Abstract
Water motion at protein surfaces is fundamental to protein structure, stability, dynamics, and function. By using intrinsic tryptophans as local optical probes, and with femtosecond resolution, it is possible to probe surface-water motions in the hydration layer. Here, we report our studies of local hydration dynamics at the surface of the enzyme Staphylococcus nuclease using site-specific mutations. From these studies of the WT and four related mutants, which change local charge distribution and structure, we are able to ascertain the contribution to solvation by protein side chains as relatively insignificant. We determined the time scales of hydration to be 3–5 ps and 100–150 ps. The former is the result of local librational/rotational motions of water near the surface; the latter is a direct measure of surface hydration assisted by fluctuations of the protein. Experimentally, these hydration dynamics of the WT and the four mutants are also consistent with results of the total dynamic Stokes shifts and fluorescence emission maxima and are correlated with their local charge distribution and structure. We discuss the role of protein fluctuation on the time scale of labile hydration and suggest reexamination of recent molecular dynamics simulations.
Keywords: protein hydration, femtosecond dynamics, protein fluctuation, selective mutation
From the laboratories of the senior authors of this study (A.H.Z. and D.Z.) (1–11), there has been a series of reports regarding the time and length scales of the water layer around protein surfaces. These studies were for proteins subtilisin Carlsberg (2), monellin (3), phospholipase A2 (5), melittin (9), and human serum albumin (8, 10). A theoretical model was developed to take into account the exchange with bulk water (4, 12), and the dynamics are consistent with molecular dynamics (MD) simulations of residence times (13–16) on time scales from femtoseconds to picoseconds. Earlier NMR studies have reported hydration dynamics (residence times) in the subnanosecond regime (17–20), but, more recently, a claim has been made that water motions at protein surfaces are ultrafast compared with bulk water, only slowing down by a factor of two to three (21, 22). This <10-ps range would imply that the observed long-time hydration dynamics in tens of picoseconds are due to protein side-chain relaxation (22, 23). In our earlier studies (6), we addressed in detail this issue and the reasons for dominance of hydration dynamics. To quantify the contribution of side-chain motions to total solvation on the time scale of hydration, we must carefully alter the local structure while maintaining the same tryptophan site.
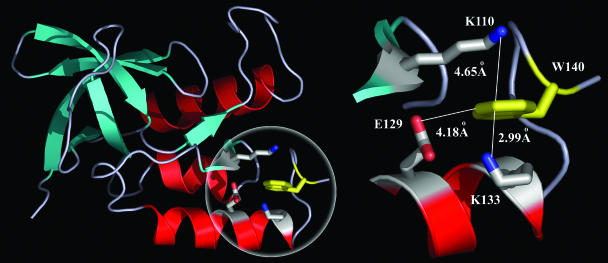
In this contribution, we report the effect of mutation (four mutants on three site selections and the WT) on hydration of the enzyme Staphylococcus nuclease (SNase). Fig. 1 shows the x-ray structure of the protein, consisting of three α-helices and a five-stranded β-barrel with a total of 149 amino acids (24). The only single tryptophan residue (W140) has one edge exposed to the surface and inserts inside the protein to form a hydrophobic cluster at the C terminus, which was found to be essential to protein foldability, stability, and activity (25). Three surface charged residues, K110, K133, and E129 (Fig. 1), surround W140 within 7 Å. Using an alanine scan, we replaced each charged residue with hydrophobic alanine one at a time by site-directed mutagenesis. We also mutated K110 into the polar residue cysteine. We measured the Stokes shifts, solvation dynamics, and local rigidity of the WT and the mutants, using the intrinsic W140 as the molecular optical probe and with femtosecond temporal resolution. From these results, we determined the hydration dynamics (from their correlation functions) and their dependence on charge distribution.
Fig. 1.
Protein structure with sites of hydration. (Left) X-ray crystallographic structure of WT SNase (Protein Data Bank ID code 1SNO). The single tryptophan W140 (in yellow) is sandwiched between K110 and K133, with one edge exposed to the protein surface. (Right) The local configuration around W140 with three charged residues (K110, K133, and E129) in close proximity (<5 Å).
Results and Discussion
Fluorescence Stokes Shifts and Femtosecond-Resolved Transients.
