Abstract
Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA-2) is a key determinant in the EBV-driven B-cell growth transformation process. By activating an array of viral and cellular target genes, EBNA-2 initiates a cascade of events which ultimately cause cell cycle entry and the proliferation of the infected B cell. In order to identify cellular target genes that respond to EBNA-2 in the absence of other viral factors, we have performed a comprehensive search for EBNA-2 target genes in two EBV-negative B-cell lines. This screen identified 311 EBNA-2-induced and 239 EBNA-2-repressed genes that were significantly regulated in either one or both cell lines. The activation of most of these genes had not previously been attributed to EBNA-2 function and will be relevant for the identification of EBNA-2-specific contributions to EBV-associated malignancies. The diverse spectrum of EBNA-2 target genes described in this study reflects the broad spectrum of EBNA-2 functions involved in virus-host interactions, including cell signaling molecules, adapters, genes involved in cell cycle regulation, and chemokines.
Epstein-Barr virus (EBV) is a DNA tumor virus that is associated with the pathogenesis of endemic Burkitt's lymphoma, Hodgkin's lymphoma, and posttransplant lymphoma. In vitro EBV infects and growth transforms primary B cells, leading to the continuous proliferation of lymphoblastoid cell lines (LCLs) in culture. EBV nuclear antigen 2 (EBNA-2) is one of the first genes expressed upon EBV infection and essential for the growth transformation process. By activating an array of primary viral and cellular target genes, EBNA-2 initiates the transcription of a cascade of secondary events that cause B-cell activation, cell cycle entry, and maintenance of LCLs (9, 16, 28, 59). In LCLs, EBNA-2 is typically coexpressed with other viral factors (EBV-associated latent membrane proteins [LMP1, -2A, and -2B], EBNA-1, -3A, -3B, -3C, and -LP, and the BamHIA rightward transcripts) that are collectively called latent antigens. This expression pattern of EBV transcription, accompanied by a specific cellular gene expression pattern, is called latency III. Important primary targets of EBNA-2 in LCLs are the proto-oncogene c-myc and the viral LMP1 gene, which contribute to the initiation of the secondary target gene cascade (6, 12, 24, 47, 57, 60). The mechanism by which EBNA-2 activates viral target genes has been intensively studied. All viral promoters that are activated by EBNA-2 share a binding site for the C-promoter binding protein CBF1. CBF1, also named RBP-J and RBP-Jκ in mice or Suppressor of Hairless in Drosophila melanogaster, is a ubiquitously expressed, sequence-specific cellular DNA binding protein that serves as a DNA adaptor protein for cellular and viral factors (17, 37). The recruitment of corepressor complexes by CBF1 can result in target gene repression, while the recruitment of transactivators like the viral EBNA-2 protein causes gene activation (17). The cellular equivalent of EBNA-2 is the activated Notch receptor. Notch proteins are transmembrane receptors that are activated upon ligand binding by proteolytic cleavage. This multistep process generates an intracellular Notch fragment that binds to CBF1 and activates the transcription of CBF1-dependent target genes (42). The constitutively active nuclear protein EBNA-2 shortcuts this pathway and might thus execute Notch-like functions. The extent to which Notch and EBNA-2 functions overlap is still an open question. Well-known targets of Notch are the basic helix-loop-helix (bHLH) proteins HES-1, -5, and -7 and HERP1/Hey2/Hesr2/HRT2/CHF1/Gridlock, HERP2/Hey1/Hesr1/HRT1/CHF2, and HERP3/HeyL/HesR3/HRT3 (3, 20, 21, 39, 43, 44). CD21, CD23, CCR7, BATF, tumor necrosis factor alpha (TNF-α), c-myc, RUNX3, and interleukin-18 receptor are activated by EBNA-2 (4, 5, 10, 23, 24, 49, 60, 66). Target genes that have been shown to be shared by both transactivators are CD21, CD23, BATF, TNF-α and c-myc (14, 19, 62). Both EBNA-2 and activated Notch can also repress immunoglobulin M (IgM) transcription (22, 62). The mechanism by which EBNA-2 represses IgM transcription is partially independent of CBF1 (40). Compared to the broad list of target genes described for Notch, the panel of EBNA-2 target genes is still limited. In this report, we describe the results of a comprehensive screen for EBNA-2 target genes based on two EBV-negative cell lines that express a conditional estrogen-responsive EBNA-2 protein. The results of this screen were confirmed for a selected group of target genes by real-time reverse transcription-PCR (RT-PCR) in the initial cell lines as well as additional EBNA-2 conditional systems and in the context of infected B cells. CBF1-dependent gene expression patterns were defined by analyzing target gene expression in CBF1-negative, somatic knockout B-cell lines.
MATERIALS AND METHODS
Plasmids.
The plasmid probe for HES-5 was a gift from R. Kageyama (Kyoto, Japan). All other plasmids (for HES-1 [HU3_p983D0695D2], HES-7 [IMAGp998C1511261Q1], HERP1 [IMAGp998H221275Q], HERP2 [IMAGp956A2118Q], and HERP3 [HU_p983C07119D2]) are distributed by the RZPD, Berlin, Germany.
Cell lines and B cells.
DG75, BL41K3, BJABK3, SM295 (clone D6), SM296 (clone D3), SM224.9, and ER/EB2-5 have been described previously (2, 27, 28, 40). AH276.1 is a stable transfectant of SM224.9 expressing tandem affinity purification-tagged CBF1. BL41 is an EBV-negative Burkitt's lymphoma cell line carrying a t(8;14) translocation (38). BJAB is an EBV-negative lymphoblastoid B-cell line (31). All cell lines were cultivated in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The hormone binding domain of the estrogen receptor (ER/EBNA-2) was induced by the addition of 1 μM β-estradiol to the cell culture medium. The medium for ER/EB2-5 was constantly supplemented with β-estradiol (1 μM) for the expansion of cell cultures. Human primary B cells were purified from adenoids by T-cell rosetting and infected with culture supernatants of B95.8 cells.
