Abstract
Some genes present in only certain strains of the genetically diverse gastric pathogen Helicobacter pylori may affect its phenotype and/or evolutionary potential. Here we describe a new 16.3-kb segment, 7 of whose 16 open reading frames are homologs of type IV secretion genes (virB4, virB7 to virB11, and virD4), the third such putative secretion gene cluster found in H. pylori. This segment, to be called tfs3, was discovered by subtractive hybridization and chromosome walking. Full-length and truncated tfs3 elements were found in 20 and 19%, respectively, of 94 strains tested, which were from Spain, Peru, India, and Japan. A tfs3 remnant (6 kb) was found in an archived stock of reference strain J99, although it was not included in this strain's published genome sequence. PCR and DNA sequence analyses indicated the following. (i) tfs3's ends are conserved. (ii) Right-end insertion occurred at one specific site in a chromosomal region that is varied in gene content and arrangement, the “plasticity zone.” (iii) Left-end insertion occurred at different sites in each of nine strains studied. (iv) Sequences next to the right-end target in tfs3-free strains were absent from most strains carrying full-length tfs3 elements. These patterns suggested insertion by a transposition-like event, but one in which targets are chosen with little or no specificity at the left end and high specificity at the right end, thereby deleting the intervening DNA.
Helicobacter pylori is a genetically diverse gastric pathogen that chronically infects more than half of all people worldwide, typically for decades. It is a major cause of gastritis and peptic ulcer disease and is an early risk factor for gastric cancer (for reviews, see references 19, 25, and 40). Comparison of full genome sequences of two strains (26695 and J99) showed that some 6 to 7% of genes of one strain were absent from the other and vice versa (3, 48). Similarly, subtractive hybridization and microarray experiments indicated that 5% or more of genes known from strains 26695 and J99 were absent from the genomes of many other strains (2, 45) and that they, in turn, contain genes not found in the fully sequenced genomes. Some two-thirds of H. pylori's strain-specific genes have no predicted function, and most have been found to date only in this species (3, 48).
Genome sequence comparisons indicated that nearly half of H. pylori's strain-specific genes are located in its plasticity zone (3). This zone is about 45 kb long in strain J99 and 68 kb long in strain 26695, where it is split in two by a chromosomal rearrangement (3, 48). Many plasticity zone genes are transcribed, suggesting that they are functional (43). The zone contains several insertion sequences, as well as genes with protein-level homology to various recombinases, integrases, and topoisomerases. Their presence implicated specialized recombination events, including transposition, as a cause of differences among strains in gene content and arrangement. Two other genes found in the plasticity zone of one strain, 26695, potentially encode VirB4- and VirD4-type ATPases, which, in other systems interact in multiprotein assemblies to help translocate specific proteins and/or DNAs through the cell envelope (type IV secretion). Type IV secretion systems are involved in various ways in pilus formation and nucleoprotein transfer from donor to recipient bacterial or plant cells (in case of tumor induction by Agrobacterium tumefaciens), DNA transformation in H. pylori, toxin secretion, cell signaling, and intracellular survival of various pathogens (14, 18, 27, 38, 46). Each type IV secretion system is quite specific for a particular set of macromolecule (protein or nucleic acid) substrates and probably has its own distinct role.
Illustration that natural differences in gene content can affect H. pylori phenotypes came from studies of the cag pathogenicity island (PAI), a 37-kb (27 genes) DNA segment that encodes a type IV secretion system. Although the cag PAI is present in nearly all East Asian strains, it is epidemiologically associated with virulence in the West: many strains that cause only benign infections lack this gene cluster, whereas nearly all of those implicated in overt disease contain it (19). Mutational tests indicated that 17 of the 27 cag PAI-encoded proteins, including its six VirB homologs, participate in translocation of CagA protein (also encoded in this PAI) to target epithelial cells, where it is tyrosine phosphorylated and disrupts cell signaling. Fourteen of these 17 proteins help promote gastric epithelial synthesis of the proinflammatory cytokine interleukin-8 and thereby potentially damaging inflammatory responses (28). A second putative type IV secretion system, which seems to be present in essentially all H. pylori strains, is needed for competence in DNA transformation (“comB” cluster) (30).
Here we describe (i) a new gene cluster in H. pylori that, based on homology, probably encodes a type IV secretion system, the third such system found in this species; (ii) its location in the plasticity zone; (iii) its distribution among H. pylori strains; and (iv) evidence suggesting it is an unusual transposon that generates large nested deletions during insertion.
MATERIALS AND METHODS
General methods.
Standard procedures were used for H. pylori growth on brain heart infusion agar (Difco) containing 10% horse blood in a microaerobic atmosphere (1, 34). High-molecular-weight genomic DNA was isolated by a hexadecyltrimethylammonium method (9). A subtractive DNA library was made with the PCR-Select bacterial genome subtraction kit (Clontech) (2) and the strains described below.
Specific PCR was carried out in 20-μl volumes containing 5 to 10 ng of DNA, 0.25 to 0.5 U of Taq polymerase (“Biolase” Midwest Scientific, St. Louis, Mo.), 2.5 pM each primer, and 0.25 mM each deoxynucleoside triphosphate (dNTP) in a standard buffer for 30 cycles with the following cycling parameters: denaturation at 94°C for 30 s, annealing as appropriate for the primer sequence (generally 52°C) for 30 s, and DNA synthesis at 72°C for the appropriate time (1 min per kilobase). PCR products for sequencing were purified with a PCR purification kit (Qiagen, Chatsworth, Calif.) or extracted from agarose by centrifugation with Ultrafree-DA (Amicon, Millipore). DNA sequencing was carried out with a Big Dye Terminator DNA sequencing kit (Perkin-Elmer) and ABI automated sequencers (gel models 373 and 377; capillary model 3100). Direct sequencing of PCR products was done with 5 μg of PCR fragment (about 100 ng of DNA), 1 μl of primer (1.6 pM), and 4 μl of Big Dye (version 2) under the following conditions: 25 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min under oil-free conditions (Perkin-Elmer 2400). Direct sequencing on chromosomal DNA was done with 5 μl of chromosomal DNA (0.5 to 1 μg), 1 μl of primer (10 pM), and 6 μl of Big Dye (version 2) under the following conditions: 96°C for 5 min and then 90 cycles with denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min under oil-free conditions (Perkin-Elmer 2400). For capillary sequencers, the reaction mixture was cleaned through a column (Performa DTR gel filtration cartridges; Edge BioSystems) instead of ethanol precipitation. DNA sequence editing and analysis were performed with programs in the GCG package (Genetics Computer Group, Madison, Wis.) and Vector NTI (Informax, Bethesda, Md.) programs and data in H. pylori genome sequence databases (3, 48) and BLAST and Pfam (version 5.3) homology search programs (http://www.ncbi.nlm.nih.gov/blast/blast.cgi; http://pfam.wustl.edu/hmmsearch.shtml). Primer sequences used for PCR and sequencing are available from the authors on request.
