Abstract
Bcl-xL protein plays a role in breast cancer dormancy, promoting survival of cells in metastatic foci by counteracting the proapoptotic signals in the microenvironment. The aim of this study was to identify phenotypes mediated by Bcl-xL in breast cancer cells that enhance in vivo survival of clinical metastases. 435/Bcl-xL or 435/Neo human breast cancer cells were injected into the inguinal mammary gland of nude mice, and tumors, metastases in lymph node, lung, and bone, and bloodstream surviving cells were examined. Proteomic analysis identified 17 proteins that were overexpressed (more than twofold) or underexpressed (less than twofold) in metastases. A protein interaction program allowed us to functionally associate peroxiredoxin 3, peroxiredoxin 2, carbonyl reductase 3, and enolase 1, suggesting a role for cellular responses to oxidative stress in metastasis organ selection. The prediction included proteins involved in redox systems, kinase pathways, and the ATP synthase complex. Furthermore, the interaction of redox proteins with enolase 1 suggests a connection between glycolysis and antioxidant pathways, enabling achievement of a high metastatic activity. In conclusion, Bcl-xL mediates a phenotype in which redox pathways and glycolysis are coupled to protect breast cancer metastatic cells during transit from the primary tumor to the metastatic state.
Metastasis is a biological process that is a part of breast cancer progression. The metastatic phenotype includes the ability to migrate from the primary tumor, survive in blood or lymphatic circulation, invade distant tissues, and establish distant metastatic nodules. It is currently believed that the metastatic cascade involves a series of interrelated events including some that tumor cells use to withstand severe proapoptotic pressures from host-cell cytokines and growth factors.1–4
Because the disseminating tumor cells are freely and ubiquitously distributed by the hematogeneous or lymphatic system, the arrival to many organs is not sufficient for the development of secondary tumors. Unless a growing tumor colony is established at a new site, the metastatic process is not fulfilled because the local organ microenvironment at the site where the tumor cells are lodged determines whether metastases emerge or not.5
Tumor dormancy describes a prolonged quiescent state during which metastasis progression is not clinically detected.6 It has been suggested that dormancy might be related to anti-angiogenic factors that indirectly promote apoptosis and thus oppose proliferative tendencies.7 Indeed, metastatic cells may be present after surgery but remain dormant due to an inability to induce angiogenesis, or to change the balance between other growth-inducing/inhibiting factors in the tumor microenvironment, a circumstance that may also determine the length of the period between dissemination and the appearance of clinical metastases.8,9 How cells that have been selected at the primary site for acquisition of self-sufficiency can suppress these activities and remain dormant for years is an enigma. It has been suggested that tumor cells may disseminate in a far less progressed genomic state than previously thought, acquiring genomic aberrations typical of metastatic cells thereafter.10 Other groups have found that solid tumors carrying a gene-expression signature were mostly associated with metastasis and poor clinical outcome, suggesting that the metastatic potential of human tumors is encoded in the bulk of a primary tumor and that tumors likely to metastasize are fundamentally different.11–13
The molecular and cellular mechanisms responsible for the metastatic phenotype in breast cancer involve, among other factors, a number of gene products that participate in apoptosis.14–17 Anti-apoptotic genes have a role in displacing the balance between death and proliferation factors toward growth, possibly shortening the period between dissemination and the appearance of clinical metastasis.8
Bcl-xL expression in breast cancer cells increases metastatic activity. This might result from resistance to apoptosis against cytokines, increasing cell survival in circulation, and enhancing anchorage-independent growth.18 We have described that overexpression of anti-apoptotic Bcl-xL plays a role in breast cancer dormancy, promoting survival of cells in metastatic foci by counteracting the proapoptotic signals in the microenvironment and favoring the successful development of metastases in specific organs,19 preferentially lodging in peripheral lymph nodes.20 Thus, by selecting the organ-specific most adaptive phenotype, anti-apoptotic proteins might be a hallmark of metastasis and resistance to therapies.21 The aim of this work is to provide insight into the metastasis phenotype of breast cancer cells overexpressing Bcl-xL, which results in a short dormancy period in several organs.
The systematic large scale profiling of protein expression by proteomic methods allows the molecular characterization of cellular events associated with cancer progression.22 We have used two-dimensional gel electrophoresis (2DE), with subsequent identification of specific proteins of interest by mass spectrometry, to define phenotypes that in vivo enhance the ubiquity of clinical metastasis of Bcl-xL-overexpressing cells. On the assumption that the progenitor of the later-arising metastasis must be present among those cells that have disseminated to distant sites, our analysis included the orthotopic tumor, lymph node, lung and bone metastasis, and the previously originated blood stream survival cells (BSCs).18
We have identified 17 proteins that are either overexpressed (more than twofold) or underexpressed (less than twofold) in metastases with regard to the tumor. Furthermore, using recently developed protein-protein interaction prediction algorithms we have proposed interactors for proteins for which no reliable interactions could be inferred from experimental data. We have used protein-protein interaction databases and in silico prediction approaches to find cell functions relevant to the 17 identified proteins and which could guide additional research into expression patterns and metabolic and signal pathways.
Materials and Methods
Human Breast Carcinoma Cell Cultures and Transfections
MDA-MB 435 cell cultures (435) were maintained in 1:1 (v/v) mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 medium supplemented with 10% fetal bovine serum, 1 mmol/L piruvate, and 2 mmol/L l-glutamine in 5% CO2-95% air at 37°C in a humidified incubator. Transfections of pSFFV-Neo Bcl-xL or pSFFV-Neo were performed with Lipofectin (Life Technologies, Inc., Gaithersburg, MD). Selection of 435/Bcl-xL and 435/Neo cells started 48 hours after transfection using 500 μg/ml of neomycin G418 (Life Technologies). The pool of transfectants was used to avoid the possibility of clonal variations.
We generated orthotopic primary tumors in 7-week-old athymic nude BALB/c female mice by inoculation of 1 × 106 cells in 0.05 ml of medium without serum in the right inguinal mammary gland (i.m.f.p.), following previous protocols.18 Tumors grew at day 45 from cell injection, and animals were put down and organs removed, weighed, and examined for metastasis at day 110. Metastatic involvement was explored in paraffin sections by hematoxylin and eosin (H&E) stain. Lung metastasis from tumors overexpressing Bcl-xL reached more than 3 mm in diameter, whereas only small metastases of ≤3 mm were found in the lungs of 435/Neo-injected mice. Monolayers of metastatic cells were obtained from trypsin-treated histocultures (metastasis fragments of ≈1 mm3) maintained until growth in medium supplemented with 20% fetal bovine serum, in the presence of 500 μg/ml of neomycin G418. Because lymph node and bone metastasis appeared in mice from Bcl-xL clones but not in mice from 435/Neo control cells, metastatic variants were established from bone, lung, and lymph node metastasis from mice carrying 435/Bcl-xL tumors, and lung metastasis from mice carrying 435/Neo tumors.
Previously, we had obtained and characterized survival cells in the blood stream (BSCs).18 Briefly, a mixture of cells 3:1, 435/Neo and 435/Bcl-xL, respectively, was injected into the tail vein of nude mice. We assessed differences in survival rates between the two clones by comparing the original mixture and cell populations recovered from the lungs. Mice were killed at 15 minutes, 2, 4, 8, and 16 hours, and recovered cells were expanded in culture and the DNA was hybridized with a radiolabeled 0.8-kb fragment of Bcl-xL to analyze the Bcl-xL cDNA in survival clones. We used for experiments as a BSC the cell variant recovered from lungs 4 hours after injection into the tail vein of nude mice.
