression), or 3+ (strong staining). The evaluation and scoring of the cores of the tumors from the patients representing different breast tumor classifications and grades incorporated in the TMAs were conducted by a pathologist (R.G.R.). The results were viewed by two pathologists. Mitoses grade in tumor cells was determined according to the standard of Bloom, Richardson, and Elston.
Cell culture, plasmid transfection, luciferase reporter assays, and Matrigel culture.
Cyclin D1−/− cells derived from cyclin D1−/− mice (64), p21CIP1−/− fibroblast cells (provided by L. Augenlicht) and p27KIP1−/− fibroblast cells (a gift from A. Koff) were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum in a humidified atmosphere with 5% CO2 at 37°C. Other cells were cultured as recommended by the American Type Culture Collection and maintained in humidified atmosphere with 5% CO2 at 37°C (59). Transfections were performed by using Superfect transfection reagent (QIAGEN, Valencia, CA) according to the manufacturer's protocol using 1.5 μg of the reporter, 50 to 300 ng of expression plasmids, or equal molar amounts of control vector. The transfection efficiency was normalized by cotransfection with 0.2 μg of pRL-CMV plasmid (Promega, Madison, WI) and measured with Promega's dual-luciferase reporter assay system according to the manufacturer's protocol. Control small interfering RNA (siRNA) was purchased from Santa Cruz. siRNAs for human DACH1 were designed and synthesized by Ambion, Inc. (gene accession number AF 356492). The target sequences were AAGGCCTCCTAAGAGGACTCA and AAGGACTTCGAGACCCTCTAC. DACH1-specific siRNAs or control siRNAs (100 or 200 nM) were transfected according to the Oligofectamine protocol (Invitrogen). Transfection efficiency was monitored by no-silencing fluorescein siRNA from QIAGEN. Statistical analyses were performed by using the Student t test. For three-dimensional culture, 1,000 cells mixed with 1:3 diluted growth factor reduced Matrigel matrix were seeded into BD biocoated cell culture inserts (24-well) with Matrigel basement membrane matrix from BD Bioscience (catalog no. 354447) and photographed after growth for 3 to 14 days. For confocal image analyses, MCF10A cells were mixed with 1:1-diluted matrigel and seeded into four-well chamber according to a previously described protocol (18).
Immunoprecipitation, immunoblotting, and Northern blot.
293T cells were used for the detection of protein-protein interaction in vivo, and immunoprecipitation-Western blotting was conducted as previously described using anti-Flag M2 antibody (Sigma) and immunoblotting with antibodies to CREB, c-Jun, or Flag (Santa Cruz Biotechnology). The guanine nucleotide dissociation inhibitor antibody (i.e., GDI) (35) was used as an internal control for protein abundance. For Northern blot analysis, total RNA was isolated by using TRIzol reagent. Cyclin D1 cDNA was randomly labeled using NEBlot Phototope kit, and the membrane was detected by Phototope-Star detection kit (New England Biolabs).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (22). The human cyclin D1 promoter-specific primers used were as follows: AP-1 site, 5′-CTGCCTTCCTACCTTGACCA-3′ and 5′-TGAAGGGACGTCTACACCCC-3′; and CRE site, 5′-GCCCCCCTCCCGCTCCCATT-3′ and 5′-TGGGGCTCTTCCTGGGCAGC-3′. The sequences as a negative control in the cyclin D1 promoter were 5′-TTTCGGAAGCGTTTTCCC and 5′-AGCGCGTTCATTCAGGAA. 293T cells were transiently cotransfected with expression plasmids encoding wild-type or mutant DACH1, with the cyclin D1 promoter constructions, −1745 or cyclin D1 −1745-AP-1/CRE mutant reporter, and then cultured for 36 h. The cells were cross-linked with formaldehyde buffer for 10 min at 37°C, and the procedure was performed as described earlier (22).
Nude mice study.
Ponasterone A was purified from taxus chinensis, and 100 μg of purified ponasterone A was introduced into 21-day slow release pellets produces by Innovative Research of American (Sarasota, FL) (5). Ponasterone A or placebo pellets were implanted subcutaneously into 4- to 6-week-old athymic female nude mice purchased from NCI. The following day, 2 × 106 MDA-MB-231 cells, stably expressing DACH1 or vector control, were injected subcutaneously, and the tumor growth was measured twice weekly by digital caliper. Ponasterone A or placebo pellets were implanted every 21 days.
Statistics.
The Student t test and the Wilcoxon-Mann-Whitney test were used to analyze the significance of expression of DACH1. Kaplan-Meier survival curves were plotted and analyzed with the Cox proportional hazards regression. The statistical significance of difference between curves was tested by using the generalized Wilcoxon test.
RESULTS
DACH1 inhibition of colony formation and tumor growth in vivo.
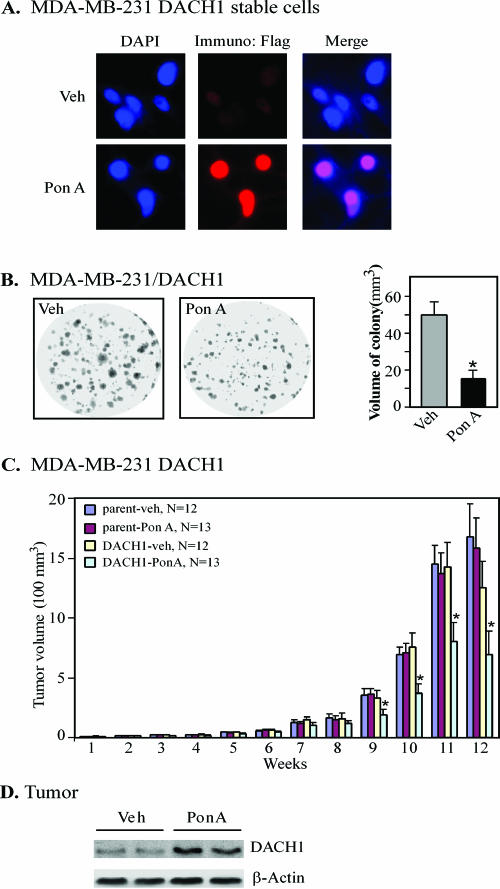
Recent studies examining the genetic signaling repressed by DACH1 identified clusters of genes promoting cellular proliferation, growth, and survival (62). To examine the function of DACH1 in regulating breast epithelial cell growth in vivo, stable cell lines inducibly expressing DACH1 under the control of the ecdysone-regulated system were constructed in MDA-MB-231 cells. Ponasterone A induced expression of the Flag-tagged DACH1 expression vector as determined by immunohistochemistry and by Western blotting at 12 to 96 h after ponasterone A addition (Fig. 1A and see Fig. S1A in the supplemental material). The numbers and sizes of colonies were reduced >70% by DACH1 (Fig. 1B). To examine the effect of DACH1 on breast tumor growth in vivo, the ponasterone-inducible stable lines were implanted in nude mice, and a comparison was made between the sustained release pellets of ponasterone A and the control placebo pellets, with greater than 12 separate animals in each group. Ponasterone A pellets induced sustained expression of an ecdysone-regulated β-galactosidase reporter mouse (data not shown) (5). The induction of DACH1 expression reduced the growth of breast tumors in mice by 50% (Fig. 1C), with an inhibition of growth by DACH1 first detected at 9 weeks postimplantation (329 ± 66 mm3 versus 186 ± 27 mm3, P < 0.05). Western blot analysis of tumors isolated from mice demonstrated the induction of DACH1 protein in mice treated with ponasterone A pellets compared to mice treated with placebo control pellets (Veh) (Fig. 1D).
FIG. 1.