Gene knockout mutations in tfs3 were made by PCR (without cloning) using a kanamycin resistance cassette (51), essentially as described previously (16). A comB deletion allele, in which the comB7-to-comB10 segment was replaced with a chloramphenicol resistance cassette, was kindly provided by K. Ogura (unpublished observations). These mutant alleles were introduced into H. pylori by electroporation and selection for resistance to kanamycin or chloramphenicol, as appropriate.
Natural transformation to test for competence was carried out with 16S ribosomal DNA (rDNA) containing the Aus108 tetracycline resistance (tet) allele (AGA to TTC in tetracycline binding site) (20, 50). Bacteria from about 1/4 of a confluent plate culture were transferred to fresh agar medium and incubated for 24 h. Exponentially growing cells from this plate were collected with a loop (generally from half or all of the plate), spread in a small circle (1 cm in diameter) on fresh medium, and incubated for 2 h. A 5-μl aliquot containing about 0.5 to 1 μg of a 16S rDNA PCR fragment containing the tet allele (20) was mixed with this patch of growth and incubated for 4 h, and then the cells were distributed over the entire plate and incubated for 20 h. Cells were then harvested and transferred to fresh medium containing 2 μg of tetracycline per ml and incubated to select Tetr transformants (3 to 6 days).
Bacterial strains.
The H. pylori strains used for subtractive library construction were PeCan18B from a Peruvian gastric cancer patient (tester) and a pool of strains SJM180A, SJM184A, and SJM189A from Peruvian gastritis patients (driver). An earlier analysis starting with another subtractive clone from this set of strains led to the finding of insertion sequence ISHp608 (35). All Peruvian strains were cultured from biopsies from persons with gastroduodenal complaints at the Servicio Universitario de Apoyo, Universidad Peruana Cayetano Heredia (UPCH), in Lima. Biopsies were obtained with written informed consent under protocols approved by the UPCH Human Studies Committee. Most other H. pylori strains used here were from the Berg laboratory collection and have been described in detail elsewhere (11, 15, 35, 41). The ethnicities of the patients and clinical disease associations are as follows. The 12 Peruvian gastric cancer strains and the 12 Peruvian gastritis strains were from persons of primarily Amerindian ancestry living in the shanty town of San Juan de Miraflores, in Lima, Peru. Eighteen of 22 Indian strains were from middle or lower middle class urban (12 strains) or rural (6 strains) residents of West Bengal; all were from patients with peptic ulcer disease. The other four strains were from Chennai (South India): three from patients with peptic ulcer and one from a patient with dyspepsia. Twelve of the 24 Japanese strains were from gastric cancer patients, and the other 12 were from gastritis patients.
Nucleotide sequence accession number.
The 20,457-nucleotide (nt) sequence we determined from strain PeCan18B was deposited in GenBank under accession no. AF487344. tfs3 extends from nt 253 to 18397. A total of 1,833 nt of this sequence (from nt 1752 to 3584) consists of ISHp608 (35), which was not found in tfs3 elements in other strains. A 6,983-nt sequence found in reference strain J99, 5,795 nt of which (from nt 905 to 6699) was not included in its published genome sequence, was deposited in GenBank under accession no. AY128679. (The tfs3 component extends from nt 460 to 6699.) A 1,968-nt sequence from strain CPY6081, which includes the gene designated “jhp926like” and the putative empty site for tfs3 insertion, was deposited in GenBank under accession no. AY128680.
RESULTS
Discovery of tfs3.
The studies presented here define a 16-kb putative transposable element (Fig. 1A; detailed in Table 1), seven of the open reading frames (ORFs) of which encode homologs of core type IV secretion proteins. This element will be called “tfs3” to connote that it is the third putative type IV secretion system found in H. pylori. These studies began with a subtractive hybridization, carried out to search for sequences in an H. pylori strain from a Peruvian patient with gastric cancer (PeCan18B) that were not present in strains from three Peruvians with more benign infections. One subtractive clone exhibited 28% protein identity to a virB11 homolog in the H. pylori cag PAI (hp525 in strain 26695 sequence), which is implicated in CagA protein translocation, induction of proinflammatory cytokine synthesis, and virulence (28, 48). It was more closely related (64% DNA identity, 55% protein identity, and 74% protein similarity) to another virB11 homolog (hp1421) whose role in H. pylori is not known. We decided to examine the genomic context of this third virB11 homolog and its distribution among H. pylori strains, based on our interest in genome evolution and VirB11-mediated secretion processes.
FIG. 1.
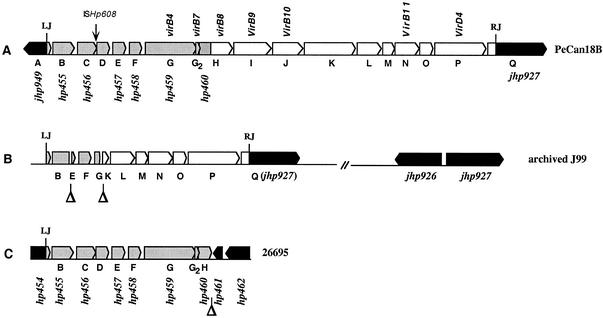
Organization of tfs3 in H. pylori. The homologies diagrammed here are detailed in Table 1. (A) tfs3 in strain PeCan18B. The tfs3 segment extends from nt 253 to 18397 of the 20,457 nt deposited under GenBank accession no. AF487344 (Details of PCR and genomic sequencing used to determine this sequence are available from the authors upon request.) ISHp608 (35) is 60 nt from the 3′ end of orfC in PeCan18B but was not found in any of 37 other tfs3 elements tested. LJ, left junction; RJ, right junction. (B) Organization of the tfs3 remnant in archived strain J99. This remnant extends from nt 460 to 6699 of the 6,983 nt deposited under GenBank accession no. AY128679. Two copies of jhp927 were found in J99: one connected to jhp926 (as published in the full genome sequence in reference 3) and another connected to the right end of tfs3 (present data) (GenBank accession no. AY128679 [absent in the J99 full genome sequence data]). (C) Map of truncated tfs3 element of strain 26695. This remnant corresponds to nt 253 to 7622 of tfs3 in strain PeCan18B (GenBank accession no. AF487344). orfD (94 codons) and orfG2 (47 codons) were not annotated as ORFs in the 26695 genome sequence, but are present in its DNA sequence.