Microarray analysis of gene expression.
Total cellular RNA (10 μg) was labeled and hybridized to Affymetrix HG-U133A 2.0 GeneChip according to the manufacturer's instructions. Affymetrix CEL files were processed for global normalization and the generation of expression values by using the robust multiarray average algorithm in the R affy package (www.bioconductor.org). Absent, marginally expressed, and present calls were defined by Affymetrix MAS 5.0 software.
Scaling factors ranged from 0.67 to 1, and the average background was below 70. The list of significantly regulated genes was achieved by applying the significance analysis of microarrays two-class algorithm (65) of the samr package for R between uninduced and β-estradiol-induced cells for each cell line individually (three replicates for BJABK3 and two for BL41K3). The 1% false discovery rate level for differentially regulated genes exceeded 784 and 1,150 probe sets for BJABK3 and BL41K3, respectively. Further filtering and data preparation were performed with Spotfire DecisionSite software (Spotfire, Sommerville, MA). The analysis of overrepresented genes was performed using the program EASE, version 2.0 (18).
Primers for PCR.
Primer pairs for RT-PCR were selected by Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). All pairs were chosen to support amplification across intron borders, with a single exception, which was CDK5R1. Primers were AATCGGATCACCAATGGAAC and CAGCTGGCTGGGTTGTATG for CD21, ATGCAGGTCTCCACTGCTG and TTCTGGACCCACTCCTCAC for CCL3, TTGCCACCAATACCATGAAG and ACCACAAAGTTGCGAGGAAG for CCL4, ATGCACAGAACAAGCCACAG and ACCCACAGTTTGGTCAAAGA for CDK5R1, GATCCCACAAATGGAAGTGG and ATCGTCATCCAGTCGAAAGC for SAMSN1, CGTGACCGAGTTCATGTGTC and TTATTCATGCCTGGGTAGGG for FGR, ACATTTCTTCGGCATTCTGC and CAACACGCACTTGATGGAAC for RHOH, GTTAATCAACTGGCCCATGC and TCTTTCCATGTGCTGTCTGC for MFN1, AGGACACCACGGTGAAGAAG and GTGAAGGCCCTTGAAGACTG for DNASE1L3, TGAGGGATGGTACCTTATGACC and ACAGCCCAGCATACACTGC for SLAMF1, TCTCACTGCACGATGAGAGG and TATGCCCGAATGGAAGAGTC for RAPGEF2, TGGCCTGCAGTACTCAACTG and TCGGAACCTTGATCTTGTCC for ABHD6, and CGGCTACCACATCCAAGGAA and CTGGAATTACCGCGGCT for 18S rRNA.
Real-time RT-PCR.
Total RNA was extracted from 1 × 107 cells using peqGOLD TriFast (PEQLAB) according to the manufacturer's protocol and treated with DNase to remove genomic DNA (RQ1 RNase-Free DNase; Promega). cDNA was synthesized from 1 μg of RNA by using avian myeloblastosis virus reverse transcriptase and random hexanucleotide mixtures as primers (First Strand cDNA synthesis kit for RT-PCR; Roche).
Relative quantification of the transcripts by real-time PCR was performed with a LightCycler, and the data were processed by LightCycler software, version 4.05 (Roche). A total of 1/20 of the cDNA was amplified using the LightCycler FastStart kit according to the manufacturer's protocol (Roche). Cycling conditions were 1 cycle of 95°C for 10 min and 38 cycles of denaturation (95°C for 0 s), annealing (60°C for 10 s), and extension (72°C for 25 s). All PCR products were examined by melting curve analysis (95°C for 0 s, 65°C for 10 s, and 99°C for 0 s; acquisition mode continuous), and the expected PCR fragment size was verified by agarose gel electrophoresis.
To correct for differences in reaction efficiencies, a standard curve was generated for each gene by using the serial dilutions as templates for amplification and plotting the crossing points versus the known dilutions. All data were normalized for the relative abundance of the 18S rRNA transcript. The abundance of each target transcript could thus be compared across all of the different RNA samples tested.
Northern blotting.
A total of 10 μg of total cellular RNA was separated on 1.2% formaldehyde agarose gels, blotted onto nylon membranes in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and cross-linked to the membranes by UV treatment. The membranes were prewashed with 0.1× SSC and 1% sodium dodecyl sulfate (SDS) for 30 min, prehybridized in Church buffer (0.5 M NaP04, pH 7.1, 7% SDS, 1 mM EDTA) for 1 hour, hybridized with 32P-labeled denatured-DNA probes in Church buffer (2.5 ng labeled DNA probe/ml) overnight, washed in 2× SSC and 1% SDS, and exposed to X-ray films.
Microarray data accession numbers.
Normalized expression values were deposited in the Gene Expression Omnibus repository as series number GSE 4525 (www.ncbi.nlm.nih.gov/geo/).
RESULTS
Microarray screen for EBNA-2 target genes.
Two cell lines, BL41K3 and BJABK3, were chosen to perform a comprehensive screen for EBNA-2 target genes. Both cell lines are EBV-negative B-cell lines and express a chimeric EBNA-2 protein fused to ER/EBNA-2. The cells are propagated in the absence of hormone (27, 29). Estrogen stimulation activates ER/EBNA-2, which in turn activates well-known EBNA-2 target genes like CD21. The c-myc gene responds differentially in the two cell lines. BL41K3 is a Burkitt's lymphoma-derived cell line carrying a t(8;14) translocation juxtaposing the immunoglobulin heavy chain locus and the c-myc oncogene. In BL41K3, IgM and c-myc are corepressed. In contrast, BJABK3 cells are derived from a lymphoblastoid B-cell line that does not carry this translocation. BJABK3 cells repress IgM expression and moderately induce c-myc (22, 40). Thus, for those genes that are coregulated in both cell lines, a contribution of Myc to target gene modulation by EBNA-2 can be excluded.