Sample Preparation
Cultured cells derived from metastasis and primary tumors were used for proteomic analyses. Cells (1 × 106) were plated onto a 75-mm flask in complete medium for 48 hours, starved for 24 hours in serum-free medium, and replaced by complete medium for 24 hours more. Cells were rinsed in ice-cold phosphate-buffered saline (PBS) before trypsinization with 0.05% trypsin and 0.5 mmol/L ethylenediamine tetraacetic acid. Variants in which a floating subpopulation was growing were preserved at each pass, by recovering the PBS wash before trypsin treatment. The cell pellets were frozen in N2 and stored at −80°C until electrophoresis. For this purpose, 1 × 106 cells were solubilized in 200 μl of lysis buffer containing 8 mol/L urea, 4% CHAPS (w/v), and 40 mmol/L Tris, and then homogenized by passing through a 25-gauge needle. Insoluble material was removed by centrifugation at 13,000 rpm for 10 minutes at 4°C. Protein concentration was determined by BCA protein assay reagent (Pierce, Rockford, IL).
Two-Dimensional Gel Electrophoresis (2DE)
Immobilized pH 4 to 7 gradient (IPG) 18-cm strips, (Immobiline DryStrip 4-7 NL; Amersham Pharmacia Biotech AB, Uppsala, Sweden) were rehydrated with 350 ml of rehydration solution (8 mol/L urea, 4% CHAPS, 1% IPG buffer, 13 mmol/L dithiothreitol), in which 200 μg of protein were applied. Isoelectric focusing was performed using a IPGphor apparatus (Amersham Pharmacia Biotech) for a total of 80 kV/hour, 50 mA/strip at 20°C, according to the manufacturer’s instructions, in a first-dimension isoelectric focusing system (Amersham Pharmacia Biotech). Immediately after being focused, IPG strips were equilibrated for 15 minutes in 6.5 mmol/L dithiothreitol, 50 mmol/L Tris, 6 mol/L urea, 30% glycerol, and 2% sodium dodecyl sulfate, followed by 13.5 mmol/L iodoacetamide, 50 mmol/L Tris, 6 mol/L urea, 30% glycerol, and 2% sodium dodecyl sulfate, 15 minutes before running in the second dimension, which was performed on 12.5% polyacrylamide gels of 18 × 20 cm in an electrophoresis tank Protean II (Bio-Rad, Hercules, CA). For protein identification (see below) preparative 2DE gels loaded with 300 μg of protein were run following the same protocol. After electrophoresis, gels were fixed and stained using a silver-staining low-fixation protocol compatible with mass spectrometry, dried between cellophane sheets, scanned in a densitometer, and processed with the Molecular Analyst Software (Bio-Rad, Richmond, CA) into Adobe Photoshop.
Analysis of Gel Images
The digitalized 2DE gel images were normalized and comparatively analyzed with the PDQuest program (version 7.0; Bio-Rad, Hercules, CA). Four gels from each specimen, in two independent experiments, were analyzed and the mean gel was used for comparisons. Normalization was assessed using the formula: normalized spot quantity = raw spot quantity × scaling factor (106 ppm) × normalization factor (total density in gel image). Significant changes were ratios <0.5, underexpressed, or >2, overexpressed, with regard to the referent sample. Molecular weight (MW) and isoelectric point (pI) were determined after comparison with reference gels in the SWISS-2DPAGE (Expasy) database.
Protein Fingerprinting by Matrix-Assisted Laser Desorption Ionization/Time of Flight (MALDI-TOF) Mass Spectrometry
Protein spots were selected according to the silver stain signal without overlap. The selected spots were excised from 2DE silver-stained gels and submitted to trypsin in-gel digestion as described before23 with minor modifications. After reduction and alkylation the gel pieces were swollen in 10 μl of a digestion buffer containing 50 mmol/L NH4HCO3 and 5 ng/μl of trypsin (modified porcine, sequencing grade; Promega, Madison, WI) in an ice bath. After 45 minutes 30 μl of 50 mmol/L NH4HCO3 were added and the digestion allowed to proceed overnight at 37°C. The samples were acidified using 5% formic acid and 10 to 20 μl of the supernatant containing tryptic peptides purified in a Poros R2 column24 (Applied Biosystems, Framingham, MA) packed in a constricted GelLoader Tip (Eppendorf, Hamburg, Germany) previously washed with 10 μl of acetonitrile and equilibrated with 10 μl of 5% formic acid. The sample was washed with 10 μl of 5% formic acid and eluted directly to the MALDI plate with 0.8 μl of matrix solution (20 mg/ml α-cyano-4-hydroxycinnamic acid in 70% in 0.1% formic acid). Peptide mass fingerprinting spectra were recorded in a Voyager STR MALDI-TOF (Applied Biosystems) in positive reflector mode with delayed extraction. The spectra were analyzed using the program m/z (Proteometrics, New York, NY). Protein identification was performed against a nonredundant database (NCBI) using the MASCOT program (http://www.matrixscience.com).
Experimental Validation of Specific Proteins Expressed in Metastasis
Western Blot Analysis
Cells from exponential cultures were lysed in 200 μl of RIPA buffer (50 mmol/L Tris, 150 mmol/L NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% sodium deoxycholate). Sample volumes were adjusted to contain 50 μg of protein as determined by the BCA protein assay reagent (Pierce), and electrophoresed in a 12% polyacrylamide gel. The separated proteins were transferred to polyvinylidene difluoride membranes (Immobilon-p; Millipore Corp., Bedford, MA) and nonspecific protein-binding sites blocked by a 5% solution of nonfat dried milk with 0.01% Tween 20 in PBS. The following antibodies (Abs) were used: polyclonal rabbit Ab specific for human Bcl-x (S-18) at 1/1000 dilution (Santa Cruz Biotechnology, Santa Cruz, CA); polyclonal rabbit Ab anti-stathmin 1 (OP18) at 1/1500 dilution (Sigma, St. Louis, MO); monoclonal Ab anti-peroxiredoxin 3 (AOP-1) at 1/8000 dilution (Sigma); polyclonal rabbit Ab anti-peroxiredoxin 2 at 1/1000 dilution (LabFrontier, Seoul, Korea); polyclonal goat Ab anti-enolase 1 (eno α) at 1/500 dilution (Santa Cruz Biotechnology); polyclonal rabbit Ab anti-glutathione transferase (GST P1-1) at 1/1000 dilution (Calbiochem, CN Biosciences Ltd., Nottingham, UK); a monoclonal anti-tyrosine 3/tryptophan 5-monooxygenase, 14-3-3-ε (clone 12) at 1/500 dilution (BD Biosciences, Pharmingen, Franklin Lakes, NJ). Peroxidase-conjugated goat anti-rabbit secondary antibody 1/2000 (Amersham), or anti-mouse secondary antibody 1/2000 (Pierce, Perbio Science Ltd., Cheshire, UK), or anti-goat secondary antibody 1/3000 (Santa Cruz Biotechnology) were used as corresponded in each case. Immunoreactive bands were visualized on CL-x posure Film (Pierce) using the Super Signal West-Pico (Pierce). MWs were established using Benchmark prestained MW markers (Invitrogen, San Diego, CA). An anti-human actin monoclonal antibody 1/2000 (Sigma) was also used as an internal standard for densitometric analysis of X-ray film. Densitometry analysis with the Quantity One program was performed using the quantity of a band that it is the sum of the intensities of all of the pixels within the band boundary multiplied by the area of each pixel.
Immunohistochemistry
Paraffin-embedded tissue was cut into 6-μm sections, deparaffinized, and rehydrated. On antigen retrieval with citrate buffer (pH, 6), we incubated with the following primary Abs: anti-stathmin 1 (OP18) at 1/500 dilution (Sigma); anti-peroxiredoxin 3 (AOP-1) at 1/500 dilution (Sigma), and anti-enolase 1 (enoα) at 1/100 dilution (Santa Cruz Biotechnology). Bound antibody was visualized with biotinylated anti-IgG (Vector Laboratories, Burlingame, CA) after diaminobenzidine staining and counterstaining with hematoxylin in an Olympus BX60 (Olympus Optical Co., Ltd., Tokyo, Japan) and digitalized in a digital camera Spot using the Spot 4.2 software (Diagnostic Instruments, Inc., Sterling Heights, MI).