Inducible DACH1 expression inhibits colony formation and tumor growth in vivo. (A) Immunohistochemical stain for the Flag epitope of DACH1 in MDA-MB-231 stable ponasterone-inducible DACH1 cell lines showing nuclear colocalization with DAP1 stain. (B) Ponasterone-inducible MDA-MB-231 stable cells were seeded onto six-well plates and stained with crystal violet after 14 days growth. Colony volumes (mean ± the standard error of the mean [SEM], in mm3) are shown on the right (*, P < 0.01). (C) Tumor growth of ponasterone-inducible MDA-MB-231 stable cells in athymic female nude mice (n > 12 separate animals in each group; *, P < 0.05). Mice were treated with the ponasterone A pellets or control pellets (vehicle), and the sizes of the tumors were assessed weekly. (D) Representative Western blot analysis of DACH1-inducible MDA-MB-231 cells from animals treated with vehicle or ponasterone A pellets.
DACH1 inhibition of breast epithelial cell DNA synthesis requires the DACH DS domain.
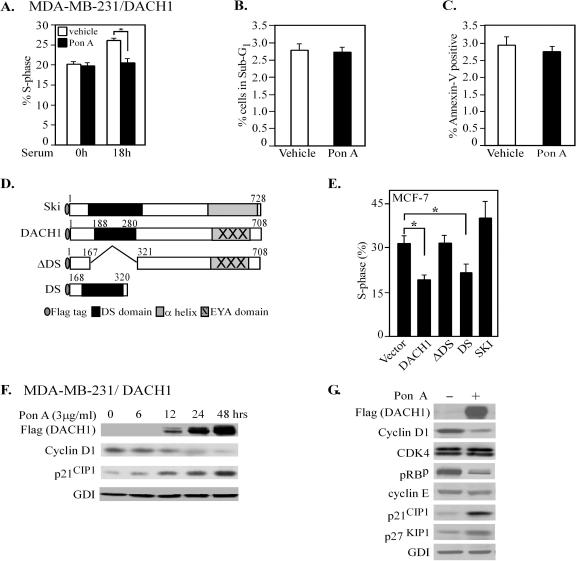
To investigate the mechanism by which DACH1 blocked MDA-MB231 cellular growth, we examined the effect of DACH1 on DNA synthesis and cellular apoptosis. Although serum starvation does not arrest DNA synthesis in MDA-MB231 cells (47), the addition of serum produces an induction of the proportion of cells in the S phase of the cell cycle (Fig. 2A). Expression of DACH1 blocked serum-induced DNA synthesis (Fig. 2A) without affecting the fraction of apoptotic cells, inferred through analysis of the sub G1 population and annexin V staining of the cells (Fig. 2B and C). MDA-MB-231 cells harbor mutation in the K-Ras gene. MCF-7 cells express amplification of c-Myc. To determine whether DACH1 was capable of inhibiting DNA synthesis in MCF-7 cells, cells were transfected with DACH1 expression vectors. To identify the domains of DACH1 involved in regulating DNA synthesis, a series of domain mutants were examined (Fig. 2D). MCF-7 cells cotransfected with DACH1 or its mutants were assessed by flow cytometry. DACH1 reduced the proportion of cells in the DNA synthetic (S) phase by 17% (Fig. 2E). One region of structural homology between DACH1 and the Ski/Sno proto-oncogene family (26) is predicted to form an α-helical structure (33) (DACH DS domain) that may convey general or specific DNA-binding activity (31). Deletion of the conserved DS domain abrogated inhibition of DNA synthesis. Expression of the related Ski protein did not inhibit DNA synthesis in MCF-7 cells. The inability of the ΔDS domain construct to inhibit DNA synthesis compared to the DS domain alone expression vector was not due to reduced expression of the ΔDS domain compared to the DS domain construct, since the ΔDS domain was expressed ∼3-fold better than the DS domain alone (see Fig. S1B in the supplemental material).
FIG. 2.
DACH1 inhibits DNA synthesis through the DS domain. (A) MDA-MB-231 cells expressing inducible DACH1 were characterized for the effects of DACH1 expression on serum-induced DNA synthesis. The effect of DACH1 expression on S phase or apoptosis assessed by Sub-G1 (B) and annexin V stain (C) is shown after 18 h of serum addition. The data are means ± the SEM for >5 separate experiments. (D) Schematic representation of DACH1 expression vectors. (E) Cell cycle analysis of MCF-7 cells transfected with DACH1 expression vectors. The percent MCF-7 cells in S phase is shown compared to the vector control. The data are mean change in S phase ± the SEM of DACH1-transfected MCF-7 cells (n = 4). (F) Dose-dependent induction of DACH1 by ponasterone A and the corresponding cyclin D1 and p21CIP1 expression in MDA-MB-231 cells. (G) Cell-cycle-related protein abundance as determined by Western blotting in MDA-MB-231 cells after 48 h of ponasterone A treatment.
Phosphorylation and inactivation of the pRb protein and the cyclin D1 gene product play a key role in DNA synthesis of MDA-MB231 cells. To investigate the candidate target cell cycle proteins regulated by DACH1, responsible for the inhibition of DNA synthesis, Western blot analysis was conducted. Cells were treated with ponasterone A to induce expression of DACH1, and serial time points were analyzed. DACH1 induction was observed at 12 h, contemporaneous with a reduction in cyclin D1 and an induction of p21CIP1. The abundance of the protein loading control GD1 was unchanged (Fig. 2F). To determine target genes involved in blocking serum-induced DNA synthesis, cells were serum starved and, after induction of DACH1 with ponasterone A, the cells were treated with serum for 18 h. After 24 h of DACH1 expression, the cyclin D1 levels were reduced 60% while increasing the abundance of p21CIP1 and p27KIP1 (Fig. 2G). A phospho-specific antibody directed to the Ser780 site phosphorylation of the pRb protein showed a 60% reduction in pRb phosphorylation. Cyclin E and cdk4 levels were unchanged (Fig. 2G).
DACH1 inhibits DNA synthesis and oncogene-induced invasive phenotype induced by Ras or Myc through distinct pathways.
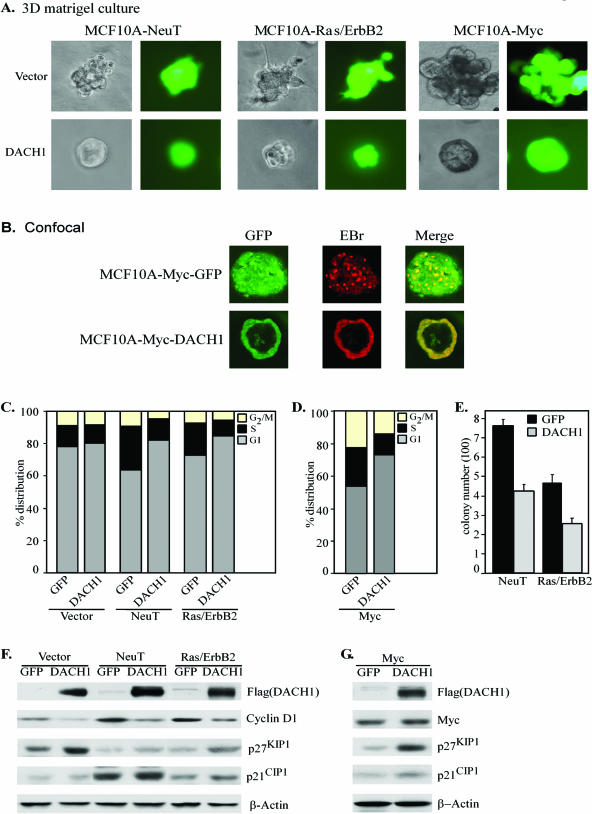
The creation of genetically defined human cancer cells has defined with greater precision the regulatory pathways contributing to the tumorigenic phenotype (19, 23). MCF10A cells are a spontaneously immortalized human luminal epithelial cell line (56). When grown on basement membrane, the cultures of MCF10A cells form growth-arrested three-dimensional structures, termed acini. These structures, comprised of a single layer of polarized epithelial cells surrounding a hollow lumen, resemble mammary acini and undergo morphological changes in response to transforming oncogenes (50, 56). The morphology of MCF10A cellular colony growth in three-dimensional Matrigel culture is characteristic for specific oncogenic pathways (9, 52). To define which oncogenic signaling pathways are inhibited by DACH1, MCF10A cells were transduced with expression vectors encoding oncogenic ErbB2 (NeuT), Ras, and ErbB2 (Ras/ErbB2) or c-Myc. These transformed cells were then transduced with viral expression vectors encoding DACH1 (MSCV-IRES-GFP or MSCV-DACH1-IRES-GFP) and selected through fluorescence-activated cell sorting (FACS) of GFP-expressing cells. Analyses were conducted of morphology in Matrigel (Fig. 3A and B), cell cycle distribution (Fig. 3C and D), colony formation ability (Fig. 3E), and cell cycle control proteins abundance determined by Western blotting (Fig. 3F and G). MCF10A cells form acinus-like spheroids. MCF10A-NeuT cells grown in Matrigel formed complex acinus-like structures (39). The activation of ErbB2 in MCF10A cells is known to induce this multiacinar phenotype through excessive cellular proliferation and changes in apicobasal polarization (17, 44). DACH1 transduction of oncogenic NeuT transformed MCF10A cells abolished the formation of these multicellular structures, reversing the phenotype to the spherical morphology of parental MCF10A (Fig. 3A). The MCF10A-Ras/ErbB2 colonies consisted of multicellular spheroids in which individual cells protruded long spikes. DACH1 transduction of these Ras/ErbB2 transformed MCF10A cells reversed the phenotype and abolished the formation of these spike structures (Fig. 3A). MCF10A-c-Myc cells grew as multicellular acinus-like structures, and DACH1 transduction resulted in the formation of well-polarized spheroids with lumen formation (Fig. 3A and B). Collectively, these studies demonstrate that DACH1 inhibits oncogene-induced morphological abnormalities in MCF10A cells.