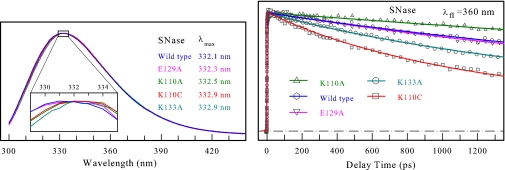
The steady-state fluorescence emission of the WT and mutants is shown in Fig. 2, together with the transients gated at the red-side emission (360 nm). All emission peaks are at ≈332.5 ± 0.5 nm, and no significant emission shift by the mutation of charged residues was observed. These striking results reveal that the three neighboring charged residues make a negligible contribution to solvation of the excited W140. These observations are consistent with the general trend that the maximum of tryptophan emission mainly depends on the location (i.e., the extent of its exposure to surface water) and not on its neighboring residues (26). Thus, the observed Stokes shift cannot be due to neighboring residues' (with charges) solvation, indicating relatively immobile charged side chains. The Stokes shift is dominantly due to hydration. From the x-ray structure (Fig. 1), we find that W140 is sandwiched between K110 and K133 (4.65 and 2.99 Å, respectively), forming two cation–π interactions. K133 is only 3.50 Å from E129, resulting in formation of a salt bridge. This unique structural motif with these strong electrostatic interactions is probably the origin of local protein rigidity around W140, which is consistent with our observation, as discussed below.
Fig. 2.
Emission spectra and transients of the proteins. (Left) Normalized steady-state fluorescence spectra of WT SNase and four mutants, E129A, K110A, K110C, and K133A. The mutant K110A has the strongest fluorescence intensity, and K110C has the weakest one because of the quenching by C110. Note that no significant emission shifts were induced by the mutation of charged residues. (Right) Normalized femtosecond-resolved fluorescence transients of W140 from WT SNase and all four mutants at the red-side emission (360 nm).
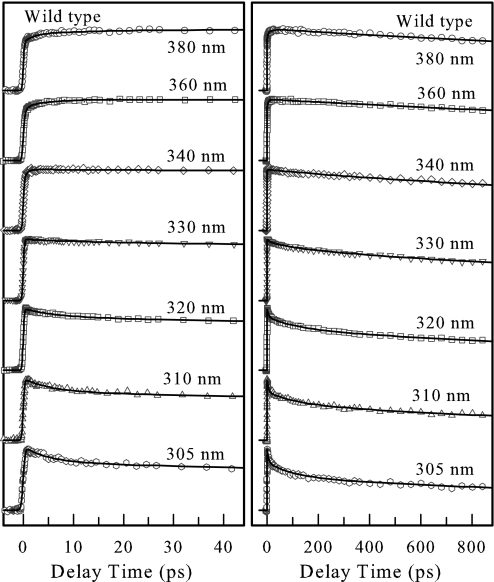
Fig. 3 shows the femtosecond-resolved fluorescence transients of W140 in the WT for several typical wavelengths, from the blue to red side, and for >10 gated emissions. The overall decay dynamics is significantly slower than that of aqueous tryptophan in a similar buffer solution (7). Clearly, the ultrafast decay components (<1 ps) observed in tryptophan solution were not observed at the blue side for the protein. Besides the lifetime contributions, solvation components for all blue-side transients are well represented by a double-exponential decay with time constants ranging from 4.9 to 12 ps and from 102 to 130 ps. For the red-side emission, the rise occurs in the range of 1.0–4.8 ps and is clearly present in all transients. For the four mutants, K133A, K110C, K110A, and E129A, besides the different lifetime emission contributions, the transients showed similar solvation patterns from the blue side to the red side (see Fig. 6, which is published as supporting information on the PNAS web site). Among the four mutants, K110A has the shortest decay time for the two solvation components, followed by E129A. The mutants K133A and K110C have similar temporal behaviors as the WT.
Fig. 3.
Normalized femtosecond-resolved fluorescence transients of W140 from WT SNase on short (Left) and long (Right) time scales with a series of gated fluorescence emissions. Note that no ultrafast decay components (<1 ps) were observed.
Solvation Correlation Functions.
Using the methodology we recently developed (7), we constructed the overall and lifetime-associated, femtosecond-resolved emission spectra (FRES) for the WT and all four mutants. By fitting these FRES to a lognormal function, we deduced the femtosecond-resolved overall emission maxima νs and lifetime-associated emission maxima νl (see Fig. 7, which is published as supporting information on the PNAS web site). The obtained total dynamic Stokes shifts are 850 ± 50 cm−1 for all of the proteins, and the emission maxima at t = 0, νs (0) are all near 321 nm, consistent with a previous report (27) giving the emission peak of several proteins as 320 nm at 2 K and 300-nm excitation. MD simulations of the Stokes shift of tryptophans in four proteins also gave an emission maximum of ≈320 nm at t = 0 (28). The possible contribution of vibrational relaxation (9–11) is negligible, and thus the observed total Stokes shift is dominantly from the local solvation.
