Abstract
To achieve accurate aminoacylation of tRNAs with their cognate amino acids, errors in aminoacylation are corrected by the “editing” mechanism in several aminoacyl-tRNA synthetases. Phenylalanyl-tRNA synthetase (PheRS) hydrolyzes, or edits, misformed tyrosyl-tRNA with its editing domain in the β subunit. We report the crystal structure of an N-terminal fragment of the PheRS β subunit (PheRS-βN) from the archaeon, Pyrococcus horikoshii, at 1.94-Å resolution. PheRS-βN includes the editing domain B3/4, which has archaea/eukarya-specific insertions/deletions and adopts a different orientation relative to other domains, as compared with that of bacterial PheRS. Surprisingly, most residues constituting the editing active-site pocket were substituted between the archaeal/eukaryal and bacterial PheRSs. We prepared Ala-substituted mutants of P. horikoshii PheRS for 16 editing-pocket residues, of which 12 are archaea/eukarya-specific and four are more widely conserved. On the basis of their activities, Tyr-adenosine was modeled on the B3/4-domain structure. First, the mutations of Leu-202, Ser-211, Asp-234, and Thr-236 made the PheRS incorrectly hydrolyze the cognate Phe-tRNAPhe, indicating that these residues participate in the Tyr hydroxy group recognition and are responsible for discrimination against Phe. Second, the mutations of Leu-168 and Arg-223, which could interact with the tRNA 3′-terminal adenosine, reduced Tyr-tRNAPhe deacylation activity. Third, the mutations of archaea/eukarya-specific Gln-126, Glu-127, Arg-137, and Asn-217, which are proximal to the ester bond to be cleaved, also reduced Tyr-tRNAPhe deacylation activity. In particular, the replacement of Asn-217 abolished the activity, revealing its absolute requirement for the catalysis.
Keywords: aminoacyl-tRNA synthetase, archaea, proofreading, x-ray crystallography
Accurate translation of the genetic code strongly depends on the specific couplings of cognate amino acid and tRNA pairs by aminoacyl-tRNA synthetases (aaRSs; refs. 1 and 2). Based on the two evolutionarily unrelated catalytic cores, the 20 aaRSs are divided into two classes (I and II), each consisting of 10 enzymes (3, 4). An aaRS generates an aminoacyl-tRNA by two reaction steps: the synthesis of an aminoacyl-adenylate from an amino acid and ATP, and the subsequent transfer of its aminoacyl moiety to the 3′-end of the tRNA.
Aminoacylation specificity generally depends on the discrimination of the cognate amino acid from the noncognate ones at the aminoacylation active site (aminoacylation site). However, several aaRSs fail to discriminate against structurally/chemically related but noncognate amino acids at the aminoacylation site and catalyze the misactivation and/or mischarging of tRNA with them. To overcome this problem, these aaRSs hydrolyze the misactivated aminoacyl-adenylate (pretransfer editing; ref. 5) or the misaminoacylated tRNA (posttransfer editing; see ref. 6) by using an extra “editing domain.” Three class I aaRSs, the isoleucyl- (7), leucyl- (8), and valyl- (9) tRNA synthetases, have a homologous domain that catalyzes the editing reactions. On the other hand, a posttransfer editing mechanism has been reported for the class II alanyl- (10), threonyl- (11), prolyl- (12), and phenylalanyl (13) tRNA synthetases [AlaRS, ThrRS, ProRS, and PheRS, respectively]. The editing domains in class II aaRSs are diverse, and four structural types have been described so far. AlaRS and ProRS have the “HxxxH” domain (11, 14) and the YbaK-like domain (12, 15, 16), respectively. Bacterial and eukaryal ThrRSs have the HxxxH domain (11, 14), whereas archaeal ThrRS has the N2A domain (17, 18). Recently, the B3/4 domain of the noncatalytic β subunit of PheRS was found to be responsible for editing Tyr-tRNAPhe (13, 19).
PheRS is a tetrameric enzyme consisting of two α/β heterodimers [(αβ)2]. A phylogenetic analysis of PheRS sequences revealed that PheRS family members can be classified into two types, the bacterial and archaeal/eukaryal types (20). Most studies on the PheRS editing mechanism have been carried out with bacterial enzymes. The crystal structure of PheRS from Thermus thermophilus is the only available PheRS structure (21–25). The PheRS structures complexed with its substrates revealed that the aminoacylation site resides in the α subunit (PheRS-α), and that the α and β subunits both interact with the tRNA. A mutational analysis of Escherichia coli PheRS located the editing site in the B3/4 domain of the β subunit (PheRS-β; ref. 13). Two amino acid residues, His-261 and Glu-334 (the numbering is according to the T. thermophilus sequence), were shown to be crucial for efficient editing activity. Recent structural work on the T. thermophilus PheRS complexed with l-tyrosine provided the structural details underlying the recognition of the Tyr moiety of the editing substrate by the editing pocket of the B3/4 domain (25).