FIG. 3.
DACH1 inhibits DNA synthesis induced by NeuT, Ras/ErbB2 or Myc. (A) The immortalized MCF10A cells were transformed with NeuT, Ras/ErbB2, or c-Myc. Cells were transduced with MSCV-IRES-GFP or MSCV-DACH1-IRES-GFP and selected through GFP FACS for analysis. Morphology of three-dimensional growth in Matrigel at 10 days was depicted by phase-contrast and fluorescence microscopy of acinar structures. (B) Confocal image of GFP and ethidium bromide staining after fixation. Transformed MCF10A cells grow as a solid ball in which the central acinar structure is filled with proliferating cells. The expression of DACH1 reverts the oncogenic morphology with a return to the nontransformed MCF10A phenotype with polarized epithelium and an acinar-like lumen structure. (C and D) Cell cycle distribution. (E to G) Colony formation assay results (E) and cell cycle control proteins as determined by Western blotting of transformed MCF10A cells with the indicated antibodies (F and G).
DACH1 inhibited NeuT, Ras/ErbB2, and c-Myc-induced DNA synthesis as assessed by a reduction in the proportion of cells distributed within the S phase of the cell cycle (Fig. 3C and D). DACH1 reduced primarily the S phase but also the G2/M fraction of c-Myc-expressing MCF10A cells. MCF10A cells transformed by oncogenic NeuT or Ras/ErbB2 gain the capacity to sustain contact-independent growth and form colonies in soft agar. DACH1 blocked colony formation induced by either NeuT or Ras/ErbB2 (Fig. 3E). To determine the components of the cell cycle regulated by DACH1 that may contribute to the inhibition of cellular growth, Western blot analyses were conducted of the oncogene-transduced MCF10A cells. We compared the effect of transducing the MCF10A cells to the effect of control vector. In randomly cycling cells, the abundance of cyclin D1 encodes a rate-limiting step in DNA synthesis (46, 68). In parental MCF10A, cyclin D1 abundance was reduced by DACH1, and p27KIP1 was induced (Fig. 3F) without a significant change in S-phase distribution. In NeuT or Ras/ErbB2-transformed MCF10A cells, cyclin D1 and p21CIP1 abundance was induced, a finding consistent with previous studies in which Raf induced cyclin D1 in MCF10A cells (50). DNA synthesis was inhibited by DACH1, associated with a reduction in cyclin D1 and induction of p27KIP1. c-Myc repressed p21CIP1 and p27KIP1, and DACH1 partially derepressed p21CIP1 and p27KIP1 abundance but did not affect cyclin D1 expression (Fig. 3G). The analysis of DACH1 in oncogene-transduced MCF10A cells indicates that DACH1 consistently reduces oncogene-induced expression of cyclin D1. Collectively, these studies show that DACH1 inhibits oncogene-induced DNA synthesis, malignant morphological changes in Matrigel, and contact-independent growth as assessed by colony formation.
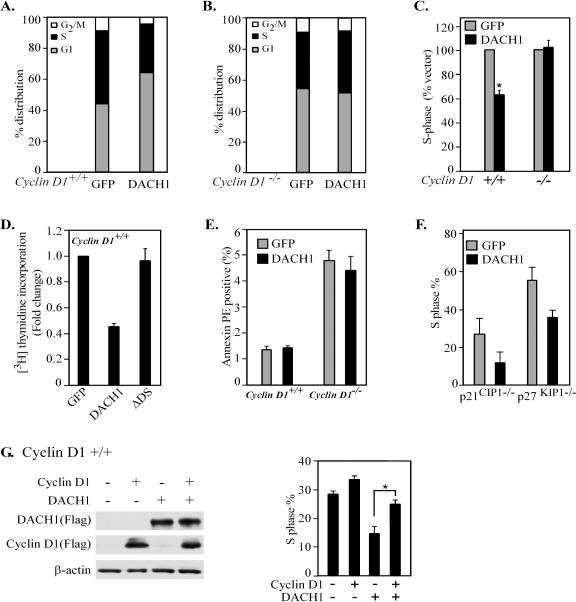
In view of the finding that DACH1 inhibited Ras and oncogenic ErbB2 induced cyclin D1 expression, we investigated the requirement for cyclin D1 in DACH1-mediated inhibition of DNA synthesis. To examine further the role of cyclin D1, p21CIP1 and p27KIP1 in DACH1-mediated cell cycle arrest, 3T3 cells were derived from mouse embryonic fibroblasts (MEFs) of mice with deletions of either the cyclin D1, the p21CIP1, or the p27KIP1 gene, and a comparison made with MEFs from sibling controls of the same strain background for each line. The cell cycle distribution was assessed by using cells transduced with MSCV-IRES-GFP or MSCV-DACH1-IRES-GFP through GFP FACS sorting. Western blot analysis confirmed the expression of DACH1. GDI was used as a protein loading control (see Fig. S2 in the supplemental material). DACH1 inhibited DNA synthesis in cyclin D1+/+ but not cyclin D1−/− cells assessed by either FACS analysis or tritiated thymidine uptake, and the inhibition of DNA synthesis by DACH1 required the DACH1 DS domain (Fig. 4A to D). Apoptosis rates were unaffected by DACH1 in either cyclin D1+/+ or cyclin D1−/− cells (Fig. 4E). Deletion of p21CIP1 or p27KIP1 altered the relative distribution of cells within the cell cycle. The proportion of cells in S phase inhibited by DACH1 in 3T3 cells with either the p21CIP1 or p27KIP1 gene deleted (Fig. 4F) was similar to that of the parental control cells. These studies indicate that cyclin D1, but not p21CIP1 or p27KIP1, are required for DACH1-mediated inhibition of DNA synthesis in 3T3 fibroblasts. To further test the role of cyclin D1 in DACH1-induced growth arrest, cyclin D1+/+ or cyclin D1−/− cells were infected with Flag-tagged cyclin D1, DACH1, or both cyclin D1 and DACH1, and Western blotting confirmed the expression of exogenous cyclin D1 and DACH1. Cell cycle analyses demonstrated the rescue of DACH1-mediated growth inhibition upon the expression of cyclin D1 (Fig. 4G).
FIG. 4.