BL41K3 and BJABK3 were cultured in estrogen-supplemented medium for 24 h, and gene expression profiles were established for unstimulated and stimulated cells. For BJABK3 and BL41K3, three and two independent estrogen stimulations, respectively, were performed. RNA was harvested and processed for the hybridization of the HGU133A 2.0 Affymetrix array. This array carries 22,277 independent probe sets, which represent 18,400 transcripts and related variants, including 14,500 well-characterized human genes. In both cell lines, 51% (±1.8%) of the probe sets scored as present before and after EBNA-2 activation, indicating that no massive overall change in transcription levels was induced.
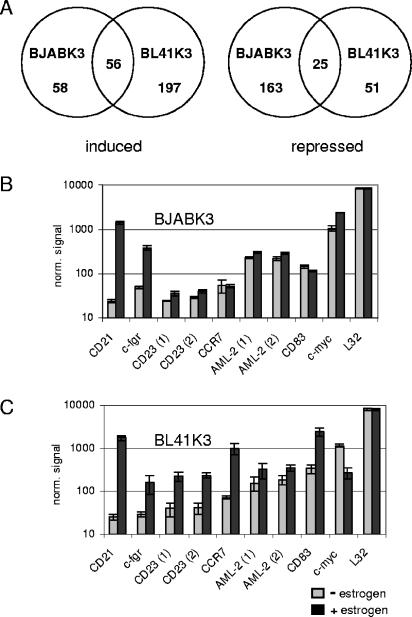
A total of 550 probe sets were significantly (q value of ≤1%) and, on average, at least twofold induced or repressed in either cell line. A total of 58 or 197 transcripts were induced in BJABK3 or BL41K3, respectively, while 56 were upregulated in both cell lines. A total of 239 transcripts were downregulated: 163 in BJABK3, 51 in BL41K3, and 25 in both cell lines (Fig. 1A).
FIG. 1.
Activation of EBNA-2 in BJABK3 or BL41K3 causes the induction or repression of 550 probe sets. Total cellular RNA was extracted from BJABK3 or BL41K3 cells before or 24 h post-estrogen activation of EBNA-2 and used for microarray analyses of mRNA expression levels. (A) Venn diagrams illustrate the distribution of 550 probe sets, which were significantly either upregulated or repressed by EBNA-2 at least twofold (q value of ≤1%). Triplicate (B) or duplicate (C) results of individual probe sets upon BJABK3 (B) or BL41K3 (C) microarray analyses are shown for EBNA-2-induced genes, which have been described in the literature. Gray bars show the mean normalized (norm.) signal intensities before estrogen (−), while black bars correspond to the signal 24 h post-estrogen (+) induction. Error bars indicate standard deviations.
Within the group of EBNA-2-induced targets, several had been described before and are now confirmed by our results. These were CD21, c-fgr, CD23, CCR7, AML-2/Runx3, and CD83 (Fig. 1B and C) (5, 10, 32, 40, 61, 66). CD21 and c-fgr belonged to the group that was induced more than 3.5-fold in both cell lines. All other genes responded more weakly or differently in the two cell lines. C-myc showed the expected differential expression levels upregulated in BJABK3 and repressed in BL41K3. The expression of the ribosomal protein L32 gene was unaffected by estrogen treatment of the cells.
Confirmation of EBNA-2 target genes by real-time RT-PCR in BJABK3 and BL41K3.
In order to confirm the results of the Affymetrix screen and compare the relative expression levels in additional B-cell lines, we performed quantitative real-time RT-PCR analyses on a selected group of 12 target genes that were induced by EBNA-2 at least 3.5-fold (q value of ≤1%) in both cellular systems. This group of 12 target genes included two well-known EBNA-2 target genes, CD21 and c-fgr, which served as controls to prove successful ER/EBNA-2 induction throughout all experiments (Table 1).
TABLE 1.