On the other hand, the structural basis of the amino acid editing by the archaeal/eukaryal type of PheRSs has remained obscure, because no 3D structure is available from this version. According to sequence analysis, the archaeal/eukaryal type differs from the bacterial type in the following points. First, the archaeal/eukaryal type is shorter than the bacterial type by ≈250 aa, because the former lacks the B2 and C-terminal B8 domains that are conserved in the latter. Second, the B3/4 domain of the archaeal/eukaryal type shares no obvious sequence similarity with the bacterial counterpart containing the editing site. Despite these remarkable differences, yeast PheRS reportedly exhibits posttransfer editing activity against the misformed Tyr-tRNAPhe, and the B3/4 domain was presumed to be involved in the reaction (19, 26, 27).
Here we report the crystal structure of an N-terminal fragment of PheRS-β from the archaeon, Pyrococcus horikoshii. This structure includes the editing domain, and in combination with a detailed mutational analysis, provides insights into the substrate recognition mechanism for editing by archaeal/eukaryal PheRS.
Results
Structure of P. horikoshii PheRS-βN.
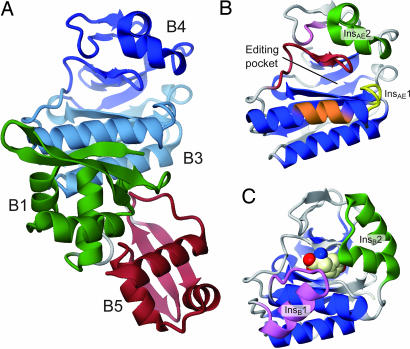
The β subunit of P. horikoshii PheRS-β consists of 556 amino acid residues and is presumed to be composed of four domains, B1, B3/4, B5, and B6/7. Because the linker connecting the B5 and B6/7 domains is highly susceptible to proteolysis (data not shown), the full-length protein was cleaved into two halves during crystallization. Only the N-terminal fragment (residues 1–348, PheRS-βN) was crystallized. Our attempts to obtain cocrystals of PheRS-β and tyrosine (or a Tyr-adenylate analog) were unsuccessful. The PheRS-βN structure was determined at a resolution of 1.94 Å (Fig. 1A). The asymmetric unit contains three nearly identical PheRS-βN molecules, and each of the refined models contains amino acid residues 2–348, comprising the complete B1, B3/4, and B5 domains.
Fig. 1.
Crystal structure of P. horikoshii PheRS-βN. (A) The overall structure of PheRS-βN is shown as a ribbon model. Green, B1 domain (residues 2–75); light blue, B3 subdomain (residues 84–149, 223–276); blue, B4 subdomain (residues 150–222); and red, B5 domain. (B and C) Ribbon models of the B3/4 domain from P. horikoshii (B) and T. thermophilus (C) PheRS. Blue, the central core region. (B) Yellow, InsAE1; green, InsAE2; orange, “φxxQ(E/D)xφ” motif; pink, “FxPL” motif; red, “SφPPφINxxx(T/S)xRφ” motif. (C) Purple, InsB1; green, InsB2. The l-tyrosine bound in the T. thermophilus PheRS-β editing pocket is shown as a cpk model. In the present paper, all graphic figures were produced by using CueMol (R. Ishitani) and POV-Ray (http://www.povray.org).
The B1 and B5 domains of PheRS-βN (Fig. 1A, green and red) superimpose well on those of T. thermophilus PheRS (rmsds for the Cα atoms are 1.7 and 1.6 Å, respectively). The B3/4 domain (Fig. 1 B and Fig. 6, which is published as supporting information on the PNAS web site) is composed of a central three-stranded antiparallel β-sheet sandwiched between a seven-stranded curved β sheet and two α helices (Fig. 1B, blue). It can be divided into the B3 and B4 subdomains, and the putative editing pocket is formed at the subdomain interface. The pocket is composed of α4-InsAE1-β6, β8-β9, and β11-β12-β13 and measures ≈18 Å in depth and ≈7 Å in diameter. The domain architecture is basically the same as that of the T. thermophilus enzyme, and they are superimposable with an rmsd of 1.9 Å for the Cα atoms (Fig. 1C). However, they differ from each other in their specific insertions/deletions (Fig. 6) and their orientations.
The B3/4 domain of T. thermophilus PheRS-β contains the bacteria-specific insertion InsB1 (residues 228–235; Fig. 1C, purple), which interacts with the B1 and B2 domains. It also possesses the helical unit InsB2 (residues 340–362; Fig. 1C, green), which is part of the Tyr-binding site. These elements are missing in P. horikoshii PheRS-βN. On the other hand, the B3/4 domain of P. horikoshii PheRS-βN possesses two additional elements, InsAE1 (residues 135–139; Fig. 1B, yellow) and InsAE2 (residues 177–198; Fig. 1B, green), which form a sidewall of the pocket, instead of InsB2.