TABLE 1.
Description of tfs3 from strain PeCan18B
| ORF (aa) | Region of homology (aa) | Homolog | Length (aa) | Protein comparison (%identity/%similarity) | %DNA | Description (reference) | ORFs in tsf3a |
|---|---|---|---|---|---|---|---|
| A (78>) | jhp949 (98% DNA), left of tsf3, 2 transmembrane helices | ||||||
| B (262) | 49-134 | jhp941 | 331 | 96/96 | 99 | Integrase-resolvase | hp455 (97% DNA) |
| C (164) | hp456 (87% DNA), ISHp608 at 145/146 aa | ||||||
| D (94) | 6-94 | hp15 | 93 | 57/74 | 64 | Hypothetical | hp456-57 intergenic region (98% DNA), 3 transmembrane helices |
| E (87) | 1-87 | hp16 | 87 | 60/81 | 67 | Hypothetical | hp457 (99% DNA), 1 transmembrane helix |
| 1-87 | hp442 | 88 | 38/56 | Hypothetical, plasticity zone | |||
| 6-87 | Cjp52 | 80 | 27/49 | C. jejuni, pVir (10) | |||
| F (78) | hp458 (100% DNA) | ||||||
| G (858) | 1-837 | hp17 | 787 | 46/64 | comB4 | hp459 (99% DNA), virB4 homolog, 2 transmembrane helices | |
| 11-843 | hp441 | 807 | 37/58 | virB4, plasticity zone | |||
| 22-848 | Cjp53 | 822 | 31/53 | virB4 in C. jejuni, pVir | |||
| 211-697 | jhp917 | 475 | 36/57 | virB4-2, plasticity zone | |||
| 708-833 | jhp918 | 140 | 34/55 | virB4-3, plasticity zone | |||
| 38-717 | hp544 | 983 | 21/39 | cag23 (virB4), cag PAI | |||
| G2 (47) | 5-33 | orf2 | 37 | 41/61 | comB7 in P1 (30) | hp459-60 intergenic region (98% DNA), virB7 homolog | |
| 7-32 | Cjp54 | 42 | 48/52 | virB7 in C. jejuni, pVir | |||
| H (352) | 116-336 | hp38 | 245 | 43/66 | comB8 | hp460 (5′ half) (98% DNA), virB8 homolog (3′ half), 1 transmembrane helix | |
| 39-345 | jhp921 | 328 | 36/56 | Putative, plasticity zone | |||
| 122-343 | Cjp01 | 225 | 34/57 | virB8 in C. jejuni, pVir | |||
| 4-337 | hp439 | 366 | 32/52 | Hypothetical, plasticity zone | |||
| I (551) | 153-551 | hp39m | 326 | 53/67 | comB9 | virB9 homolog, 1 transmembrane helix | |
| 142-545 | jhp922m | 511 | 37/52 | Conjugation, plasticity zone | |||
| 87-546 | Cjp02 | 356 | 30/50 | virB9 in C. jejuni, pVir | |||
| J (402) | 13-143 | jhp36 | 376 | 26/32 | 69 | comB10 + | virB10 homolog, 2 transmembrane helices |
| 156-384 | 77/83 | ||||||
| 27-385 | Cjp03 | 378 | 36/54 | virB10 in C. jejuni, pVir | |||
| K (735) | 7-508 | jhp945 | 668 | 34/54 | Putative, plasticity zone | 1 transmembrane helix | |
| L (320) | No homology, 1 transmembrane helix | ||||||
| M (92) | No homology | ||||||
| N (314) | 6-310 | hp1421 | 304 | 53/72 | virB11-12 | virB11 homolog | |
| 12-309 | Cjp05 | 317 | 40/59 | virB11 in C. jejuni, pVir | |||
| 27-302 | hp525 | 330 | 28/50 | virB11, cag PAI | |||
| O (172) | No homology | ||||||
| P (747) | 449-612 | hp1006 | 177 | 62/79 | traG (virD4-2), plasticity zone | virD4 homolog, 4 transmembrane helices | |
| 14-636 | Cjp06 | 628 | 37/56 | virD4 in C. jejuni, pVir | |||
| 150-604 | hp524 | 748 | 23/46 | virD4, cag PAI | |||
| Q (701>) | jhp927 (90% DNA) + jhp928 (93% DNA), tfs3 right junction is in 9th codon |
See Fig. 1 for the overall structure of tsf3. orfA and orfQ are adjacent to tsf3 in PeCan18B (orfQ in PeCan18B is a fusion of jhp927 and jhp928) orfB through orfP are within tsf3.
tfs3 sequence.