Bioinformatics
We used the STRING (search tool for the retrieval of interacting genes/proteins25) to find interactors of the proteins identified by mass spectrometry. STRING contains protein-protein interactions that have been predicted using different methods such as gene fusion,26 phylogenetic profiles,27 or gene neighborhood.28 For each of these predictions, STRING gives a confidence score, which measures how likely it is that the prediction is a true interaction. Furthermore, we have developed an algorithm that predicts interactions between proteins by propagating known interactions in one species to ortholog proteins in another species (ie, assuming protein-protein interologs).29 For this algorithm we have used the protein-protein interactions contained in the data base of interacting proteins.30
Using interactions from STRING (at a confidence level of 400) and the protein-protein interologs found by our algorithm, we built a protein-protein interaction network for each of the metastatic organs studied in this work. Taking as root proteins of the network those that in the metastatic organ were under- or overexpressed with respect to the tumor, we obtained networks that attempt to describe the relationship between proteins relevant to metastasis in that organ. We then grouped proteins by their cellular function and extracted those functions that were connecting at least two root proteins. These cellular functions and the root proteins that they connected provided us with clues about processes that are potentially involved in metastasis.
Results
Analysis of Bcl-xL-Mediated Protein Expression Changes in Breast Cancer Metastasis
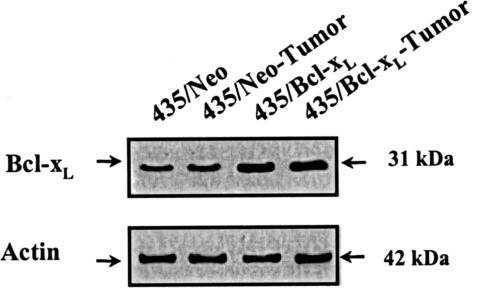
For these experiments we used MDA-MB 435 cells transfected with the anti-apoptotic gene Bcl-xL or the control 435/Neo cells. The parental cells expressed low levels of Bcl-xL protein, and the transgene protein expression was twofold higher than in the control cells. Comparative analyses of Bcl-xL protein expression by Western blot showed stable overexpression of these genes in 435/Neo and 435/Bcl-xL tumor cells (Figure 1).
Figure 1.
Western blot analyses from 435/Neo and 435/Bcl-xL cells. Whole cell lysates containing 50 μg of total protein were loaded, separated by PAGE, and blotted to nitrocellulose membranes. The indicated proteins were detected using specific primary antibodies and visualized using HRP-conjugated secondary antibodies.
The differential expression of proteins was analyzed in quadruplicate 2DE gels from two independent experiments, after normalizing and quantifying spot volumes by means of the PDQuest program. We were able to identify an average of 250 spots in each gel. Seventy-one protein expression changes were found between breast tumors induced with either 435/Bcl-xL or 435/Neo cells. Differential protein expression between 435/Bcl-xL metastatic and 435/Bcl-xL tumor cells involved 126 modified proteins. We found 75 proteins whose expression changed in 435/Bcl-xL lung metastasis with regard to 435/Bcl-xL tumor: 31 were altered only in lung metastatic cells; 7 were found modified in lung and lymph node metastasis, 16 in lung and bone metastasis; and 21 in lung, lymph node, and bone metastasis (Figure 2A).
Figure 2.
Proteins differentially expressed in metastasis with regard to the tumor. A: Representative diagrams with the comparative proteome analysis of 435/Bcl-xL lung, 435/Bcl-xL lymph node, and bone metastases with regard to 435/Bcl-xL tumor. We show the number of protein expression changes, and the location of identified proteins by MALDI-TOF in each metastasis. Some of them are present in all metastases; others are bone, lymph node, or lung organ-specific proteins. B: Comparison of protein changes between lung metastasis in control mice, 435/Neo lung, and lung metastasis in mice with tumors overexpressing Bcl-xL, 435/Bcl-xL lung. The number of changes with regard to their respective tumor is represented, as well as the number of changes shared by both metastases.
Bone metastasis had the most different expression profile with regard to the tumor, with 82 protein changes. In contrast, 42 proteins were differentially expressed in lymph node metastasis when compared to the tumor. On the other hand, only 39 modified proteins were found in 435/Neo lung metastasis with regard to 435/Neo tumor. Thus, lung metastasis from 435/Bcl-xL cells had more phenotypic changes with regard to their tumor than lung metastasis from control cells (Figure 2B).
Identification of Bcl-xL-Mediated Protein Expression Changes in Metastasis
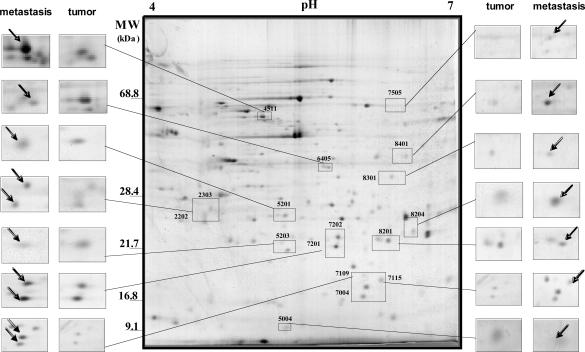
Seventeen proteins displaying differential spots in 2DE gels from either tumor cells or metastases (Figure 3) could be identified by MALDI-TOF peptide fingerprint analysis, from a total of 29 picked out. These proteins are listed in Table 1, with their corresponding MW, pI, and recognized function (according to SwissProt database), as well as references to their relevance to breast cancer, primarily derived from experiments using array technologies.
Figure 3.
Image of a silver-stained 2DE gel with 200 μg of protein. The gel is oriented with acidic proteins to the left and basic proteins to the right. High molecular weight proteins are near the top whereas low molecular weight proteins are near the bottom. Proteins that were identified by excision followed by mass spectrometry are indicated with spot numbers. Differentially expressed proteins between tumor and metastasis are enlarged with the boxes.
Table 1.
Identities of Differentially Expressed Proteins in Metastasis from MDA-MB 435/Bcl-xLBreast Tumors
| Spot no.* | Protein identity | Acc. no. | pI | MW (kd) | Function | Differential expression in breast cancer† |
|---|---|---|---|---|---|---|
| 2202 | Multicatalytic endopeptidase complex ζ chain | P28066 | 4.7 | 26.58 | Proteolysis | Su et al, 2001; Perou et al, 1999; Ross et al, 2000 |
| 2303 | Tyrosine 3/tryptophan 5 monoxygenase | NP_006752 | 4.6 | 29.33 | Transduction | Vercoutter-Edouart et al, 2001 |
| 4511 | ATP synthase β chain | P06576 | 5.3 | 56.53 | Synthesis of ATP | Chen et al, 2001 |
| 5004 | β-tubulin co-factor A | O75347 | 5.3 | 12.90 | Complex assembly | Perou et al, 1999 |
| 5201 | Ran-BP1 (ran-binding protein 1) | P34022 | 5.2 | 23.74 | GTPase activating | |
| 5203 | My 032 protein | AF061735 | 6.6 | 15.82 | ||
| 6405 | F-actin capping protein α-1 subunit | P52907 | 5.5 | 33.07 | Complex assembly | Su et al, 2001; Ross et al, 2000 |
| 7004 | Stathmin 1 | P16949 | 5.8 | 17.29 | Transcription | Perou et al, 1999; Curmi et al, 2000; Brattsand, 2000; Bieche et al, 1998 |
| 7109 | Familial Als mutant G37r | IAZV_A | 5.9 | 16.12 | Redox | Chandel, 2000 |
| 7115 | Nonmetastatic cells 1 protein nm-23 | P15531 | 5.8 | 17.31 | Transcription | Su et al, 2001; Nakopoulou et al, 1999; Kapranos, 1996 |
| 7201 | Peroxiredoxin 2 | 1QMV_A | 5.4 | 21.91 | Redox | Su et al, 2001; Perou et al, 1999; Ross et al, 2000; Husbeck et al, 2001; Berggren et al, 2001; Harvey et al, 2001 |
| 7202 | Glutathione transferase | P09211 | 5.4 | 23.57 | Glutathione transferase | Perou et al, 1999; Ross et al, 2000; Dialyna et al, 2001; Ghalia et al, 2000; Huang et al, 2000 |
| 7505 | Aminoacylase 1 | Q03154 | 5.8 | 46.08 | Hydrolysis | Su et al, 2001; Perou et al, 1999; Ross et al, 2000 |
| 8201 | Hypothetical protein MGC15429 | NP_116139 | 5.9 | 22.45 | ||
| 8204 | Peroxiredoxin 3 | P30048 | 7.1 | 28.05 | Redox | Su et al, 2001; Perou et al, 1999; Ross et al, 2000 |
| 8301 | Carbonyl reductase 3 | O75828 | 5.8 | 31.23 | Redox | |
| 8401 | Enolase 1 | P06733 | 7.1 | 47.78 | Glycolysis | Su et al, 2001; Perou et al, 1999; Ross et al, 2000 |
Spots were excised from 2DE gels and peptide mass spectrum from gel trypsin digestion was obtained by MALDI-TOF mass spectrometry.