The orientation of the B3/4 domain relative to the B1 and B5 domains significantly differs between the P. horikoshii and T. thermophilus PheRS-βs. A superposition of the B1 and B5 domains revealed that the P. horikoshii B3/4 domain is rotated by ≈30° as compared with the T. thermophilus counterpart, probably because P. horikoshii PheRS-β lacks the bacteria-specific InsB1 (Fig. 7 A and B, which is published as supporting information on the PNAS web site). In T. thermophilus, the B3/4 domain interacts with the B1 domain through InsB1, whereas in P. horikoshii, it interacts through the α4 and α7 helices. The superposition also suggests that the 3′ end of the tRNA can be translocated to the putative editing pocket of the P. horikoshii B3/4 domain (Fig. 7 C and D). These findings indicate that the B3/4-domain orientation relative to the tRNA 3′ end differs between the archaeal/eukaryal and bacterial types.
The Editing Pocket.
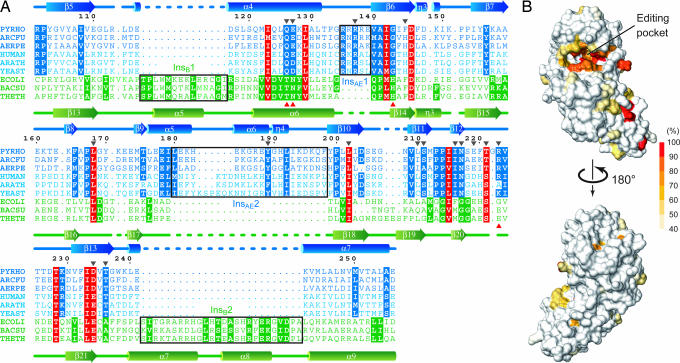
The P. horikoshii PheRS-βN structure provided structural guidelines to compare the B3/4 domains between the archaeal/eukaryal and bacterial types and thus facilitated the alignment of the corresponding amino acid residues (Fig. 2A). The structure-based alignment revealed that the editing domain sequences are highly diverse between the two types, except for a few conserved residues. In the archaeal/eukaryal type, 21 amino acid residues in the putative editing pocket are well conserved (Fig. 2B).
Fig. 2.
Sequence conservation of archaeal/eukaryal PheRS-βs. (A) Structure-based alignment of the B3/4 domain sequences. Three representative sequences are shown for each of archaeal (blue), eukaryal (light blue), and bacterial (green) PheRS-βs. The residues conserved in the entire PheRS-β family, those specific to the archaeal/eukaryal type, and those specific to the bacterial type are highlighted in red, blue, and green, respectively [residues with >50% conservation according to a similarity matrix (Gonnet PAM 250) are highlighted]. Black frames indicate type-specific insertions. Positions with introduced Ala substitutions and those postulated to be involved in hydrolysis in E. coli PheRS are indicated by black and red arrows, respectively. PYRHO, P. horikoshii; ARCFU, Archaeoglobus fulgidus; AERPE, Aeropyrum pernix; HUMAN, Homo sapiens; ARATH, Arabidopsis thaliana; YEAST, S. cerevisiae; ECOLI, E. coli; BACSU, Bacillus subtilis; and THETH, T. thermophilus. (B) Conservation mapping of archaeal/eukaryal PheRS-β. Extent of conservation of corresponding residues is mapped on the surface model of P. horikoshii PheRS-βN.
Among these conserved amino acid residues, Ile-123, Ile-142, Leu-168, Leu-202, Ile-215, Thr-221, and Asp-234 are also conserved in the bacterial type and correspond to the T. thermophilus residues Val-246, Met-260, Leu-286, Ile-300, Val-316, Ser-322, and Glu-334, respectively. In the T. thermophilus PheRS·tyrosine complex structure, the Met-260, Ile-300, and Val-316 side chains are buried inside, whereas the Val-246, Leu-286, and Glu-334 side chains, as well as the main-chain parts of Met-260 and Val-316, interact with (or are in the vicinity of) the bound tyrosine molecule (25).
On the other hand, the rest of the amino acid residues (Gln-126, Glu-127, Arg-137, Ala-141, Gly-143, Pro-167, Tyr-189, Ser-211, Pro-213, Pro-214, Ile-216, Asn-217, Arg-223, and Thr-236) are unique to the archaeal/eukaryal type. They belong to one of four archaea/eukarya-specific sequence signatures: the “φxxQ(E/D)xφ” motif (residues 123–129, where φ stands for a hydrophobic residue; Fig. 1B, orange), the “RxR” motif (InsAE1, residues 135–137; Fig. 1B, yellow), the “FxPL” motif (residues 165–168; Fig. 1B, pink), and the “SφPPφINxxx(T/S)xRφ” motif (residues 211–224; Fig. 1B, red). It is remarkable that T. thermophilus Thr-249, Asn-250, His-261, and Glu-323, which were postulated to be involved in the hydrolysis of the misformed product (25), are all replaced in the archaeal/eukaryal type (corresponding to Gln-126, Glu-127, Gly-143, and Arg-223, respectively, in P. horikoshii; Fig. 2A).
