Abstract
Functional genomics approaches, which use combined computational and expression-based analyses of large amounts of sequence information, are emerging as powerful tools to accelerate the comprehensive understanding of cellular metabolism in specialized tissues and whole organisms. As part of an ongoing effort to identify genes of essential oil (monoterpene) biosynthesis, we have obtained sequence information from 1,316 randomly selected cDNA clones, or expressed sequence tags (ESTs), from a peppermint (Mentha x piperita) oil gland secretory cell cDNA library. After bioinformatic selection, candidate genes putatively involved in essential oil biosynthesis and secretion have been subcloned into suitable expression vectors for functional evaluation in Escherichia coli. On the basis of published and preliminary data on the functional properties of these clones, it is estimated that the ESTs involved in essential oil metabolism represent about 25% of the described sequences. An additional 7% of the recognized genes code for proteins involved in transport processes, and a subset of these is likely involved in the secretion of essential oil terpenes from the site of synthesis to the storage cavity of the oil glands. The integrated approaches reported here represent an essential step toward the development of a metabolic map of oil glands and provide a valuable resource for defining molecular targets for the genetic engineering of essential oil formation.
Keywords: functional genomics, isoprenoid biosynthesis, monoterpene biosynthesis
Essential oil plants have been valued historically for their medicinal, culinary, and fragrance properties and include members of the genus Mentha (mint), which produce some of the most widely used essential oils (1). The essential oil of peppermint is biosynthesized and stored in specialized anatomical structures, termed peltate glandular trichomes, on leaf surfaces (2, 3). Given the commercial value of these oils, the processes involved in their biosynthesis and secretion are attractive targets for genetic engineering. The characteristic flavor and aroma components of mint oils are monoterpenes (4). However, because of the complexity of the monoterpene biosynthetic pathway and the difficulties in purifying the responsible enzymes (5), obtaining gene probes from the corresponding target proteins by classical biochemical approaches has presented a considerable challenge.
The partial sequencing of anonymous cDNA clones [expressed sequence tags (ESTs)] has become a rapid and cost-effective means of gaining information about gene expression and coding capacity of Arabidopsis thaliana (6) and rice (7). In addition, a few groups have analyzed ESTs expressed in specialized tissues and organs to assist in the identification of new genes involved in specialized pathways, such as those of developing rice endosperm (8), isolated guard cells of Brassica campestris (9), wood-forming tissues of poplar (10), and immature xylem of Loblolly pine (11).
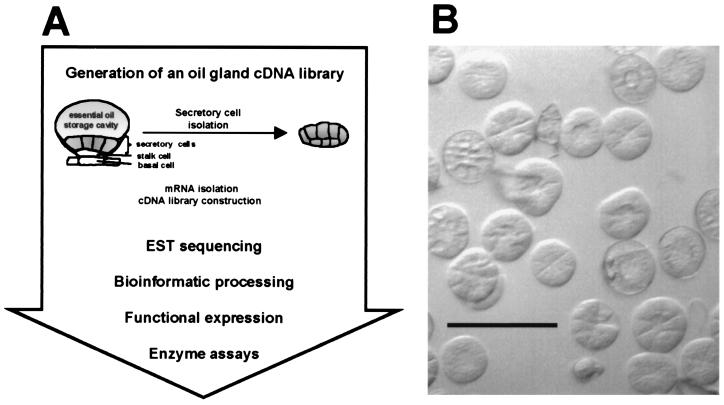
Here we report on a functional genomics approach (Fig. 1A) directed toward the characterization of genes involved in essential oil formation in peppermint (Mentha x piperita), that was initiated with the construction of an oil gland secretory cell cDNA library. The secretory cells of the oil glands responsible for essential oil biosynthesis can be isolated in high yield from leaves (3). These isolated cells are capable of de novo biosynthesis of monoterpenes from primary carbohydrate precursors (12), and they have been shown to be highly enriched in the enzymes of monoterpene biosynthesis (13). By using a modification of a published protocol (14), mRNA from isolated peppermint secretory cells was obtained and subsequently used to generate a cDNA library. A modest effort to sequence ESTs from this enriched cDNA library (≈1, 300 acquisitions) led by bioinformatic processing of the data to candidate genes putatively involved in essential oil biosynthesis and secretion, that, by heterologous expression in Escherichia coli, allowed functional characterization of the corresponding recombinant proteins.
