Abstract
DEAD-box proteins are characterized by nine conserved motifs. According to these criteria, several hundreds of these proteins can be identified in databases. Many different DEAD-box proteins can be found in eukaryotes, whereas prokaryotes have small numbers of different DEAD-box proteins. DEAD-box proteins play important roles in RNA metabolism, and they are very specific and cannot mutually be replaced. In vitro, many DEAD-box proteins have been shown to have RNA-dependent ATPase and ATP-dependent RNA helicase activities. From the genetic and biochemical data obtained mainly in yeast, it has become clear that these proteins play important roles in remodeling RNP complexes in a temporally controlled fashion. Here, I shall give a general overview of the DEAD-box protein family.
INTRODUCTION
Nucleic acids can be present in single-stranded, double-stranded or even multiple-stranded forms. The advantages of a double-stranded molecule with strands of opposite polarity have been known since the discovery of the double-stranded DNA molecule (1). However, the possibility of finding a matching partner can be of importance not only for DNA but also for RNA. This can be true for extended double-stranded RNA molecules as found in viruses, for local secondary structures as in ribosomes and for short RNA–RNA interactions, as in pre-mRNA splicing or RNA-mediated silencing. The caveat of double-stranded nucleic acids is that at some point they may need to be unwound if the sequence information of the nucleic acid needs to be deciphered or to be used for an alternative sequence-specific binding event. Therefore, an obligatory complement of double-stranded nucleic acids is the presence of enzymes that can unwind these helical molecules, i.e. helicases. Since the two strands are held together by base pairing, helicases require energy for unwinding. Text books discuss in detail helicases required for initiation and elongation of DNA replication, but only rarely helicases that are involved in the separation of RNA strands. Nevertheless, genes encoding helicases make up a considerable portion of the coding information of a eukaryotic genome (2) and many of these helicases have a preference or even an exclusive requirement of RNA molecules. Several reviews on different aspects of DEAD-box proteins have been published in recent years (3–12). Here, I shall give a general overview of the DEAD-box field as it stands today.
WHAT IS THE DEAD-BOX FAMILY?
One of the earliest descriptions of an RNA helicase was the report that incubation of globin mRNA with the translation initiation factor eIF4A and ATP changed the susceptibility of the mRNA to nucleases (13). Thus, eIF4A altered the structure of the mRNA in such a way, that the RNase digestion pattern changed. This change was dependent on a source of energy in the form of ATP. The translation initiation factor eIF4A could therefore be considered as a helicase that melts (local) secondary structures and makes the RNA accessible to nucleases. Since then, many RNA helicases involved in a variety of cellular processes have been described.
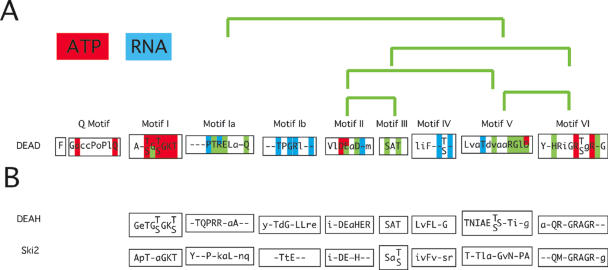
In 1988, Gorbalenya et al. (14) defined a group of NTPases and showed that they had several common sequence elements. This analysis, together with the description of a number of proteins involved in RNA metabolism (p68, SrmB, MSS116, vasa, PL10, mammalian eIF4A, yeast eIF4A) resulted, based on the sequence of eIF4A, in the birth of the DEAD-box protein family (15). Today, the alignment of all annotated sequences in SwissProt from all species reveal nine conserved sequence motifs with very little variation (15,16) (Figure 1). The simultaneous presence of these motifs is a criterion for inclusion of a protein within the family, although an enzymatic activity has been demonstrated only for a limited number. Motif II (or Walker B motif) has the amino acids D-E-A-D, which gave the name to the family. This motif, together with motif I (or Walker A motif), the Q-motif and motif VI, is required for ATP binding and hydrolysis (16–19). Motifs Ia and Ib, III, IV and V have been characterized less well but may be involved in interaction with RNA (20) and in intramolecular rearrangements necessary for remodeling activity (Figure 1).
Figure 1.
A schematic presentation of the conserved motifs of the DEAD-box family. (A) Consensus sequence of the DEAD-box family. Residues identified in the structure of the Vasa protein (70) to interact with ATP (red), RNA (blue) or involved in intra-protein interactions (green) are highlighted. (B) Consensus sequences of the DEAH-box and Ski2 family. The consensus sequences (capital letters represent amino acids conserved at least 80%, lower case letters represent amino acids that are conserved 50–79%) are taken from Tanner and Linder (10).
Proteins related to eIF4A in sequence can be found in all eukaryotic cells and in most eubacteria and archaebacteria. The genome of the yeast Saccharomyces cerevisiae encodes 25 DEAD-box proteins (21,22). Interestingly, it has two genes (TIF1 and TIF2) encoding exactly the same eIF4A protein, and it encodes two related proteins Ded1 and Dbp1. The deletion of DED1 is lethal, whereas the deletion of DBP1 is not lethal under normal laboratory conditions. However, overexpression of Dbp1 can suppress the lethal deletion of DED1 (23), indicating (but not proving) a functional redundancy. A comparison with another fungal species, Ashbya gosypii, which is considered to be the free living eukaryote with the smallest genome (24), reveals all the DEAD-box proteins found in S.cerevisiae, with the exception of Dbp1 and Prp28 (involved in pre-mRNA splicing, see below), and with only one eIF4A copy. Thus, the DEAD-box proteins of A.gosypii could represent the minimal number of such proteins required for a free-living eukaryote.
In multicellular eukaryotes, several additional DEAD-box proteins can be found. A search in the human genome revealed 38 DEAD-box proteins (Table 1), which can tentatively be classified into 32 subfamilies. These subfamilies have been defined by iterative blast searches against the SwissProt/trEMBL databases, using all human DEAD-box proteins. Approximately 250 best scoring sequences from each blast search were then used for a ClustalW analysis to identify related sequences. In some cases, where twohuman or two yeast proteins clustered together, the members of the putative subfamily from other model organisms were analyzed further to determine whether there were one or two proteins within this subfamily. If other model organisms had only one protein, the subfamily was defined as such. However, if most model organisms had also two representatives within the putative subfamily, the subfamily was divided into two. An example is the separation of the Ddx3/Ded1 and Vasa subfamilies. Drosophila and other multi-cellular eukaryotes have two or more DEAD-box protein related to Ded1 or Vasa. However, with the exception of the yeast S.cerevisiae, unicellular eukaryotes have only one of these proteins (25) and therefore these proteins have been divided in two subfamilies. Another example would be the subfamily of proteins homologous to the yeast Dbp5 protein. In the human genome three proteins, Ddx19A, Ddx19B and Ddx25, are very similar to Dbp5 and are therefore being included in the same subfamily. It is clear, that this definition of subfamilies is somehow arbitrary and should be regarded as a working tool to compare proteins and predict functions. In some cases cross species complementation could be demonstrated (26,27) but in any case, experiments are needed to characterize these subfamilies further. The Ddx7 (28) protein has no homologs in other mammals and a tblast against the human genome does not report any significant similarity. It is therefore excluded from the list presented here. According to the criteria defined above, 11 human DEAD-box proteins have no direct or obvious counterpart in yeast (Ddx1, Ddx4/vasa, Ddx20/DP103, Ddx21/RNA helicase Gu-alpha, Ddx28, Ddx50/RNA helicase Gu-beta, Ddx41/abstrakt, Ddx42, Ddx43, Ddx53, Ddx59). Although it may be expected that the human genome contains more DEAD-box proteins than the simple budding yeast, it may seem surprising that three DEAD-box proteins present in yeast (Dbp3, Mss116, Mrh4) have no obvious counterpart in humans. The DEAD-box proteins Mss116 and Mrh4 have been shown to be required for gene expression in yeast mitochondria (29,30). It is tempting to speculate that these proteins are simply not required in human mitochondria, because the structural organization of human mitochondrial genes is different from that of yeast mitochondrial genes, which harbor many introns. In contrast, Ddx28, may be involved in mitochondrial gene expression in human mitochondria, insofar as it shows nuclear and mitochondrial localization (31,32). The yeast Dbp3 protein is involved in ribosome biogenesis and it is one of the rare DEAD-box proteins that are not essential for growth under normal laboratory conditions (33). In contrast to eukaryotes, bacterial genomes encode far fewer DEAD-box proteins and some bacterial species seem not to encode DEAD-box proteins at all (5,8). Today, searches in SwissProt reveal ∼205 annotated sequences and >700 different entries in SwissProt and trEMBL. Based on the activity of eIF4A and on the sequence alignments, it is thought that the members of the DEAD-box family have similar biochemical activities.