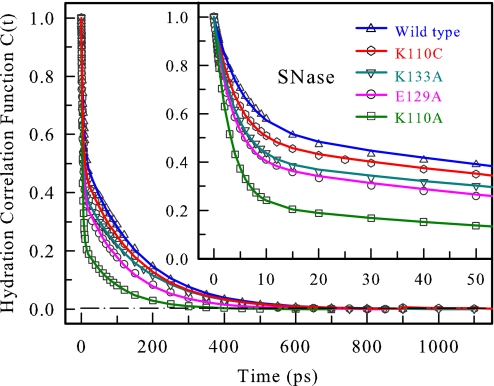
Using c(t) = [νs(t) − νl(t)]/[νs(0) − νl(0)] (7), we constructed all solvation correlation functions, and the obtained results are shown in Fig. 4 and summarized in Table 1. All solvation correlation functions can be represented by a double-exponential decay. For the WT, the time scales are 5.1 ps with 46% of the total amplitude and 153 ps (with 54% of the total amplitude); for K110C, 4.2 ps (51%) and 149 ps (49%); for K133A, 3.9 ps (59%) and 157 ps (41%); for E129A, 3.5 ps (60%) and 124 ps (40%); for K110A, 3.1 ps (77%) and 96 ps (23%). Overall, all four mutants show faster temporal behaviors than the WT, and all of the second long solvation times are within ≈100–150 ps. Note that no ultrafast solvation dynamics in <1 ps were observed for the proteins studied here.
Fig. 4.
Hydration correlation functions probed by tryptophan W140 for WT SNase and four mutants, K110C, K133A, E129A, and K110A. Inset shows the correlation functions in the short time range. Note the similarity of the time scales and the difference in amplitudes.
Table 1.
Results obtained from the hydration correlation functions c(t) of WT SNase and four mutant proteins
| SNase | τ1 | τ2 | c1 | c2 |
|---|---|---|---|---|
| WT | 5.1 | 153 | 0.46 | 0.54 |
| K110C | 4.2 | 149 | 0.51 | 0.49 |
| K133A | 3.9 | 157 | 0.59 | 0.41 |
| E129A | 3.5 | 124 | 0.60 | 0.40 |
| K110A | 3.1 | 96 | 0.77 | 0.23 |
All of the hydration correlation functions were fitted with c(t) = c1e−t1/τ1 + c2e−t2/τ2, where c1+ c2 = 1. The time constants are in picoseconds.
Hydration vs. Charged Side-Chain Solvation.
The constructed solvation correlation function is the response of the local environment around W140 to the sudden dipole change from the ground state to the excited state. In principle, the response results from both surrounding water molecules and neighboring protein polar/charged residues (and protein peptide bonds). If the second long-time solvation came only from protein side-chain motions (22, 23) (in this case, from three neighboring charged residues), we should observe significantly different solvation time scales and amplitudes of the second solvation components. However, for the mutants K133A and K110C, we observed a long relaxation time (149 and 157 ps) similar to the WT (153 ps). Overall, the obtained solvation dynamics for the four mutants become faster with the decrease in the local charge distribution around W140 (Fig. 4), consistent with MD simulations that show that water has longer residence times around charged residues than near hydrophobic side chains (15, 29). The observed long time scale of ≈100–150 ps also is consistent with our recent observation in the melittin tetramer and human serum albumin (E and F isomers); in both cases (9, 10), the probe tryptophan has a similar emission maxima (≈332 nm) and charge surroundings as in SNase. Thus, the obtained solvation correlation functions essentially reflect the local hydration dynamics at the protein surfaces.
For SNase, W140 has one edge exposed to the surface. Because of the local strong electrostatic interactions of the cation–π stacking/salt bridge and the tight packing of the hydrophobic cluster near the C terminus, the local structure is more rigid and W140 is less mobile. We studied the femtosecond-resolved rotational dynamics of W140 by the measurement of anisotropy changes with time for all of the proteins (see Fig. 8, which is published as supporting information on the PNAS web site). We observed that all anisotropy dynamics dominantly decay in nanoseconds with a small decay component at the early time. The long nanosecond dynamics represents the whole-protein tumbling motion. The early small decays, on the time scales from 62 to 464 ps, result from the local wobbling motions with semiangles of 11–19° (30). The time scales of 62–464 ps do not correlate with those of solvation dynamics (Fig. 4). All of these findings suggest a relatively small local fluctuation; the protein structure around W140 does not undergo large conformation changes in the time window of our measurements (≈1.3 ns). These results from anisotropy dynamics are consistent with those obtained above from site-selected mutagenesis. The conclusion is indeed consistent with the results of the steady-state fluorescence emission maxima (i.e., the dynamic Stokes shifts of the WT and four mutant proteins are independent of the three charged residues K110, K133, and E129). However, these findings are contrary to those of recent MD simulations, which indicate that, for this protein (SNase), K110 and K133 effects combine to make a very large contribution to the total Stokes shift (28).