The set of genes containing this new virB11 homolog in strain PeCan18B was sequenced by primer walking from the original subtractive clone, with the intent to continue until genes known from published sequences were encountered. The first such genes were jhp927 and then jhp928 (known from strain J99) on the right, and hp460 and then hp459 (known from strain 26695) on the left (Fig. 1A). PCR tests with DNAs from 94 representative H. pylori strains (see “Geographic distribution,” below) identified jhp927 in 22 of 37 strains that contained any tfs3 genes and in 28 of 57 strains that seemed to be tfs3 free. This pattern was taken as initial evidence that jhp927 is to the right of tfs3, not within it. In contrast, PCR tests for hp459 and hp460 indicated that they were present in each of 37 strains that contained tfs3 sequences, but not in any of 57 strains that lacked them. These data suggested that hp459 and hp460 were part of tfs3 and that tfs3 was truncated in strain 26695. We therefore continued sequencing leftward and checked sequences obtained by PCR using DNAs from a panel of other representative H. pylori strains at each step. The results suggested that tfs3 extended leftward through orfB (hp455) and the 3′ end of hp454. Of 20,457 nt sequenced (GenBank accession no. AF487344), 16,312 nt were assigned to the tfs3 element itself; another 1,833 nt consisted of ISHp608 (35), which was inserted near the 3′ end of orfC. The leftmost 252 nt and rightmost 2,060 nt were considered flanking sequences, just outside the left and right ends, respectively, of tfs3. Seven of tfs3's 16 ORFs encoded homologs of VirB proteins that act in type IV secretion: four that form a membrane-traversing transporter channel (VirB7, VirB8, VirB9, and VirB10) and three that are cytoplasmic membrane-associated ATPases and that efficiently move their cognate macromolecular substrates to and through the channel (VirB4, VirB11, and VirD4) (Fig. 1A and Table 1). We also note that orfB (hp455), at the left end of tfs3, was nearly identical in the first third of its length to part of H. pylori gene jhp941, another segment of which exhibits high protein-level homology to members of the XerC/XerD-type integrase-recombinase-transposase family.
Geographic distribution.
A set of 94 H. pylori strains from four geographic regions (Japan, Peru, Spain, and India) were tested by PCR for 15 tfs3 ORFs. One-fifth of the strains carried an apparently full-length tfs3 cluster, as defined in relation to the sequence in strain PeCan18B (Fig. 1A), and another one-fifth of the strains contained just part of this segment (most often the left half) (Table 2). No correlation of tfs3 carriage with disease status was observed: tfs3 was equally present in strains from Peruvian and Japanese patients with gastric cancer and with gastritis. Similarly, no correlation with cag PAI status was seen with Spanish strains (half of which carried and half of which lacked the cag PAI).
TABLE 2.
Geographic distribution of tfs3 elementsa
| Country | No. of strains tested | No. (%) containing tfs3
|
|
|---|---|---|---|
| Full | Partial | ||
| Japan | 24 | 4 (17) | 7 (29) |
| Peru | 24 | 6 (25) | 4 (17) |
| Spain | 24 | 6 (25) | 1 (4) |
| India | 22 | 3 (14) | 6 (27) |
| Total | 94 | 19 (20) | 18 (19) |
This distribution was determined by PCR with primers specific for each of 15 ORFS in tfs3 (see Fig. 1). Primer sequences are available from the authors upon request.
tfs3 in strain J99.
DNA from an archived stock of reference strain J99 had been chosen initially as a negative control for tests of the geographic distribution of tfs3 elements, because no tfs3 genes had been included in this strain's published genome sequence (GenBank accession no. AE001439). Our PCR tests indicated, however, that parts of tfs3 were present in the strain J99 genome. Our DNA sequencing then identified a 6.2-kb segment consisting of 741-, 402-, and 5,097-nt segments from the left end, interior, and right end, respectively, of full-length tfs3 (GenBank accession no. AY128679). This structure can be ascribed to deletion of 1.3 kb between sites in orfB and orfE and deletion of 8.7 kb between sites in orfG and orfK (Fig. 1B).
We found orfP of this tfs3 remnant in strain J99 next to jhp927 (as in strain PeCan18B), although another gene, jhp926, had been placed next to jhp927 in the published J99 sequence. Direct PCR tests identified both connections (both jhp926- and tfs3-jhp927), and DNA sequencing showed that a 750-nt segment containing the jhp926-jhp927 junction was identical to that in the published genome sequence. The implication that J99 contained two copies of jhp927 was confirmed by Southern blot hybridization (data not shown). Further PCR tests of DNAs from two single-colony isolates from this J99 stock identified the same two connections to jhp927. Thus, archived J99 had a duplication of the region containing jhp927; it was not a mixture of two strains with different jhp927 connections.
The structure of tfs3 and its context were examined further by using an aliquot of the genomic DNA that had been provided years earlier to Genome Therapeutics Corporation for genome sequencing and that was kindly given to us by T. L. Cover. PCR tests of this DNA revealed the internally deleted copy of tfs3 and the tfs3-hp927 and jhp926-jhp927 connections noted above. However, this DNA also seemed to contain full-length tfs3, based on PCR tests for each tfs3 ORF. These results imply that the DNA used for J99 genome sequencing and the original J99 culture may have contained two closely related strains: one with full-length tfs3 and one with the tfs3 remnant. Presumably, only the strain with the remnant was archived or survived long-term frozen storage.
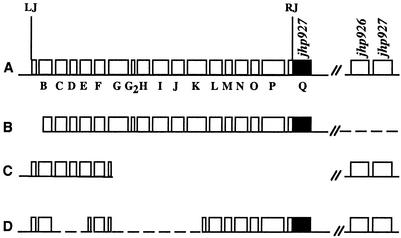
Limited divergence among H. pylori isolates recovered from individual patients, reminiscent of that found in DNA from the original J99 culture, had been reported sporadically in the past (26, 34, 53) and emerged again with further studies of H. pylori that were recovered from patient J99 by re-endoscopy and culture 6 years after his initial endoscopy (31). This yielded a complex population of strains that were closely related to, but not identical to, the original J99 strain (31). Our PCR analysis of 14 of these new isolates (DNAs kindly provided by D. Israel) identified three main variants with respect to tfs3 region markers (diagrammed as variants A, B, and C in Fig. 2), none of which was perfectly matched to the tfs3 remnant in archived strain J99 (line D in Fig. 2).
FIG. 2.
Structures of tfs3 regions in J99 variants from patient J99, recovered in the second endoscopy (31) 6 years after isolation of archived strain J99. Gene designations here (B, C, D, etc.,) are as in Fig. 1, and are annotated in Table 1. RJ and LJ, right and left junctions, respectively, of tfs3 with flanking sequences. (A) Variant A. Shown is the full-length tfs3 region, as in strain PeCan18B, containing two copies of jhp927 (clones A1, A7, A9, C5, C6, C8, and D1) (31). (B) Variant B. Variant B is similar to variant A, except it lacks the jhp926-jhp927 segment and has deletion or rearrangement at tfs3's left end (clones A3, A5, C10, C12, and D3). (C) Variant C. Variant C is a truncated tfs3, lacking tfs3 sequences that extend rightward from the 5′ end of orfG (hp459) and also jhp927 (clones Ca1 and C2). (D) Variant D. tfs3 remnant in archived stock of strain J99 found here (Fig. 1B).
tfs3 ends, flanking sequences, and site of insertion.