Summary of results obtained for 12 genes induced by EBNA-2 at least 3.5-fold
| Gene or gene producta | Gene IDb | Molecular or biological function(s) | Result for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affymetrixc
|
Real-time RT-PCRd
|
||||||||||||
| BL41K3 | BJABK3 | BL41K3 | BL41 | BJABK3 | BJAB | SM295 | SM296 | CBF1neg. | ER/EB2-5 | EBV | |||
| Abhydrolase domain-containing 6 protein (ABHD6) | 57406 | Aromatic compound metabolism, lipase activity | 4.4 | 4.8 | 3.6 | 1.8 | 3.7 | 1.2 | 2.1 | 1.1 | 10.4 | 78.1 | |
| CD21 (CR2) | 1380 | B-cell receptor cosignaling/complement receptor 2 (CR2)/EBV receptor | 70.4 | 350.8 | 139.2 | 1.5 | 59.8 | 1.2 | +++ | 4.4 | +++ | ||
| Chemokine ligand 3 (CCL3); macrophage inflammatory protein 1-alpha (MIP1α) | 6348 | Lymphocyte trafficking | 34.9 | 624.8 | 44.9 | 0.4 | 73.5 | 0.7 | 41.1 | 1.0 | + | 8.2 | 11.8 |
| Chemokine ligand 4 (CCL4); macrophage inflammatory protein 1-beta (MIP1β) | 6354 | Lymphocyte trafficking | 10.2 | 109.5 | 9.1 | 1.0 | 16.4 | 1.0 | 66.0 | 1.3 | 80.7 | <2 | |
| Regulatory subunit 1 (p35) of cdk5 (CDK5R1) | 8851 | Neuronal specific activator of cyclin-dependent kinase 5 | 21.7 | 8.2 | 10.4 | 1.7 | 8.2 | 1.9 | +++ | 1.1 | + | 11.7 | 7.1 |
| DNase I-like 3 (DNASE1L3) | 1776 | Apoptotic DNase activity, hypermutation | 4.7 | 522.9 | 18.1 | 1.2 | 4.4 | 1.4 | 37.9 | 0.3 | 30.9 | <2 | |
| c-fgr (FGR) | 2268 | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog/tyrosine kinase | 6.2 | 22 | 17.5 | 1.2 | 7.8 | 0.9 | 1 | 1.3 | <2 | +++ | |
| Mitofusin1 (MFN1) | 55669 | Mitochondrial fusion | 5.7 | 5.0 | 2.1 | 1.7 | 5.09 | 1.6 | 4.2 | 1.2 | <2 | 4.4 | |
| Rap guanine nucleotide exchange factor 2 (RAPGEF2) | 9693 | Ras activation | 4.8 | 3.1 | 3.9 | 0.2 | 4.0 | 1.4 | 4.0 | 2.2 | + | <2 | 8.7 |
| ras homolog gene family, member H (RHOH) | 399 | Inhibitor of Rho GTPases | 8.4 | 5.3 | 3.4 | 1.1 | 6.8 | 0.9 | 4.6 | 1.3 | + | <2 | <2 |
| HACS1/SH3-SAM adaptor (SAMSN1) | 64092 | SH3-SAM adaptor protein | 5.7 | 9.3 | 2.4 | 2.2 | 7.8 | 2.2 | +++ | 1.1 | 28.3 | 16.5 | |
| CD150; signaling lymphocytic activation molecule 1 (SLAMF1) | 6504 | Lymphocyte activation, receptor for measles virus | 33.6 | 3.8 | 25.8 | 1.1 | 4.3 | 1.2 | +++ | 0.9 | 3.5 | 10.0 | |
Genes that are induced at least 3.5-fold in BJABK3 and BL41K3 after estrogen induction.
Unique gene ID according to the National Center for Biotechnology Information. ID, identification number.
Induction (n-fold) 24 h post-estrogen stimulation according to the mean of the Affymetrix screen.
Induction (n-fold) 24 h post-estrogen stimulation according to real-time RT-PCR data. +++, strong upregulation from levels which are below the detection limits; +, significantly upregulated in CBF1-negative (CBF1neg.) DG75 cells.
For each of these 12 genes, transcript abundances were referred to a calibrator sample generated by using the PCR product of the respective gene as a template for amplification. This allowed the comparison of transcript abundances and the relative induction levels obtained by RT-PCR for each gene between different cell lines.
All original Affymetrix data sets are provided in Excel sheet format, including expression levels, a score for the detection call, mean values of the triplicates, standard deviations, and the relative induction or repression for both cell lines (see http://www.gsf.de/kmolbi/group3_frameset.htm).
For all 12 genes, the induction by EBNA-2 could be confirmed by RT-PCR in BL41K3 and BJABK3 (see http://www.gsf.de/kmolbi/group3_frameset.htm). In general, relative induction rates and relative expression levels, both strong and weak, correlated well when the two techniques were compared, while absolute induction levels differed. Since cDNA synthesis protocols differed for both techniques, experimental procedures and gene-specific efficiencies might account for these minor discrepancies. Control experiments were performed in order to test whether target gene activation upon estrogen treatment was dependent on EBNA-2. To this end, the parental BL41 and BJAB cells were treated with estrogen and transcript levels were determined. We detected 2.2-fold changes in response to estrogen at most. Importantly, the activation by EBNA-2 significantly exceeded these potential estrogen effects in all cases (see http://www.gsf.de/kmolbi/group3_frameset.htm).
The contribution of CBF1 to EBNA-2 target gene induction and repression.
Next, the contribution of CBF1 signaling to the expression of these 12 target genes by ER/EBNA-2 was analyzed (see http://www.gsf.de/kmolbi/group3_frameset.htm). We recently described a CBF1 somatic knockout B-cell line, which was generated by the inactivation of the CBF1 gene in the Burkitt's lymphoma cell line DG75 (40). ER/EBNA-2 transfectants of DG75 wild-type cells (SM295) and of CBF1-negative DG75 cells (SM296) were estrogen stimulated, and gene expression was studied by real-time RT-PCR. FGR was expressed at high levels before ER/EBNA-2 activation but not induced by EBNA-2 in the cellular background of DG75, while the remaining 11 target genes were activated. Analysis of these 11 target genes in CBF1-negative SM296 cells revealed that RAPGEF2 responded weakly but also reproducibly in the absence of CBF1. The activation of the remaining 10 target genes was strictly dependent on CBF1, indicating that CBF1 was absolutely required for the activation of the majority of target genes.
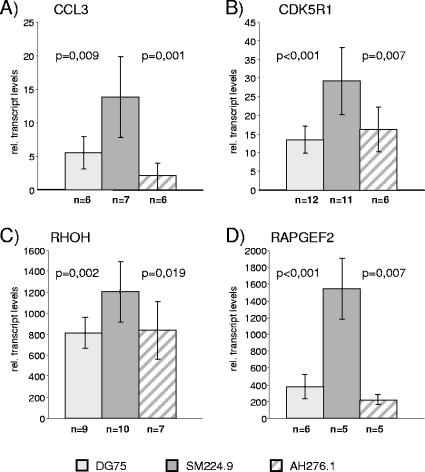
In order to detect the contribution of CBF1 to the repression of these 12 modulated target genes, basal expression levels were compared in wild-type and CBF1-negative DG75 cells before EBNA-2 induction. CCL3, CDK5R1, RHOH, and RAPGEF2 expression was increased at least twofold in CBF1-negative cells (SM296) compared to that in CBF1-positive cells (SM295) independent of EBNA-2 activation (0 h) (see http://www.gsf.de/kmolbi/group3_frameset.htm). Thus, these four genes were candidates for CBF1-repressed genes. In order to exclude trivial reasons like clonal variation as an explanation for these results, we employed three additional cell lines in our studies: the parental cell line DG75, the corresponding CBF1-negative knockout cell line SM224.9, and a transfectant of SM224.9 into which tandem affinity purification-tagged CBF1 had been reintroduced (AH276.1). Real-time RT-PCR analysis of the four candidate CBF1-repressed genes revealed elevated transcript levels in CBF1-negative cells (SM224.9) as opposed to the CBF1-positive counterparts (DG75 and AH276.1) (Fig. 2). For all other genes, the baseline expression before EBNA-2 activation was below the detection levels or unchanged in the absence of CBF1.