Molecular Mechanism of Surface-Water Hydration.
To summarize, the studies of mutations, steady-state fluorescence, anisotropy measurements, and biphasic behavior of dynamic Stokes shifts exclude charged side-chain solvation as a significant contributor. As discussed in ref. 6, in all previous work reported from our laboratories, the robustness of the temporal behavior for different proteins of different sequence and structure supported the above conclusion, which is now reexamined with site-selected mutagenesis of the same protein. We did not observe a significant ultrafast component (<1 ps) for any of the proteins studied here, contrary to the initial dynamics obtained from recent MD simulations (13–15, 23). Given that we were able to resolve the 200-fs solvation dynamics of tryptophan in bulk water (1, 7), we should have been able to detect subpicosecond dynamics present in the proteins studied. The absence of subpicosecond solvation suggests that the force field used in the MD simulations might be too flexible and should be reexamined to account for relevant interactions between the water network and protein.
Previous MD simulations gave a biphasic distribution of residence times with two discrete time scales (13–15), which show remarkable similarity with our obtained hydration dynamics. Two distinct residence times represent two different ways for water to escape hydration sites. The fast route is through consecutive hopping by breaking and making of the neighboring hydrogen bonds, and the slow path is through direct exchange with bulk water by disrupting the local water structure: hence, the distribution in residence times (31). Along the slow path, water has longer residence times at concave surfaces than around convex patches, and around charge residues than near hydrophobic side chains (32). Our recent MD simulations of nonequilibrium excited-state trajectories of tryptophan in proteins give a similar dynamic pattern of two distinct time scales in addition to a subpicosecond component (T. Li, A. Hassanali, D.Z., and S. Singer, unpublished data). The initial dynamics occurs in a few picoseconds and represents local water librational/rotational motions as discussed before. The long-time solvation is also observed in tens of picoseconds from local water collective motions but is strongly assisted by local protein fluctuations. Only if the local protein structural rigidity is lost by conformational changes does the protein contribute to the total solvation energy in a significant way (33). It should be noted that static spectral shifts reflect the internal and static Stark perturbation, which attractively or repulsively alters the energy levels at equilibrium, whereas dynamical solvation measures the fluctuations in energy as a function of time (6).
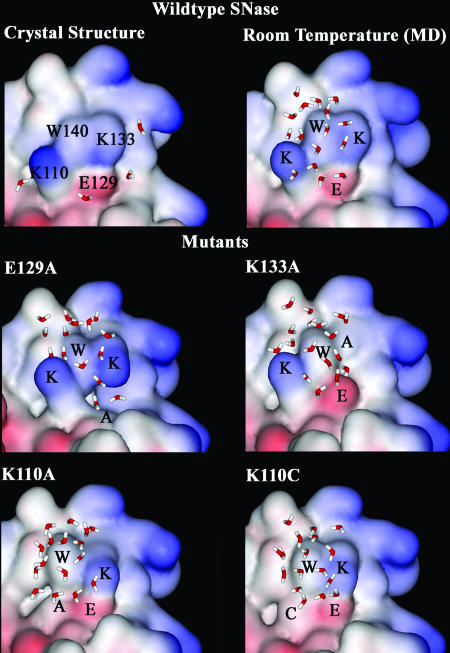
Fig. 5 shows a series of surface maps around W140 for the WT and four mutants with the local surface topography and neighboring residues. The x-ray structure at 1.7-Å resolution shows around W140 four surface-water molecules sticking to three charged residues and one water molecule buried inside the protein. As also shown by recent MD simulations of water penetration in SNase (34), the buried water molecule bridges three residues, the side chain of W140 (NE1–O distance of 2.87 Å) and two backbone oxygen atoms of V104 (O–O distance of 2.89 Å) and A109 (O–O distance of 2.6 Å), by three hydrogen bonds and has a residence time that is longer than the simulation time window of 10 ns. Another recent 7-ns MD simulation (35) of surface-water properties reveals a complex hydration structure with a pronounced pentagon–pentagon ring distribution in the hydration layers that depends highly on the surface topography. Around the hydrophobic atoms, clathrate-like cage structures parallel to the protein surface were observed. Near the polar atoms, the ring structures, perpendicular to the surface, have one or two hydrogen bonds with the polar atoms. From our recent 2-ns MD simulations, we found that within 5 Å from the O atom of H2O to the indole moiety (including H), there are ≈17 water molecules around the ground-state W140. We did not observe significant changes of total water molecules for the different mutants, implying nearly similar polar water environments, consistent with our observation of the same Stokes shifts for all mutants and the WT.