Thus, the editing pocket of archaeal/eukaryal PheRS-β drastically differs from the bacterial counterpart at the amino acid sequence level, although their editing domains share a similar global fold. This suggests there may be differences in the substrate recognition and/or catalytic mechanisms between the two types of editing domains.
Tyrosine Mischarging by PheRS Mutants.
We prepared 16 Ala-substituted PheRS mutants. Mutations were introduced to Gln-126, Glu-127, Arg-137, Phe-145, Leu-168, Tyr-189, Leu-202, Leu-210, Ser-211, Ile-216, Asn-217, Glu-219, Thr-221, Arg-223, Asp-234, and Thr-236 (Q126A, Ε127A, R137A, F145A, L168A, Y189A, L202A, L210A, S211A, I216A, N217A, E219A, T221A, R223A, D234A, and T236A, respectively; Fig. 2A) in the B3/4 domain of the β subunit. All of these residues are constituents of the editing pocket, and their side chains could be proximal to the editing substrate (Fig. 2B). In addition, we prepared PheRS with the replacement of Ala-141 with Trp (A141W), which would plug the editing pocket and prevent substrate binding. All enzymes were prepared in the background of the A463G mutation in the α subunit (αA463G). This mutation is equivalent to the αA294G mutation in E. coli PheRS and is expected to enlarge the aminoacylation site to enhance tyrosine misactivation (13, 28). The αA463G mutation has no effect on the editing activity of P. horikoshii PheRS, because the activities of the αA463G/βWT (WT) and αWT/βWT enzymes were the same in all experiments performed in the present study (data not shown).
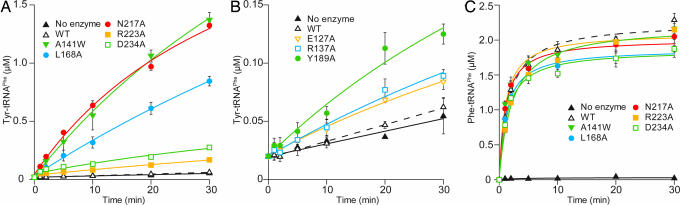
First, we tested whether these mutant PheRS enzymes misaminoacylate tRNAPhe with tyrosine (Fig. 3A and B). Significant Tyr-tRNAPhe formation was observed for the E127A, R137A, A141W, L168A, Y189A, N217A, R223A, and D234A mutants. On the other hand, the WT enzyme (Fig. 3B) and the other mutant enzymes examined (data not shown) displayed no detectable tyrosine mischarging. This result suggests that Glu-127, Arg-137, Leu-168, Tyr-189, Asn-217, Arg-223, and Asp-234 in the β subunit contribute to deacylation of the misformed product by PheRS.
Fig. 3.
Tyr-tRNAPhe formation by the P. horikoshii PheRS mutants. (A and B) Tyr-tRNAPhe formation by mutant PheRSs. The graphs in A and B represent the same experiment (note the different y axis scales). (C) Phe-tRNAPhe formation by mutant PheRSs. Values shown are the means of three independent experiments, with error bars representing ± 1 SD.
The A141W mutant resulted in the largest misincorporation of tyrosine; the mischarging activity of the N217A mutant is comparable to that of the A141W mutant. On the other hand, the tyrosine mischarging level was moderate for the L168A, R223A, and D234A mutants and relatively low for the E127A, R137A, and Y189A mutants. Note that the overall aminoacylation activities of these mutants are comparable to that of the WT enzyme (Fig. 3C and data not shown), indicating that mutations in the editing domain have no effect on aminoacylation at the aminoacylation site.
Mutants Deficient in Posttransfer Editing.