DACH1 inhibition of DNA synthesis requires cyclin D1. (A to F) DACH1 regulation of cell cycle distribution by FACS analysis (A to C and F) or DNA synthesis using [3H]thymidine uptake (D) and apoptosis using annexin V staining in 3T3 cells derived from the MEFs of mice with either the cyclin D1 (A to E) or the p21CIP1or p27KIP1 (F) gene deleted compared to sibling controls of the same strain background. Cell cycle distribution was assessed using cells transduced with MSCV-IRES-GFP or MSCV-DACH1-IRES-GFP through GFP FACS sorting. Western blotting confirmed the expression of DACH1 with GDI as the loading control (see Fig. S2 in the supplemental material). (G) cyclin D1+/+ cells were transduced with expression vectors encoding Flag epitope-tagged cyclin D1 or DACH1. Western blot analysis demonstrated the expression of DACH1 and cyclin D1. FACS analyses demonstrated that cyclin D1 rescues the inhibition of DNA synthesis by DACH1.
DACH1 repression of the cyclin D1 promoter requires the AP-1/CRE DNA-binding site.
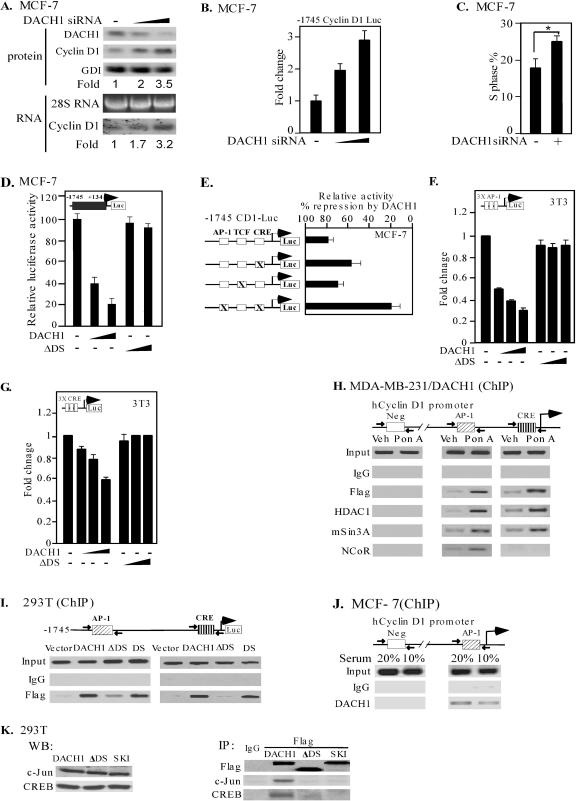
DACH1 inhibited DNA synthesis in the breast cancer epithelial cell lines MCF-7 and MDA-MB-231 and in NIH 3T3 cells. We sought to determine whether physiological levels of DACH1 regulated cyclin D1 abundance. The administration of siRNA directed to endogenous DACH1 in MCF-7 cells reduced DACH1 abundance and induced cyclin D1 protein and mRNA abundance (∼3-fold), indicating physiological levels of DACH1 repress cyclin D1 (Fig. 5A). Since DACH1 expression inhibited MCF-7 cell DNA synthesis (Fig. 2) and the abundance of cyclin D1 is rate limiting in DNA synthesis of MCF-7 cells (68), the role of DACH1 in regulating cyclin D1 was next assessed in these cells. To examine the role of endogenous DACH1 in regulating the cyclin D1 promoter, DACH1 siRNA was compared to control siRNA for its effect in regulating the cyclin D1 promoter. DACH1 siRNA induced the full-length cyclin D1 promoter threefold (Fig. 5B). Consistent with previous studies demonstrating that cyclin D1 abundance is rate limiting in MCF-7 cell DNA synthesis (40), DACH1 siRNA induced the S phase in MCF-7 cells (Fig. 5C). The cyclin D1 promoter was repressed in a dose-dependent manner by the expression of DACH1, and deletion of the DACH1 DS domain abrogated repression of the cyclin D1 promoter (Fig. 5D). In contrast to DACH1, the related Ski protein did not inhibit cyclin D1 promoter activity (not shown). Mutation of the AP-1 and CRE site of the cyclin D1 promoter reduced DACH1 repression by 80%, suggesting that these sequences together played a key role in repression by DACH1 (Fig. 5E). DACH1 repressed cyclin D1 promoter activity in 293T and NIH 3T3 cells and again repression required both the AP-1 and the CRE sites (data not shown). The cyclin D1 promoter AP-1 and CRE sites serve as both basal level and inducible enhancer elements; therefore, experiments were conducted to determine whether these elements were sufficient for repression by DACH1. The CRE and AP-1 sites were sufficient for repression by DACH1, and repression required the DACH1 DS domain (Fig. 5F and G).
FIG. 5.
DACH1 recruits HDAC to the AP-1/CRE site of the cyclin D1 promoter in the context of local chromatin. (A) MCF-7 cells treated with DACH1 or control siRNA. Endogenous cyclin D1 protein abundance and cyclin D1 mRNA level were detected; GDI was used as a loading control for Western blot analyses, and 18S RNA was used as a loading control for Northern blot analyses. (B) Cyclin D1 promoter activity assessed in MCF-7 cells transfected with DACH1 siRNA. Promoter activity is shown as the means ± the SEM for six separate transfections. (C) Mean S-phase analyses of DACH1 siRNA transfected MCF-7 cells. (D and E) Cyclin D1 promoter activity in cells transfected with increasing amounts of expression vector encoding DACH1 or a DACH1 mutant with the DS domain deleted (ΔDS) in MCF-7 cells. The vector control value was set as 100. Cyclin D1 promoter point mutants were compared for repression by DACH1. The data (means ± the SEM) are shown as the percent repression compared to the vector for eight separate experiments. (F and G) Heterologous reporters encoding the cyclin D1 AP-1 and CRE sites linked to the luciferase reporter gene were assessed for regulation by DACH1 in 3T3 cells. (H) ChIP assay using oligonucleotides directed to DNA sequences of the endogenous human cyclin D1 promoter AP-1 or CRE sites or a control DNA sequence at −3060. MDA-MB-231 cells expressing inducible Flag-tagged DACH1 are shown treated with PonA or vehicle. (I) 293T cells cotransfected with −1745 cyclin D1 promoter and Flag-tagged wild-type or mutant DACH1 expression vectors. ChIP assays were conducted using oligonucleotides directed to DNA sequences of the human cyclin D1 AP-1 or CRE sites. (J) ChIP analyses of endogenous DACH1 binding to the human cyclin D1 promoter comparing the −3060 region (negative) and the cyclin D1 promoter AP-1 site. (K) 293T cells were transfected with control or DACH1 or Ski expression vector and analyzed by Western blotting to detect c-Jun and CREB. Immunoprecipitation was conducted with antibodies directed to the Flag epitope of the DACH1, ΔDS, and Ski expression vectors and immunoprecipitation analyzed for c-Jun or CREB.
To determine the mechanisms by which DACH1 repressed the cyclin D1 promoter, ChIP assays were conducted to examine the occupancy of DACH1 at the endogenous human cyclin D1 promoter in human breast cancer cells. Comparison was made between the inducible DACH1 stable cell line and the vehicle control. Using oligonucleotides directed to the endogenous human cyclin D1 AP-1 and CRE sites, ChIP assays conducted with the anti-Flag antibody demonstrated the presence of DACH1 in the context of the local chromatin structure of the endogenous human cyclin D1 promoter upon the induction of DACH1 expression (Fig. 5H). HDAC1, mSin3A, and NCoR were recruited by DACH1 to form a multimeric repressor complex at the AP-1 site of the endogenous human cyclin D1 promoter. Amplification was not observed using oligonucleotides directed toward sequences within the −3060 region of the cyclin D1 promoter. No amplification was observed using either the vehicle control cells or immunoglobulin G in the ponasterone-inducible DACH1 stable cell line (Fig. 5H). To determine which region of the DACH1 protein was necessary for the formation of a DNA-associated complex at the cyclin D1 promoter in the context of its local chromatin structure, 293T cells were transfected with the human cyclin D1 luciferase promoter and wild-type or mutant DACH1 expression vectors. ChIP assay using the Flag antibody demonstrated that both wild-type DACH1 and the DS domain alone were recruited to the AP-1 and the CRE site, whereas the DS domain deleted mutant was defective in recruitment to the cyclin D1 promoter (Fig. 5I). Since DACH1 regulated DNA synthesis and cyclin D1 expression and promoter activity, we sought to determine whether endogenous DACH1 bound the cyclin D1 promoter in the context of local chromatin. Endogenous DACH1 was associated with the cyclin D1 promoter AP-1 site but was not identified at the −3060 region. Serum addition did not significantly alter DACH1 occupancy in ChIP assays (Fig. 5J).