Fig. 5.
Protein surface maps of the WT and four mutants showing the local topography and neighboring protein residues around W140. There are four water molecules sticking to three surface charged residues (K110, E129, and K133) near the probe W140 in the x-ray structure. The water molecules within 5 Å from the O atom of H2O to the indole ring are shown from our 2-ns single-trajectory MD simulations at 295 K in aqueous solution. Note that the number of water molecules is nearly the same for all of the proteins but the structures are different. The structural landscapes are different, especially for E129A and K110A, with ruptures at the surfaces near W140.
When tryptophan is excited to the 1La state, the local equilibrium is shifted, and the system is in a nonequilibrium state. Our observed hydration dynamics, with two distinct time scales, reflect the temporal evolution of two types of motions from the initial nonequilibrium configuration to new equilibrated state around the excited tryptophan. The initial hydration dynamics, occurring in 3–5 ps, represents the librational/rotational motions of these surrounding water molecules, slowing down by a factor of three to five compared with similar bulk-water motions around tryptophan, which occur in <1 ps (1, 7). We clearly observed a correlation of the initial fast time scale and amplitude with the local charge distribution (Table 1). For example, we observed the fastest hydration dynamics, in 3.1 ps, for the mutant K110A with the largest amplitude (77%); for K110C, we obtained a time scale of 4.2 ps with 51%; for the WT (K110), we have the initial dynamics of 5.1 ps with 46%. Thus, from the hydrophobic (A110) to the polar (C110) and to the charged residue (K110), we observed a lengthening of the time scale, consistent with stronger interactions of local water with charges. Furthermore, we observed a decrease, not an increase, in the amplitude, suggesting that the charged/polar residue does not directly contribute to the solvation energy. These results reveal that the initial relaxation is from the local surface-water motions and that the observed variations in hydration dynamics in the mutations reflect the alternation of the local landscape and the change of the neighboring chemical identity (Fig. 5).
The observed slow water dynamics in 100–150 ps represent the long-time collective motions, reflecting a dynamic structural change of the local water molecules. The time scale to reach the new structural configuration depends on the local protein–water interactions. Upon the sudden change of the dipole moment of W140 in the protein, besides initial librational/rotational motions, at least two local structural motions are expected: one is the alignment of the local water network/cluster to the new excited-state (1La) dipole moment, and the other is the increase of water molecules around W140 due to the larger excited-state dipole moment (Δμ ≈ 5 debye units). Because of the longer time scale (tens of picoseconds), these overall structural changes are coupled with the protein fluctuation. These fluctuations assist in structural rearrangement of surface water and its exchange with bulk water. As such, the connection of residence time to hydration is incomplete without knowledge of fluctuations. Thus, the long-time hydration dynamics, defined here as the rate for water structure to reach the new equilibrated state of minimal free energy, is an integrated process determined by the local interactions with the protein (Fig. 5) and assisted by its fluctuations. The protein–water coupling motions enable surface-water molecules to make structural arrangements (solvation), but these small protein fluctuations themselves do not make direct contributions to total solvation.
Conclusion
The studies reported here, using site-selected mutagenesis of the enzyme SNase, indicate that surface protein hydration occurs on the picosecond time scale with a biphasic behavior, consistent with observations made previously for a number of other proteins that we have studied. The fastest component (a few picoseconds) is due to the librational/rotational motions of water near the surface, whereas the longest one (tens of picoseconds) is due to the water molecules coupled to the surface and structurally modified by protein fluctuations. The evidence for hydration, in contrast to the dominance of side-chain solvation, comes from a number of observations discussed above and in ref. 6 in studies of different proteins, but here the evidence comes from studies of the same protein but with different side chains of different charges and structures. It would be fortuitous, and it seems unlikely, that mutations at three different sites would exactly cancel out the effect of hydration and protein solvation to give nearly the same Stokes shifts and correlation functions. For another protein, the insignificant contribution of side-chain solvation was recently examined in experiments involving the native and denatured states (36).