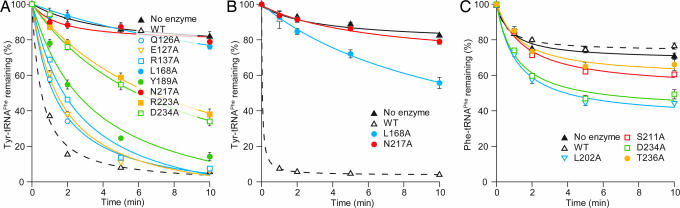
Next, we measured Tyr-tRNAPhe deacylation activity by these mutant PheRSs. Tyr-tRNAPhe was prepared by using the A141W mutant, which has the highest mischarging activity. All of the mutant PheRSs that misaminoacylated tRNAPhe with tyrosine also exhibited impaired deacylation activities. A severe defect in Tyr-tRNAPhe hydrolysis was observed for the L168A and N217A mutants (Fig. 4A). We performed the deacylation experiment with a 10-fold higher enzyme concentration for these two mutant PheRSs (Fig. 4B). Under these conditions, the L168A mutant showed a trace level of deacylation activity, whereas the deacylation level of the N217A mutant was comparable to that of nonenzymatic hydrolysis. This suggests that Asn-217 is essential for posttransfer editing by P. horikoshii PheRS. On the other hand, the deacylation deficiency was relatively moderate for the R223A and D234A mutants and small for the Q126A, E127A, R137A, and Y189A mutants (Fig. 4A). It is interesting that the F145A, L202A, L210A, S211A, T221A, and T236A mutants showed increased Tyr-tRNAPhe deacylation activities (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 4.
Posttransfer editing by the P. horikoshii PheRS mutants. (A) Tyr-tRNAPhe deacylation by the WT and mutant PheRSs. Reactions were performed with 10 nM PheRSs. (B) The same experiment as in A, but with a 10-fold higher concentration (100 nM) of the L168A and N217A mutants. (C) Misediting activities toward Phe-tRNAPhe.
Some residues investigated here are expected to be involved in Tyr recognition. Mutations of such residues would impair strict Tyr recognition and would allow the editing domain to recognize/hydrolyze an improper substrate, such as the cognate Phe-tRNAPhe. Therefore, we tested whether the present mutant PheRSs were able to hydrolyze the cognate Phe-tRNAPhe. As compared with WT, the L202A, S211A, D234A, and T236A mutants showed significant misediting activities (Fig. 4C), strongly suggesting that Leu-202, Ser-211, Asp-234, and Thr-236 are responsible for the discrimination against Phe.
Discussion
Categorization of the Residues Crucial for Editing by Archaeal/Eukaryal PheRS.
We determined the crystal structure of the P. horikoshii PheRS-β editing domain and constructed a structure-based alignment of the PheRS editing-domain sequences. By a mutational analysis based on structural data, we identified the residues important for the editing of misformed Tyr-tRNAPhe by the archaeal/eukaryal type of PheRSs.
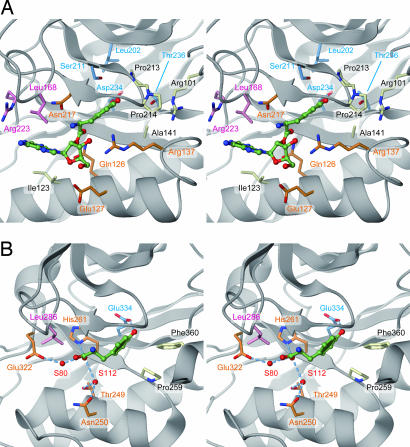
To examine the roles of the important residues in the editing activity, we created a plausible model of the P. horikoshii PheRS editing domain bound with the posttransfer substrate. In the model, the Tyr moiety was placed in the P. horikoshii PheRS editing pocket by superposing it on the T. thermophilus editing domain with the bound tyrosine molecule. The tRNA 3′-terminal adenosine moiety (A76) was attached to the tyrosine to produce the 2′-tyrosylated adenosine (Tyr-A76), as previously proposed for the T. thermophilus enzyme (25), and was energy-minimized (Fig. 5). Based on the model, amino acid residues important for P. horikoshii PheRS editing activity can be categorized into three groups: (i) residues close to the Tyr hydroxy group (Leu-202, Ser-211, Asp-234, and Thr-236; Fig. 5A, light blue), (ii) residues close to the A76 moiety (Leu-168 and Arg-223; Fig. 5A, pink), and (iii) residues close to the Tyr-A76 ester bond (Gln-126, Glu-127, Arg-137, and Asn-217; Fig. 5A, orange).
Fig. 5.
Model of Tyr-tRNAPhe recognition by archaeal/eukaryal PheRS-β. (A) The editing pocket of P. horikoshii PheRS-β. The posttransfer substrate (Tyr-A76, green) was placed on the basis of the structure of T. thermophilus PheRS complexed with tyrosine. The residues shown are proposed to participate in Tyr-A76 recognition. Light blue, residues close to the Tyr hydroxy group; pink, residues close to the A76 moiety; orange, residues close to the Tyr-A76 ester bond; white, residues not mutated. (B) The T. thermophilus PheRS editing pocket, bound with tyrosine for comparison. Water molecules are depicted by red spheres. Dashed lines indicate hydrogen-bonding interactions.
Specific Interaction with the Tyrosyl Moiety.