Figure 1.
Functional genomics approach to evaluate ESTs from a peppermint oil gland secretory cell cDNA library. (A) Flow diagram of program. (B) Light micrograph of oil gland secretory cell clusters isolated from peppermint leaves (Bar = 200 μm).
Materials and Methods
Plant Material, mRNA Isolation, and Generation of an Oil Gland Secretory Cell cDNA Library.
Peppermint plants (Mentha x piperita L. cv. Black Mitcham) were propagated and grown under greenhouse conditions as previously described (3). Leaves (15–20 g; <10 mm in length) were excised and soaked in an ice-cold imbibition buffer containing 1 mM aurintricarboxylic acid, 5 mM thiourea, and 2 mM DTT, for 1 h. Oil gland secretory cells were then isolated by using a modification of the glass bead abrasion method (3) with an extraction buffer (pH 6.6) consisting of 200 mM sorbitol, 10 mM sucrose, 25 mM Mopso, 0.5 mM sodium phosphate, 1% (wt/vol) polyvinylpyrrolidone-40, 0.6% (wt/vol) methyl cellulose, 1 mM aurintricarboxylic acid, 5 mM thiourea, and 2 mM DTT. The isolated secretory cells were next frozen in liquid N2, powdered with a mortar and pestle, and RNA was extracted by using a modified protocol based on a previously published method (14). This method involves extraction at pH 7.0 with 8 M guanidine⋅HCl containing 20 mM Mes, 20 mM EDTA, 50 mM ethanethiol, and 1% (wt/vol) polyvinylpyrrolidone-40, followed by phenol and chloroform extractions to remove proteins, acidic precipitation of RNA to remove DNA, and, finally, ethanol (10%, vol/vol) precipitation of polysaccharides. RNA yield and quality were assessed by formaldehyde-agarose gel electrophoresis (15) and by in vitro translation by using the wheat germ system (Promega, L4140) followed by gel electrophoresis of the resulting labeled proteins. mRNA was isolated by two rounds of oligo(dT)-cellulose column chromatography (Pharmacia Biotech, no. 27–9258-02), and the quality was evaluated again by using the in vitro translation assay. cDNA synthesis from 5 μg of purified mRNA and library construction were carried out with a commercial kit (Stratagene, λZAPII cDNA library construction kit no. 1200454). To estimate the average cDNA insert size, an aliquot of the cDNA library was plated, 96 individual clones were in vivo excised according to the manufacturer's instructions, plasmid DNA of these clones was isolated and used as template for PCR amplication by using T3 and T7 primers, and the resulting amplicons were analyzed by agarose gel electrophoresis. To acquire the 5′-termini corresponding to truncated cDNA clones, 5′-rapid amplification of cDNA ends was carried out by using the Marathon cDNA amplification system (CLONTECH, K1802–1) with purified mRNA from oil gland secretory cells as template. The resulting amplicons were cloned into the TOPO TA cloning vector (Invitrogen, K4550–01), and the inserts were sequenced from both ends.
Nucleotide Sequencing.
pBluescript SK(−) phagemids were excised from the λZAPII vector according to the in vivo mass excision protocol of the manufacturer (Stratagene). Plasmid DNA was isolated from randomly selected colonies by using a commercial kit (Qiagen, no. 16151) and a 48-well vacuum manifold (Qiagen, no. 19503). Taq polymerase cycle sequencing reactions were performed by using FS terminator chemistry (Perkin–Elmer) and T3 primer. For selected clones, the complete sequence of both strands was obtained by using T3, T7, and gene-specific primers. Automated sequencing was carried out with a model 373 sequencer (Applied Biosystems).
Sequence Analysis.