Since DACH1 was recruited in a DNA sequence-specific manner to the cyclin D1 AP-1/CRE site and is not thought to bind a defined DNA sequence directly, we sought to determine whether DACH1 formed a complex with AP-1 proteins in cultured cells. Immunoprecipitation was conducted with the Flag antibody using cell extracts derived from 293T cells transfected with DACH1 or Ski, normalized for equal amounts of c-Jun and CREB by Western blotting (Fig. 5K). Equal amounts of DACH1 and c-Jun were coprecipitated with the Flag antibody; however, only DACH1 coprecipitated c-Jun and CREB. Deletion of the DACH1 DS domain abolished binding to c-Jun and CREB (Fig. 5K). These findings are consistent with a model in which DACH1 binds c-Jun and is recruited to an AP-1 site in the context of its local chromatin structure, requiring the DS domain of DACH1 to recruit the corepressors HDAC1, NCoR, and mSin3A to repress cyclin D1 gene expression.
DACH1 expression is regulated during mammary gland development and reduced in metastatic breast cancer.
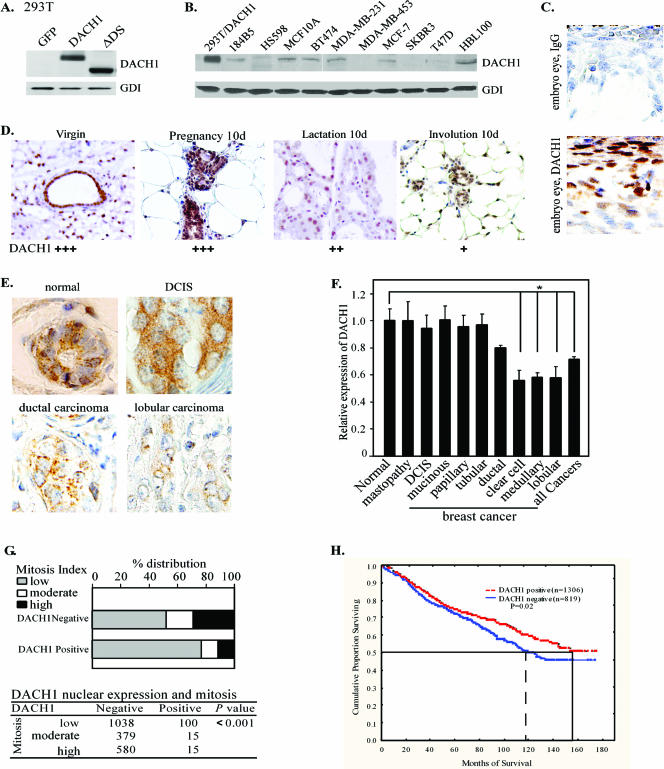
In view of the finding that endogenous DACH1 inhibited cyclin D1 expression and that DACH1 expression inhibited DNA synthesis in breast cancer cell lines, we investigated the distribution and physiological regulation of DACH1 in human and murine breast epithelium. DACH1 expression was examined in breast epithelium and breast cancer cell lines by immunohistochemical staining and Western blotting with a DACH1-specific antibody. The specificity of the antibody was confirmed through Western blotting of 293T cells transfected with expression vectors encoding the DACH1 wild-type or the DACH1 ΔDS domain (Fig. 6A). Immunoreactive DACH1 was detected in MCF-7, MCF-10A, and MDA-MB-231 cells and several other breast cancer cell lines (Fig. 6B). As a form of positive control for immunohistochemical staining, DACH1 was identified in the embryonic eye as previously described (8) (Fig. 6C). To determine whether Dach1 expression is regulated during normal physiological changes in the breast, Dach1 immunohistochemistry was assessed during murine mammary gland development and lactation. Dach1 expression was detectable in normal virgin mammary gland, increased at 10 days pregnancy, and decreased in lactation (day 10) and involution (day 10) (Fig. 6D). The homeodomain protein Six1, which functions in developmental networks including eye development in Drosophila, is expressed in mammary carcinoma but is absent or detected at low levels only in normal mammary tissues (15, 21). Six1 expression, although detectable in breast cancers, was weakly expressed in the normal murine mammary epithelium (see Fig. S3 in the supplemental material), as recently shown (15). The expression of Six1 was dramatically increased during pregnancy, coinciding with high expression of cyclin D1. We tested whether Six1 was capable of inducing cyclin D1. HEK293T cells were transiently transfected with the cyclin D1 promoter luciferase reporter and expression vectors encoding either Six1 or DACH1. A dose-dependent activation of the cyclin D1 promoter by Six1 and repression by DACH1 were observed (see Fig. S3B in the supplemental material), a finding consistent with recent findings that Six1 induces cyclin D1 promoter in a rhabdomyosarcoma cell line (65). Collectively, These findings are consistent with a model in which the relative abundance of the Eya/Six complex are under physiological control and may together contribute to the relative abundance of cyclin D1 in a given cell type.
FIG. 6.
DACH1 expression in human breast cancer cell lines, mouse mammary gland, and breast carcinoma tissues. (A) 293T cells were transiently transfected with control or DACH1 expression vector and analyzed by Western blotting with a DACH1 specific antibody. (B) Western blot for DACH1 abundance of breast epithelial cell lines, breast cancer cell lines, and control extracts of 293T cells transfected with DACH1 (lane 1). GDI was used as a loading control. (C) DACH1 expression in mouse embryonic eye was used as a positive control. (D) DACH1 immunohistochemistry during the murine mammary gland development of the virgin, at pregnancy (day 10), at lactation (day 10), and at involution (day 10). (E) Representative DACH1 expression in normal breast epithelium, ductal carcinoma in situ, or invasive human breast cancer samples using DACH1 specific antibodies. (F) Analysis of DACH1 expression in subcategory of human breast tissue microarray (*, P < 0.05). (G) Relationship of DACH1 expression to mitosis index. (H) DACH1 expression and prognosis of human breast carcinoma (n = 2,125 patients).
In order to clarify the DACH1 expression profile during the development of breast cancer and its prognostic significance, we analyzed DACH1 expression in a breast tissue microarray containing normal mammary epithelial cells, mastopathy, carcinoma in situ, and over 2,100 breast cancer samples for which detailed clinical-pathological characteristics and follow-up information were available. The results indicated that DACH1 was expressed in normal mammary epithelial (n = 81) but was significantly reduced in breast cancer (n = 2117). Similar DACH1 immunostaining was seen in mastopathy (n = 17) and ductal carcinoma and lobular carcinoma in situ (n = 63) compared to normal breast tissue (Fig. 6E; see also Fig. S4 in the supplemental material). The separate analysis of different tumor subtypes revealed significantly reduced expression in clear cell carcinoma or cribriform carcinoma (n = 75), lobular carcinoma (n = 300), and medullary carcinoma (n = 58) (Fig. 6F). DACH1 expression was inversely related with tumor diameter (P < 0.001), local tumor stage (P < 0.0001), and nodal metastasis (P = 0.04). Importantly, nuclear expression of DACH1 correlated inversely with mitosis (P < 0.001). The finding that DACH1 expression was reduced in invasive breast cancer compared to ductal carcinoma in situ prompted us to examine additional breast cancer tissue arrays that allowed direct comparison of invasive versus noninvasive breast cancer. Reduced DACH1 expression in invasive human breast cancer was confirmed with two additional distinct DACH1 antibodies on two additional distinct arrays (see Fig. S5 and S6 in the supplemental material). Semiquantitative immunostaining for DACH1 of invasive breast cancers (n = 124) demonstrated less intense and predominantly cytoplasmic, rather than nuclear, staining compared to either the normal breast epithelium (n = 23) or ductal carcinoma in situ (n = 19) (see Fig. S6 in the supplemental material). Within the invasive breast cancer group, immunostaining was less intense among patients with lymph node involvement (see Fig. S6 in the supplemental material). Analysis of the prognostic significance of DACH1 expression in breast cancers demonstrated that DACH1 expression is associated with favorable survival time in all cancer samples (Fig. 6G). Kaplan-Meier survival curves showed significantly better survival for DACH1-positive versus DACH1-negative patients (P = 0.02). This finding was confirmed by Cox proportional hazards regression, with only DACH1 as a predictor of survival. In univariable modes, DACH1-positive patients were estimated to have an ∼20% lower mortality than DACH1-negative patients (hazard ratio = 0.81, P = 0.005). Thus, DACH1 levels are regulated during normal mammary gland development in the epithelial cells, with reduced expression in invasive human breast cancer. When examined collectively across all breast cancer types, DACH1 abundance is significantly inversely correlated with patient survival.