Table 1.
A tentative assignment of yeast and human DEAD-box protein subfamilies
| Human | SwissProt | Alias | Function | Reference | Yeast | SwissProt | Function | Reference |
|---|---|---|---|---|---|---|---|---|
| DDX1 | Q92499 | DEAD-box protein-retinoblastoma | Amplified in retinoblastoma, cellular co-factor of HIV-1 Rev, nucleolar | (105,143,144) | — | — | ||
| DDX2A | P60842 | eIF4A I | Translation initiation | (9) | Tif1 | P10081 | Translation initiation | (145) |
| Tif2 | ||||||||
| DDX2B | Q14240 | eIF4A II | ||||||
| DDX3Y | O15523 | DBY | (146,147) | Ded1 | P06634 | Translation initiation, re-mRNA splicing, mRNA export | (90,125,126,148) | |
| Dbp1 | P24784 | |||||||
| DDX3X | O00571 | DDX3, mDEAD3 | Similar to mouse PL10, Xenopus An3, and Drosophila Bel; required for Rev-dependent export of intron-containing HIV-1 RNA, nucleolar | (105,149,150) | ||||
| DDX4 | Q9NQI0 | vasa | Translation initiation, imilar to Drosophila vasa that interacts with eIF5B | (151,152) | — | — | — | — |
| DDX5 | P17844 | p68, HLR1 | transcription, pre-mRNA splicing, mRNA stability and ribosome biogenesis, nucleolar | (105,153,154) | Dbp2 | P24783 | ribosome biogenesis, interacts with Upf1 and is involved in NMD | (155) |
| DDX17 | Q92841 | p72 | nucleolar | (105,156) | ||||
| DDX6 | P26196 | p54, RCK | Oncogene RCK, translation initiation of c-myc mRNA, nuclear assembly of stored mRNP particles, mRNA masking in analogy to clam homolog | (137,138,157–159) | Dhh1 | P39517 | Assists decapping, Required for mRNA storage, | (135,160) |
| DDX10 | Q13206 | nucleolar | (105,161) | Dbp4 | P20448 | Ribosome biogenesis | (162) | |
| DDX17 | See subfamily DDX5/DDX17 | |||||||
| DDX18 | Q9NVP1 | MrDb, mRNA export | Nucleolar, Myc-regulated | (105,163) | Has1 | Q03532 | Ribosome biogenesis | (108) |
| DDX19A | Q9NUU7 | DEAD box protein | (53) | Dbp5 | P20449 | mRNA export | (52,111) | |
| DDX19B | Q9UMR2 | |||||||
| DDX25 | Q9UHL0 | GRTH | Gonadotropin-regulated testicular RNA helicase | (164) | ||||
| DDX20 | Q9UHI6 | DP103, Gemin3, survival of motor neurons (SMN)-interacting protein | Spliceosomal snRNP biogenesis | (165,166) | — | — | — | — |
| DDX21 | Q9NR30 | Nucleolar RNA helicase II, Nucleolar RNA helicase Gu Gu-alpha | Ribosomal RNA production, co-factor for c-Jun-activated transcription | (105,167–169) | — | — | — | — |
| DDX50 | Q9BQ39 | RNA helicase Gu-beta DDX21B according to Abdelhaleem et al. | Localizes to nuclear speckles containing splicing factor SC35 Co-factor for c-Jun-activated transcription, nucleolar | (61,105,170,171) | — | — | — | — |
| DDX23 | Q9BUQ8 | Pre-mRNA splicing | (172) | Prp28 | P23394 | pre-mRNA splicing | (72,173,174) | |
| DDX24 | Q9GZR7 | nucleolar | (105,175) | Mak5 | P38112 | Ribosome biogenesis | (176) | |
| DDX25 | See subfamily DDX19A/DDX19B/DDX25 | |||||||
| DDX27 | Q96GQ7 | Nucleolar | (105) | Drs1 | P32892 | Ribosome biogenesis | (177) | |
| DDX28 | Tr_Q9NUL7 | MDDX28 | Mitochondrial and nuclear localization | (31) | — | — | — | — |
| DDX31 | Q9H8H2 | Nucleolar | (105) | Dbp7 | P36120 | Ribosome biogenesis | (178) | |
| DDX39 | O00148 | URH49 | Pre-mRNA splicing and export | (118) | ||||
| BAT1 | Q13838 | UAP56 | (179–181) | Sub2 | Q07478 | Pre-mRNA splicing and export | (85,115,117,182) | |
| DDX41 | Q9UJV9 | DEAD-box protein abstrakt homolog | (183,184) | — | — | — | — | |
| DDX42 | Tr_Q86XP3 | SF3b125 DEAD-box protein | Pre-mRNA splicing, splicing | (185) | — | — | — | — |
| DDX43 | Tr_Q9NXZ2 | Displays tumor-specific expression | (186) | — | — | — | — | |
| DDX53 | Tr_Q6NVV4 | CAGE | CAGE is expressed in a variety of cancers but not in normal tissues except testis, | (187) | — | — | — | — |
| DDX46 | Tr_Q7L014 | Pre-mRNA splicing | (185) | Prp5 | P21372 | Pre-mRNA splicing | (46,47,188) | |
| DDX47 | Q9H0S4 | Co-transfection of GABARAP and DDX47 cDNA into a tumor cell line induces apoptosis, nucleolar localization | (105,106) | Rrp3 | P38712 | Ribosome biogenesis | (189) | |
| DDX48 | P38919 | NMP265/NUK34, eIF4A III | DDX48 is a component of the EJC; has also been found in proteomic studies of the nucleolus | (98,105,190) | Fal1 | Q12099 | Ribosome biogenesis | (100) |
| DDX49 | tr_Q9Y6V7 | nucleolar | (105) | Dbp8 | Ribosome biogenesis | (191) | ||
| DDX50 | See subfamily DDX21/DDX50 | |||||||
| DDX51 | Tr_Q8IXK5 | Nucleolar | (105) | Dbp6 | P53734 | Ribosome biogenesis | (192) | |
| DDX52 | Q9Y2R4 | nucleolar | (105,106) | Rok1 | P45818 | Ribosome biogenesis | (193) | |
| DDX53 | See subfamily DDX43/DDX53 | |||||||
| DDX54 | Q8TDD1 | DP97 | nucleolar | (105,106,194) | Dbp10 | Q12389 | Ribosome biogenesis | (195) |
| DDX55 | Tr_Q8NHQ9 | Nucleolar associates with nucleoplasmic 65S preribosomal particles,nucleolar | (105) | Spb4 | P25808 | Ribosome biogenesis | (196) | |
| DDX56 | Q9NY93 | noH61, DDX21 | (105,197) | Dbp9 | Q06218 | Ribosome biogenesis | (198) | |
| DDX59 | tr_Q8IVW3 | — | — | — | — | |||
| — | Dbp3 | P20447 | Ribosome biogenesis | (33) | ||||
| — | MSS116 | P15424 | Mitochondrial gene expression | (30,199) | ||||
| — | Mrh4 | P53166 | Mitochondrial function | (29) | ||||
The yeast DEAD-box proteins have been described previously (21). The human subfamilies have been determined with the help of Abdelhaleem et al. (2003), a search for DDX genes in SwissProt, a search in the human gene nomenclature search site (www.gene.ucl.ac.uk/nomenclature/), and by running a blast search using yeast eIF4A against the initio proteins of the human genome (http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html). Representative samples (∼250 sequences) from the blast searches using every individual human DEAD-box protein defined above was used for a second round of blast analysis for confirmation and for ClustalW analysis at EBI and a tentative tree has been established by using the TreeView (Rod page, http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) program. Proteins related to DDX2A and DDX2B, DDX3Y and DDX3X, DDX5 and DDX17, DDX19A and DDX19B and DDX25, DDX21 and DDX50, DDX39 and BAT1, DDX43 and DDX53, form each one subfamily, respectively. Based on this analysis and the absence of any significant match in a blast with the human genome, the DDX7 entry (28) has been removed from the list. References are given for information but are by far not exhaustive. More information on RNA helicases can be found on http://www.helicase.net and http://www.medecine.unige.ch/~linder/RNA_helicases.html.