FIG. 2.
Expression of CCL3, CDK5R1, RHOH, and RAPGEF2 is repressed by CBF1. Total cellular RNAs of DG75, SM224.9, and AH276.1 were tested for the expression of CCL3, CDK5R1, RHOH, and RAPGEF2 by real-time RT-PCR. The results are given as mean values (n = number of independent experiments). The relative (rel.) transcript levels for all genes refer to a gene-specific calibrator template. Standard deviations are given as error bars, and P values were calculated by an unpaired two-sided Student t test.
Target gene activation in EBV-positive B cells.
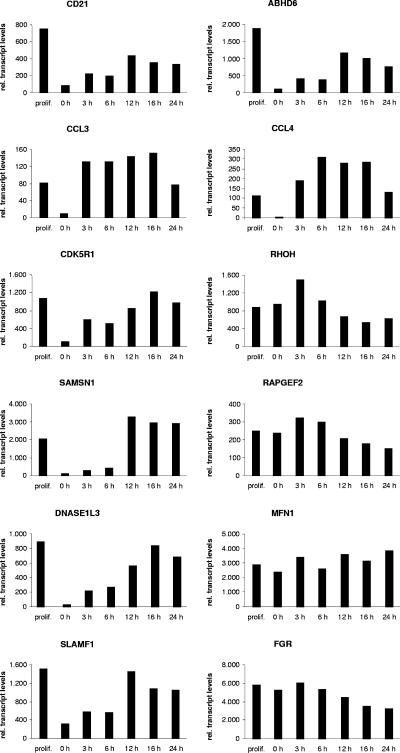
In order to validate the 12 candidate EBNA-2 target genes in the context of an EBV-positive cellular background, expression was analyzed in ER/EB2-5 cells by real-time RT-PCR (Fig. 3). ER/EB2-5 cells are EBV-transformed B cells expressing ER/EBNA-2, which proliferate and express EBNA-2 target genes in only estrogen-conditioned growth medium (28). Estrogen deprivation of the growth medium leads to cell cycle arrest, which can be reversed by estrogen restimulation.
FIG. 3.
Induction of target genes in ER/EB2-5 cells. Total cellular RNAs of proliferating ER/EB2-5 cells, growth-arrested ER/EB2-5 cells (0 h) which had been cultivated in the absence of estrogen for 3 days, or estrogen-restimulated cells were harvested at the indicated time points, and gene expression was tested by real-time RT-PCR. The results are given as relative transcript levels referring to a gene-specific calibrator template.
Total cellular RNA was isolated from ER/EB2-5 cells (i) cultivated in the presence of estrogen, (ii) depleted from estrogen for 3 days, and (iii) restimulated with estrogen for different time periods. Importantly, all 12 genes were expressed in proliferating ER/EB2-5 cells when EBNA-2 was active (Fig. 3). Upon estrogen depletion, the expression levels of CD21, CCL3, CDK5R1, SAMSN1, DNASE1L3, SLAMF1, ABHD6, and CCL4 were significantly diminished. Reinduction could already be monitored 3 h post-EBNA-2 activation. In contrast, the expression levels of RHOH, RAPGEF2, MFN1, and FGR were maintained or only marginally modulated in response to estrogen. Thus, the expression levels of the latter genes are controlled by other viral or cellular factors independent of EBNA-2 in the cellular context of ER/EB2-5 cells.
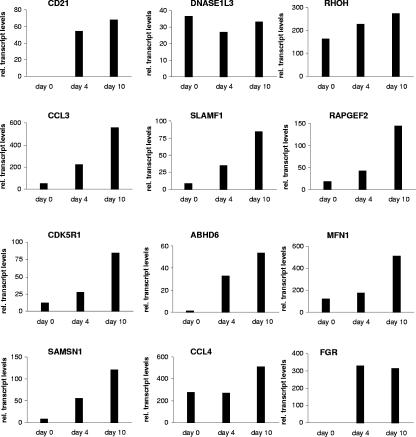
Target genes induced upon infection of primary B cells by EBV infection.
All results presented so far were obtained by using established cell culture systems based on the ER/EBNA-2 chimeric protein. In order to exclude potential artifacts of the estrogen-inducible system, primary B cells infected with EBV-carrying wild-type EBNA-2 were analyzed for target gene expression. To this end, purified primary B cells were infected with B95.8 viral supernatants and the expression levels of all 12 candidate target genes were tested before infection as well as 4 and 10 days postinfection of short-term B-cell cultures (Fig. 4). All 12 target genes were expressed 4 and 10 days postinfection. Transcript levels of CCL3, CD21, CDK5R1, SAMSN1, FGR, MFN1, SLAMF1, RAPGEF2, and ABHD6 were elevated at least twofold on day 4 and further elevated on day 10. DNASE1L3 expression levels were low and did not rise postinfection. For CCL4, RHOH initial transcript levels were already elevated before infection and the induction by virus infection was only marginal (1.5- to 1.9-fold).