Sequences were edited manually to remove contaminants originating from the vector and to discard poor quality 3′-sequence. Sequence comparisons against the GenBank nonredundant protein database were performed by using the blastx algorithm (16). A match was declared when the score was higher than 120 (optimized similarity score), with 65% sequence identity over a minimum of 30 deduced amino acid residues. EST sequences were then grouped, where appropriate, into sequence clusters by using the tigr assembler (17). In addition, the sequences of each contig were aligned by using the fragment assembly program of the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, WI; based on ref. 18), and consensus sequences were generated with 90% identity over a minimum of 40 nucleotides.
Recombinant Protein Expression.
For candidate genes putatively involved in isoprenoid biosynthesis, five methods for heterologous expression in E. coli were used. In general, bacteria harboring a plasmid contruct were grown in LB medium supplied with appropriate antibiotics at 37°C to an OD600 of 0.2–0.5. After addition of a chemical inducer (for details, see below), cells were incubated for an additional 12–20 h at 15–20°C. Low-level expression, without subcloning, was achieved from pBluescript SK(−) (Stratagene, no. 212206) under control of the lac promoter (E. coli SOLR cells as a host, Stratagene, no. 200298), which was induced by addition of 1.0–2.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). If purification of the recombinant protein was required, expression under control of the araBAD promoter from the pBAD TOPO TA expression vector (Invitrogen, K4300–01) was induced by arabinose (0.1–1 mM) addition (E. coli OneShot TOP10 cells as a host, Invitrogen, no. C4040–10); this approach allowed a convenient one-step purification of the corresponding His-tagged recombinant protein by using Ni2+ affinity-binding columns. For high-level expression, the vector pSBET (19) was used, from which the expression of transgenes is regulated by the T7 phage promoter and is induced by IPTG (0.1–1 mM) addition. This vector supplements a tRNA for the arginine triplets AGA and AGG, which are of low abundance in E. coli genes but are frequently found in plant genes. In more recent work, tRNAs for rare codons were successfully complemented by the use of E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene, no. 230245), which contain extra copies of the argU, ileY, and leuW tRNA genes encoding tRNA species that recognize the arginine codons AGA and AGG, the isoleucine codon AUA, and the leucine codon CUA. Thus, the pSBET vector was replaced by the pET-3 vector (Novagen, no. 69409–3) in this application; this vector also features the T7 phage promoter for control of expression. It should be noted that the high-level expression of plant proteins in E. coli often results in the formation of inclusion bodies; however, sufficient soluble protein is usually produced to give measurable enzyme activity. For the heterologous expression of cytochrome P450 monooxygenases, the corresponding genes were subcloned into the pCWOri+ vector (20). Expression from this vector under control of the lacUV5 promoter was induced by addition of 0.2–1.0 mM IPTG. Protocols for the isolation of bacterial cells, enzyme extraction, and activity assays have been previously described (21–25).
Results and Discussion
Generation of an Oil Gland Secretory Cell cDNA Library.
Essential oil (monoterpene) biosynthesis and accumulation in mint is specifically localized in epidermal oil glands (12, 26). Disk-like clusters of oil gland secretory cells were obtained in high yield from young peppermint leaves by an improved protocol based on the glass-bead abrasion technique (3). Microscopic examination indicated that >99% of the glandular disks isolated contained eight secretory cells each (Fig. 1B). This distribution of glandular structures is in contrast to the developmental distribution of two-celled (≈6%), four-celled (≈14%), and eight-celled (≈80%) peltate glandular trichomes on the surface of young leaves (<10 mm in length) (G. Turner and R.C., unpublished results); this fortuitous enrichment in eight-celled structures most active in essential oil biosynthesis is likely a physical consequence of the heavily cutinized lateral wall of the supporting stalk cell (Fig. 1A) that may render the interface of the stalk cell–secretory cell junction most susceptible to cleavage (G. Turner, J. Gershenzon, and R.C., unpublished results).