THE DEAD-BOX FAMILY IS DISTINCT, BUT NOT ALONE
Bioinformatic searches have revealed related proteins that share some motifs with the DEAD-box family, but have other distinguishing motifs that are conserved within their own family (34). The related proteins belong to the DEAH and Ski2 families, which together with the DEAD-box family are often referred to as the DExD/H families. However, based on their sequences, the families are clearly distinct, despite the similarities they share (Figure 1B). In other words, no protein has been found so far that could belong to two families, as judged from the conserved motifs. This could simply be explained by a co-evolution of the different motifs within one family. However, another perspective is that the different families serve different purposes in RNA metabolism in a cell. In this respect, it is interesting to note that biochemical and structural analyses have revealed certain similarities amongst members of the various families, but also differences. For example both DEAD-box and DEAH-box proteins are stimulated by RNA in their NTPase activity, but DEAD-box proteins use only ATP, whereas DEAH box proteins are more promiscuous in their NTP usage (16,35).
WHAT DO WE KNOW ABOUT THE BIOCHEMICAL ACTIVITIES OF DEAD-BOX PROTEINS?
In comparison to the enormous number of DEAD-box proteins present in protein databases, only few RNA helicases from the DEAD-box family have been characterized biochemically (36). As expected from the presence of the Walker A and Walker B motifs typical for NTPases, DEAD-box proteins show ATPase activity. Normally, this activity is dependent on RNA, although in some instances an RNA-independent activity has been reported (36). Further experiments are needed to determine whether these differences are intrinsic to the analyzed proteins themselves, or dependent on the purification of the proteins. In general, stimulation of the ATPase activity is not dependent on a particular RNA species. Indeed, in many cases such as in the scanning process of the 40S ribosomal subunit in translation initiation or in mRNA export from the nucleus, sequence specificity for the substrate would be in contradiction to its function. This implies that their specificity relies on the interaction with other RNP components. In the case of eIF4A it has been shown, for a long time, that its RNA-dependent ATPase activity is stimulated by eIF4B, although the molecular details of this stimulation are still not known (37). More recently the stimulation of the activity of eIF4A by eIF4H and eIF4G has also been described previously (38,39). In the case of eIF4G, it has been suggested that eIF4G forms a ‘soft clamp’ that stabilizes eIF4A in a closed active conformation (39). Interestingly eIF4A can also be stimulated by pateamine A, a natural marine product that inhibits translation initiation and decreases the interaction of eIF4G and eIF4A (40,41). In contrast to these examples of stimulation by other proteins, in the case of proteins from the bacterial DbpA subfamily, a large stimulation by a hairpin structure of the 23S rRNA can be observed (42–44). This stimulation is dependent on a C-terminal domain that contains an RNA recognition fold motif (45). To a lesser extend, the yeast Prp5 protein, involved in pre-mRNA splicing, is stimulated in its activity by the snRNA U2 (46,47). It is noteworthy that Prp5 interacts with components of the U2 RNP (48). It is likely that, for other DEAD-box proteins, other stimulating or regulatory conditions/environments will be found in the near future [e.g. eIF4AIII, below, and Dbp8 (49)].
DEAD-box proteins are often referred to as RNA helicases. This implies that the proteins unwind, in an energy-dependent manner, double-stranded RNA molecules. Such an activity has indeed been demonstrated for several DEAD-box proteins (50–69). In most cases, however, unwinding activity is limited to short duplexes, indicating that it is not processive. Two simple explanations can be offered. First, recombinant proteins out of their biological context may not be efficient or processive. This is also true for proteins that are considered to have highly processive activities, such as the DNA polymerase that requires a clamping factor to become processive in its activity. The second explanation would be that indeed the DEAD-box proteins are not processive even in vivo, since they do not need to unwind lengthy double-stranded structures. In this scenario, which is at present the most likely one, their requirement would be a local action to unwind a limited double-stranded RNA or dissociate a protein from the RNA (see below), to allow further steps in a process to occur. The recently published data on the structure of the Drosophila Vasa protein with non-hydrolyzable ATP and an RNA substrate are clearly consistent with this view (70). In this structure, the Vasa protein bends the bound RNA in such a way that a double-stranded nucleic acid would be partially unwound (71). Clearly, the destabilization of the double-stranded RNA by virtue of the binding of the helicase to the double-stranded substrate, would suggest a non-processive and local dissociation activity (71).
Following the idea of a local dissociation activity, it has been shown recently, by genetic and biochemical experiments, that DEAD-box proteins are able to dissociate proteins from RNA molecules. Genetic experiments demonstrated that mutations in the genes encoding DEAD box proteins Prp28 and Sub2 can be suppressed by mutations in genes encoding proteins that are part of RNPs (72,73). In the case of Prp28, it has been shown that mutations in the U1 snRNA or the U1-C protein bypass the requirement of Prp28 (72). Similarly, deletion of Mud2 bypasses the requirement of Sub2 (73) and it has been shown recently that a mutation in the export factor Mex67 can suppress a mutation in DBP5 (74). These results suggest that either these DEAD-box proteins can directly dissociate RNA–protein complexes or modify RNA structures that stabilize RNA–protein interactions. How this applies to the structure of Vasa, remains to be determined.
Thus, DEAD-box proteins are modulators of RNP complexes [see also (6)]. This modulating function is dependent on the presence of RNA, since the ATPase activity of most, if not all, DEAD-box proteins is dependent or largely stimulated by the presence of RNA. In order to limit the activity in time and space, RNA helicases may only transiently associate with an RNP complex. However, they also may be part of a complex for a certain period as found in proteomic studies of successive intermediates in pre-ribosomal particles (75–83). In this case it is likely that a conformational change, induced by the binding or dissociation of another subunit of the complex, brings the RNA substrate in such a position as to activate the ATPase activity of the DEAD-box protein. The DEAD-box protein would then induce a further conformational change in the RNP structure. This change will in turn modify the structure in such a way that it might no longer be a substrate for this particular RNA helicase. This would be an easy and elegant way to limit the activity of DEAD-box proteins and to provide a force for a unidirectional development of an RNP complex.
BIOLOGICAL FUNCTIONS OF DEAD-BOX PROTEINS
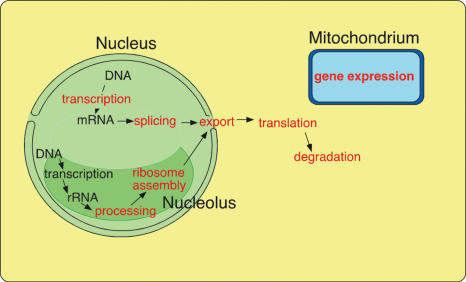
DEAD-box proteins have been described to be necessary for, or involved in, many different processes of RNA metabolism. In eukaryotic cells, in particular, these range from the transcription to the degradation of RNA, and include pre-mRNA splicing, mRNA export, ribosome biogenesis, translation initiation and gene expression in organelles (Figure 2).
Figure 2.
Schematic presentation of cellular processes that require DEAD-box proteins in eukaryotic cells.