PCR and sequencing further identified the left and right tfs3 ends and empty sites in strains lacking tfs3 and gave insights into insertion specificity. First, we tested whether the putative tfs3 right-end-jhp927 junction of strain PeCan18B was typical or unique. PCR using orfP- and jhp927-specific primers resulted in a product of the expected size from 25 of 41 strains with full-length tfs3 elements that were tested. DNA sequencing of PCR products from five such strains and comparison with the published strain J99 genome sequence identified tfs3's right end at the ninth codon of jhp927 (Fig. 3, right junction). Eight of the remaining 16 tfs3-containing strains yielded PCR products with primers specific for the tfs3 right end and jhp931 instead of jhp927. Products from six of these eight strains were of one size class (variant 1a in Table 3), and DNA sequences from three of them suggested that these variants had arisen by deletion involving a direct repeat of 22 nt, ATTTTAAAAGGAGCAACAAATG, present near the tfs3 right end and again near the jhp931 start; a second variant end (variant 1b) was ascribed to a shorter direct repeat, GCAACC(G/C)AA, at the tfs3 right end and upstream of jhp931; and a third variant (1c) did not seem to be associated with repeat sequences.
FIG. 3.
Empty site in the 5′ end of jhp927 and left and right tfs3 junctions. tfs3 sequences are in uppercase, and flanking sequences are in lowercase. (A) The empty site is the site of tfs3 right-end insertion in codon 9 of jhp927 (probable translation start is underlined). This start is 33 codons downstream from the start assigned in strain J99 due to the presence of one extra nucleotide in the annotated jhp927 sequence (3). The underline designates the first in-frame ATG in these 10 strains. PCR and sequence analysis indicated that jhp927 is adjacent to jhp926 in strains J99, SpainB43, SpainB49, PeCan9A and India-Chennai2 and is adjacent to jhp926like (GenBank accession no. AY128680) in strains SpainB50, JapCPY2362, JapHU131, JapCPY6081, India39A, and SpainB70. (B) Right junction sequences of the predominant type (Table 3). (C) Left junctions of tfs3. The sequences are adjacent to the following: in PeCan18B, to jhp949; in archived J99, to the intergenic region of jhp941-jhp942; in SJM148(Peru), to a site 38 nt upstream of a 5S ribosomal DNA gene; in PeCan10B, to an unknown sequence (no homology in database); in SpainB84, to a weak homolog of hp1383 and jhp1422 (restriction modification system S subunit, 55 to 66% protein identity and similarity); in 26695, to the 3′ end of hp454; in JapHU87, to hp513; in JapCPY1672, to the jhp644-jhp645 intergenic region; and in India77A, to a jhp1297 homolog.
TABLE 3.
tfs3 right junction types
| Country | Total no. of strains | No. of strains with complete tfs3 | No. (%) of strains with right junction typea
|
||||
|---|---|---|---|---|---|---|---|
| 1 | Variant
|
Unidentified | |||||
| 1a | 1b | 1c | |||||
| Japan | 24 | 4 (17) | 3 (75) | 0 | 0 | 0 | 1 (25) |
| Peru | 24 | 6 (25) | 5 (83) | 0 | 0 | 1 (17) | 0 |
| Spain | 24 | 6 (25) | 3 (50) | 3 (50) | 0 | 0 | 0 |
| India | 121 | 25 (21) | 14 (56) | 3 (12) | 1 (4) | 0 | 7 (28) |
| Total | 193 | 41 (21) | 25 (61) | 6 (15) | 1 (2) | 1 (2) | 8 (20)b |
Predominant type 1 is described in the legend to Fig. 3. Variant types 1a and 1b contain short deletions that can be ascribed to slippage between direct repeats (see text). Variant type 1c contains a 23-nt deletion from the right end of tfs3 to a site 110 nt from the 3′ end of jhp930.
About 20% of strains did not amplify with available primers, suggesting that they might contain other rearrangements. However, chromosomal sequencing of three such strains identified a tfs3-jhp927 connection (as in Fig. 3) in each case.
The lack of amplification with orfP- and jhp927- or jh931-specific primers in cases of eight tfs3-containing strains (“unidentified” column in Table 3) was attributable to chromosome rearrangement or to sequence divergence affecting primer binding sites. Direct sequencing of genomic DNAs from three of these eight strains identified a typical tfs3-jhp927 connection in each case. This outcome further supports the inference that the tfs3 right end inserts preferentially at the ninth codon of jhp927.
We next tested the possibility that jhp926 and jhp927 might be adjacent to one another unless disrupted by tfs3 insertion. PCR tests of strains that contained jhp927 but not tfs3 identified this connection in only 11 of 41 strains tested. Primer walking was carried out to identify a sequence adjacent to the 5′ end of jhp927 by using genomic DNA from 1 of the 30 other strains. The sequence obtained was distantly related to jhp926 and will be called “jhp926like” (34% protein identity, 43% protein similarity, no significant DNA identity; GenBank accession no. AY128680). Further PCR tests identified a jhp926like-jhp927 connection in 28 of the remaining 29 strains that carried jhp927 but not canonical jhp926. The jhp926like sequence can be considered a divergent allele of jhp926 based on (i) its connection to jhp927, (ii) the inability to find any strain among 94 tested that contained both jhp926like and jhp926, and (iii) the protein-level homology noted above. Both jhp926 and jhp926like alleles were found in Peruvian, Spanish, and Indian populations, although jhp926like was twice as common as canonical jhp926 itself; only jhp926like (9 strains) was found among the 24 Japanese strains tested. Neither jhp926 nor jhp926like was found next to the tfs3 left end in any of 37 strains containing partial or complete tfs3 elements, nor were these genes found at all in the genomes of 16 of 19 strains with apparently full-length tfs3 elements; the three exceptions contained duplication of jhp927, equivalent to that in strain J99. Eight of 18 strains with only partial tfs3 elements (mainly the left end) also lacked jhp926 sequences, and the other 10 contained either jhp926 (1 strain) or jhp926like (9 strains) connected to jhp927.