To protect RNA from degradation during the secretory cell isolation procedure, the RNase inhibitor aurintricarboxylic acid (27) and the phenol oxidase inhibitor thiourea (28) were added to the leaf imbibition and secretory cell isolation buffers. The RNA isolation method of Logemann et al. (14) was modified by adding polyvinylpyrrolidone-40 to the buffer to bind phenolics released during the process and by including a polysaccharide precipitation step by using 10% ethanol (29). Quality assessment of isolated RNA by agarose gel electrophoresis and by in vitro translation indicated that these modifications allowed the preparation of intact mRNA from oil glands. Notably, about 20 distinct protein bands, in addition to a faint smear of other labeled proteins, were observed on SDS/PAGE analysis of the in vitro translation mixture, indicating the presence of several highly abundant mRNA species (data not shown). A λZAPII cDNA library was then constructed from purified oil gland mRNA according to the manufacturer's instructions (Stratagene). The average insert size, based on PCR analysis of 96 individual clones, was estimated to be about 1,400 bp. For sequencing and expression, the cDNA clones were in vivo excised as pBluescript SK(−) phagemids in E. coli host strain SOLR.
Nucleotide Sequencing, Fragment Assembly, and Database Searching.
A total of 1,316 individual clones were randomly picked and sequenced from the 5′-terminus, of which 1,250 clones gave high-quality sequencing results. After deletion of vector sequences and poor quality 3′-sequences and editing of artefacts, the average readable coding sequence was determined to be approximately 560 bp. Of the 1,250 ESTs evaluated, 66% showed significant homology to sequences with assigned putative functions in public databases by using the blastx search algorithm (16). Fragment assembly of these sequences revealed 109 clusters of contiguous sequence, consisting of two or more ESTs, whereas 747 sequences remained as single ESTs with no significant homology to any other sequence in the data set (corresponding to a total of 856 unique transcripts). About 66% of the ESTs belonged to sequence clusters ranging from low redundancy (2–4 sequences per contig, with 69 contigs detected) and medium redundancy (5–10 sequences per contig, with 25 contigs detected), to high redundancy (11–77 sequences per contig, with 13 contigs detected).
Selection and Expression of Candidate Genes.
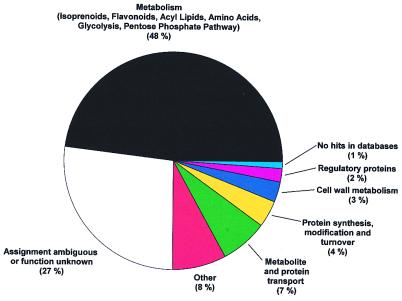
The ESTs were classified into eight functional groups according to their sequence characteristics (Fig. 2). About 48% of the clones from the oil gland secretory cell cDNA library coded for enzymes involved in primary metabolism (glycolysis, pentose phosphate pathway, amino acid metabolism, and acyl lipid metabolism) and secondary metabolism (isoprenoid and flavonoid biosynthesis). Another group of ESTs of high abundance were represented by putatively assigned genes coding for enzymes of metabolite and protein transport (7%). These results, together with the observed complete absence of genes involved in photosynthesis and carotenoid biosynthesis, confirmed that the preparation of oil gland secretory cells [which contain only photosynthetically inactive leucoplasts (30)] was devoid of contaminating photosynthetic leaf tissue.
Figure 2.
Functional classification of ESTs from the peppermint oil gland secretory cell cDNA library. Secondary metabolism accounts for about 35% of total metabolism.