FIG. 4.
Induction of target genes upon EBV infection. Total cellular RNA was harvested from purified primary B cells before and 4 and 10 days after B95.8 infection. Gene expression was analyzed by real-time RT-PCR. The results are given as relative (rel.) transcript levels referring to a gene-specific calibrator template.
Notch target genes activated by EBNA-2.
An extensive list of Notch target genes has been described for diverse biological systems. Notch target genes, including a list of genes with diverse functions, such as receptor tyrosine-protein kinase, pre-T-cell antigen receptor alpha, Deltex1, Notch-related ankyrin receptor protein, IκBα, NF-κB2, PU.1 or cyclin D1, and p21/WAF-1/CIP1 or Nodal, have been described previously (8, 11, 33, 34, 45, 48, 52, 53, 55, 58). The genes NRARP and deltex1 were not represented by probe sets on array U133A 2.0. Cyclin D1, Nodal, erB-2, PU.1, BATF, and pre-T-cell antigen receptor alpha scored as absent or only marginally expressed on the array and were not induced by EBNA-2. NF-κB2 scored as present in most samples, but no significant change of expression was induced by EBNA-2. Both IκBα and p21/WAF-1/CIP1 were expressed but (0.5- to 0.6-fold) downregulated by EBNA-2.
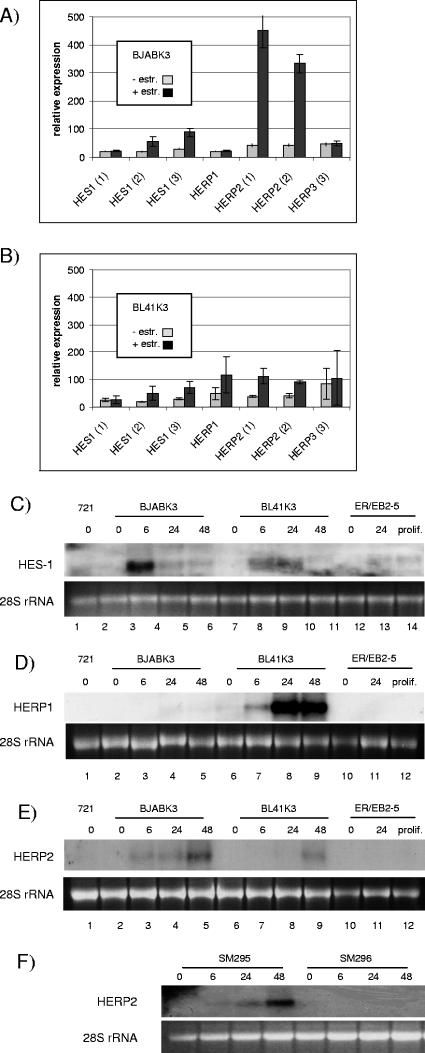
Next we focused our studies on members of the bHLH proteins: HES-1, -5, and -7 and HERP1, -2, and -3, which are well-established Notch target genes (Fig. 5) (3, 20, 21, 39, 43, 44). Only four of these six genes, HES-1 and HERP1, -2, and -3, were represented on the Affymetrix U133A 2.0 array by multiple probe sets, and the induction of HES-1 and HERP1 and -2 was validated by Northern blot analyses (Fig. 5). Hes-1 and HERP1 were not induced in DG75-derived cell lines. Surprisingly, the degree of induction differed considerably between B-cell lines and did not follow any general scheme.
FIG. 5.
Activation of basic helix-loop-helix genes by EBNA-2. Total cellular RNA of BJABK3 (A) and BL41K3 (B) was isolated before estrogen (−) and 24 h post-estrogen (+) treatment, and expression profiles were established by hybridization of the U133A 2.0 Affymetrix array. Results are given as the mean value of triplicates for HES-1, HERP1, -2, and -3. Results of all available probe sets are provided. Error bars indicate standard deviations. Different B-cell lines treated with estrogen for the indicated time periods were tested for HES-1 (C), HERP1 (D), and HERP2 (E and F) expression by Northern blot analysis. Equal loading of the lanes is visualized by ethidium bromide staining of the 28S rRNA for each blot. prolif., proliferation.
HERP3 scored as absent on the microarray and could not be detected by Northern blot analysis (data not shown). Neither HES-5 nor HES-7 was detected by real-time RT-PCR or Northern blotting, suggesting that these two Notch targets are not expressed in these B-cell lines. None of the bHLH genes were induced by estrogen in ER/EBNA-2-negative parental cell lines (data not shown). In 721, an established EBV-positive LCL, and proliferating ER/EB2-5 cells, expression levels for all bHLH genes were below or close to the detection limit.
EBNA-2-repressed genes.
Table 2 lists genes which are significantly repressed by EBNA-2 by at least twofold (q value of ≤1%) in both cell lines. These 18 genes, represented by 25 probe sets, cover a broad array of functions relevant to signal transduction, cellular metabolism, and transcriptional regulation. The repression by EBV of IGLL1, MEF2B, CD79B, and CD52 has been reported previously (6, 7). The repression of these genes can now be assigned at least partially to EBNA-2 functions. Since CD52 is a target for antibody-based chemotherapy of lymphoid malignancies, potential therapeutic implications might arise from our findings (15). The repression of T-cell leukemia I oncogene (TCL1A) by EBNA-2 was unexpected since TCL1A has recently been described as an EBV-induced gene (30). Our results suggest that EBNA-2 activation represses TCL1A but is counteracted by other viral factors in EBV-positive Burkitt's lymphoma cell lines.
TABLE 2.