Transcription
Recently several RNA helicases of the DEAD-box family have been described to be involved in transcription [see the contribution by Fuller-Pace (4)].
Pre-mRNA splicing
Splicing of pre-mRNAs has become a paradigm for the analysis of the function of DEAD/DExH proteins. Although the removal of an intron by two transesterification reactions is energetically neutral, the splicing reaction requires ATP. This could be explained by temporal modification reactions, such as phosphorylation, or by active remodeling of the spliceosome. Indeed the formation of the spliceosome, the rearrangements within the spliceosome during the splicing reactions and the final release of the product, as well as the recycling of the components, require the rearrangement of five large RNP complexes (snRNPs U1, U2, U4, U5 and U6). Proteomic approaches of the spliceosome suggest >200 proteins involved in this process (84). Part of these rearrangements need to occur rapidly and in a controlled fashion, and therefore most likely require an energy input, which, at least partially, may be attributed to the function of DEAD-box proteins. In yeast, three DEAD-box proteins have been shown to be required for in vivo splicing [Sub2, Prp28 and Prp5 (85)]. In higher eukaryotes, p68 was shown to be involved in constitutive and alternative mRNA splicing (86,87), and its homolog p72 has been implicated in alternative splicing (88). Other proteins, such Ded1p (see below), may also be involved in splicing (89,90), although their role in this process has not been established definitely. In addition to the known DEAD-box proteins, other DExD/H proteins are required for splicing to occur, namely the DEAH proteins Prp2, Prp16, Prp22, Prp43 and the Ski2-like protein Brr2 (85,91–97). Interestingly, DEAD-box proteins are required for establishment of a functional spliceosome, whereas DEAH-box proteins are (indirectly) required for the transesterification reactions, the release of the mRNA, and the recycling of the spliceosome components.
In addition to these ‘classical’ splicing DEAD/H-box proteins, the proteomic approaches of spliceosomes from higher eukaryotes also revealed the presence of other DEAD-box proteins such as homolog of the Drosophila abstrakt, eIF4AIII, Ddx35 and Ddx9 (84). The eIF4AIII protein has been shown to be an important component of the exon-junction complex (EJC) (98). In the case of eIF4AIII it has been reported recently that its ATPase activity is inhibited by the presence of another component of the EJC (99). Interestingly, a homologous protein, Fal1, from yeast is involved in ribosome biogenesis (100).
Ribosome biogenesis
Pre-ribosomal complexes with well over 100 transacting factors, including small nucleolar RNAs (snoRNAs) and many proteins of different activities (101,102), represent another example of a complex and highly dynamic RNP. Many NTPases have been shown to be involved in ribosome biogenesis in prokaryotes and eukaryotes. Beside many DEAD-box proteins, DEAH-box proteins, a Ski2-like RNA helicase (Dob1/Mtr4), and AAA proteins, are required for ribosome biogenesis. Whereas the number varies in prokaryotes and is relatively small [i.e. 0 in Borrelia burgdorferi; 3 in Escherichia coli, (5)], 14 DEAD-box proteins have been shown by genetic experiments to be required for ribosome biogenesis in S.cerevisiae (21,103,104). Most of these DEAD-proteins from S.cerevisiae have counterparts in higher eukaryotes, indicating that their requirement is conserved. This is further supported by the fact that most of the human DEAD-box proteins homologous to those required in ribosome biogenesis in yeast can be detected in proteomic approaches of human nucleoli (105,106). One of the rare exceptions is Dbp3, required for the MRP RNase assisted cleavage at A3 (33). This protein is highly conserved amongst fungi, but has no obvious counterpart in higher eukaryotes, as judged from blast searches and ClustalW analyses (Table 1). Interestingly, Dbp3, together with Dbp7, is not absolutely essential for ribosome biogenesis in yeast. However, in contrast to their bacterial DEAD-box counterparts (5), the other proteins involved in ribosome biogenesis in yeast are essential for cell viability. Moreover, they are highly specific and cannot be replaced by each other, even when overexpressed.
Ribosome biogenesis is an ideal playground for DEAD-box proteins. Eukaryotic ribosomes are composed of 4 rRNAs and 78 proteins (102). Three of the mature rRNA species are transcribed as a large pre-rRNA and are, during the assembly reaction, processed to the three mature 18S, 5.8S and 25S rRNAs. In addition to the processing reactions, the 18S and 25S rRNA are modified by pseudouridylation and methylation. These modifications are guided by snoRNAs (107) that are complementary to the rRNA. Thus, DEAD-box proteins could play roles in reorganizing the pre-ribosomal complexes by dissociating snoRNAs from the pre-rRNA, to allow new and mutually exclusive RNA–RNA interactions to occur. Moreover, as in pre-mRNA splicing, DEAD-box proteins may be involved in RNP remodeling by altering RNA–protein interactions [see contribution by (6)]. Whereas all DEAD-box proteins involved in ribosome biogenesis in yeast have been characterized genetically, little is known about these proteins in higher eukaryotes. Nevertheless, most of them have been found in proteomic analyses of nucleoli, the cradle of ribosomes (105,106).
Genetic analyses of ribosome biogenesis and of the DEAD-box proteins required for this process in yeast indicate functions of RNA helicases by dead-end products that can be detected. By this criterion, six DEAD-box proteins are required for early cleavages and affect synthesis of the small ribosomal subunit, whereas eight DEAD-box proteins are required for the synthesis of the large ribosomal subunit. Nevertheless, this analysis of dead-end products in ribosome biogenesis may not reflect appropriately the actual function of DEAD-box proteins. It is likely that the absence of a protein of this family does not induce an immediate defect in ribosome biogenesis or assembly, but only a delayed processing/assembly defect. Moreover, a strong defect may mask a weaker effect in a completely different step. As an example, it is intriguing that Has1 is mainly found associated with pre-60S particles, but the genetic analysis reveals a clear 40S deficit (108). It is therefore important to characterize interacting partners of these enzymes. Genetic screens, such as the search for synthetic lethal interactions, suppressor analyses and complex purification, will certainly help to go in this direction (109,110).
Nuclear export
In eukaryotic cells transcription and translation occur in separate compartments. Therefore the mature mRNA needs to be exported, through nuclear pores, from the nucleus to the cytoplasm. A defect in the yeast Dbp5/Rat8 gene results in accumulation of poly(A) mRNA in the nucleus, clearly indicating a role in mRNA export (52,111). In a beautiful experiment, it has been shown that a mutation in the Nup159 protein of the nuclear pore complex leads to a cytoplasmic localization of Dbp5, rather than at the nuclear rim (53). These data suggest a role of Dbp5 on the cytoplasmic side of the nuclear pore. In addition, recent data show that Dbp5 localizes to Balbiani ring of Chironomus tentans (112) and demonstrate genetic and physical interactions of yeast Dbp5 with the transcription machinery (113). This suggests that Dbp5 needs to be loaded on the mRNA early, travels along to the nuclear pore, where it is required for export. A genetic interaction between mex67 and dbp5 suggests that Dbp5 is required for the release of Mex67 (74). Another DEAD-box protein, Uap56/Bat1 in higher eukaryotes and Sub2 in yeast (that are in reality DECD proteins), has also been shown to be required for export of mRNA, in addition to its role in pre-mRNA splicing (114–117). Interestingly this splicing factor is also required for export of mRNAs that do not contain introns, arguing against the simple scenario that Sub2 remains bound to the message after splicing. Thus, Uap56/Bat1/Sub2 proteins play two roles in the life of an mRNA. Intriguingly, two highly homologous (89% identity) DECD proteins, Ddx39 and Bat1, are encoded by the human genome (118).