The key to understanding surface hydration is the time scales involved. In a simple model (for reviews, see refs. 4, 6, 12, and 37), the time scale is related to hydrogen-bond exchanges in layered water, consistent with residence times obtained from several MD simulations (6, 13). Such a picture can explain slow hydration also in micelles (37). However, on time scales of a few picoseconds, proteins are “rigid,” whereas on time scales of tens of picoseconds, they fluctuate. This separation of the time scales is relevant not only for hydration but also for other protein interactions, such as molecular recognition (38). The fluctuations assist solvation by allowing for restructuring in the new nonequilibrium state (in this case, the structure of water around tryptophan's dipole). They themselves do not contribute directly to solvation by minimizing energy, as evidenced by the robustness of hydration time scales and Stokes shifts independent of local charge and structural changes around tryptophan. Large-amplitude side-chain motions may occur, but they are on somewhat longer time scales and may involve conformation changes; such changes contribute to solvation (33).
Because it directly represents interactions with the protein, unlike libration or distant rotation, the longer picosecond hydration discussed here is significant for biological function (for recent reviews, see refs. 39 and 40). The picosecond time scale, as pointed out in ref. 6, is ideal for many biological processes involved in molecular recognition, reactivity, and conformational intactness. The robustness in longer-time hydration for proteins of different sequence and structure cannot be ignored. In earlier NMR work, it was shown in a series of papers (for reviews, see refs. 17–20) that the hydration of proteins and DNA occurs in subnanoseconds, and it was the work from our laboratories (for review, see refs. 6 and 9) that pointed out that the time scale of surface hydration is much shorter than the time scales reported by NMR: namely, from 1 ps to tens of picoseconds (biphasic) with femtosecond resolution. However, in more recent work published from one of the same NMR groups (21–23), it was ascertained that hydration occurs in a few picoseconds. Similarly, in earlier MD simulations (see, for example, ref. 13), the time scales varied from picosecond to nanosecond residence times, depending on the strength of hydrogen bonding and the location of water molecules, but with a force field and linear response theory, the claim now is that water is much more labile, although fast and slow (10%) components were present, with the latter assigned to protein solvation (23). As discussed above, the search for the large ultrafast (<1 ps) dynamics predicted by MD simulations was unsuccessful. It is possible that the flexibility imposed by MD modeling is reducing all time scales and increasing amplitudes of subpicosecond components by order(s) of magnitude. Protein fluctuations, including side chains, cannot be blind to local water hydration that is responsible for solvation and is structurally evident through x-ray and mutation studies. It is concluded that the picture based on MD simulations and emphasized in several recent publications (22, 23) needs to be revisited, taking into consideration the approximations made regarding force field, linear response, and the exact meaning of hydration for the coupled protein–water network. Protein fluctuation-assisted interactions with surface water are an integral part of hydration dynamics, particularly at longer times.
Methods
Femtosecond Methods.
All experimental measurements were carried out by using the femtosecond-resolved fluorescence up-conversion apparatus described in refs. 11 and 41. Briefly, the pump laser was set at 290 nm, and the energy was typically attenuated to ≈140 nJ before being focused into the motor-controlled moving sample cell. The fluorescence emission was collected by a pair of parabolic mirrors and mixed with a gating pulse (800 nm) in a 0.2-mm β-barium borate (BBO) crystal through a noncollinear configuration. The up-converted signal ranging from 218 to 292 nm was detected. The instrument response time under the current noncollinear geometry is between 400 and 500 fs as determined from the up-conversion signal of Raman scattering by water at ≈320 nm. Most measurements were carried out at the magic-angle (54.7°) condition. For fluorescence anisotropy measurements, the pump-beam polarization was rotated to become either parallel or perpendicular to the BBO acceptance axis to obtain the parallel (I‖) and perpendicular (I⊥) signals, respectively. These transients were used to construct the time-resolved anisotropy: r(t) = (I‖ − I⊥)/(I‖ + 2I⊥).
Protein Site-Selected Mutagenesis and Preparation.
The four mutants of SNase, E129A, K133A, K110A, and K110C, were prepared by the method of Kunkel as described in ref. 42. Protein expression and purification were performed by following the procedure described in ref. 43. The obtained proteins were finally dissolved in a buffer of 25 mM sodium phosphate and 100 mM NaCl at pH 7. The protein concentration used in femtosecond-resolved studies was 200–300 μM. The fluorescence emission was measured by using a SPEX FluoroMax-3 spectrometer. To ensure that no change of the protein quality occurred during data acquisition, we measured the protein fluorescence emission before and after experiments. We also kept the sample in a rotating cell to minimize possible photobleaching.
Mutant Lifetime.
The lifetimes of all five proteins were determined by gating the relaxed-state emission at the red side of the 360-nm emission. The obtained transients are shown in Fig. 2 Right. For the WT, the transient was best fitted by a double-exponential decay with time constants of 1.2 and 5.6 ns, consistent with previous frequency-domain measurements (44). Because the time interval between two consecutive pump excitations was 2 ms and each transient was averaged over >1 h, all fluctuating protein configurations were probably sampled on such long times. Thus, the apparent two lifetimes do not mean that only two static configurations are present in solution but that two types of temporal configurations are present; the system is dynamically heterogeneous on a time scale longer than a few nanoseconds.