EBNA-2-repressed genes
| Gene or gene producta | Gene IDb | Molecular or biological function(s) |
|---|---|---|
| A2A adenosine receptor (ADORA2A) | 135 | G protein-coupled receptor |
| Arachidonate 5-lipoxygenase-activating protein (ALOX5AP) | 241 | Inflammatory response |
| CD24 | 934 | Humoral immune response/small cell lung carcinoma cluster 4 antigen |
| CD52 (CAMPATH-1 antigen) | 1043 | Target for antibody-based chemotherapy |
| CD79B | 974 | BCR associated, signal transduction |
| Chimerin (CHN2) | 1124 | Intracellular signaling cascade |
| DKFZP586A0522 protein (DKFZP586A0) | 25804 | |
| FCH and double SH3 domains 2 (FCHSD2) | 9873 | Adaptor for signal transduction |
| cDNA: FLJ23572 fis, clone LNG12403 (AK027225) | ||
| Major histocompatibility complex class II, DPa1 (HLA-DPA1) | 3113 | Antigen presentation |
| Immunoglobulin lambda-like polypeptide 1 (IGLL1) | 3543 | B-cell development/BCR signaling |
| Lysosome-associated multispanning membrane protein-5 (LAPTM5) | 7805 | B-cell differentiation |
| Lipin 1 (LPIN1) | 23175 | Adipocyte differentiation |
| Leucine zipper, putative tumor suppressor 1 (LZTS1) | 11178 | Transcription, negative regulation of cell cycle |
| MADS box transcription enhancer factor 2 (MEF2B) | 4207 | Regulation of transcription |
| Cyclic AMP-dependent protein kinase inhibitor g (PKIG) | 11142 | Negative regulation of protein kinase activity |
| Ras association (RalGDS/AF-6) domain family 2 (RASSF2) | 9770 | Neuropeptide signaling pathway |
| T-cell leukemia/lymphoma 1A (TCL1A) | 8115 | Cell growth and/or maintenance |
| Kell blood group precursor (XK) | 7504 | Membrane transport |
Genes that scored present before estrogen induction and were repressed at least twofold in BJABK3 and BL41K3 after estrogen induction.
Unique gene ID according to the NCBI. ID, identification number.
Previously we reported that EBNA-2 represses IgM transcription (22, 40). Eleven individual probe sets represented the IgM gene locus. However, upon closer inspection, only five probe sets (215621_s_at, 222285_at, 213674_x_at, 209374_s_at, and 212827_at) mapped to chromosome 14 only, while the other six were complementary to additional sites on chromosome 2, 15, or 16 (217320_at, 211637_x_at, 216491_x_at, 211642_at, 211632_at, and 214916_x_at). While the polyspecific probe sets responded inconsistently, the five monospecific probe sets indicated repression of 0.9- to 0.4-fold (see http://www.gsf.de/kmolbi/group3_frameset.htm). Since for immunoglobulin lambda joining 3 and immunoglobulin heavy constant gamma 1, multiple polyspecific probe sets also generated inconsistent results, we excluded the corresponding results in this study.
DISCUSSION
In healthy hosts, EBV establishes a balance which permits lifelong persistence, based on tightly controlled viral gene expression patterns and immune surveillance of the host. When posttransplant patients are treated with immunosuppressive drugs to prevent graft rejection, highly malignant EBV-positive posttransplant lymphoproliferative disease (PTLD) can arise. These PTLDs typically express a latency III viral gene transcription pattern that is controlled by EBNA-2 (35, 54). In a specific subset of cells in infectious mononucleosis and PTLD, EBNA-2 can also be expressed in the absence of LMP1 (35, 36). In addition, EBNA-2 is expressed in EBV-positive central nervous system lymphomas of AIDS patients (1).
In the context of the viral genome, EBNA-2 induces a cascade of events that relieve the resting status of the peripheral B cell and initiate and maintain B-cell activation, cell cycle entry, and proliferation. A complex transcriptional program is established by primary and secondary target genes, which hamper the identification of primary EBNA-2 functions. In addition, EBNA-2 and LMP1 target genes are not necessarily mutually exclusive, as exemplified by the activation of the EBNA-2 target genes CCR7, CD83, SAMSN1, and c-myc by LMP1 (6, 12, 13). Obviously, signaling by both viral factors frequently converges on the same targets.
In this study, targets of viral factors like LMP1 were totally excluded and did not conceal those events caused by EBNA-2 directly. Since BJABK3 and BL41K3 regulate c-myc in response to EBNA-2 in a differential manner, genes induced in both systems are Myc independent.
In response to EBNA-2 activation, gene expression profiles did not change globally, while dramatic alterations of specific gene sets were observed. Within this study, we focused on transcripts activated by EBNA-2, while transcripts repressed by EBNA-2 are briefly described but have not yet been extensively investigated. Among the genes repressed by EBNA-2 were components of the B-cell receptor complex like CD79B and the surrogate lambda light chain. If this EBNA-2 function is mimicked by activated Notch, it might explain T- versus B-cell fate decisions in response to activated Notch as well as the toxicity of activated Notch in a diverse set of B-cell lines (51, 62, 69). Some of the EBNA-2-repressed genes, such as the antiapoptotic gene TCL1A, might contribute to the recently described antagonism of EBNA-2 and Myc, which can lead to EBNA-2 loss in endemic Burkitt's lymphoma cell lines (25, 26, 50).
A more profound analysis of EBNA-2-repressed genes needs to be performed, in order to (i) test whether these genes are repressed by EBNA-2 in the context of viral infection, (ii) elucidate the relevance of these target genes for the pathogenesis of EBV-associated diseases, and (iii) understand the mechanism by which EBNA-2 executes this function. bHLH proteins are transcriptional repressors and thus attractive candidate mediators of repression. EBNA-2 induced HES-1, HERP1, or HERP2 in at least one of the two cell lines. HES-1 was strongly induced in BJABK3, HERP1 strongly in BL41K3, and HERP2 strongly in BJABK3 and DG75. Obviously the cellular background has a strong and specific impact on target gene activation by EBNA-2. Interestingly, mutually exclusive expression of HERP1 and HERP2 has been described before and might account for the differential expression patterns observed in the different B-cell lines (20).