In silico validation was made taking into account experimental results referred to breast cancer and metastasis.
As shown in Table 2, 12 of these identified proteins were significantly overexpressed (more than twofold with regard to the tumor) in 435/Bcl-xL lung metastasis: multicatalytic endopeptidase complex ζ-chain (twofold), tyrosine 3/tryptophan 5-monoxygenase activation protein (48-fold), ATP synthase β-chain (twofold), My 032 protein (twofold), stathmin 1 (fourfold), familial ALS mutant G37r (fourfold), nonmetastatic cells 1 protein nm-23 (fivefold), peroxiredoxin 2 (threefold), glutathione transferase (twofold), aminoacylase 1 (fivefold), peroxiredoxin 3 (threefold), and enolase 1 (sixfold).
Table 2.
Differentially Expressed Proteins (More than or Less than Twofold) in BSC Cells and Metastasis
| Spot no. | Protein identity | 435/Bcl-xL tumor* | 435/Neo lung† | 435/Bcl-xL BSC‡ | 435/Bcl-xL lung§ | 435/Bcl-xL node 1§ | 435/Bcl-xL bone 1§ |
|---|---|---|---|---|---|---|---|
| 2202 | Multicatalytic endopeptidase complex ζ chain | 0.45 | 1.27 | 0.04 | 2.30 | 2.48 | 2.60 |
| 2303 | Tyrosine 3/tryptophan 5-monoxygenase | 0.03 | 0.36 | 0.73 | 48.20 | 32.90 | 20.20 |
| 4511 | ATP synthase β chain | 0.68 | 1.50 | 1.20 | 2.05 | 3.00 | 0.60 |
| 5004 | β-tubulin co-factor A | 0.63 | 1.50 | 0.03 | 1.20 | 0.70 | 0.06 |
| 5201 | Ran BP1 (ran-binding protein) | 1.35 | 1.43 | 0.04 | 0.38 | 1.04 | 0.94 |
| 5203 | My 032 protein | 0.73 | 1.78 | 0.30 | 2.10 | 1.08 | 1.29 |
| 6405 | F-actin capping protein α-1 subunit | 1.60 | 2.37 | 0.61 | 1.04 | 0.80 | 0.29 |
| 7004 | Stathmin 1 | 0.23 | 1.03 | 0.03 | 3.70 | 1.70 | 1.40 |
| 7109 | Familial Als mutant G37r | 0.32 | 1.69 | 0.81 | 4.00 | 3.90 | 1.80 |
| 7115 | Nonmetastatic cells 1 protein nm-23 | 0.19 | 1.14 | 0.65 | 4.60 | 1.80 | 2.80 |
| 7201 | Peroxiredoxin 2 | 0.70 | 1.50 | 1.45 | 2.49 | 1.80 | 1.00 |
| 7202 | Glutathione transferase | 0.68 | 1.60 | 0.24 | 2.04 | 1.80 | 0.74 |
| 7505 | Aminoacylase 1 | 0.02 | 0.90 | 9.33 | 4.86 | 5.88 | 1.49 |
| 8201 | Hypothetical protein MGC15429 | 0.90 | 1.23 | 0.38 | 1.40 | 1.30 | 0.16 |
| 8204 | Peroxiredoxin 3 | 0.83 | 1.12 | 0.85 | 2.90 | 2.12 | 0.79 |
| 8301 | Carbonyl reductase 3 | 1.09 | 1.06 | 0.26 | 0.60 | 0.68 | 0.38 |
| 8401 | Enolase 1 | 0.70 | 1.20 | 11.07 | 6.17 | 2.15 | 2.17 |
Differentially expressed proteins in 435/Bcl-xL tumor as regards 435/Neo tumor.
Differentially expressed proteins in 435/neo-lung metastasis as regards 435/Neo tumor.
Differentially expressed proteins in 435/Bcl-xL BSC as regards 435/Bcl-xLtumor.
Differentially expressed proteins in lung, lymph node, and bone metastasis as regards 435/Bcl-xL tumor.
In contrast, only seven of these proteins were overexpressed in lymph node metastasis: multicatalytic endopeptidase complex ζ-chain (threefold), tyrosine 3/tryptophan 5-monoxygenase activation protein (33-fold), ATP synthase β-chain (threefold), familial ALS mutant G37r (fourfold), aminoacylase 1 (sixfold), peroxiredoxin 3 (twofold), and enolase 1 (twofold). Only three proteins were overexpressed in bone metastasis: multicatalytic endopeptidase complex ζ-chain (threefold), tyrosine 3/tryptophan 5-monoxygenase activation protein (20-fold), nonmetastatic cells 1 protein nm-23 (threefold), and enolase 1 (twofold).
Protein underexpression with regard to the tumor was more frequent in bone metastasis. We found less β-tubulin co-factor A (0.01-fold), F-actin capping protein α-1 subunit (0.3-fold), hypothetical protein MGC15429 (0.1-fold), and carbonyl reductase 3 (0.3-fold). In contrast, only Ran BP1 protein was underexpressed (0.4-fold) in 435/Bcl-xL lung metastasis. In lymph node metastasis we did not find underexpression of these proteins relative to the tumor. Thus, the phenotype evolving in lung metastasis was quite different from that of bone metastasis.
In contrast to 435/Bcl-xL lung metastasis, neither of the above described changes were found in lung metastasis from 435/Neo cells, where the only observed modifications, with regard to its tumor, were overexpression of F-actin capping protein α-1 subunit (twofold), and underexpression of tyrosine 3/tryptophan 5-monoxygenase activation protein (0.4-fold). Furthermore, neither of the 435/Bcl-xL metastasis-related proteome alterations were found in 435/Bcl-xL tumor cells with regard to 435/Neo tumor cells, thus expression changes induced by Bcl-xL must be emerging during tumor development and modulated throughout metastatic steps.
Survival in the Blood Stream (BSC) versus Organ-Specific Changes
Because protein expression profiles varied between metastatic sites, we analyzed the expression of proteins in cells living in bloodstream and rescued from lung 4 hours after intravenous injection.18 This allowed us to discern whether changes in protein expression resulted from cross-talk between metastatic cells and the different environments that finally select the phenotype, or belonged to subpopulations living in tumors selected during progression to metastasis.
The expression of aminoacylase 1 and enolase 1 in BSCs with regard to 435/Bcl-xL tumor cells increased by 9- and 11-fold, respectively. Moreover, no over- or underexpression was found in the rest of proteins. Thus, the enrichment of both proteins is a phenotype selected in the bloodstream and maintained in the following steps of the metastatic process.
Some organ-specific changes were already present in BSCs, such as underexpression of β-tubulin co-factor A (0.03-fold), hypothetical protein MGC15429 (0.4-fold), and carbonyl reductase 3 (0.3-fold), present in bone metastasis; or Ran BP1 (0.04-fold) present only in lung metastasis. Conversely, other organ-specific proteins were not enough represented in BSCs but did occur in metastatic foci, such as My 032 protein and stathmin 1 (twofold and fourfold, respectively) in lung cells, and F-actin capping protein α-1 subunit (0.3-fold) in bone cells.