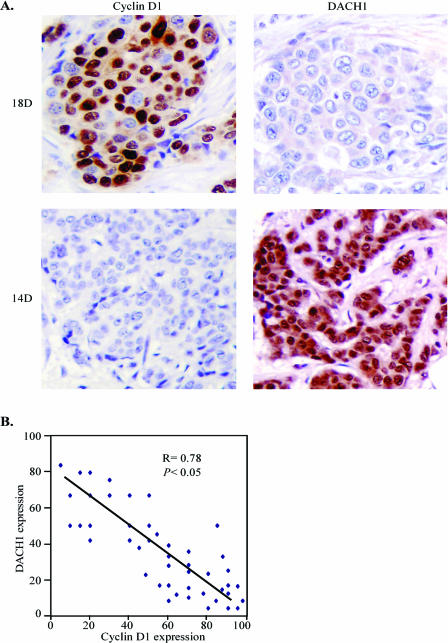
Since DACH1 repressed cyclin D1 expression in cultured cells, we examined the relationship between DACH1 and cyclin D1 expression by costaining for both proteins in human breast cancer samples. The expression of DACH1 and cyclin D1 was detected by immunohistochemistry. The semiquantitative analyses of 46 breast cancer tissues revealed that nuclear staining of DACH1 was inversely correlated with the expression of cyclin D1 (Fig. 7).
FIG. 7.
Relative expression of DACH1 and cyclin D1. (A) Representative immunostaining of cyclin D1 and DACH1 in paired breast cancer tissues. (B) Inverse correlation of cyclin D1 and DACH1 in breast cancers (n = 46).
DISCUSSION
Studies in Drosophila have identified a genetic network consisting of the eyeless, sine oculis (so), DNA-binding homeodomain factor, and the eyes absent (eya) and dachshund (dac) cofactors. Remarkably, each of these genes is sufficient for eye determination and function together in a molecular network during development. The mechanism by which Dachshund regulates cellular proliferation and differentiation was previously unknown. In the present studies, Dach1 expression was regulated during normal murine mammary gland development, and DACH1 was expressed in human breast cancer with reduced expression in metastatic breast cancer. DACH1 inhibited both ErbB2 and c-Myc-induced oncogenic morphogenesis in three-dimensional basement membrane cultures. DACH1 blocked contact-independent growth of human breast cancer cells in vivo. DACH1 inhibited ErbB2- and c-Myc-induced DNA synthesis without affecting cellular apoptosis. DACH1-dependent inhibition of DNA synthesis involved cyclin D1 and p21CIP1. Together, these studies demonstrate that the Drosophila cell fate determination network directly intersects the genetic determinants of cell cycle control through cyclin D1/p21CIP1.
Cyclin D1 is sufficient for mammary tumorigenesis in transgenic mice, collaborates in oncogenesis through pRb inactivation, and functions as a common downstream target induced by oncogenic signals (48, 64). In the present study, cyclin D1 abundance was repressed by DACH1. Reduction in DACH1 by siRNA induced cyclin D1 abundance and S-phase progression, suggesting that endogenous DACH1 is a physiological regulator of cyclin D1 in human breast cancer epithelial cells. Inhibition of DNA synthesis by DACH1 was abrogated by genetic deletion of cyclin D1, demonstrating the cyclin D1 gene is a functional genetic target of DACH1 function in mammalian cells. DACH1 reduced the proportion of MCF-7 cells in S phase. DACH1 inhibition of DNA synthesis, cyclin D1 abundance, and promoter activity required the conserved DS domain. Cyclin D1 abundance is limiting in the induction of MCF-7 cell S-phase progression, DNA synthesis, and cellular proliferation (46, 68). The cyclin D1 gene is known to encode a labile, growth factor- and oncogene-inducible protein required for mammary tumorigenesis induced by Ras and ErbB2. The present study extends these findings, demonstrating that cyclin D1 functions at a genetic interface with the pathways regulating cell fate determination.
The present study identified a novel mechanism by which DACH1 regulates gene expression. Although a direct DNA-binding capacity for DACH1 has not been identified, DACH1 is known to form part of a DNA-binding complex, with the So/Six homeodomain proteins providing the DNA specific binding capacity (37). We now show that DACH1 was recruited within chromatin complexes, with DNA sequence-specific binding capabilities provided by c-Jun or CREB. These findings raise the possibility that DACH1 may function as one of the previously hypothesized c-Jun repressor binding proteins (60). The DS domain of DACH1, which is predicted to form a helix-turn-helix structure, was required for recruitment to AP-1/CRE sites. Proteins conveying intrinsic or associated histone deacetylase (HDAC) activity (NCoR, Sin3A, and HDAC1) were also recruited with DACH1 to the sites of DACH1-dependent repression. The present study extends the mechanisms by which the cell fate determination factor complex regulates gene transcription by demonstrating the ability of DACH1 to use AP-1/CRE binding proteins as a molecular scaffold to recruit HDAC-containing repressor complexes to these sites. These findings are important in identifying the molecular mechanisms by which DACH1 may regulate the molecular genetic targets previously identified by high-density microarray analysis (62).
The three-dimensional culture of MCF10A on a reconstituted basement membrane results in polarized growth-arrested acinar-like spheroids, which are thought to recapitulate aspects of glandular architecture in vivo (18, 30). Oncogenic disruption of this morphogenetic process has identified distinct changes induced by different oncogenes. DACH1 reversed the ErbB2- and c-Myc-induced morphological changes of MCF10A cells in Matrigel. Furthermore, DACH1 inhibited DNA synthesis in cell lines harboring distinct activating mechanisms, through mutation in K-Ras (MDA-MB-231) or amplification of c-Myc (MCF-7 cells) (32). DACH1 inhibition of MCF10A-c-Myc-induced DNA synthesis correlated with the induction of p21CIP1, consistent with previous findings that c-Myc promotes growth in part through p21CIP1 (14) and Smad-dependent mechanisms (20).
c-Jun contributes to cellular proliferation and tumorigenesis (53). c-Jun promotes G1 phase cell cycle progression, and immunoneutralizing antibodies to c-Jun inhibit DNA synthesis (11). MEFs deficient in c-Jun exhibit a severe defect in proliferation due to extension of the G1 transition time (49). DACH1 repressed AP-1 activity, associating with c-Jun in the context of local promoter chromatin. DACH1 purified whole-cell extracts contained AP-1 binding activity, and DACH1 coprecipitated c-Jun through the DS domain. DACH1 repression of cyclin D1 expression, DNA synthesis, and cell survival are consistent with previous findings that AP-1 induces cyclin D1 transcription (4, 12, 61). Cyclin D1 contributes to cell survival since cyclin D1−/− MEF cells display both reduced rates of G1 phase progression and increased cellular apoptosis (3). DACH1, but not the related Ski, blocked serum-induced DNA synthesis in both mammary epithelial cells and in fibroblasts, demonstrating dissociable functions of these homologous proteins. The current model suggests DACH1, like Sno/Ski and NCoR (41, 57, 63), is recruited with DNA-binding proteins. DACH1, but not Ski, repressed the transcription of c-Jun. Recruitment to distinct DNA-binding proteins through the DS domain may contribute to the transcriptional specificity of the DACH1 versus Ski complexes (41).