We obtained a similar transient for the E129A mutant, indicating negligible excited-state quenching by the charged residue E129. For the K110A mutant, a nearly single-exponential decay was observed with a lifetime of 8.7 ns, indicating minor dynamic quenching by immobile K133 and consistent with a strong cation–π interaction at 2.99 Å. Without any quenching in proteins, tryptophan emission has a typical lifetime of ≈9 ns (26). For the K133A mutant, the transient becomes faster, and the lifetimes shorten to 0.9 and 4.5 ns, suggesting a noticeable quenching by K110 due to a “loose” cation–π interaction at 4.65 Å. The mutant K110C has the shortest lifetimes, 0.8 and 3.5 ns, and has stronger quenching than K133A due to the quenching by C110 through an electron-transfer mechanism (45). These obtained lifetimes correlate with neighboring chemical identities (46), separation distances, and observed fluorescence intensities and were used in the data analyses above. It should be noted that although changes in intensities and lifetimes reflect quenching, these quenching dynamics are on the nanosecond time scale, and the blue-side emission wavelengths are more sensitive to solvation, which is typically on the picosecond time scale (11, 47).
Supplementary Material
Acknowledgments
We thank Prof. Patrik R. Callis for catalysis of the initial collaboration for the work with W.E.S. and for constructive comments on the manuscript, Bradford Bullock for help with preparing protein samples, Prof. Sherwin Singer for helpful discussion, Tanping Li for efforts in preparing Fig. 5, and Dr. Wenyun Lu for initial help with the experiments. This work was supported by Petroleum Research Fund Grant PRF-42734-G4, the Packard Foundation Fellowship (to D.Z.), National Science Foundation grants (to D.Z. and A.H.Z.), and National Institutes of Health Grant NCRR COBRE P20 RR15569 (to W.E.S.).
Abbreviations
- SNase
Staphylococcus nuclease
- MD
molecular dynamics.
Footnotes
The authors declare no conflict of interest.
References
- 1.Zhong D, Pal SK, Zhang D, Chan SI, Zewail AH. Proc Natl Acad Sci USA. 2002;99:13–18. doi: 10.1073/pnas.012582399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pal SK, Peon J, Zewail AH. Proc Natl Acad Sci USA. 2002;99:1763–1768. doi: 10.1073/pnas.042697899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peon J, Pal SK, Zewail AH. Proc Natl Acad Sci USA. 2002;99:10964–10969. doi: 10.1073/pnas.162366099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal SK, Peon J, Bagchi B, Zewail AH. J Phys Chem B. 2002;106:12376–12395. [Google Scholar]
- 5.Zhao L, Pal SK, Xia T, Zewail AH. Angew Chem Int Ed. 2004;43:60–63. doi: 10.1002/anie.200352961. [DOI] [PubMed] [Google Scholar]
- 6.Pal SK, Zewail AH. Chem Rev. 2004;104:2099–2123. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 7.Lu W, Kim J, Qiu W, Zhong D. Chem Phys Lett. 2004;388:120–126. [Google Scholar]
- 8.Kamal JKA, Zhao L, Zewail AH. Proc Natl Acad Sci USA. 2004;101:13411–13416. doi: 10.1073/pnas.0405724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu W, Zhang L, Kao Y-T, Lu W, Li T, Kim J, Sollenberger GM, Wang L, Zhong D. J Phys Chem B. 2005;109:16901–16910. doi: 10.1021/jp0511754. [DOI] [PubMed] [Google Scholar]
- 10.Qiu W, Zhang L, Okobiah O, Yang Y, Wang L, Zhong D, Zewail AH. J Phys Chem B. 2006;110:10540–10549. doi: 10.1021/jp055989w. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Kao Y-T, Qiu W, Wang L, Zhong D. J Phys Chem B. 2006. in press. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya SM, Wang ZG, Zewail AH. J Phys Chem B. 2003;107:13218–13228. [Google Scholar]
- 13.Levitt M, Sharon R. Proc Natl Acad Sci USA. 1988;85:7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocchi C, Bizzarri AR, Cannistraro S. Phys Rev E. 1998;57:3315–3325. [Google Scholar]