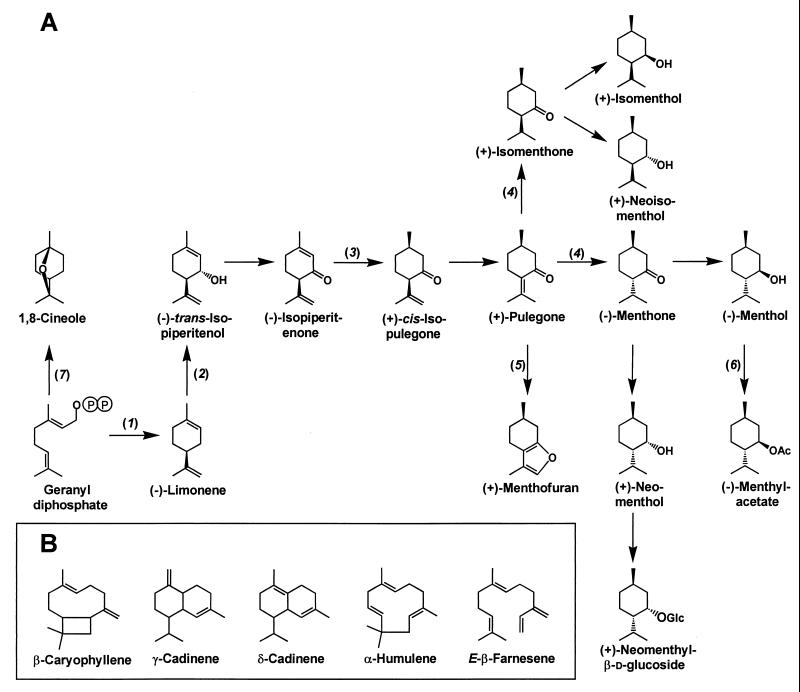
To limit the number of ESTs for expression studies, attention was first focused on clones with similarity to known genes of isoprenoid (monoterpene and sesquiterpene) biosynthesis (e.g., prenyl transferases and terpene synthases) and on those with sequence properties predicted for other genes involved in isoprenoid biosynthesis (e.g., transketolases, cytochrome P450 monooxygenases, and oxidoreductases) (Fig. 3). In addition, more extensive sequence analyses were conducted with selected candidate genes (e.g., putative genes for kinases, glucosyl transferases, and acyl transferases) by searching the Prosite [http://www.expasy.ch/prosite (31)], ProDom [http://protein.toulouse.inra.fr/prodom.html (32)], and Promise [http://bioinf.leeds.ac.uk/promise (33)] databases.
Figure 3.
(A) Outline of monoterpene biosynthesis in peppermint glandular trichomes. The enzymes indicated are: (−)-limonene synthase (1), (−)-limonene-3-hydroxylase (2), (−)-isopiperitenone reductase (3), (+)-pulegone reductase (4), (+)-menthofuran synthase (5), (−)-menthol acetyltransferase (6), and 1,8-cineole synthase (7). (B) Structures of selected sesquiterpenes of peppermint oil.
Full-length clones corresponding to truncated versions of candidate genes were obtained by a 5′-rapid amplification of cDNA ends method. For functional expression in E. coli, five different methods were considered, and the most promising protocol for a particular class of enzyme was then selected. For initial characterization, the pBluescript SK(−) vector was used. For cases in which an affinity-based purification of the recombinant protein was desirable to eliminate competing enzyme activities of the host, genes were subcloned into the pBAD TOPO TA vector (Invitrogen) and expressed as His-tag fusion proteins. For high-level expression, genes were subcloned into pSBET (19), a plasmid that supplements for a tRNA for arginine codons that rarely occur in E. coli but are commonly found in plant genes. Recently, the use of BL21-CodonPlus cells (Stratagene) has become a convenient alternative to supply tRNAs for rare arginine, leucine, and isoleucine codons; this approach allows substitution of the pET-3 vector (Novagen) for pSBET. Cytochrome P450 monooxygenases were expressed by using the pCWOri+ vector (20).
Genes of Essential Oil Biosynthesis.
In mint oil glands, the classical mevalonate pathway for the biosynthesis of isopentenyl diphosphate (IPP), the central intermediate of isoprenoid biosynthesis, is blocked (34); not a single gene of this pathway was among the ESTs sequenced. This specialization facilitated the isolation and characterization of genes of the alternative 1-deoxyxylulose-5-phosphate (DXP) pathway for IPP biosynthesis (35, 36), including DXP synthase (22), DXP reductoisomerase (23) and isopentenyl monophosphate kinase (37) (Table 1). In addition, a putative homologue of 4-diphosphocytidyl-2-C-methylerythritol synthase (38) was acquired in the EST dataset, and confirmation by the functional expression of this gene is in progress.
Table 1.