Translation initiation
The translation initiation factor eIF4A was the first DEAD-box protein described to have a RNA-dependent ATPase activity (119). Several reviews have been published about eIF4A (9,25) and I summarize here only its essentials. The translation initiation factor eIF4A is a very abundant protein (120,121). It is part of the cap-binding complex eIF4F but is also present in a free form. Its biochemical activities are greatly stimulated by eIF4B, eIF4H and eIF4G (37–39,122). It has been proposed that eIF4A helps to unwind secondary structures in the 5′-untranslated region (5′-UTR), which are inhibitory for the scanning process of the small ribosomal subunit (123). Experimental evidence supporting this hypothesis has been reported in an in vitro translation system with increasing secondary structures in the 5′-UTR (20) and by the analysis of cell cycle defects in Schizosaccharomyces pombe (124). Interestingly, however, a mRNA substrate with the initiator AUG positioned 8 nt downstream of the cap structure is still absolutely dependent on eIF4A in a yeast in vitro translation system (17).
The laboratory of Tien-Hsien Chang and our laboratory have demonstrated that another DEAD-box protein, Ded1, is also required for translation initiation in vivo and in vitro (125,126). Although its precise role in translation initiation remains elusive, the laboratory of John McCarthy has shown, by testing mRNAs with 5′-UTR of various lengths, that Ded1 plays a role in the scanning process in vivo and in vitro (127). In multicellular organisms, yet another DEAD-box protein, Vasa, has been shown to play an important role in translation initiation via its interaction with IF2 (128). Interestingly, Vasa is highly related to Ded1, but is absent in fungi, in accordance with its role in embryonic development.
Degradation
Elegant studies demonstrated the requirement for DEAD-box proteins in RNA degradation in E.coli (5,129,130). In eukaryotes, RNA degradation occurs mainly via the multisubunit exosome, assisted by RNA helicases of the Ski2 family (131–134). However, no DEAD-box protein seems to be directly required for the progression of the exosome. The Dhh1 protein plays an essential role in mRNA degradation through its implication in decapping of the mRNA (135,136). Interestingly, proteins from the same subfamily play important functions in masking mRNAs in higher eukaryotes (13) (137,138).
Organelle gene expression
In yeast, two DEAD-box proteins are required for mitochondrial gene expression, Mss116 and Mrh4. The Mss116 protein was shown to be involved in mitochondrial splicing. However, a strain with no mitochondrial introns still required Mss116 for growth on non-fermentable carbon sources (139). Intriguingly, overexpression of Mss116 does suppress the absence of a completely unrelated helicase, Suv3, which is involved in mitochondrial RNA turnover (140). The Mrh4 protein was isolated as a low-copy suppressor of a point mutation in the mitochondrial aI5γ intron, although a block in the splicing reaction could not be observed in a Δmrh4 strain (29). As mentioned above, both these DEAD-box proteins have no direct homolog in humans, which could be related to differences in the mode of mitochondrial gene expression in yeast and humans. A human DEAD-box protein, Ddx28, has been reported to localize to the mitochondria (31). However, its function is not known.
DEAD-box proteins from Trypanosomes have also been shown to be required for editing (141). Interestingly these proteins have homologs only in another kinetoblast organism: Leishmania. It is thus tempting to speculate that these proteins are required for the guide RNA assisted editing of mitochondrial RNA, characteristic for these organisms (142).
Altogether, the emerging picture of DEAD-box proteins depicts a large family of proteins that possess a non-processive dissociation activity. This activity is particularly used in many RNA metabolic processes in eukaryotic cells. It is likely that in these processes, the DEAD-box proteins are important place-holders or check-point proteins, allowing processes to proceed efficiently in one direction and connected with previous or following steps in the RNA metabolism machinery.
DISCUSSION AND OPEN QUESTIONS
Eukaryotic cells have a large number of DEAD-box proteins, most of which are essential, as judged from genetic experiments in yeast. From our actual knowledge of these proteins, we can deduce that they are involved in many if not all steps of RNA metabolism. Although they present a high degree of similarity within the core region, which is responsible for the enzymatic activities of these proteins, they are all highly specific for their function and cannot be replaced by each other.
Despite intensive studies in many laboratories, the exact role of these proteins remains elusive. Although we know that they are required for the dynamics of RNP complexes, such as the establishment of the spliceosome, the biogenesis of ribosomes, export of mRNA through the nuclear pore or the presence of EJCs on spliced mRNAs, their exact function remains unclear. Do they need their enzymatic activity for an active remodeling of RNP structures, or do they play a more passive role and use the enzymatic activity to leave the complex to make place for new interactions within the RNP complex?
Yet another enigma is their regulation. As far as we know, DEAD-box proteins require the presence of RNA for stimulation of their enzymatic activity. Also, it is clear that the Q-motif plays an important role in ATP recognition and thus in regulation. Nevertheless, the activity needs to be tightly controlled in time and space. Presumably, it is regulated by the interaction with specific RNA sequences or other proteins of the RNP complexes.
Finally, some DEAD-box proteins may function as a sort of ‘check point’ control. In a dynamic RNP assembly, such as the spliceosome or the ribosome, the cell needs to control the correct functionality of these super-machines to avoid erroneous splicing or protein synthesis. In this view, a particular DEAD-box protein can only be activated if the intermediate structure is correct. If the structure is not correct, the synthesis has to wait or be abandoned.
Acknowledgments
The author is very grateful to Frances Fuller-Pace, Beate Schwer, Josette Banroques and an unknown referee for very helpful comments on the manuscript. The author is particularly grateful to all his present and past collaborators and to Costa Georgopoulos for continuous support. Progress in the field has been made possible by many friendly and collaborative interactions. Work in the laboratory is supported by the Swiss National Science Foundation, Novartis Foundation, Roche Research Foundation, E. & L. Schmidheiny Foundation and the Canton of Geneva. Funding to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Watson J.D., Crick F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Shiratori A., Shibata T., Arisawa M., Hanaoka F., Murakami Y., Eki T. Systematic identification, classification, and characterization of the open-reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. YEAST. 1999;15:219–253. doi: 10.1002/(SICI)1097-0061(199902)15:3<219::AID-YEA349>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Fuller-Pace F.V. DExD/H-box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl460. doi:10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iost I., Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl500. doi:10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankowsky E., Bowers E. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl410. doi:10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeang K.-T., Yedavalli V. Role of RNA Helicases in HIV-1 replication. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl398. doi:10.1093/nar/gkl398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nature Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 9.Rogers G.W., Jr, Komar A.A., Merrick W.C. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 10.Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 11.Owttrim W.G. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34:3220–3230. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weston A., Sommerville S. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray B.K., Lawson T.G., Kramer J.C., Cladaras M.H., Grifo J.A., Abramson R.D., Merrick W.C., Thach R.E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 14.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schnier J., Slonimski P.P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 16.Tanner N.K., Cordin O., Banroques J., Doère M., Linder P. The Q motif. A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 17.Blum S., Schmid S.R., Pause A., Buser P., Linder P., Sonenberg N., Trachsel H. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1992;89:7664–7668. doi: 10.1073/pnas.89.16.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pause A., Méthot N., Svitkin Y., Merrick W.C., Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4A in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Cruz J., Kressler D., Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 22.Linder P., Gasteiger E., Bairoch A. A comprehensive web resource on RNA helicases from the baker's yeast Saccharomyces cerevisiae. YEAST. 2000;16:507–509. doi: 10.1002/(SICI)1097-0061(200004)16:6<507::AID-YEA549>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Jamieson D.J., Beggs J.D. A suppressor of yeast spp81/ded1 mutations encodes a very similar putative ATP-dependent RNA helicase. Mol. Microbiol. 1991;5:805–812. doi: 10.1111/j.1365-2958.1991.tb00753.x. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 25.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 26.Tseng-Rogenski S.S., Chong J.L., Thomas C.B., Enomoto S., Berman J., Chang T.H. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 2003;31:4995–5002. doi: 10.1093/nar/gkg712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnstone O., Deuring R., Bock R., Linder P., Fuller M.T., Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Kitajima Y., Yatsuki H., Zhang R., Matsuhashi S., Hori K. A novel human homologue of a dead-box RNA helicase family. Biochem. Biophys. Res. Commun. 1994;199:748–754. doi: 10.1006/bbrc.1994.1292. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt U., Lehmann K., Stahl U. A novel mitochondrial DEAD box protein (Mrh4) required for maintenance of mtDNA in Saccharomyces cerevisiae. FEM Yeast Res. 2002;2:267–276. doi: 10.1016/S1567-1356(02)00109-5. [DOI] [PubMed] [Google Scholar]