The docking model of the P. horikoshii PheRS editing domain and the posttransfer substrate suggests the Tyr ring could be accommodated in a hydrophobic pocket composed of Ala-141, Ile-142 (main chain), Gly-143, Pro-213, and Pro-214 (Fig. 5A, white). Ala-141, Gly-143, Pro-213, and Pro-214 are the archaea/eukarya-specific residues, and the interior of the pocket is shaped somewhat differently from that in T. thermophilus PheRS-β. This indicates that the orientation/conformation of the bound tyrosine is not exactly the same between the archaeal/eukaryal and bacterial types.
On the other hand, the Tyr hydroxy group could interact with Asp-234 (Fig. 5A, light blue), which is an acidic residue conserved throughout the entire PheRS-β family. The Ala substitution of this residue decreased Tyr-tRNAPhe hydrolysis activity by P. horikoshii PheRS. Consistently, the corresponding acidic amino acid residue is reportedly crucial for efficient editing by the E. coli and S. cerevisiae PheRSs (13, 19). In the structure of T. thermophilus PheRS complexed with tyrosine, the corresponding residue, Glu-334, forms a hydrogen bond with the Tyr hydroxy group (25). In the present study, we showed that the Asp-234 mutation allows P. horikoshii PheRS to hydrolyze Phe-tRNAPhe, evidence demonstrating that an acidic residue plays a crucial role in discrimination against Phe.
Misediting of Phe-tRNAPhe was also observed for PheRS variants with L202A, S211A, and T236A substitutions in the β subunit. Ser-211 and Thr-236 are archaea/eukarya-specific residues. They are close to Asp-234 and together form a local hydrophilic environment that would be favorable for accommodating the Tyr hydroxy group (Fig. 5A, light blue). The misediting activities of the S211A and T236A mutants suggest that the hydrophilic environment might also be important to exclude the hydrophobic Phe side chain. Leu-202 is the conserved hydrophobic residue in the PheRS-β family, and it contacts the Asp-234 and Ser-211 side chains. Leu-202 might indirectly contribute to the amino acid specificity of the editing site, probably by constraining the conformations of Asp-234 and Ser-211 (Fig. 5A, light blue).
It is interesting that the mutant PheRSs substituted at Phe-145, Leu-202, Leu-210, Ser-211, Thr-221, and Thr-236 in the β subunit display hyperactive Tyr-tRNAPhe deacylation. These residues constitute one half of the binding pocket for Tyr-A76 in our model. Substitutions of these residues would impair the substrate interaction because of the decreased complementarity. It is possible that the hyperactivity of the mutant enzymes might be explained by an increased rate of product release. The above-mentioned L202A, S211A, and T236A mutants are not only hyperactive in the canonical Tyr-tRNAPhe deacylation but also deficient in discrimination against Phe.
Interaction with the Adenosine Moiety.
Leu-168 in P. horikoshii PheRS-β is absolutely conserved in the family, and it protrudes from the tip of the β8-β9 hairpin (residues 161–176). Leu-168, together with Ile-123, Ile-216, Glu-219, and Arg-223, constitutes the opening of the editing pocket, which could accommodate the A76 portion of the editing substrate. The corresponding Leu residue in T. thermophilus (Leu-286) was proposed to be involved in the interaction with A76 (25). We demonstrated that the Ala substitution of Leu-168 in P. horikoshii PheRS-β causes a severe defect in PheRS editing activity (Figs. 3B and 4 A and B). Thus, the conserved Leu residue was confirmed to play a pivotal role in editing (Fig. 5A, pink).
Although the reduction in editing activity by the R223A mutation was moderate relative to that by the L168A mutation, Arg-223 might also interact with the adenosine. Arg-223 corresponds to T. thermophilus Glu-323, which points toward the inside of the pocket and therefore may participate in the cleavage of Tyr-A76 (25). In contrast, the Arg-223 side chain points away from the pocket and would be far from the Tyr-A76 ester bond. If Arg-223 underwent a conformational change upon substrate binding, then its guanidinium group could interact with the ester bond. Therefore, another possibility is that Arg-223 directly contributes to catalysis.
The Editing Active Site.
It is remarkable that the amino acid residues close to the Tyr-A76 ester bond dramatically differ between the archaeal/eukaryal and bacterial types of PheRS-βs. In the T. thermophilus PheRS·tyrosine structure, the α-carboxy group of the bound tyrosine is surrounded by Thr-249, Asn-250, His-261, and Glu-323. These residues were postulated to be involved in hydrolysis, because their arrangement is similar to that in peptidyl-tRNA hydrolase (25). In contrast, none of these residues is conserved in P. horikoshii PheRS-β. Our model suggests that Gln-126, Glu-127, Arg-137, and Asn-217 are close to the ester bond (Fig. 5A, orange). This type of catalytic site is not present in any known hydrolase structure (29).