The left end of tfs3 had been identified provisionally at 79 nt upstream of the 3′ end of hp454, based on divergence between sequences from strains PeCan18B and 26695 (Fig. 3). Consistent with this, primer walking along tfs3 in archived J99 showed that 76 nt matching the 3′ end of hp454 were joined to a segment that seemed to correspond to the intergenic region between jhp941 and jhp942. However, none of these three connections (jhp949-orfB in PeCan18B, jhp941/2 intergenic region-orfB in J99, hp454 [5′ end]-orfB in 26695) was found in any of 37 other tfs3-containing strains tested by PCR. Left-end junction sequences were determined by primer walking from hp455 by using three additional strains containing full-length tfs3 elements and three others containing left-end remnants of tfs3. In each case, the 79-nt segment from hp454's 3′ end was joined to a different sequence (Fig. 3). These results established again that the left end of tfs3 was about 79 nt from hp454's 3′ end, that this end was well conserved, and that it could be joined to many target sites.
Test for possible tfs3 function.
The closest homologs of four tfs3 genes were comB genes of H. pylori (comB7 to comB10, Table 1), which are needed for competence in DNA transformation (30). Accordingly, three mutant derivatives of strain PeCan18B were constructed to test if these tfs3 genes might participate in natural transformation, such that the corresponding comB genes would be redundant. The three derivatives contained (i) a deletion of the entire comB7-to-comB10 region and replacement with a cat (chloramphenicol resistance) gene, (ii) a deletion of the entire tfs3 segment and replacement with an aphA (kanamycin resistance) gene, and (iii) replacement of both gene clusters by cat and aphA genes. These three derivatives were then used as recipients for natural transformation using a 16S rDNA PCR fragment that confers tetracycline resistance (due to mutation in the tetracycline binding site) (20). Tetr transformants were obtained at frequencies of about 10−6 by using the PeCan18B wild type and its tfs3 deletion derivative as recipients, but at ≤10−9 when using PeCan18B derivatives with the comB gene deletion alone or in combination with the tfs3 deletion. Thus, these data did not support the idea that tfs3 virB homologs contribute to transformability.
Tests for tfs3 in several additional strains gave no support for models in which tfs3 might have a critical role in general bacterial conjugation, in intracellular entry or survival, or in mouse colonization. First, no tfs3 sequences were found in HPK5, a strain that seemed to serve as a donor in conjugation (36). Second, no tfs3 sequences were found in strain G27, which is quite invasive (4), and, as noted above, only remnant tfs3 elements were found in strains J99 and 26695, two other strains that also enter epithelial cells quite well (37). Third, neither of the two special and widely used mouse-colonizing strains, SS1 or X47-2AL, was found to contain tfs3 sequences.
DISCUSSION
We have described a new gene cluster in H. pylori, designated tfs3, 7 of whose 16 genes are protein-level homologs of genes involved in type IV secretion. Four of these genes are transmembrane pore genes (virB7, virB8, virB9, and virB10), and three code for ATPases that move cognate macromolecule substrates to and through the pore with specificity and efficiency (virB4, virB11, and virD4). None of the nine other tfs3 ORFs had homologs in current databases, but transmembrane motifs encoded in four of them also suggest possible type IV secretion involvement. Full-length and truncated tfs3 elements were each found in about one-fifth of H. pylori strains—obtained from Peru, Spain, India, and Japan. In all cases analyzed, they were inserted in the plasticity zone, a genomic region rich in insertions and deletions. This is the third type IV secretion gene complex found in H. pylori, but the first found to be in a putative transposable element (discussed below) or in H. pylori's special plasticity zone.
tfs3's role in H. pylori is not known. The closest known homologs of some tfs3 genes are needed in H. pylori for competence in natural transformation; more distant homologs form part of the cag PAI and contribute to virulence. There are also striking homologies with genes in a virulence plasmid of Campylobacter jejuni that are needed for this pathogen's intracellular growth and competence for transformation (10). Our mutational test did not support a possible tfs3 involvement in transformation, nor was an association of tfs3 carriage and overt disease detected: tfs3 was either entirely absent or was present only in truncated form in several strains that can persist in cultured epithelial cells and was absent from a strain that acts as a donor in conjugation. In terms of other possible roles, H. pylori strains are diverse phenotypically, especially in their interactions with host cells and tissues during chronic infection (23, 39, 44), and it is thus tempting to imagine that tfs3-encoded proteins may help secrete macromolecules involved in some of these interactions.
An unusual tfs3 derivative with two internal deletions was found in an archived copy of reference strain J99, and both this vestigial element and full-length tfs3 were found in an aliquot of J99 DNA that had been provided for genome sequencing, but no tfs3 genes were reported in J99's full genome sequence. This discrepancy might be explained by supposing that a J99 variant, putatively free of tfs3, was used to complete the genome sequence and that inconsistent sequences were omitted from the final contig. (This project was started by one company and finished by another several years later [3, 6].) Another explanation invokes difficulty in tfs3 segment cloning, possibly related to its genes for membrane proteins, and then in silico assembly of a circular contig without tfs3 and one jhp927 region, due to the duplicated jhp927 region.
The principle that H. pylori infections can be mixed and can involve clusters of related strains (26, 34, 53) was well illustrated by the study by Israel et al. (31) of H. pylori isolates recovered from “patient J99” at the second endoscopy, 6 years after the one that had yielded the original reference strain. We found three tfs3 variants in 14 new J99-related isolates, none of which completely matched the tfs3 element found in the archived strain. This was in accord with the diversity Israel et al. found at other loci. Possible explanations for such variant strains include (i) mutant or recombinant formation in the years between the first and second endoscopies (as postulated in reference 31); (ii) the strain's being present at the time of the first endoscopy, but not recovered by culture; and (iii) the strain being transmitted to the patient from other family members during the intervening 6 years.