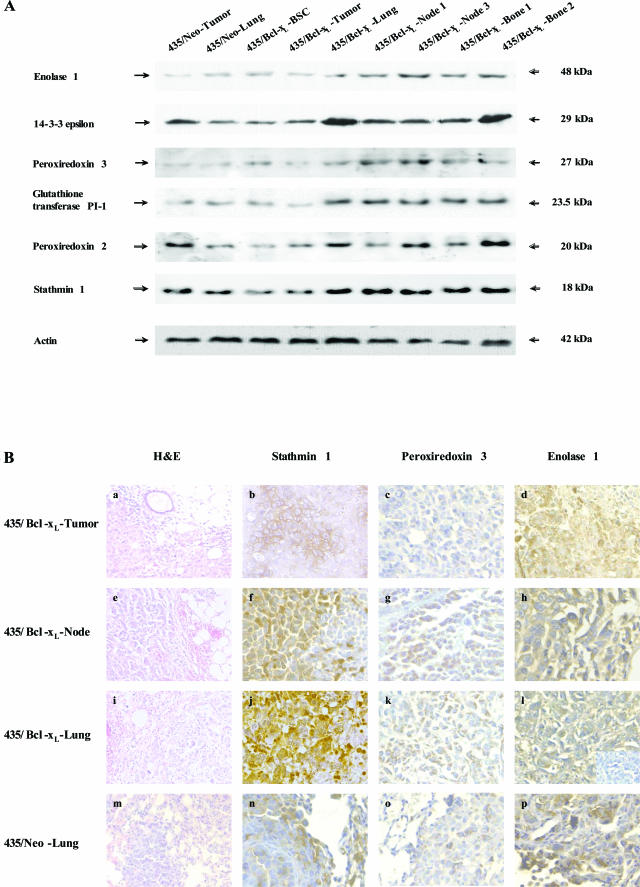
Finally, changes common to all metastases but not evident in BSCs were multicatalytic endopeptidase complex ζ-chain and tyrosine 3/tryptophan 5-monoxygenase activation protein (14-3-3-ε). We used Western blot analyses with specific antibodies against some of these proteins to assess with an alternative technique the different expression between tumor, BSCs, and metastases (Figure 4A). The analyses confirmed the overexpression (more than twofold) of enolase 1 and glutathione transferase in BSCs, and in all metastases from 435/Bcl-xL cells with regard to the tumor; the increased expression of peroxiredoxin 2 and stathmin 1 in 435/Bcl-xL lung metastasis with regard to tumor and BSCs; and the overexpression of 14-3-3-ε in all metastasis with regard to the tumor and BSCs (Table 3).
Figure 4.
Specific expression of proteins in cells and tissues. A: Western blot analyses from metastatic variants. Whole cell lysates containing 50 μg of total protein from control lung metastasis, 435/Bcl-xL-BSC, and 435/Bcl-xL metastatic variants from lung, lymph nodes (node and node 3), and bones (bone and bone 2), were loaded, separated by PAGE, and blotted to nitrocellulose membranes. The indicated proteins were detected using specific primary antibodies and visualized using HRP-conjugated secondary antibodies. Anti-human actin monoclonal antibody was used as an internal standard. B: Immunohistochemistry of paraffin-embedded tissues obtained from mice, in which breast cancer cells were injected i.m.f.p. a, e, i, m: H&E staining of each tissue are shown, visualized by light microscopy. Immunolocation of stathmin 1, peroxiredoxin 3, and enolase 1 expression, respectively, are shown: in breast tumors from mice injected with 435/Bcl-xL cells (b–d); in lymph node metastasis (f–h) and lung metastasis (j–l) from these mice; and lung metastasis obtained from mice whose tumors were induced with 435/Neo cells (n–p). The inset in l shows the negative control for enolase 1 staining using an unrelated primary antibody. Original magnifications: ×20 (a, e, i, m, and inset in l); ×40 (b–d, f–h, j–l, n–p).
Table 3.
Ratio of Protein Expression Using the Quantity One Program to Analyze the Western Blot (Figure 4A)
| Protein identity | 435/Bcl-xL tumor* | 435/Neo lung† | 435/Bcl-xL BSC‡ | 435/Bcl-xL lung§ | 435/Bcl-xL node§ | 435/Bcl-xL node 3§ | 435/Bcl-xL bone§ | 435/Bcl-xL bone 2§ |
|---|---|---|---|---|---|---|---|---|
| 14-3-3-ε | 0.48 | 0.43 | 0.76 | 2.86 | 2.00 | 1.86 | 1.97 | 2.62 |
| Stathmin 1 | 0.87 | 0.81 | 0.50 | 3.62 | 1.61 | 1.74 | 1.13 | 1.40 |
| Peroxiredoxin 2 | 1.03 | 1.18 | 0.85 | 2.42 | 1.88 | 1.50 | 1.29 | 1.58 |
| Glutathione transferase | 0.34 | 0.96 | 1.93 | 4.23 | 2.71 | 2.72 | 2.54 | 2.41 |
| Peroxiredoxin 3 | 2.02 | 0.88 | 1.17 | 1.86 | 2.43 | 2.23 | 1.56 | 0.98 |
| Enolase 1 | 1.11 | 1.78 | 2.29 | 5.03 | 3.62 | 3.63 | 2.08 | 2.81 |
Differentially expressed proteins in 435/Bcl-xL tumor as regards 435/Neo tumor.
Differentially expressed proteins in 435/neo-lung metastasis as regards 435/Neo tumor.
Differentially expressed proteins in 435/Bcl-xL BSC as regards 435/Bcl-xL tumor.
Differentially expressed proteins in lung, lymph node, and bone metastasis as regards 435/ Bcl-xL tumor.
To assess the influence of the culture in protein expression we performed immunohistochemistry in experimental metastatic tissue obtained by injecting cells into the intra-mammary fat pad (i.m.f.p.). Immunohistochemistry confirmed the 90% expression of enolase 1 in metastatic lung and lymph node cells, in contrast to 40% in lung metastasis control, and 60% in 435/Bcl-xL tumor. Peroxiredoxin 3 was expressed in 80 to 60% in 435/Bcl-xL lung and lymph node metastatic cells, showing a cytoplasm staining compatible with its mitochondria location. In contrast 20% of 435/Neo lung metastatic cells and 435/Bcl-xL tumor cells were peroxiredoxin 3-positive. Stathmin 1 had similar nuclear and cytoplasmic expression in 435/Bcl-xL lung and lymph node metastasis (70%) with 20% of cells with nuclear location, in contrast to the cytoplasmic expression in lung metastasis control (10%) and in 435/Bcl-xL tumor (30%) in which cells are regrouped (Figure 4B).
An in Silico Proposal of Interacting Proteins
The bioinformatic analysis revealed several potential human-protein interactors within the 17 proteins (root proteins hereafter) for which an organ-specific change in expression was experimentally found. These protein-connectors were clustered according to functional criteria: 1) cell division; 2) DNA-repair, synthesis, and translation; 3) redox; 4) structure and motion (ie, dynein, tubulin, actin, etc); 5) ATP synthase; 6) kinase-phosphatase; 7) calcium metabolism; 8) protein folding stability; 9) Krebs cycle; and 10) notch signaling. Proteins for which no function or human homologue was found were ignored.
The largest set of putative interactors was found linking proteins involved in lung metastasis, which could be expected from the fact that most root proteins exhibited differential expression in lung metastasis. Table 4 lists proteins bridging two or more proteins with organ-specific changes in expression. For all organs a central connection with a proteasome subunit, P28066, was found. It is also noteworthy that calmodulin (P62158), involved in calcium metabolism, connects P28066 and NDP kinase A, a metastasis-associated protein, P15531.
Table 4.