The mutant PheRS with an Ala substitution of Asn-217 in the β subunit showed the largest decrease in Tyr-tRNAPhe hydrolysis activity among the mutants tested. The level of deacylation by this mutant enzyme is as low as that of nonenzymatic deacylation, indicating the essential role of this archaea/eukarya-specific Asn residue in catalysis. In bacterial PheRS-βs, a Gly residue is conserved at the corresponding position (Gly-318 in T. thermophilus). Archaea/eukarya-specific Gln-126 and Glu-127, corresponding to Thr-249 and Asn-250 in T. thermophilus, are also proximal to the ester bond. Mutational analysis revealed that Gln-126 and Glu-127 contribute to editing efficiency, although these residues do not seem to be essential for catalysis.
Arg-137, belonging to InsAE1 in P. horikoshii PheRS-β, was shown to be important for efficient editing. Arg-137 is one of the major constituents of the pocket interior, and its side chain is close to the α-amino group of the tyrosyl moiety in our model. The substitution of Tyr-189 from InsAE2, which stacks on Arg-137, also reduced PheRS editing activity. Tyr-189 seems to be required to stabilize the conformation/orientation of Arg-137, and thus the archaea/eukarya-specific insertions may cooperatively contribute to efficient editing.
Materials and Methods
Preparation of P. horikoshii PheRS-β.
The P. horikoshii PheRS-β gene was cloned into the vector pET-11a (Novagen, San Diego, CA). Selenomethionine (SeMet)-substituted PheRS-β was expressed in E. coli B384(DE3) at 37°C. The protein was purified by heat treatment (90°C, 14 min), followed by chromatography on a TOYOPEARL SuperQ-650M column (Tosoh, Tokyo), a Resource Q column (GE Healthcare, Little Chalfont, Buckinghamshire, U.K.), a Bio-Scale CHT10-I column (Bio-Rad, Hercules, CA), and a HiLoad 16/60 Superdex 200 column (GE Healthcare). The purified protein was concentrated to 36 mg/ml in 20 mM Tris·HCl buffer (pH 8.0) containing 200 mM NaCl, using a Vivaspin 10-kDa cutoff concentrator (Vivascience, Goettingen, Germany), and was stored at 4°C.
Crystallization, Data Collection, Structure Determination, and Refinement.
Crystallization was done by the microbatch method. A drop was prepared by mixing 1 μl of protein solution (36 mg/ml) with 1 μl of precipitant solution, containing 100 mM sodium citrate (pH 5.6) and 16.5% PEG 20k, and was covered by 15 μl of a 1:1 mixture of silicone and paraffin oils (Hampton, San Diego, CA). After incubation at 20°C for 1 wk, only the N-terminal fragment, PheRS-βN, was crystallized (see Results). Crystals were soaked in a cryoprotectant solution containing 100 mM sodium citrate (pH 5.6), 18.2% PEG 20k, and 20% glycerol, and were flash-frozen with liquid nitrogen for data collection.
The data set was obtained from the frozen SeMet-containing crystal at 100 K at the Photon Factory beamline NW12 of the High-Energy Accelerator Research Organization (KEK, Tsukuba, Japan) and was processed with HKL2000 (30). The crystal belongs to the monoclinic space group C2, with unit-cell parameters of a = 142.8, b = 82.0, c = 139.1 Å, β = 116.79°. The structure was solved by the multiwavelength anomalous dispersion method, by using SOLVE/RESOLVE (31, 32). The refinement was carried out by using REFMAC5 (33) and was completed with Rwork = 18.8% and Rfree = 23.1%. The stereochemistry of the model was assessed with PROCHECK (34). The data collection and structure refinement statistics are summarized in Table 1, which is published as supporting information on the PNAS web site.
Sequence Analysis.
See Supporting Text, which is published as supporting information on the PNAS web site.
Preparation of PheRS-α/β Mutants.
The P. horikoshii PheRS-α and -β genes were each subcloned, and single amino acid substitutions were introduced with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The coexpression system of the N-terminally His-tagged PheRS-α and untagged PheRS-β is based on the pETDuet-1 vector (Novagen). PheRS (α/β)2 was overexpressed in E. coli BL21(DE3) at 37°C and was purified by heat treatment (85°C, 10 min) followed by Ni-Sepharose (GE Healthcare) chromatography.
Aminoacylation and Deacylation Assays.