The diversity in gene content among H. pylori strains stimulates interest in how DNA is gained and lost during H. pylori evolution. tfs3's lower G+C content (35% versus 39% for the H. pylori genome overall) suggests transfer from another bacterial species, as does its presence in only a subset of H. pylori strains. tfs3 entry into H. pylori gene pools might have occurred long ago, because tfs3 is common among strains from both East Asia and the West. Alternatively, it might have been transferred into H. pylori recently and could be increasing in abundance by lateral DNA transfer and insertion into new genomes (“molecular drive”) (21, 22) at rates that also reflect any effects of this element on bacterial fitness.
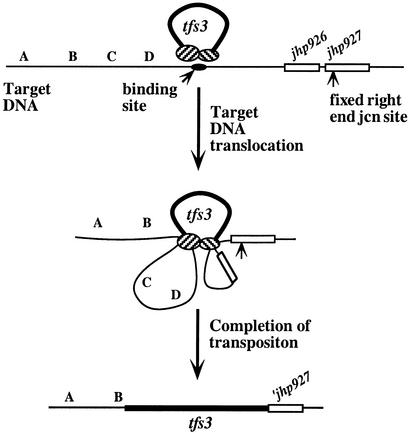
In terms of possible acquisition mechanisms, our analyses showed (i) that left and right ends of full-length tfs3 elements were conserved; (ii) that tfs3's right end was joined to the same target site in jhp927 in most H. pylori strains; whereas (iii) its left end could be joined to many different target sequences; and (iv) that the jhp926 or jhp926like genes to the left of tfs3's fixed insertion site were missing from most strains with full-length tfs3. These findings fit with a formal model of clean insertion into a fixed site in jhp927, followed by formation of leftward deletions, equivalent to those generated by elements such as IS1, Tn5, and γδ (32, 49, 52, 54). Because such deletions are rare in other systems, however, we prefer a model in which the deletions observed here result directly from tfs3 insertion and the joining of its right and left ends to different chromosomal sites. Although none of the proteins or specific DNA sites needed for tfs3 transposition are known, we are attracted to a model that entails “motorized” tracking of recipient DNA through the transposition complex (Fig. 4), prompted in part by knowledge of how type I restriction endonucleases act (13, 29, 42, 47). Insertion would begin with binding of a tfs3-transposition protein complex to a specific target site. Adjacent recipient DNA would then be pulled by the complex from both sides, perhaps coupled to ATP hydrolysis, until signals for DNA cleavage and tfs3 end joining are encountered. Signals for cleavage might be inherent in the DNA sequence itself, as is seen with RecBC and related recombination enzymes (5, 7, 17), or it might consist of other proteins that the transposition complex encounters along the recipient DNA, as is seen with type I restriction endonucleases (13, 29, 42, 47). Activation of latent nucleases, as in MutSLH-mediated mismatch repair (12), also merits consideration. Finally, differences between right and left junctions suggest that different signals may be used to trigger the processing of sites for left and right end joining.
FIG. 4.
Motorized transposition model for tfs3 acquisition. This model envisions that two at least subtly different sets of transposition proteins (hatched ovals) act on tfs3's two ends and then bind to a specific recognition site in target DNA (solid oval). DNAs adjacent to this recognition site are pulled progressively through the protein complex until they encounter sequences or DNA-bound proteins that tfs3 uses as signals for left-end joining to one of many potential target sites and right-end joining to its specific jhp927 site. jcn, junction.
The truncated (left-end containing) tfs3 elements can be explained by this tracking model, based on a finding that half of tfs3-free H. pylori strains lacked the jhp927 site for right-end insertion. We propose that transposition in these strains also entails transposition protein binding to the preferred target site, tracking bidirectionally along target DNA, and left-end joining to one of its many possible target sites, essentially as in Fig. 4. Right-end joining might fail, however, either because of deletion or point mutation of its specific target sequence or because of the lack of a putative protein signal. Joining of just one end would linearize the chromosome, but recircularization would be needed for viability, and this might be achieved by illegitimate recombination, which can occur by various mechanisms (8, 24, 33).
Because most deletions are either potentially deleterious or at best only neutral, transposable elements that regularly generate them would usually be lost during evolution. tfs3 may be an exception, however, able to escape such contraselection in two ways. First, if the initial target is between sites of tfs3 joining to recipient DNAs, as postulated (Fig. 4), tfs3 insertion would remove this site. This might be selected because it would lower the chance of displacing resident tfs3s by “superinfecting” tfs3 elements. Second, many deletions formed by tfs3 insertion might not be deleterious because they occur in the plasticity zone, where DNA loss and rearrangement are the norm, and flexibility and diversity in gene content may contribute to bacterial fitness in different members of the diverse human host population.
Acknowledgments
We thank Timothy Cover and Dawn Israel for providing DNAs from J99 and related H. pylori strains and stimulating discussion, Daiva Dailidiene and Keiji Ogura for mutant DNAs, Patricia Guerry for access to her C. jejuni pVir plasmid data before publication, and anonymous reviewers for valuable suggestions.
This research was supported by grants from the Public Health Service (AI38166, AI49161, DK53727, P30 DK52574, and 3 D43 TW 00910).
REFERENCES
- 1.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., A. Fradkov, L. Diatchenko, P. D. Siebert, S. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Amieva, M. R., N. R. Salama, L. S. Tompkins, and S. Falkow. 2002. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol. 4:677-690. [DOI] [PubMed] [Google Scholar]
- 5.Amundsen, S. K., and G. R. Smith. 2003. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112:741-744. [DOI] [PubMed] [Google Scholar]
- 6.Anonymous. 1995. The Gold Bug: Helicobacter pylori. Science 267:173. [Google Scholar]
- 7.Arnold, D. A., N. Handa, I. Kobayashi, and S. C. Kowalczykowski. 2000. A novel, 11 nucleotide variant of chi, chi*: one of a class of sequences defining the Escherichia coli recombination hotspot chi. J. Mol. Biol. 300:469-479. [DOI] [PubMed] [Google Scholar]
- 8.Ashizawa, Y., T. Yokochi, Y. Ogata, Y. Shobuike, J. Kato, and H. Ikeda. 1999. Mechanism of DNA gyrase-mediated illegitimate recombination: characterization of Escherichia coli gyrA mutations that confer hyper-recombination phenotype. J. Mol. Biol. 289:447-458. [DOI] [PubMed] [Google Scholar]
- 9.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A., Smith, and K. A. Struhl. 1994. Current protocols in molecular biology; supplement 27 CPMB, p. 2.4.1. Greene Publishing and Wiley Interscience, New York, N.Y.