Organ-Specific Predicted Graph of Interactions
| Clustered function | Linking Proteins* | Connects root nodes† |
|---|---|---|
| Node | ||
| DNA repair synthesis and translation | ATP-dependent DNA helicase II, 70-kd subunit | Multicatalytic endopeptidase complex, peroxiredoxin 3 |
| Kinase-phosphatase | Homeodomain-interacting protein kinase 2 | Multicatalytic endopeptidase complex, peroxiredoxin 3 |
| Notch signaling | Sel-1 homolog | ATP synthase β chain, enolase 1 |
| Bone | ||
| Calcium metabolism | Calmodulin | Nucleoside diphosphate kinase A, Multicatalytic endopeptidase complex, carbonyl reductase 3 |
| Cell division | Transducer of Cdc42-dependent actin assembly-1 | F-actin capping protein α-1 subunit, multicatalytic endopeptidase complex |
| Estructure and motion | Coatomer β′ subunit | Carbonyl reductase 3, multicatalytic endopeptidase complex |
| Krebs cycle | Citrate synthase, mitochondrial | Carbonyl reductase 3, multicatalytic endopeptidase complex |
| Lung | ||
| ATP synthase | ATP synthase α chain, mitochondrial, ATP synthase 6, ATP synthase oligomycin sensitivity conferral protein mitochondrial | ATP synthase β chain, ATP synthase D chain, mitochondrial |
| Calcium metabolism | Calmodulin | Nucleoside diphosphate kinase A, multicatalytic endopeptidase complex |
| Cell division‡ | Hypothetical protein FLJ36874 | Peroxiredoxin 2, peroxiredoxin 3 |
| Kinase-phosphatase/cell division‡ | Cdc14A3 phosphatase | Peroxiredoxin 2, peroxiredoxin 3, ATP synthase D chain, mitochondrial |
| Kinase-phosphatase | Homeodomain-interacting protein kinase 2 | Multicatalytic endopeptidase complex, peroxiredoxin 2, peroxiredoxin 3 |
| Kinase-phosphatase | Ras-related protein Rab-22A | Nucleoside diphosphate kinase A, aminoacylase 1 |
| DNA repair synthesis and translation | ATP-dependent DNA helicase II, 70-kd subunit, SMARCA3 protein | Multicatalytic endopeptidase complex, peroxiredoxin 2, peroxiredoxin 3 |
| Structure and motion | DNCI1 protein | Peroxiredoxin 2, peroxiredoxin 3 |
| Structure and motion | Importin α-7 subunit | Nucleoside diphosphate kinase A, peroxiredoxin 2, peroxiredoxin 3 |
| Notch signaling | Sel-1 homolog | ATP synthase β chain, enolase 1 |
| Protein fold stability | Heat-shock protein 105 kd | Peroxiredoxin 2, peroxiredoxin 3 |
| Redox | DNA-directed RNA polymerase I | Peroxiredoxin 2, peroxiredoxin 3 |
| Ubiquitination | Ubiquitin carboxyl-terminal hydrolase 8, SMARCA3 protein | Peroxiredoxin 2, peroxiredoxin 3 |
Proteins predicted as links (with a predicted interaction between two root nodes) are indicated and grouped according to their principal cell function.
Proteins identified by 2DE with organ-specific changes of expression in metastasis are indicated in the connects root nodes column.
Function is postulated by homology.
Proteins with functions such as cell-division, protein kinase, and DNA-repair, synthesis and translation appear significantly involved in connecting proteins with differential expression in all organs. It is worth mentioning that ATP-dependent DNA helicase II (P12956) interacts with two root proteins with changed expression in lymph node and lung metastases. The root proteins are P28066 and different types of thioredoxin-dependent peroxide reductase (peroxiredoxin, P32119 and P30048), involved in redox regulation of the cell and protecting radical-sensitive enzymes from oxidative damage.
In lung metastasis there are more proteins linking the two types of peroxiredoxin P32119 and P30048. Among them it is worth to point out: 1) O15160, another peroxiredoxin; 2) Q8N9M6, assigned to cell division function (see above); and 3) P40818 (ubiquitin thiolesterase 8) and Q86YA5 (SMARCA3), two proteins involved in ubiquitination and containing domains for DNA-binding, helicase C activity, and a ring zinc finger. Among the proteins found to link proteins with altered expression in lung metastasis, many are kinases, usually related to signal cascades: Q9UBV2, a homologue of Sel-1; HIPK2 (Q9H2X6), which phosphorylates homeodomain transcription factors; Rab22 (Q9UL26), in the GTPase family; and Cdc14A3 phosphatase (O60728), which is a Tyr-phosphatase that may play a role in cell division.
The functions exerted by metastatic cells have been validated in silico. It has to be understood that these links are proposed through predictions based on homology because no real experiments have been performed with these proposed human proteins. Therefore, only the implication on a cluster of related proteins should be considered as potential real connections. This limits the prediction to few clusters: for proteins involved in a redox system (putatively affecting other functions involved in DNA helicase and cell division and ubiquitination), the set of kinases (which putatively could implicate some signal transduction), and the ATP synthase complex. Taking into account known proteins and their interactions, we suggest a model of breast cancer metastasis progression of Bcl-xL-overexpressing tumors, in which Bcl-xL could mediate changes in metabolic pathways of breast cancer cells, with the result of ubiquity and organ specificity of a progressively selected phenotype from the survival in the blood stream to organ-specific clinical metastasis (Figure 5).
Figure 5.
Hypothesis of an adaptor phenotype mediated by Bcl-xL in breast cancer progression by connecting several metabolic pathways that might allow the organ-specific metastasis development. Overexpression of anti-apoptotic Bcl-xL promotes survival advantage of cells in metastatic foci by counteracting the proapoptotic signals in the microenvironment and favoring the successful development of metastasis in specific organs.17 The shortened dormancy of Bcl-xL metastatic cells might be induced by the selection of a phenotype through cell survival in the blood stream (BSC) created by inducing resistance to apoptosis against cytokines, increasing cell survival in circulation, and enhancing anchorage-independent growth.16 We have identified aminoacylase 1 and enolase 1 overexpression as a common phenotype in 435/Bcl-xL BSCs and metastatic variants in lymph nodes and lung (putatively affecting other functions involved on Notch signaling and kinase activity). The organ-specific growth in lung might be as a result of the interaction between metabolic routes involved in a redox system as well as peroxiredoxin 3 and 2, and glutathione transferase (putatively affecting other functions involved in DNA helicase and cell division and ubiquitination), with kinase activity thought bridging proteins that link two proteins, ATP synthase β-chain and enolase 1. The low expression of these proteins in bone metastasis might be related to an increased dormancy of cells, with demonstrated lung and lymph node tropism59 that reach the bone, in which organ selective growth might be mediated by changes in carbonyl reductase 3 linked to structural proteins. Therefore, we hypothesize that a common core related to the redox system, involving peroxiredoxins and carbonyl reductase 3 that connects several structural proteins, proteins implied in ubiquitination, in DNA binding, cell division, and kinase-phosphatase activity, could determine in each foci the development of clinical metastasis (arrows summarize the changes in protein levels for metastatic variants in each location).
Discussion
Although the molecular mechanisms that enable individual cells to escape from the tumor, migrate, and survive, remain essentially unknown, they are likely to represent independent activities that, having otherwise deleterious effects in normal cells, may coincide in tumor cells to promote metastasis.31 The Bcl-2 family of proteins constitutes a critical intracellular checkpoint in the intrinsic pathways of apoptosis regulated by the permeabilization of mitochondrial membranes. Expression of anti-apoptotic members Bcl-2 and Bcl-xL blocks cell death after multiple physiological and pathological stimuli.32 Indeed, metastatic cells might become resistant to treatment by modifying expression levels or function of proteins involved in apoptosis-signaling pathways.33 The aim of this study was to identify phenotypes selected by Bcl-xL-overexpressing cells that in vivo enhance the ubiquity of clinical metastasis and shorter dormancy.
Proteomic profiling of metastatic human breast cancer cells by 2DE and mass spectrometry identified a Bcl-xL-mediated phenotype that harbors cells from the primary tumor to the metastatic state and appears to be useful for shortening dormancy with the result of clinical metastasis in many organs. Several proteins that function in redox system are overexpressed (familial Als mutant G37r, peroxiredoxin 2, peroxiredoxin 3) or underexpressed (carbonyl reductase 3), particularly in lung metastasis from 435/Bcl-xL cells. The presence of these proteins has been reported in some tumor cells and tissues,34–36 validating their implications in cancer. Furthermore, the protein-protein interaction program shows a common core related to the redox system, involving peroxiredoxins and carbonyl reductase, that connects several structural proteins, proteins implicated in ubiquitination, in DNA binding, cell division, kinase-phosphatase activity, and ATP synthase complex. These results suggest that cellular response to oxidative stress is a mechanism implicated in the selective growth of cells at the metastatic foci.