P. horikoshii tRNAPhe was transcribed in vitro. The aminoacylation reaction was performed at 65°C in 100 mM Hepes-Na buffer (pH 7.2) containing 30 mM KCl, 10 mM MgCl2, 1.4 μg/ml BSA, 5 μM tRNAPhe, 4 mM ATP, 30 μM [14C]-amino acid (tyrosine or phenylalanine), and 100 nM PheRS. The deacylation reaction was performed at 37°C in 100 mM Hepes-Na buffer (pH 7.2) containing 30 mM KCl, 10 mM MgCl2, 1.4 μ/ml BSA, 200 nM [14C]-aminoacyl-tRNA (Tyr-tRNAPhe or Phe-tRNAPhe), and PheRS (10 or 100 nM in the Tyr-tRNAPhe deacylation; 200 nM in the Phe-tRNAPhe deacylation). Phe-tRNAPhe and Tyr-tRNAPhe were prepared by using the αWT/βWT and αA463G/βA141W PheRSs, respectively. In each experiment, an aliquot of the reaction was removed at various incubation times, and the reaction was quenched with trichloroacetic acid. The radioactivity of the acid-insoluble fraction was quantitated by scintillation counting. For optimization of the enzyme reaction efficiencies, we examined the temperature (37–65°C) and PheRS concentration (10–300 nM).
The Docking Model.
See Supporting Text.
Supplementary Material
Acknowledgments
We thank Dr. T. Yanagisawa for supporting data collection at the Photon Factory. This work was supported by grants-in-aid for Science Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the RIKEN Structural Genomics/Proteomics Initiative; and the National Project on Protein Structural and Functional Analyses of MEXT.
Abbreviations
- aaRS
aminoacyl-tRNA synthetase
- PheRS
phenylalanyl-tRNA synthetase
- PheRS-β
phenylalanyl-tRNA synthetase β-subunit.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2CXI).
References
- 1.Freist W, Sternbach H, Cramer F. Eur J Biochem. 1988;173:27–34. doi: 10.1111/j.1432-1033.1988.tb13962.x. [DOI] [PubMed] [Google Scholar]
- 2.Martinis SA, Plateau P, Cavarelli J, Florentz C. Biochimie. 1999;81:683–700. doi: 10.1016/s0300-9084(99)80126-6. [DOI] [PubMed] [Google Scholar]
- 3.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 4.Cusack S. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 5.Eldred EW, Schimmel PR. J Biol Chem. 1972;247:2961–2964. [PubMed] [Google Scholar]
- 6.Fersht AR. Biochemistry. 1977;16:1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 7.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 8.Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grotli M, et al. Mol Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 9.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 10.Tsui WC, Fersht AR. Nucleic Acids Res. 1981;9:4627–4637. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dock-Bregeon A, Sankaranarayanan R, Romby P, Caillet J, Springer M, Rees B, Francklyn CS, Ehresmann C, Moras D. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 12.Beuning PJ, Musier-Forsyth K. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy H, Ling J, Irnov M, Ibba M. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beebe K, Ribas De Pouplana L, Schimmel P. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahel I, Korencic D, Ibba M, Soll D. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. J Biol Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- 17.Korencic D, Ahel I, Schelert J, Sacher M, Ruan B, Stathopoulos C, Blum P, Ibba M, Soll D. Proc Natl Acad Sci USA. 2004;101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beebe K, Merriman E, Ribas De Pouplana L, Schimmel P. Proc Natl Acad Sci USA. 2004;101:5958–5963. doi: 10.1073/pnas.0401530101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy H, Ling J, Alfonzo J, Ibba M. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 20.Woese CR, Olsen GJ, Ibba M, Soll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosyak L, Reshetnikova L, Goldgur Y, Delarue M, Safro MG. Nat Struct Biol. 1995;2:537–547. doi: 10.1038/nsb0795-537. [DOI] [PubMed] [Google Scholar]
- 22.Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M. Structure (London) 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 23.Reshetnikova L, Moor N, Lavrik O, Vassylyev DG. J Mol Biol. 1999;287:555–568. doi: 10.1006/jmbi.1999.2617. [DOI] [PubMed] [Google Scholar]
- 24.Fishman R, Ankilova V, Moor N, Safro M. Acta Crystallogr D. 2001;57:1534–1544. doi: 10.1107/s090744490101321x. [DOI] [PubMed] [Google Scholar]
- 25.Kotik-Kogan O, Moor N, Tworowski D, Safro M. Structure (Cambridge, UK) 2005;13:1799–1807. doi: 10.1016/j.str.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Lin SX, Baltzinger M, Remy P. Biochemistry. 1983;22:681–689. doi: 10.1021/bi00272a024. [DOI] [PubMed] [Google Scholar]
- 27.Lin SX, Baltzinger M, Remy P. Biochemistry. 1984;23:4109–4116. doi: 10.1021/bi00313a015. [DOI] [PubMed] [Google Scholar]
- 28.Ibba M, Kast P, Hennecke H. Biochemistry. 1994;33:7107–7112. doi: 10.1021/bi00189a013. [DOI] [PubMed] [Google Scholar]
- 29.Porter CT, Bartlett GJ, Thornton JM. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R, MacArthur M, Moss D, Thornton J. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.