- 10.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg, D. E., R. H. Gilman, J. Lelwala-Guruge, K. Srivastava, Y. Valdez, J. Watanabe, J. Miyagi, N. S. Akopyants, A. Ramirez-Ramos, T. H. Yoshiwara, S. Recavarren, and R. Leon-Barua. 1997. Helicobacter pylori populations in individual Peruvian patients. Clin. Infect. Dis. 25:996-1002. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell, L. J., K. P. Bjornson, D. J. Allen, and P. Modrich. 2001. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem. 276:34339-34347. [DOI] [PubMed] [Google Scholar]
- 13.Bourniquel, A. A., and T. A. Bickle. 2002. Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84:1047-1059. [DOI] [PubMed] [Google Scholar]
- 14.Cao, T. B., and M. H. Saier, Jr. 2001. Conjugal type IV macromolecular transfer systems of gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147:3201-3214. [DOI] [PubMed] [Google Scholar]
- 15.Chalkauskas, H., D. Kersulyte, I. Cepuliene, V. Urbonas, D. Ruzeviciene, A. Barakauskiene, A. Raudonikiene, and D. E. Berg. 1998. Genotypes of Helicobacter pylori in Lithuanian families. Helicobacter 3:296-302. [PubMed] [Google Scholar]
- 16.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chedin, F., and S. C. Kowalczykowski. 2002. A novel family of regulated helicases/nucleases from gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43:823-834. [DOI] [PubMed] [Google Scholar]
- 18.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cover, T. L., D. E. Berg, M. J. Blaser, and H. L. T. Mobley. 2001. H. pylori pathogenesis, p. 509-558. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, New York, N.Y.
- 20.Dailidiene, D., M. T. Bertoli, J. Miciuleviciene, A. K. Mukhopadhyay, G. Dailide, M. A. Pascasio, L. Kupcinskas, and D. E. Berg. 2002. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob. Agents Chemother. 46:3940-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doolittle, W. F., and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601-603. [DOI] [PubMed] [Google Scholar]
- 22.Dover, G., and W. F. Doolittle. 1980. Modes of genome evolution. Nature 88:646-647. [DOI] [PubMed] [Google Scholar]
- 23.Dubreuil, J. D., G. Del Giudice, and R. Rappuoli. 2002. Helicobacter pylori interactions with host serum and extracellular matrix proteins: potential role in the infectious process. Microbiol. Mol. Biol. Rev. 66:617-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrlich, S. D., H. Bierne, E. d'Alencon, D. Vilette, M. Petranovic, P. Noirot, and M. Michel. 1993. Mechanisms of illegitimate recombination. Gene 135:161-166. [DOI] [PubMed] [Google Scholar]
- 25.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 26.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer, W., R. Haas, and S. Odenbreit. 2002. Type IV secretion systems in pathogenic bacteria. Int. J. Med. Microbiol. 292:159-168. [DOI] [PubMed] [Google Scholar]
- 28.Fischer, W., J. Puls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 29.Garcia, L. R., and I. J. Molineux. 1999. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by EcoKI. Proc. Natl. Acad. Sci. USA 96:12430-12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 31.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jilk, R. A., J. C. Makris, L. Borchardt, and W. S. Reznikoff. 1993. Implications of Tn5-associated adjacent deletions. J. Bacteriol. 175:1264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 34.Kersulyte, D., H. Chalkauskas., and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 35.Kersulyte, D., B. Velapatiño, G. Dailide, A. K. Mukhopadhyay, Y. Ito, L. Cahuayme, A. J. Parkinson, R. H. Gilman, and D. E. Berg. 2002. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 184:992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuipers, E. J., D. A. Israel, J. G. Kusters, and M. J. Blaser. 1998. Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J. Bacteriol. 180:2901-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwok, T., S. Backert, H. Schwarz, J. Berger, and T. F. Meyer. 2002. Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70:2108-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15:1725-1752. [DOI] [PubMed] [Google Scholar]
- 39.Mahdavi, J., B. Sonden, M. Hurtig, F. O. Olfat, L. Forsberg, N. Roche, J. Angstrom, T. Larsson, S. Teneberg, K. A. Karlsson, S. Altraja, T. Wadstrom, D. Kersulyte, D. E. Berg, A. Dubois, C. Petersson, K. E. Magnusson, T. Norberg, F. Lindh, B. B. Lundskog, A. Arnqvist, L. Hammarstrom, and T. Boren. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mobley, H. L. T., G. L. Mendez, and S. L. Hazell (ed.). 2001. Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 41.Mukhopadhyay, A. K., D. Kersulyte, J.-Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray, N. E. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64:412-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Mégraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 68:6240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhen, M., S. Eriksson, M. Clements, S. Bergström, and S. J. Normark. 2003. The basis of persistent bacterial infections. Trends Microbiol. 11:80-86. [DOI] [PubMed] [Google Scholar]
- 45.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 47.Studier, F. W., and P. K. Bandyopadhyay. 1988. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl. Acad. Sci. USA 85:4677-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 49.Tomcsanyi, T., C. M. Berg, S. H. Phadnis, and D. E. Berg. 1990. Intramolecular transposition by a synthetic IS50 (Tn5) derivative. J. Bacteriol. 172:6348-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trieber, C. A., and D. E. Taylor. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trieu-Cuot, P., G. Gerbaud, T. Lambert, and P. Courvalin. 1985. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 4:3583-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turlan, C., and M. Chandler. 1995. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 14:5410-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Ende, A., E. A. Rauws, M. Feller, C. J. Mulder, G. N. Tytgat, and J. Dankert. 1996. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology 111:638-647. [DOI] [PubMed] [Google Scholar]
- 54.Wang, G., R. W. Blakesley, D. E. Berg, and C. M. Berg. 1993. pDUAL: a transposon-based cosmid cloning vector for generating nested deletions and DNA sequencing templates in vivo. Proc. Natl. Acad. Sci. USA 90:7874-7878. [DOI] [PMC free article] [PubMed] [Google Scholar]