- 15.Makarov VA, Andrews BK, Smith PE, Pettitt BM. Biophys J. 2000;79:2966–2974. doi: 10.1016/S0006-3495(00)76533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay S, Chakraborty S, Balasubramanian S, Bagchi B. J Am Chem Soc. 2005;127:4071–4075. doi: 10.1021/ja042847r. [DOI] [PubMed] [Google Scholar]
- 17.Otting G, Liepinsh E, Wüthrich K. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 18.Denisov VP, Halle B. J Mol Biol. 1995;245:682–697. doi: 10.1006/jmbi.1994.0055. [DOI] [PubMed] [Google Scholar]
- 19.Wüthrich K, Billeter M, Güntert P, Luginbühl P, Riek R, Wider G. Faraday Discuss. 1996;103:245–253. [Google Scholar]
- 20.Denisov VP, Halle B. Faraday Discuss. 1996;103:227–244. doi: 10.1039/fd9960300227. [DOI] [PubMed] [Google Scholar]
- 21.Modig K, Liepinsh E, Otting G, Halle B. J Am Chem Soc. 2004;126:102–114. doi: 10.1021/ja038325d. [DOI] [PubMed] [Google Scholar]
- 22.Halle B. Philos Trans R Soc London B. 2004;359:1207–1224. doi: 10.1098/rstb.2004.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson L, Halle B. Proc Natl Acad Sci USA. 2005;102:13867–13872. doi: 10.1073/pnas.0504181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truckses DM, Somoza JR, Prehoda KE, Miller SC, Markley JL. Protein Sci. 1996;5:1907–1916. doi: 10.1002/pro.5560050917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano S, Kamikubo H, Yamazaki Y, Kataoka M. Proteins. 2005;58:271–277. doi: 10.1002/prot.20333. [DOI] [PubMed] [Google Scholar]
- 26.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Plenum; 1999. [Google Scholar]
- 27.Scott TW, Campbell BF, Cone RL, Friedman JM. Chem Phys. 1989;131:63–79. [Google Scholar]
- 28.Vivian JT, Callis PR. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bizzarri AR, Cannistraro S. J Phys Chem B. 2002;106:6617–6633. [Google Scholar]
- 30.Steiner RF. In: Topics in Fluorescence Spectroscopy. Lakowicz JR, editor. Vol 2. New York: Plenum; 1991. pp. 1–51. [Google Scholar]
- 31.Tarek M, Tobias DJ. Phy Rev Lett. 2002;88:138101. doi: 10.1103/PhysRevLett.88.138101. [DOI] [PubMed] [Google Scholar]
- 32.Luise A, Falconi M, Desideri A. Proteins. 2000;39:56–67. doi: 10.1002/(sici)1097-0134(20000401)39:1<56::aid-prot6>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Hassanali AA, Li T, Zhong D, Singer SJ. J Phys Chem B. 2006;110:10497–10508. doi: 10.1021/jp0601926. [DOI] [PubMed] [Google Scholar]
- 34.Damjanovic A, Garcia-Moreno B, Lattman EE, Garcia AE. Proteins. 2005;60:433–449. doi: 10.1002/prot.20486. [DOI] [PubMed] [Google Scholar]
- 35.Smolin N, Winter R. J Phys Chem B. 2004;108:15928–15937. [Google Scholar]
- 36.Shaw AK, Sarkar R, Banerjee D, Hintschich S, Monkman A, Pal SK. J Photochem Photobiol A. 2006. in press. [Google Scholar]
- 37.Bagchi B. Chem Rev. 2005;105:3197–3219. doi: 10.1021/cr020661+. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Baskin JS, Xia T, Zewail AH. Proc Natl Acad Sci USA. 2004;101:18000–18005. doi: 10.1073/pnas.0408379102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy Y, Onuchic JN. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 40.Raschke TM. Curr Opin Struct Biol. 2006;16:152–159. doi: 10.1016/j.sbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Saxena C, Sancar A, Zhong D. J Phys Chem B. 2004;108:18026–18033. [Google Scholar]
- 42.Kunkel TA. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byrne MP, Manuel RL, Lowe LG, Stites WE. Biochemistry. 1995;34:13949–13960. doi: 10.1021/bi00042a029. [DOI] [PubMed] [Google Scholar]
- 44.Eftink MR, Gryczynski I, Wiczk W, Laczko G, Lakowicz JR. Biochemistry. 1991;30:8945–8953. doi: 10.1021/bi00101a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Barkley MD. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 46.Callis PR, Liu T. J Phys Chem B. 2004;108:4248–4259. [Google Scholar]
- 47.Xu JH, Toptygin D, Graver KJ, Albertini RA, Savtchenko RS, Meadow ND, Roseman S, Callis PR, Brand L, Knutson JR. J Am Chem Soc. 2006;128:1214–1221. doi: 10.1021/ja055746h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.