Cells living under aerobic conditions, as might be the case of lung metastasis, need to protect themselves against the damage caused by reactive oxygen species, arising from either the incomplete reduction of oxygen or exposure to external agents.37 Peroxiredoxins are mitochondrial or cytoplasmic enzymes that catalyze the destruction of peroxides and use cellular thiol to regenerate their active state.38 It has been reported that peroxiredoxin 3 protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis.39 Experiments using mitochondria-specific fluorescent probes demonstrate that peroxiredoxin 3 is essential for maintaining mitochondrial mass and membrane potential in transformed rat and human cells.40
The protein-protein interaction program showed a relationship between enolase 1 and aminoacylase 1 proteins, both cytoplasmic, with a high reliability score, suggesting that glucose and amino group metabolism could be associated to metastatic activity in terms of functional gain. Furthermore, the interaction of both with redox system proteins suggests that glycolysis metabolism and antioxidant pathways might be connected to the achievement of metastatic activity.
Enolase 1 is an enzyme that converts 2-phospho-d-glycerate to phosphoenol-pyruvate in the glycolysis pathway. The gene α enolase encodes a nuclear shorter monomeric structural protein by an alternative translation start, which has been found to bind to an element in the c-myc promoter. It is expressed in mammary gland and in breast carcinoma with endocrinal cells.41 Aminoacylase 1 is a hydrolase involved in the urea cycle and metabolism of amino groups. Mutational inactivation has been found in small cell lung cancer.42 The presence of both genes has also been established in array studies in which tumor cell lines and tissues, including breast, were analyzed and different expression patterns among samples were found.34–36
Expression of Bcl-xL influences a signaling network that links changes in mitochondria metabolism to alterations in nuclear gene expression, even under nonapoptotic conditions.43 Thus, genetic predisposition at primary tumor must exist because tumor cells stably overexpressed Bcl-xL, but a higher restriction takes place in the blood stream, ie, enolase 1 and aminoacylase 1 are overexpressed in BSCs with regard to breast tumor, and they are further represented in metastases. Because Bcl-xL inhibits anoikis and increases the survival of BSCs,18 overexpression of these proteins might be a result of a selection process. Indeed, the role of Bcl-xL in metastases might be associated to its ability to prevent apoptosis by coupling oxidative phosphorylation and glycolysis.44
Some phenotypic changes appeared only in metastatic foci, such as, tyrosine 3/tryptophan 5-monoxygenase activation protein, which activate tyrosine and tryptophan hydroxylases in the presence of calmodulin protein kinases; and multicatalytic endopeptidase complex ζ-chain, a proteasome subunit, characterized by its ability to cleave peptides in an ATP/ubiquitin-dependent nonlysosomal proteolytic pathways.45 Because both are overexpressed in all metastases, these proteins could have a role in common metabolic pathways involved in shortening dormancy.
Tyrosine 3/tryptophan 5-monoxygenase activation protein belongs to the 14-3-3 family of cytoplasmic proteins that mediate signal transduction binding to phosphoserine-containing proteins. A down-regulation has been reported in breast cancer cell lines MCF-7 and MDA-MB-231, and in primary breast carcinomas as compared with normal breast epithelial cells.46 In agreement with these results, we found underexpression of this protein in 435/Bcl-xL tumor cells with regard to control tumor. Moreover, a higher expression occurred in metastases, thus emphasizing the importance of the cross-talk between metastatic cells and the microenvironment in the development of clinical metastasis. Indeed, 14-3-3 proteins prevent apoptosis by directly binding to proapoptotic proteins as Bax and Bad.47 Moreover, it interacts with CDC25 phosphatases, RAF1 and IRS1 proteins, suggesting its role in diverse biochemical activities related to cell division.45
The organ-specific phenotype of tumor cells might be as a result of the selection in both the blood stream and the lodging site: differential expression of hypothetical protein MGC15429 and β-tubulin co-factor A underexpressed in bone metastasis, and Ran BP1, a GTP-binding nuclear protein, underexpressed in lung metastasis, are still represented in blood cells with regard to the tumor. Moreover, My 032 protein, glutathione transferase, and stathmin 1 might be a phenotype positively selected in the lodging site.
Stathmin 1, an ubiquitous cytosolic phosphoprotein that binds to two α/β-tubulin heterodimers, functions as an intracellular relay integrating regulatory signals of the cellular environment.48,49 Its overexpression has been found in highly proliferative breast carcinomas.50,51 Furthermore, glutathione transferase belongs to an enzyme family that plays an important role in detoxification by catalyzing the conjugation of many hydrophobic and electrophilic compounds with reduced glutathione, with a prognostic significance on invasive breast cancer.52
Increased expression of stathmin, enolase, peroxiredoxin 2, and some 14-3-3 protein isoforms in human breast ductal carcinoma in situ with regard to the normal breast has been described.53 Moreover, a serological proteomic study has identified a 14-3-3 protein σ as a breast cancer biomarker.54 These proteins are not included in the protein expression map of the normal human breast cells, suggesting that they belong to the earliest stage of breast cancer progression.55
The prevailing paradigm of carcinogenesis suggests that epithelial cells sequentially accumulate multiple genetic and epigenetic changes. It has been shown that cells isolated from metastases are frequently more highly metastatic than the bulk population of cells from primary tumors.56 Progenitors of the later-arising metastasis must be present among those cells that have disseminated to distant sites before the primary tumor is removed, and metastatic variants can arise by genetic or epigenetic mechanisms. Therefore, a signature of metastatic genes can exist.11–13 Moreover, tumor cells may disseminate in a far less progressed phenotype and acquire the metabolic properties of cells that live under the stress of no adhesion conditions, and the metabolic properties of cells with organ-specific growth, during the eventual development of metastatic disease.10 Because disseminate cells progress independently from the primary tumor, a simple extrapolation from primary tumor data to disseminated cancer cells is impossible.
Up-regulation of Bcl-xL protein may be a marker of tumor progression. High levels of Bcl-xL expression in breast tumors correlate with poor patient survival and increased metastasis.57 We suggest that overexpression of Bcl-xL in breast cancer cells act as an adaptor protein mediating breast cancer progression by connecting several metabolic pathways with the result of a phenotype that includes several functional pathways useful to metastasis development. Because metastasis is a state that emerges from the tumor-host interface, in which there is an interactive dialog between the neoplastic cells and the local activated host cells,58 the cross-talk results in different facilitated functions in each microenvironment (metastatic foci, cells that survive in blood, and tumor cell populations), arguing for the importance of anti-apoptotic proteins in paracrine and autocrine signaling as a crucial feature to therapeutic strategies to prevent metastases.
Footnotes
Address reprint requests to Dr. Angels Sierra, Centre d’Oncologia Molecular, Institut de Recerca Oncològica-IDIBELL, Hosp. Duran i Reynals, C.S.U.B., Gran Vía, km 2,7, 08907 L’ Hospitalet de Llobregat, Barcelona, Spain. E-mail: asierra@iro.es.
Supported by the Ministerio de Sanidad y Consumo (grants FIS 01/1469 and FIS/PI041937), the EC MetaBre (contract no. LSHC-CT-2004-506049), the Spanish Ministerio de Ciencia y Tecnologia (MCyT BIO02-0369 to R.A. and B.O.), and the Fundación Ramón Areces (to R.A. and B.O.).
L.E. and B.M. contributed equally in this project.