Peppermint ESTs related to monoterpene and sesquiterpene biosynthesis
| EST identification | Total number of hits (of 1,250) | Redundancy, ‰ |
|---|---|---|
| DXP synthase | 11 | 8.8 |
| DXP reductoisomerase* | — | 0.7 |
| Isopentenyl monophosphate kinase | 2 | 1.6 |
| IPP isomerase | 2 | 1.6 |
| Geranyl diphosphate synthase (small subunit) | 4 | 3.2 |
| Geranyl diphosphate synthase (large subunit) | 4 | 3.2 |
| Farnesyl diphosphate synthase | 3 | 2.4 |
| Monoterpene synthases (total) | 37 | 29.6 |
| (−)-Limonene synthase (isoenzyme 1) | 18 | 14.4 |
| (−)-Limonene synthase (isoenzyme 2) | 12 | 9.6 |
| 1,8-Cineole synthase | 2 | 1.6 |
| Sesquiterpene synthases (total) | 12 | 9.6 |
| β-Farnesene synthase | 2 | 1.6 |
| (−)-Limonene-3-hydroxylase | 77 | 61.1 |
| (+)-Menthofuran synthase | 10 | 8.0 |
| (−)-Isopiperitenone reductase | 28 | 22.4 |
| (+)-Pulegone reductase | 14 | 11.2 |
| (−)-Menthol acetyltransferase | 4 | 3.2 |
This clone was not obtained as part of the EST sequencing project. A cDNA library screen revealed an abundance of one in 1,500 clones (0.7‰).
IPP, by the action of IPP isomerase (39), is converted to dimethylallyl diphosphate (DMAPP). By sequence comparison, two homologous ESTs for IPP isomerase were detected, the functional properties of which are now being evaluated. DMAPP serves as the starter unit for sequential condensation reactions catalyzed by prenyltransferases (40). Addition of one molecule of IPP to DMAPP yields geranyl diphosphate; this reaction is catalyzed by geranyl diphosphate synthase, a heterodimeric enzyme that was recently cloned and characterized from this oil gland cDNA library (24). Addition of a second molecule of IPP (to geranyl diphosphate) leads to the formation of farnesyl diphosphate; ESTs encoding farnesyl diphosphate synthase were detected by sequence homology. Similarly, elongation of DMAPP with a total of three molecules of IPP yields geranylgeranyl diphosphate by the action of geranylgeranyl diphosphate synthase, for which the corresponding ESTs were also detected.
These prenyl diphosphates undergo subsequent cyclization reactions, catalyzed by terpene synthases, leading to the various parent skeletons for monoterpenes (from geranyl diphosphate), sesquiterpenes (from farnesyl diphosphate) (Fig. 3B), and diterpenes (from geranylgeranyl diphosphate) (41). The peppermint oil gland terpene synthases characterized by functional expression thus far include (−)-limonene synthase and 1,8-cineole synthase, β-farnesene synthase (42) (Fig. 3) and, by similarity considerations, (−)-copalyl diphosphate synthase and (−)-ent-kaurene synthase.
The late stages of monoterpene metabolism in peppermint involve a series of secondary transformations (redox reactions, isomerizations, and conjugations) to yield numerous end products (Fig. 3A). Functional expression of full-length cDNAs derived from the corresponding EST acquisitions have yielded (−)-limonene-3-hydroxylase (25), (−)-isopiperitenone reductase, (+)-pulegone reductase, (+)-menthofuran synthase, and (−)-menthol acetyltransferase. The abundance of some ESTs was remarkably high, including those encoding DXP synthase (8.8‰), (−)-limonene synthase (LS1, 14.4‰; LS2, 9.6‰), (−)-limonene-3-hydroxylase (61.6‰), (−)-isopiperitenone reductase (22.4‰), and (+)-pulegone reductase (11.2‰), confirming the enrichment in the library of genes involved in essential oil biosynthesis.
In addition to the monoterpenes and sesquiterpenes, flavonoids form another, although quantitatively less significant, group of mint essential oil constituents (43). As inferred by sequence homology, genes encoding 4-coumarate-CoA ligase, (0.8‰), chalcone synthase (4‰), chalcone isomerase (4‰), flavonoid-3′,5′-hydroxylase (4‰), flavonol-4-reductase (8‰), flavonol sulfotransferase (2.4‰), and flavonoid O-methyltransferases (40‰) were detected, as was one EST related to the CYP93 family of cytochrome P450 monooxygenases.