Several recent findings suggest the RD pathway may regulate aberrant cellular growth and metastasis. Six1 expression is enhanced in metastatic rhabdomyosarcoma and contributes to cellular migration (66). Six6 functions as a tissue-specific repressor in association with Dach corepressors through Six6 binding sites (37), and the EWS/NOR1 translocation, implicated in extraskeletal myxoid chondrosarcoma, is repressed by Six3 (34). It has been hypothesized that Six1, which regulates DNA-damage-induced G2-phase arrest, may regulate migration through Lbx1h (66). In view of the reduction in DACH1 in metastatic human breast cancer and findings that cyclin D1 promotes cellular migration, through altering the adhesion and phosphorylation of tyrosine-phosphorylated paxillin (38, 45), it will be of interest to determine whether DACH1 contributes to cellular migration. The accumulating evidence that the RD pathway contributes to tumorigenesis through intersecting cell cycle control raises the possibility that this pathway may be a useful new therapeutic prospect.
Supplementary Material
Acknowledgments
We thank C. Chang, S. Ishii, J. Massague, G. H. Merlino, and M. G. Rosenfeld for plasmids; L. H. Augenlicht and A. Koff for cells; G. H. Nuckolls for the Dach1 antibody; and P. A. Furth for mouse mammary slides. We thank Dawn Scardino and Almeta Mathis for help with manuscript preparation.
This study was supported in part by awards from the Susan Komen Breast Cancer Foundation, the Breast Cancer Alliance, Inc., and NIH (R01CA70896, R01CA75503, R01CA86072, and R01CA86071 [R.G.P.]). Work conducted at the Kimmel Cancer Center was supported by the NIH Cancer Center Core grant P30CA56036 (R.G.P.). A.C. was supported by NIH grant EY12200, a Research to Prevent Blindness Career Development Award, and an American Cancer Society Junior Faculty Institutional Award. This project is funded, in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abate-Shen, C. 2002. Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer 2:777-785. [DOI] [PubMed] [Google Scholar]
- 2.Al-Kuraya, K., P. Schraml, J. Torhorst, C. Tapia, B. Zaharieva, H. Novotny, H. Spichtin, R. Maurer, M. Mirlacher, O. Kochli, M. Zuber, H. Dieterich, F. Mross, K. Wilber, R. Simon, and G. Sauter. 2004. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 64:8534-8540. [DOI] [PubMed] [Google Scholar]
- 3.Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundaram, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. [DOI] [PubMed] [Google Scholar]
- 4.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 5.Albanese, C., A. T. Reutens, B. Bouzahzah, M. Fu, M. D'Amico, T. Link, R. Nicholson, R. A. Depinho, and R. G. Pestell. 2000. Sustained mammary gland-directed, ponasterone A-inducible expression in transgenic mice. FASEB J. 14:877-884. [DOI] [PubMed] [Google Scholar]
- 6.Albanese, C., K. Wu, M. D'Amico, C. Jarrett, D. Joyce, J. Hughes, J. Hulit, T. Sakamaki, M. Fu, A. Ben-Ze'ev, J. F. Bromberg, C. Lamberti, U. Verma, R. B. Gaynor, S. W. Byers, and R. G. Pestell. 2003. IKKα regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol. Biol. Cell 14:585-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashton, A. W., G. Watanabe, C. Albanese, E. O. Harrington, J. A. Ware, and R. G. Pestell. 1999. Protein kinase Cδ inhibition of S-phase transition in capillary endothelial cells involves the cyclin-dependent kinase inhibitor p27Kip1. J. Biol. Chem. 274:20805-20811. [DOI] [PubMed] [Google Scholar]
- 8.Ayres, J. A., L. Shum, A. N. Akarsu, R. Dashner, K. Takahashi, T. Ikura, H. C. Slavkin, and G. H. Nuckolls. 2001. DACH: genomic characterization, evaluation as a candidate for postaxial polydactyly type A2, and developmental expression pattern of the mouse homologue. Genomics 77:18-26. [DOI] [PubMed] [Google Scholar]
- 9.Bentires-Alj, M., G. S., R. Chan, Z. C. Wang, Y. Wang, N. Imanaka, L. N. Harris, A. Richardson, B. G. Neel, and H. Gu. 2006. A role for the scaffolding adapter GAB2 in breast cancer. Nat. Med. 12:114-121. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard, C., K. Thieke, A. Maier, R. Saffrich, J. Hanley-Hyde, W. Ansorge, S. Reed, P. Sicinski, J. Bartek, and M. Eilers. 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18:5321-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bravo, R. 1990. Genes induced during the G0/G1 transition in mouse fibroblasts. Semin. Cancer Biol. 1:337-346. [PubMed] [Google Scholar]
- 12.Brown, J. R., E. Nigh, R. J. Lee, H. Ye, M. A. Thompson, F. Saudou, R. G. Pestell, and M. E. Greenberg. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 18:5609-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, R., M. Amouri, Z. Zhang, and G. Mardon. 1997. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91:893-903. [DOI] [PubMed] [Google Scholar]
- 14.Claassen, G. F., and S. R. Hann. 2000. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor-induced cell-cycle arrest. Proc. Natl. Acad. Sci. USA 97:9498-9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coletta, R. D., K. Christensen, K. J. Reichenberger, J. Lamb, D. Micomonaco, L. Huang, D. M. Wolf, C. Muller-Tidow, T. R. Golub, K. Kawakami, and H. L. Ford. 2004. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl. Acad. Sci. USA 101:6478-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debnath, J., K. R. Mills, N. L. Collins, M. J. Reginato, S. K. Muthuswamy, and J. S. Brugge. 2002. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111:29-40. [DOI] [PubMed] [Google Scholar]
- 18.Debnath, J., S. K. Muthuswamy, and J. S. Brugge. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30:256-268. [DOI] [PubMed] [Google Scholar]
- 19.Elenbaas, B., L. Spirio, F. Koerner, M. D. Fleming, D. B. Zimonjic, J. L. Donaher, N. C. Popescu, W. C. Hahn, and R. A. Weinberg. 2001. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15:50-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, X. H., Y. Y. Liang, M. Liang, W. Zhai, and X. Lin. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15 (Ink4B). Mol. Cell 9:133-143. [DOI] [PubMed] [Google Scholar]
- 21.Ford, H. L., E. N. Kabingu, E. A. Bump, G. L. Mutter, and A. B. Pardee. 1998. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc. Natl. Acad. Sci. USA 95:12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, M., M. Rao, K. Wu, C. Wang, X. Zhang, M. Hessien, Y. G. Yeung, D. Gioeli, M. J. Weber, and R. G. Pestell. 2004. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J. Biol. Chem. 279:29436-29449. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 24.Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter, and W. J. Gehring. 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125:2181-2191. [DOI] [PubMed] [Google Scholar]
- 25.Halder, G., P. Callaerts, and W. J. Gehring. 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267:1788-1792. [DOI] [PubMed] [Google Scholar]
- 26.Hammond, K. L., I. M. Hanson, A. G. Brown, L. A. Lettice, and R. E. Hill. 1998. Mammalian and drosophila dachshund genes are related to the ski proto-oncogene and are expressed in eye and limb. Mech. Dev. 74:121-131. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan, D., and R. Weinberg. 2000. The hallmarks of cancer. Cell 100: 57-70. [DOI] [PubMed] [Google Scholar]
- 28.Hermeking, H., C. Rago, M. Schuhmacher, Q. Li, J. F. Barrett, A. J. Obaya, B. C. O'Connell, M. K. Mateyak, W. Tam, F. Kohlhuber, C. V. Dang, J. M. Sedivy, D. Eick, B. Vogelstein, and K. W. Kinzler. 2000. Identification of CDK4 as a target of c-MYC. Proc. Natl. Acad. Sci. USA 97:2229-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold, S., M. Wanzel, V. Beuger, C. Frohme, D. Beul, T. Hillukkala, J. Syvaoja, H. P. Saluz, F. Haenel, and M. Eilers. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10:509-521. [DOI] [PubMed] [Google Scholar]