- 30.Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 31.Valgardsdottir R., Brede G., Eide L.G., Frengen E., Prydz H. Cloning and characterization of MDDX28, a putative dead-box helicase with mitochondrial and nuclear localization. J. Biol. Chem. 2001;276:32056–32063. doi: 10.1074/jbc.M011629200. [DOI] [PubMed] [Google Scholar]
- 32.Valgardsdottir R., Prydz H. Transport signals and transcription-dependent nuclear localization of the putative DEAD-box helicase MDDX28. J. Biol. Chem. 2003;278:21146–21154. doi: 10.1074/jbc.M300888200. [DOI] [PubMed] [Google Scholar]
- 33.Weaver P.L., Sun C., Chang T.-H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassarman D.A., Steitz J.A. Alive with DEAD proteins. Nature. 1991;349:463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka N., Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 36.Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Grifo J.A., Abramson R.D., Satler C.A., Merrick W.C. RNA-stimulated ATPase activity of eukaryotic initiation factors. J. Biol. Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 38.Korneeva N.L., First E.A., Benoit C.A., Rhoads R.E. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 39.Oberer M., Marintchev A., Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordeleau M.E., Matthews J., Wojnar J.M., Lindqvist L., Novac O., Jankowsky E., Sonenberg N., Northcote P., Teesdale-Spittle P., Pelletier J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl Acad. Sci. USA. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Low W.K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Merrick W.C., Romo D., Liu J.O. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Fuller-Pace F.V., Nicol S.M., Reid A.D., Lane D.P. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicol S.M., Fuller-Pace F.V. The ‘DEAD box’ protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl Acad. Sci. USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsu C.A., Kossen K., Uhlenbeck O.C. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S., Hu Y., Overgaard M.T., Karginov F.V., Uhlenbeck O.C., McKay D.B. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Day C.-L., Dalbadie-McFarland G., Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 47.Perriman R., Barta I., Voeltz G.K., Abelson J., Ares M., Jr ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl Acad. Sci. USA. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y.Z., Newnham C.M., Kameoka S., Huang T., Konarska M.M., Query C.C. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 2004;23:376–385. doi: 10.1038/sj.emboj.7600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Granneman S., Lin C., Champion E.A., Nandineni M.R., Zorca C., Baserga S.J. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 2006;34:3189–3199. doi: 10.1093/nar/gkl419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rozen F., Edery I., Meerovitch K., Dever T.E., Merrick W.C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers G.W.J., Richter N.J., Lima W.F., Merrick W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 52.Tseng S.S.-I., Weaver P.L., Liu Y., Hitomi M., Tartakoff A.M., Chang T.-H. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt C., von Kobbe C., Bach I.A., Pante N., Rodrigues J.P., Boscheron C., Rigaut G., Wilm M., Seraphin B., Carmo-Fonseca M., et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iost I., Dreyfus M., Linder P. Ded1p, a DEAD-box protein required for translation initation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 55.Gururajan R., Weeks D.L. An3 protein encoded by a localized maternal mRNA in Xenopus laevis is an ATPase with substrate-specific RNA helicase activity. Biochim. Biophys. Acta. 1997;1350:169–182. doi: 10.1016/s0167-4781(96)00155-8. [DOI] [PubMed] [Google Scholar]
- 56.Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 57.Okanami M., Meshi T., Iwabuchi M. Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana. Nucleic Acids Res. 1998;26:2638–2643. doi: 10.1093/nar/26.11.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang L., Diehl-Jones W., Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 59.Ladomery M., Wade E., Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 1997;25:965–973. doi: 10.1093/nar/25.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valdez B.C., Henning D., Perumal K., Busch H. RNA-unwinding and RNA-folding activities of RNA helicase II/Gu—two activities in separate domains of the same protein. Eur. J. Biochem. 1997;250:800–807. doi: 10.1111/j.1432-1033.1997.00800.x. [DOI] [PubMed] [Google Scholar]
- 61.Valdez B.C., Perlaky L., Henning D. Expression, cellular localization, and enzymatic activities of RNA helicase II/Gu(beta) Exp. Cell Res. 2002;276:249–263. doi: 10.1006/excr.2002.5538. [DOI] [PubMed] [Google Scholar]
- 62.Yu E., Owttrim G.W. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res. 2000;28:3926–3934. doi: 10.1093/nar/28.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhlmann-Schiffler H., Jalal C., Stahl H. Ddx42p—a human DEAD box protein with RNA chaperone activities. Nucleic Acids Res. 2006;34:10–22. doi: 10.1093/nar/gkj403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rocak S., Emery B., Tanner N.K., Linder P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 2005;33:999–1009. doi: 10.1093/nar/gki244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bizebard T., Ferlenghi I., Iost I., Dreyfus M. Studies on three E.coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 66.Kikuma T., Ohtsu M., Utsugi T., Koga S., Okuhara K., Eki T., Fujimori F., Murakami Y. Dbp9p, a member of the DEAD box protein family, exhibits DNA helicase activity. J. Biol. Chem. 2004;279:20692–20698. doi: 10.1074/jbc.M400231200. [DOI] [PubMed] [Google Scholar]
- 67.Li S.C., Chung M.C., Chen C.S. Cloning and characterization of a DEAD box RNA helicase from the viable seedlings of aged mung bean. Plant Mol. Biol. 2001;47:761–770. doi: 10.1023/a:1013687412020. [DOI] [PubMed] [Google Scholar]
- 68.Yan X., Mouillet J.F., Ou Q., Sadovsky Y. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell. Biol. 2003;23:414–423. doi: 10.1128/MCB.23.1.414-423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 71.Linder P., Lasko P. Bent out of shape: RNA unwinding by the DEAD-box helicase vasa. Cell. 2006;125:219–221. doi: 10.1016/j.cell.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 72.Chen J.Y.-F., Stands L., Staley J.P., Jackups R.R., Jr, Latus L.J., Chang T.-H. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 73.Kistler A.L., Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lund M.K., Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 75.Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;88:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 76.De Marchis M.L., Giorgi A., Schinina M.E., Bozzoni I., Fatica A. Rrp15p, a novel component of pre-ribosomal particles required for 60S ribosome subunit maturation. RNA. 2005;11:495–502. doi: 10.1261/rna.7200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 79.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 80.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 81.Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schafer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jurica M.S., Moore M.J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 85.Staley J.P., Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 86.Liu Z.R. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 2002;22:5443–5450. doi: 10.1128/MCB.22.15.5443-5450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guil S., Gattoni R., Carrascal M., Abian J., Stevenin J., Bach-Elias M. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol. Cell. Biol. 2003;23:2927–2941. doi: 10.1128/MCB.23.8.2927-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Honig A., Auboeuf D., Parker M.M., O'Malley B.W., Berget S.M. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 2002;22:5698–5707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jamieson D.J., Rahe B., Pringle J., Beggs J.D. A suppressor of a yeast splicing mutation (prp8–1) encodes a putative ATP-dependent RNA helicase. Nature. 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- 90.Stevens S.W., Ryan D.E., Ge H.Y., Moore R.E., Young M.K., Lee T.D., Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 91.Arenas J.E., Abelson J.N. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl Acad. Sci. USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen J.-H., Lin R.-J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 1990;18:6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 94.King D.S., Beggs J.D. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:6559–6564. doi: 10.1093/nar/18.22.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martin A., Schneider S., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 96.Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 97.Silverman E., Edwalds-Gilbert G., Lin R.J. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 98.Ferraiuolo M.A., Lee C.S., Ler L.W., Hsu J.L., Costa-Mattioli M., Luo M.J., Reed R., Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl Acad. Sci. USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ballut L., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nature Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 100.Kressler D., de la Cruz J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kressler D., Linder P., de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Venema J., Tollervey D. Ribosome biosynthesis in Saccharomyces cerevisiae. Ann. Rev. Gen. 1999;33:261–331. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 103.Bernstein K.A., Granneman S., Lee A.V., Manickam S., Baserga S.J. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol. Cell. Biol. 2006;26:1195–1208. doi: 10.1128/MCB.26.4.1195-1208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Granneman S., Bernstein K.A., Bleichert F., Baserga S.J. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell. Biol. 2006;26:1183–1194. doi: 10.1128/MCB.26.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 106.Scherl A., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kos M., Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 108.Emery B., De La Cruz J., Rocak S., Deloche O., Linder P. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2004;52:141–158. doi: 10.1111/j.1365-2958.2003.03973.x. [DOI] [PubMed] [Google Scholar]
- 109.de la Cruz J., Daugeron M.-C., Linder P. In: Yeast Gene Analysis. Brown A.J.P., Tuite M.F., editors. Vol. 26. London, UK: Academic Press Limited; 1998. pp. 269–295. [Google Scholar]
- 110.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 111.Snay-Hodge C.A., Colot H.V., Goldstein A.L., Cole C.N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao J., Jin S.B., Bjorkroth B., Wieslander L., Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002;21:1177–1187. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Estruch F., Cole C.N. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol. Biol. Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gatfield D., Le Hir H., Schmitt C., Braun I.C., Köcher T., Wilm M., Izaurralde E. The DExH/D protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 115.Jensen T.H., Boulay J., Rosbash M., Libri D. The DECD-box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- 116.Luo M.-J., Zhou Z., Magni K., Christoforides C., Rappslibere J., Mann M., Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 117.Strasser K., Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 118.Pryor A., Tung L., Yang Z., Kapadia F., Chang T.H., Johnson L.F. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic Acids Res. 2004;32:1857–1865. doi: 10.1093/nar/gkh347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Grifo J.A., Tahara S.M., Leis J.P., Morgan M.A., Shatkin A.J., Merrick W. Characterization of eukaryotic initiation factor 4A, a protein involved in ATP-dependent binding of globin mRNA. J. Biol. Chem. 1982;257:5246–5252. [PubMed] [Google Scholar]
- 120.Duncan R., Hershey J.W.B. Identification and quantification of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimentional polyacrylamide gel electrophoresis. J. Biol. Chem. 1983;258:7228–7235. [PubMed] [Google Scholar]
- 121.von der Haar T., McCarthy J.E. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 122.Richter N.J., Rogers G.W., Jr, Hensold J.O., Merrick W.C. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 1999;274:35415–35424. doi: 10.1074/jbc.274.50.35415. [DOI] [PubMed] [Google Scholar]
- 123.Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog. Nucleic Acid Res. Mol. Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- 124.Daga R.R., Jimenez J. Translational control of the cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J. Cell Sci. 1999;112:3137–3146. doi: 10.1242/jcs.112.18.3137. [DOI] [PubMed] [Google Scholar]
- 125.Chuang R.-Y., Weaver P.L., Liu Z., Chang T.-H. Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 126.de la Cruz J., Iost I., Kressler D., Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J.E. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 2004;51:987–1001. doi: 10.1046/j.1365-2958.2003.03898.x. [DOI] [PubMed] [Google Scholar]
- 128.Carrera P., Johnstone O., Nakamura A., Casanova J., Jackle H., Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- 129.Khemici V., Poljak L., Toesca I., Carpousis A.J. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc. Natl Acad. Sci. USA. 2005;102:6913–6918. doi: 10.1073/pnas.0501129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Py B., Higgins C.F., Krisch H.M., Carpousis A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 131.Anderson J.S.J., Parker R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 133.Margossian S.P., Butow R.A. RNA turnover and the control of mitochondrial gene expression. Trends Biochem. Sci. 1996;21:392–396. [PubMed] [Google Scholar]
- 134.Raijmakers R., Schilders G., Pruijn G.J. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- 135.Coller J.M., Tucker M., Sheth U., Valencia-Sanchez M.A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Minshall N., Thom G., Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Smillie D.A., Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- 139.Huang H.R., Rowe C.E., Mohr S., Jiang Y., Lambowitz A.M., Perlman P.S. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl Acad. Sci. USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Minczuk M., Dmochowska A., Palczewska M., Stepien P.P. Overexpressed yeast mitochondrial putative RNA helicase Mss116 partially restores proper mtRNA metabolism in strains lacking the Suv3 mtRNA helicase. Yeast. 2002;19:1285–1293. doi: 10.1002/yea.906. [DOI] [PubMed] [Google Scholar]
- 141.Missel A., Souza A.E., Nörskau G., Göhringer H.U. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stuart K.D., Schnaufer A., Ernst N.L., Panigrahi A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 143.Godbout R., Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc. Natl Acad. Sci. USA. 1993;90:7578–7582. doi: 10.1073/pnas.90.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 145.Blum S., Mueller M., Schmid S.R., Linder P., Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4A-dependent cell-free system. Proc. Natl Acad. Sci. USA. 1989;86:6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sekiguchi T., Iida H., Fukumura J., Nishimoto T. Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp. Cell Res. 2004;300:213–222. doi: 10.1016/j.yexcr.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 147.Ditton H.J., Zimmer J., Kamp C., Rajpert-De Meyts E., Vogt P.H. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- 148.Hayashi N., Seino H., Irie K., Watanabe M., Clark K.L., Matsumoto K., Nishimoto T. Genetic interaction of DED1 encoding a putative ATP-dependent RNA helicase with SRM1 encoding a mammalian RCC1 homolog in Saccharomyces cerevisiae. Mol. Gen. Genet. 1996;253:149–156. doi: 10.1007/s004380050307. [DOI] [PubMed] [Google Scholar]
- 149.Chung J., Lee S.-G., Song K. Identification of a human homolog of a putative RNA helicase gene (mDEAD3) expressed in mouse erythroid cells. Korean J. Biochem. 1995;27:193–197. [Google Scholar]
- 150.Yedavalli V.S., Neuveut C., Chi Y.H., Kleiman L., Jeang K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 151.Castrillon D.H., Quade B.J., Wang T.Y., Quigley C., Crum C.P. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl Acad. Sci. USA. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Johnstone O., Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 153.Hloch P., Schiedner G., Stahl H. Complete cDNA sequence of the human p68 protein. Nucleic Acids Res. 1990;18:3045. doi: 10.1093/nar/18.10.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ford M.J., Anton I.A., Lane D.P. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988;332:736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- 155.Bond A.T., Mangus D.A., He F., Jacobson A. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 2001;21:7366–7379. doi: 10.1128/MCB.21.21.7366-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lamm G.M., Nicol S.M., Fuller-Pace F.V., Lamond A.I. p72: a human nuclear DEAD box protein highly related to p68. Nucleic Acids Res. 1996;24:3739–3747. doi: 10.1093/nar/24.19.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Akao Y., Yoshida H., Matsumoto K., Matsui T., Hogetu K., Tanaka N., Usukura J. A tumour-associated DEAD-box protein, rck/p54 exhibits RNA unwinding activity toward c-myc RNAs in vitro. Genes Cells. 2003;8:671–676. doi: 10.1046/j.1365-2443.2003.00665.x. [DOI] [PubMed] [Google Scholar]