Genes of Essential Oil Transport and Secretion.
Lipid transfer protein homologues are very abundant in the peppermint oil gland cDNA library (32%). These proteins are known to transport lipophilic molecules between intracellular membranes and are secreted into the medium of plant cell cultures (44). Because essential oil biosynthesis involves three intracellular compartments (leucoplasts, endoplasmatic reticulum and cytosol) and final export to the subcuticular storage cavity, a potential role for lipid transfer proteins is suggested in the intracellular trafficking and ultimate secretion of essential oil.
Genes of Primary Metabolism.
Among the ESTs sequenced were homologues for all enzymes of glycolysis (with the exception of phosphoglycerate kinase), the total relative abundance of which was about 17‰. The relative abundance of genes related to the oxidative pentose phosphate pathway was 8.8‰. Genes of oxidative phosphorylation were represented with a relative abundance of 9.6‰ of the ESTs. These results indicate that the pathways that provide energy equivalents and redox cofactors for essential oil biosynthesis are very active. The transcript abundance for certain specific transporters, including a plastidial ADP/ATP antiporter (4‰) and a phosphoenolpyruvate/phosphate antiporter (1.6‰) as well as several unidentified putative transport proteins, suggests that the machinery for efficient translocation of metabolic intermediates and cofactors is also present.
Conclusions
The use of isolated oil gland secretory cell messages to generate a cDNA library has proven to provide an excellent source of ESTs related specifically to essential oil biosynthesis and accumulation. The functional expression of full-length forms corresponding to a significant number of target ESTs has been achieved by judicious choice of generic heterologous expression systems. This functional genomics approach has successfully accelerated gene discovery and characterization and has also provided promising targets for genetic engineering of essential oil production (45).
Acknowledgments
This work was supported in part by the United States Department of Energy, Division of Energy Biosciences, the Mint Industry Research Council, and project no. 0268 from the Agricultural Research Center, Washington State University.
Abbreviations
- DXP
1-deoxyxylulose-5-phosphate
- EST
expressed sequence tag
- IPP
isopentenyl diphosphate
Footnotes
References
- 1.Parry J W. Spices. I and II. New York: Chemical Publishing Co.; 1969. [Google Scholar]
- 2.Amelunxen F, Wahlig T, Arbeiter H. Z Pflanzenphysiol. 1969;61:68–72. [Google Scholar]
- 3.Gershenzon J, McCaskill D, Rajaonarivony J I M, Mihaliak C, Karp F, Croteau C. Anal Biochem. 1992;200:130–138. doi: 10.1016/0003-2697(92)90288-i. [DOI] [PubMed] [Google Scholar]
- 4.Gasic O, Mimika-Dukic N, Adamovic D, Borojevic K. Biochem System Ecol. 1987;15:335–340. [Google Scholar]
- 5.Alonso W R, Croteau R. Methods Plant Biochem. 1993;9:239–260. [Google Scholar]
- 6.Delseny M, Cooke R, Raynal M R, Grellet F. FEBS Lett. 1997;405:129–132. doi: 10.1016/s0014-5793(97)00184-1. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto K, Sasaki T. Plant Mol Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- 8.Liu J, Hara C, Umeda M, Zhao Y, Okita T W, Uchimiya H. Plant Mol Biol. 1995;29:685–689. doi: 10.1007/BF00041159. [DOI] [PubMed] [Google Scholar]
- 9.Kwak J M, Kim S A, Hong S W, Nam H G. Planta. 1997;202:9–17. doi: 10.1007/s004250050097. [DOI] [PubMed] [Google Scholar]
- 10.Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, et al. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allona I, Quinn M, Shoop E, Swope K, St. Cyr S, Carlis J, Riedl J, Retzel E, Campbell M M, Sederoff R, et al. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCaskill D, Gershenzon J, Croteau R. Planta. 1992;187:445–454. doi: 10.1007/BF00199962. [DOI] [PubMed] [Google Scholar]