- 30.Jacks, T., and R. A. Weinberg. 2002. Taking the study of cancer cell survival to a new dimension. Cell 111:923-925. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S.-S., R.-G. Zhang, S. E. Braunstein, A. Joachimiak, A. Cvekl, and R. S. Hegde. 2002. Structure of the retinal determination protein dachshund reveals a DNA binding motif. Structure 10:787-789. [DOI] [PubMed] [Google Scholar]
- 32.Kozma, S. C., M. E. Bogaard, K. Buser, S. M. Saurer, J. L. Bos, B. Groner, and N. E. Hynes. 1987. The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res. 15:5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozmik, Z., P. Pfeffer, J. Kralova, J. Paces, V. Paces, A. Kalousova, and A. Cvekl. 1999. Molecular cloning and expression of the human and mouse homologues of the drosophila dachshund gene. Dev. Genes Evol. 209:537-545. [DOI] [PubMed] [Google Scholar]
- 34.Laflamme, C., C. Filion, J. A. Bridge, M. Ladanyi, M. B. Goldring, and Y. Labelle. 2003. The homeotic protein Six3 is a coactivator of the nuclear receptor NOR-1 and a corepressor of the fusion protein EWS/NOR-1 in human extraskeletal myxoid chondrosarcomas. Cancer Res. 63:449-454. [PubMed] [Google Scholar]
- 35.Lee, R. J., C. Albanese, M. Fu, M. D'Amico, B. Lin, G. Watanabe, G. K. Haines III, P. M. Siegel, M. C. Hung, Y. Yarden, J. M. Horowitz, W. J. Muller, and R. G. Pestell. 2000. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20:672-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, X., K. A. Oghi, J. Zhang, A. Krones, K. T. Bush, C. K. Glass, S. K. Nigam, A. K. Aggarwal, R. Maas, D. W. Rose, and M. G. Rosenfeld. 2003. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426:247-254. [DOI] [PubMed] [Google Scholar]
- 37.Li, X., V. Perrisi, F. Liu, D. W. Rose, and M. G. Rosenfeld. 2002. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 297:1180-1183. [DOI] [PubMed] [Google Scholar]
- 38.Li, Z., C. Wang, X. Jiao, Y. Lu, M. Fu, A. A. Quong, C. Dye, J. Yang, M. Dai, X. Ju, X. Zhang, A. Li, p. Burbelo, E. R. Stanley, and R. G. Pestell. 2006. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol. Biol. Cell 26:4240-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, H., D. C. Radisky, F. Wang, and M. J. Bissell. 2004. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J. Cell Biol. 164:603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukas, J., J. Bartkova, and J. Bartek. 1996. Convergence of mitogenic signalling cascades from diverse classes of receptors at the cyclin D-cyclin-dependent kinase-pRb-controlled G1 checkpoint. Mol. Cell. Biol. 16:6917-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, K., S. L. Stroschein, W. Wang, D. Chen, E. Martens, S. Zhou, and Q. Zhou. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes Dev. 13:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mardon, G., N. M. Solomon, and G. M. Rubin. 1994. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120:3473-3486. [DOI] [PubMed] [Google Scholar]
- 43.Massague, J. 2004. G1 cell-cycle control and cancer. Nature 432:298-306. [DOI] [PubMed] [Google Scholar]
- 44.Muthuswamy, S. K., D. Li, S. Lelievre, M. J. Bissell, and J. S. Brugge. 2001. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 3:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumeister, P., F. J. Pixley, Y. Xiong, H. Xie, K. Wu, A. Ashton, M. Cammer, A. Chan, M. Symons, E. R. Stanley, and R. G. Pestell. 2003. Cyclin D1 governs adhesion and motility of macrophages. Mol. Biol. Cell 14:2005-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagano, M., A. M. Theodorus, S. W. Tam, and G. F. Draetta. 1994. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 8:1627-1639. [DOI] [PubMed] [Google Scholar]
- 47.Pervin, S., R. Singh, and G. Chaudhuri. 2001. Nitric oxide-induced cytostasis and cell cycle arrest of a human breast cancer cell line (MDA-MB-231): potential role of cyclin D1. Proc. Natl. Acad. Sci. USA 98:3583-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pestell, R. G., C. Albanese, A. T. Reutens, R. J. Lee, J. Segall, and A. Arnold. 1999. The cyclins and cyclin dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocrinol. Rev. 20:501-534. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber, M., A. Kolbus, F. Piu, A. Szabowski, U. Mohle-Steinlein, J. Tian, M. Karin, P. Angel, and E. F. Wagner. 1999. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 13:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulze, A., K. Lehmann, H. B. Jefferies, M. McMahon, and J. Downward. 2001. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 15:981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seoane, J., H. V. Le, and J. Massague. 2002. Myc suppression of the p21 (Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419:729-734. [DOI] [PubMed] [Google Scholar]
- 52.Seton-Rogers, S. E., Y. Lu, L. M. Hines, M. Koundinya, J. LaBaer, S. K. Muthuswamy, and J. S. Brugge. 2004. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 101:1257-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 54.Shen, W., and G. Mardon. 1997. Ectopic eye development in drosophila induced by directed dachsund expression. Development 124:45-52. [DOI] [PubMed] [Google Scholar]
- 55.Sherr, C. J., and J. M. Roberts. 1995. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149-1163. [DOI] [PubMed] [Google Scholar]
- 56.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D. Peterson, Jr., R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 57.Tarapore, P., C. Richmond, G. Zheng, S. B. Cohen, B. Kelder, J. Kopchick, U. Kruse, A. E. Sippel, C. Colmenares, and E. Stavnezer. 1997. DNA binding and transcriptional activation by the Ski oncoprotein mediated by interaction with NFI. Nucleic Acids Res. 25:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlach, J., S. Hennecke, K. Alevizopoulis, D. Conti, and B. Amati. 1996. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 15:6595-6604. [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe, G., A. Howe, R. J. Lee, C. Albanese, I. W. Shu, A. N. Karnezis, L. Zon, J. Kyriakis, K. Rundell, and R. G. Pestell. 1996. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc. Natl. Acad. Sci. USA 93:12861-12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss, C., S. Schneider, E. F. Wagner, X. Zhang, E. Seto, and D. Bohmann. 2003. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 22:3686-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, K., Y. Yang, C. Wang, M. A. Davoli, M. D'Amico, A. Li, K. Cveklova, Z. Kozmik, M. P. Lisanti, R. G. Russell, A. Cvekl, and R. G. Pestell. 2003. DACH1 inhibits TGF-beta signaling through binding Smad4. J. Biol. Chem. 278:51673-51684. [DOI] [PubMed] [Google Scholar]
- 63.Xu, W., K. Angelis, D. Danielpour, M. M. Haddad, O. Bischof, J. Campisi, E. Stavnezer, and E. E. Medrano. 2000. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor. Proc. Natl. Acad. Sci. USA 97:5924-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 65.Yu, Y., E. Davicioni, T. J. Triche, and G. Merlino. 2006. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 66:1982-1989. [DOI] [PubMed] [Google Scholar]
- 66.Yu, Y., J. Khan, C. Khanna, L. Helman, P. S. Meltzer, and G. Merlino. 2004. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat. Med. 10:175-181. [DOI] [PubMed] [Google Scholar]
- 67.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwijsen, R. M. L., R. Klompmaker, E. B. H. G. M. Wientjens, P. M. P. Kristel, B. van der Burg, and R. J. A. M. Michalides. 1996. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol. Cell. Biol. 16:2554-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.