References
- McCawley LJ, Matrisian LM. Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol Med Today. 2000;6:149–156. doi: 10.1016/s1357-4310(00)01686-5. [DOI] [PubMed] [Google Scholar]
- Wells A. Tumor invasion: role of growth factor-induced cell motility. Adv Cancer Res. 2000;78:31–101. doi: 10.1016/s0065-230x(08)61023-4. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- Goodison S, Kawai K, Hihara J, Jiang P, Yang M, Urquidi V, Hoffman RM, Tarin D. Prolonged dormancy and site-specific growth potential of cancer cells spontaneously disseminated from nonmetastatic breast tumors as revealed by labeling with green fluorescent protein. Clin Cancer Res. 2003;9:3808–3814. [PubMed] [Google Scholar]
- Yefenof E, Picker LJ, Scheuermann RH, Tucker TF, Vitetta ES. Cancer dormancy: isolation and characterization of dormant lymphoma cells. Proc Natl Acad Sci USA. 1993;90:1829–1833. doi: 10.1073/pnas.90.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgreen L, O’Reilly MS, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- Karrison TG, Ferguson DJ, Meier P. Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst. 1999;91:80–85. doi: 10.1093/jnci/91.1.80. [DOI] [PubMed] [Google Scholar]
- Hart IR. Perspective: tumour spread the problems of latency. J Pathol. 1999;187:91–94. doi: 10.1002/(SICI)1096-9896(199901)187:1<91::AID-PATH234>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, Bischoff J, Harich D, Schlimok G, Riethmuller G, Eils R, Klein CA. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van’t Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- Ladeda V, Adam AP, Puricelli L, Bal de Kier Joffe E. Apoptotic cell death in mammary adenocarcinoma cells is prevented by soluble factors present in the target organ of metastasis. Breast Cancer Res Treat. 2001;69:39–51. doi: 10.1023/a:1012201805486. [DOI] [PubMed] [Google Scholar]
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- Fernandez Y, Espana L, Manas S, Fabra A, Sierra A. Bcl-xL promotes metastasis of breast cancer cells by induction of cytokines resistance. Cell Death Differ. 2000;7:350–359. doi: 10.1038/sj.cdd.4400662. [DOI] [PubMed] [Google Scholar]
- Rubio N, Espana L, Fernandez Y, Blanco J, Sierra A. Metastatic behavior of human breast carcinomas overexpressing the Bcl-x(L) gene: a role in dormancy and organospecificity. Lab Invest. 2001;81:725–734. doi: 10.1038/labinvest.3780281. [DOI] [PubMed] [Google Scholar]
- Espana L, Fernandez Y, Rubio N, Torregrosa A, Blanco J, Sierra A. Overexpression of Bcl-x(L) in human breast cancer cells enhances organ-selective lymph node metastasis. Breast Cancer Res Treat. 2004;87:33–44. doi: 10.1023/B:BREA.0000041579.51902.89. [DOI] [PubMed] [Google Scholar]
- Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- Celis JE, Gromov P. Proteomics in translational cancer research: toward an integrated approach. Cancer Cell. 2003;3:9–15. doi: 10.1016/s1535-6108(02)00242-8. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 1999;34:105–116. doi: 10.1002/(SICI)1096-9888(199902)34:2<105::AID-JMS768>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, Bertin N, Chung S, Vidal M, Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- Nonn L, Berggren M, Powis G. Increased expression of mitochondrial peroxiredoxin-3 (thioredoxin peroxidase-2) protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis. Mol Cancer Res. 2003;1:682–689. [PubMed] [Google Scholar]
- Wonsey DR, Zeller KI, Dang CV. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc Natl Acad Sci USA. 2002;99:6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse GM, Ma TK. Fine-needle aspiration cytology of breast carcinoma with endocrine differentiation. Cancer. 2000;90:286–291. [PubMed] [Google Scholar]
- Cook RM, Burke BJ, Buchhagen DL, Minna JD, Miller YE. Human aminoacylase-1. Cloning, sequence, and expression analysis of a chromosome 3p21 gene inactivated in small cell lung cancer. J Biol Chem. 1993;268:17010–17017. [PubMed] [Google Scholar]
- Li C, Fox CJ, Master SR, Bindokas VP, Chodosh LA, Thompson CB. Bcl-X(L) affects Ca(2+) homeostasis by altering expression of inositol 1,4,5-trisphosphate receptors. Proc Natl Acad Sci USA. 2002;99:9830–9835. doi: 10.1073/pnas.152571899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock DS, Santore MT, Lee VY, Brunelle J, Budinger GR, Zong WX, Thompson CB, Hay N, Chandel NS. Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Mol Cell Biol. 2002;22:94–104. doi: 10.1128/MCB.22.1.94-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The 14-3-3 cancer connection. Nat Rev Cancer. 2003;3:931–943. doi: 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- Vercoutter-Edouart AS, Lemoine J, Le Bourhis X, Louis H, Boilly B, Nurcombe V, Revillion F, Peyrat JP, Hondermarck H. Proteomic analysis reveals that 14-3-3sigma is down-regulated in human breast cancer cells. Cancer Res. 2001;61:76–80. [PubMed] [Google Scholar]
- Tsuruta F, Sunayama J, Mori Y, Hattori S, Shimizu S, Tsujimoto Y, Yoshioka K, Masuyama N, Gotoh Y. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P, Brattsand G, Gullberg M. MAP4 counteracts microtubule catastrophe promotion but not tubulin-sequestering activity in intact cells. Curr Biol. 2002;12:1034–1039. doi: 10.1016/s0960-9822(02)00897-7. [DOI] [PubMed] [Google Scholar]
- Amayed P, Pantaloni D, Carlier MF. The effect of stathmin phosphorylation on microtubule assembly depends on tubulin critical concentration. J Biol Chem. 2002;277:22718–22724. doi: 10.1074/jbc.M111605200. [DOI] [PubMed] [Google Scholar]
- Curmi PA, Nogues C, Lachkar S, Carelle N, Gonthier MP, Sobel A, Lidereau R, Bieche I. Overexpression of stathmin in breast carcinomas points out to highly proliferative tumours. Br J Cancer. 2000;82:142–150. doi: 10.1054/bjoc.1999.0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattsand G. Correlation of oncoprotein 18/stathmin expression in human breast cancer with established prognostic factors. Br J Cancer. 2000;83:311–318. doi: 10.1054/bjoc.2000.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Tan PH, Thiyagarajan J, Bay BH. Prognostic significance of glutathione S-transferase-pi in invasive breast cancer. Mod Pathol. 2003;16:558–565. doi: 10.1097/01.MP.0000071842.83169.5A. [DOI] [PubMed] [Google Scholar]
- Wulfkuhle JD, Sgroi DC, Krutzsch H, McLean K, McGarvey K, Knowlton M, Chen S, Shu H, Sahin A, Kurek R, Wallwiener D, Merino MJ, Petricoin EF, III, Zhao Y, Steeg PS. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 2002;62:6740–6749. [PubMed] [Google Scholar]
- Posadas EM, Simpkins F, Liotta LA, MacDonald C, Kohn EC. Proteomic analysis for the early detection and rational treatment of cancer—realistic hope? Ann Oncol. 2005;16:16–22. doi: 10.1093/annonc/mdi004. [DOI] [PubMed] [Google Scholar]
- Page MJ, Amess B, Townsend RR, Parekh R, Herath A, Brusten L, Zvelebil MJ, Stein RC, Waterfield MD, Davies SC, O’Hare MJ. Proteomic definition of normal human luminal and myoepithelial breast cells purified from reduction mammoplasties. Proc Natl Acad Sci USA. 1999;96:12589–12594. doi: 10.1073/pnas.96.22.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Metastatic potential: generic predisposition of the primary tumor or rare, metastatic variants—or both? Cell. 2003;113:821–823. doi: 10.1016/s0092-8674(03)00468-9. [DOI] [PubMed] [Google Scholar]
- Olopade OI, Adeyanju MO, Safa AR, Hagos F, Mick R, Thompson CB, Recant WM. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J Sci Am. 1997;3:230–237. [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. Cancer’s deadly signature. Nat Genet. 2003;33:10–11. doi: 10.1038/ng0103-10. [DOI] [PubMed] [Google Scholar]
- Montel V, Huang T-Y, Mose E, Pestonjamasp K, Tarin D. Expression profiling of primary tumors and matched lymphatic and lung metastases in a xenogeneic breast cancer model. Am J Pathol. 2005;166:1565–1579. doi: 10.1016/S0002-9440(10)62372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]