- 13.Alonso W R, Rajaonarivony J I M, Gershenzon J, Croteau R. J Biol Chem. 1992;267:7582–7587. [PubMed] [Google Scholar]
- 14.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;2:15. doi: 10.1016/S0022-2836(05)80360-2. , 403–410. [DOI] [PubMed] [Google Scholar]
- 17.Sutton G G, White O, Adams M D, Kervalage A R. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- 18.Staden R. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 19.Schenk P M, Baumann S, Mattes R, Steinbiss H-H. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 20.Barnes H J. Methods Enzymol. 1996;272:3–14. doi: 10.1016/s0076-6879(96)72003-7. [DOI] [PubMed] [Google Scholar]
- 21.Colby S M, Alonso W R, Katahira E, McGarvey D J, Croteau R. J Biol Chem. 1993;268:23016–23024. [PubMed] [Google Scholar]
- 22.Lange B M, Wildung M R, McCaskill D, Croteau R. Proc Natl Acad Sci USA. 1998;95:2100–2104. doi: 10.1073/pnas.95.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange B M, Croteau R. Arch Biochem Biophys. 1999;365:170–174. doi: 10.1006/abbi.1999.1168. [DOI] [PubMed] [Google Scholar]
- 24.Burke C C, Wildung M R, Croteau R. Proc Natl Acad Sci USA. 1999;96:13063–13067. doi: 10.1073/pnas.96.23.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupien S, Karp F, Wildung M, Croteau R. Arch Biochem Biophys. 1999;368:181–192. doi: 10.1006/abbi.1999.1298. [DOI] [PubMed] [Google Scholar]
- 26.Gershenzon J, Maffei M, Croteau R. Plant Physiol. 1989;89:1351–1357. doi: 10.1104/pp.89.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzales R G, Haxo R S, Schleich T. Biochemistry. 1980;19:4299–4303. doi: 10.1021/bi00559a023. [DOI] [PubMed] [Google Scholar]
- 28.Van Driessche E, Beeckmans S, Dejaegere R, Kanarek L. Anal Biochem. 1984;141:184–188. doi: 10.1016/0003-2697(84)90443-3. [DOI] [PubMed] [Google Scholar]
- 29.Lewinsohn E, Steele C L, Croteau R. Plant Mol Biol Rep. 1994;12:20–25. [Google Scholar]
- 30.Turner G, Gershenzon J, Nielson E E, Froehlich J E, Croteau R. Plant Physiol. 1999;120:879–886. doi: 10.1104/pp.120.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann K, Bucher P, Falquet L, Bairoch A. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corpet F, Gouzy J, Kahn D. Nucleic Acids Res. 1999;27:263–267. doi: 10.1093/nar/27.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Degtyarenko K N, North A C T, Finlay J B C. Nucleic Acids Res. 1999;27:233–236. doi: 10.1093/nar/27.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCaskill D, Croteau R. Planta. 1995;197:49–56. [Google Scholar]
- 35.Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk M H, Bacher A. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 36.Lichtenthaler H K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–66. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- 37.Lange B M, Croteau R. Proc Natl Acad Sci USA. 1999;96:13714–13719. doi: 10.1073/pnas.96.24.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk M H. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos-Valdivia A C, van der Heiden R, Verpoorte R. Nat Prod Rep. 1997;14:591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- 40.Ogura K, Koyama T. Chem Rev. 1998;98:1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- 41.Bohlmann J, Meyer-Gauen G, Croteau R. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crock J, Wildung M, Croteau R. Proc Natl Acad Sci USA. 1997;94:12833–12838. doi: 10.1073/pnas.94.24.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voirin B, Bayet C. Phytochemistry. 1992;31:2299–2304. [Google Scholar]
- 44.Kader J-C. Trends Plant Sci. 1997;2:66–70. [Google Scholar]
- 45.Lange B M, Croteau R. Curr Opin Plant Biol. 1999;2:139–144. doi: 10.1016/s1369-5266(99)80028-4. [DOI] [PubMed] [Google Scholar]