- 158.Akao Y., Seto M., Yamamoto K., Iida S., Nakazawa S., Inazawa J., Abe T., Takahashi T., Ueda R. The RCK gene associated with t(11;14) translocation is distinct from the MLL/ALL-1 gene with t(4;11) and t(11;19) translocations. Cancer Res. 1992;52:6083–6087. [PubMed] [Google Scholar]
- 159.Lu D., Yunis J.J. Cloning, expression and localization of an RNA helicase gene from a human lymphoid cell line with chromosomal breakpoint 11q23.3. Nucleic Acids Res. 1992;20:1967–1972. doi: 10.1093/nar/20.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fischer N., Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Savitsky K., Ziv Y., Bar-Shira A., Gilad S., Tagle D.A., Smith S., Uziel T., Sfez S., Nahmias J., Sartiel A., et al. A human gene (DDX10) encoding a putative DEAD-box RNA helicase at 11q22-q23. Genomics. 1996;33:199–206. doi: 10.1006/geno.1996.0184. [DOI] [PubMed] [Google Scholar]
- 162.Liang W.-Q., Clark J.A., Fournier M.J. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol. Cell. Biol. 1997;17:4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Grandori C., Mac J., Siebelt F., Ayer D.E., Eisenman R.N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 164.Tang P.Z., Tsai-Morris C.H., Dufau M.L. A novel gonadotropin-regulated testicular RNA helicase. A new member of the dead-box family. J. Biol. Chem. 1999;274:37932–37940. doi: 10.1074/jbc.274.53.37932. [DOI] [PubMed] [Google Scholar]
- 165.Charroux B., Pellizzoni L., Perkinson R.A., Shevchenko A., Mann M., Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Grundhoff A.T., Kremmer E., Tureci O., Glieden A., Gindorf C., Atz J., Mueller-Lantzsch N., Schubach W.H., Grasser F.A. Characterization of DP103, a novel DEAD box protein that binds to the Epstein–Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 1999;274:19136–19144. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- 167.Valdez B.C., Henning D., Busch R.K., Woods K., Flores-Rozas H., Hurwitz J., Perlaky L., Busch H. A nucleolar RNA helicase recognized by autoimmune antibodies from a patient with watermelon stomach disease. Nucleic Acids Res. 1996;24:1220–1224. doi: 10.1093/nar/24.7.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Henning D., So R.B., Jin R., Lau L.F., Valdez B.C. Silencing of RNA helicase II/Gualpha inhibits mammalian ribosomal RNA production. J. Biol. Chem. 2003;278:52307–52314. doi: 10.1074/jbc.M310846200. [DOI] [PubMed] [Google Scholar]
- 169.Yang H., Zhou J., Ochs R.L., Henning D., Jin R., Valdez B.C. Down-regulation of RNA helicase II/Gu results in the depletion of 18 and 28 S rRNAs in Xenopus oocyte. J. Biol. Chem. 2003;278:38847–38859. doi: 10.1074/jbc.M302258200. [DOI] [PubMed] [Google Scholar]
- 170.Valdez B.C., Yang H., Hong E., Sequitin A.M. Genomic structure of newly identified paralogue of RNA helicase II/Gu: detection of pseudogenes and multiple alternatively spliced mRNAs. Gene. 2002;284:53–61. doi: 10.1016/s0378-1119(01)00888-5. [DOI] [PubMed] [Google Scholar]
- 171.Westermarck J., Weiss C., Saffrich R., Kast J., Musti A.M., Wessely M., Ansorge W., Seraphin B., Wilm M., Valdez B.C., et al. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 2002;21:451–460. doi: 10.1093/emboj/21.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Teigelkamp S., Mundt C., Achsel T., Will C.L., Luhrmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 173.Strauss E.J., Guthrie C. PRP28, a ‘DEAD-box’ protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994;22:3187–3193. doi: 10.1093/nar/22.15.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Staley J.P., Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 175.Zhao Y., Yu L., Fu Q., Chen W., Jiang J., Gao J., Zhao S. Cloning and characterization of human DDX24 and mouse Ddx24, two novel putative DEAD-Box proteins, and mapping DDX24 to human chromosome 14q32. Genomics. 2000;67:351–355. doi: 10.1006/geno.2000.6255. [DOI] [PubMed] [Google Scholar]
- 176.Zagulski M., Kressler D., Becam A.M., Rytka J., Herbert C.J. Mak5p, which is required for the maintenance of the M1 dsRNA virus, is encoded by the yeast ORF YBR142w and is involved in the biogenesis of the 60S subunit of the ribosome. Mol. Genet. Genomics. 2003;270:216–224. doi: 10.1007/s00438-003-0913-4. [DOI] [PubMed] [Google Scholar]
- 177.Ripmaster T.L., Vaughn G.P., Woolford J.L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl Acad. Sci. USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Daugeron M.C., Linder P. Dbp7p, a putative ATP-dependent RNA helicase of Saccharomyces cerevisiae is required for 60S ribosomal subunit assembly. RNA. 1998;4:566–581. doi: 10.1017/s1355838298980190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Peelman L.J., Chardon P., Nunes M., Renard C., Geffrotin C., Vaiman M., Van-Zeveren A., Coppieters W., van-de-Weghe A., Bouquet Y., et al. The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family. Genomics. 1995;26:210–218. doi: 10.1016/0888-7543(95)80203-x. [DOI] [PubMed] [Google Scholar]
- 180.Fleckner J., Zhang M., Valcarcel J., Green M.R. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 181.Lehner B., Semple J.I., Brown S.E., Counsell D., Campbell R.D., Sanderson C.M. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 182.Shi H., Cordin O., Minder C.M., Linder P., Xu R.M. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl Acad. Sci. USA. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Irion U., Leptin M., Siller K., Fuerstenberg S., Cai Y., Doe C.Q., Chia W., Yang X. Abstrakt, a DEAD box protein, regulates Insc levels and asymmetric division of neural and mesodermal progenitors. Curr. Biol. 2004;14:138–144. doi: 10.1016/j.cub.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 184.Irion U., Leptin M. Developmental and cell biological functions of the Drosophila DEAD-box protein abstrakt. Curr. Biol. 1999;9:1373–1381. doi: 10.1016/s0960-9822(00)80082-2. [DOI] [PubMed] [Google Scholar]
- 185.Will C.L., Urlaub H., Achsel T., Gentzel M., Wilm M., Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martelange V., De Smet C., De Plaen E., Lurquin C., Boon T. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res. 2000;60:3848–3855. [PubMed] [Google Scholar]
- 187.Cho B., Lim Y., Lee D.Y., Park S.Y., Lee H., Kim W.H., Yang H., Bang Y.J., Jeoung D.I. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem. Biophys. Res. Commun. 2002;292:715–726. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
- 188.Dalbadie-McFarland G., Abelson J. PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc. Natl Acad. Sci. USA. 1990;87:4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.O'Day C.L., Chavanikamannil F., Abelson J. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 1996;24:3201–3207. doi: 10.1093/nar/24.16.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chan C.C., Dostie J., Diem M.D., Feng W., Mann M., Rappsilber J., Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Daugeron M.C., Linder P. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 2001;29:1144–1155. doi: 10.1093/nar/29.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kressler D., de la Cruz J., Rojo M., Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Venema J., Bousquet-Antonelli C., Gelugne J.-P., Caizergues-Ferrer M., Tollervey D. Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol. 1997;17:3398–3407. doi: 10.1128/mcb.17.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rajendran R.R., Nye A.C., Frasor J., Balsara R.D., Martini P.G., Katzenellenbogen B.S. Regulation of nuclear receptor transcriptional activity by a novel DEAD box RNA helicase (DP97) J. Biol. Chem. 2003;278:4628–4638. doi: 10.1074/jbc.M210066200. [DOI] [PubMed] [Google Scholar]
- 195.Burger F., Daugeron M.-C., Linder P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 2000;28:2315–2323. doi: 10.1093/nar/28.12.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.de la Cruz J., Kressler D., Rojo M., Tollervey D., Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zirwes R.F., Eilbracht J., Kneissel S., Schmidt-Zachmann M.S. A novel helicase-type protein in the nucleolus: protein NOH61. Mol. Biol. Cell. 2000;11:1153–1167. doi: 10.1091/mbc.11.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Daugeron M.C., Kressler D., Linder P. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA. 2001;7:1317–1334. doi: 10.1017/s1355838201010640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Niemer I., Schmelzer C., Börner G.V. Overexpression of DEAD box protein pMSS116 promotes ATP-dependent splicing of a yeast group II intron in vitro. Nucleic Acids Res. 1995;23:2966–2972. [PMC free article] [PubMed] [Google Scholar]
- 200.Abdelhaleem M., Maltais L., Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]