Notably, for all activated target genes that were induced more than 3.5-fold in BJABK3 and BL41K3, we could confirm the primary data obtained in the Affymetrix screen by real-time RT-PCR. The majority of genes activated by EBNA-2 were exclusively induced in one cellular B-cell system, either BL41K3 or BJABK3. Activation of well-known EBNA-2 target genes was restricted to either cell line. In order to test whether the EBNA-2 target genes would also score in the cellular context of EBV-transformed B cells, the relative transcript levels were determined in a kinetic study of ER/EB2-5 as well as after EBV infection of primary B cells in culture. All 12 genes were either expressed at high levels before stimulation or induced upon ER/EBNA-2 stimulation or EBV infection, indicating that by using both cellular systems, which complement each other, we could define target genes which are relevant in EBV-positive B cells also.
A limited number of genes induced in either cell line was also validated. As a paradigm, a selected group of Notch targets, the bHLH genes, was chosen and differentially induced in both cell lines according to the Affymetrix data. By Northern blot analyses, these data could be confirmed, suggesting that the Affymetrix array data sets are reliable resources for EBNA-2 target genes, even for low-level expression or induction in a single cell line. Minor relative deviations between the Northern blot and Affymetrix data are most likely due to the different hybridization kinetics of the oligonucleotides used in the arrays and the cDNA probes used for Northern blot hybridization. When control EBNA-2-negative BL41 or BJAB cells were treated with estrogen, none of the EBNA-2 genes analyzed were induced by the estrogen treatment. This indicates that the conditional cellular systems generated highly reproducible data sets that are specific for EBNA-2 functions. All published target genes were confirmed in one or both cell lines with two exceptions: BATF and interleukin-18 receptor. TNF-α was induced 1.45-fold in BL41K3 but not induced in BJABK3. This result was partially expected since TNF-α had been described in EBV-positive cell lines as a secondary target gene and therefore might score weakly in EBV-negative cells (60). BATF and interleukin 18 receptor both scored as absent before and after EBNA-2 activation and thus might have been missed by the specific probe sets. In our hands, BATF activation by EBNA-2 is detected in Northern blot analysis 48 h post-EBNA-2 activation (data not shown). Thus, the time limits of our experiment might have biased the results and we might have missed genes which are expressed with a transient or slow kinetic.
While this paper was in preparation, a study was published (68) that described EBNA-2 target genes in the background of EBV-immortalized B cells expressing a tamoxifen-responsive EBNA-2. Our study confirmed at least twofold induction levels for the MAFF, CR2, CCL5, CD23, CMKOR1, and CDK5R1 transcripts.
Those genes, which were induced by EBNA-2 in BL41K3 and BJABK3 more than 3.5-fold, were confirmed in several additional cellular systems (Table 1). Eleven out of 12 target genes were activated in ER/EBNA-2-expressing DG75 (SM295). The activation of 10 of these 11 genes was CBF1 dependent. FGR was expressed but not induced in SM295 cells and could not been tested. In this study, RAPGEF2 responded weakly but also reproducibly in the absence of CBF1. These results might suggest that RAPGEF2 is activated by EBNA-2 by a partially CBF1-independent mechanism and expression is repressed by CBF1. Recently, CBF1-independent activation of the interleukin 18 receptor by EBNA-2 was demonstrated (49). The situation is perhaps reminiscent of LMP1 promoter activation by EBNA-2, which also is partially CBF1 independent (67). However, among the 12 genes studied, RAPGEF2 was an exception, being an ensemble-predicted gene that has not yet been functionally analyzed in humans. Since for the remaining 10 genes, activation was strictly dependent on CBF1, the overall conclusion drawn from our results is that CBF1 is the major rate-limiting factor for EBNA-2 activation of cellular target genes as shown before for the viral C and LMP2A promoters.
Since CBF1 has been previously described to be a repressor of target gene activation, we compared the transcript levels in wild-type DG75 and CBF1-negative cell lines in order to find potentially CBF1-repressed genes. For CCL3, CDK5R1, RHOH, and RAPGEF2, transcript levels were upregulated in the absence of CBF1. However, repression was by no means a consistent feature of EBNA-2 target genes and had a minor impact on relative expression levels. We have previously shown that DG75 cells carry only one functional CBF1 allele and express CBF1 protein at twofold-reduced levels. However, heterozygous CBF1 knockout mice do not have an apparent phenotype (46). According to our own estimates based on recombinant CBF1 as a reference, at least 1 × 105 CBF1 molecules per cell are present in DG75 (K. Henning, unpublished data). In summary, CBF1 is not a low-abundance protein and the intracellular CBF1 concentration in DG75 is unlikely to be rate limiting.
We thus would like to suggest an alternative scenario for CBF1 action. EBNA-2 might not only use CBF1 as a simple adaptor but in addition may facilitate CBF1/DNA complex formation. If CBF1/DNA complex formation were facilitated by EBNA-2 or cellular factors in the context of specific promoters, we would anticipate that the corresponding genes are not actively repressed by CBF1. Recently, it has been demonstrated that CBF1 binding to the promoter of CD23 and to the FcRH5 promoter is enhanced by EBNA-2 (41). This finding might support the latter scenario, which implies that target genes actively repressed by CBF1 carry specific or high-affinity binding sites.
The group of EBNA-2 target genes described in this study is heterogeneous in function, including cell signaling molecules, adapters, genes involved in cell cycle regulation, and chemokines. The diverse spectrum of EBNA-2 target genes described in this and previous studies reflects the broad spectrum of EBNA-2 functions involved in virus-host interactions, such as chemokines like CCL3 and CCL4 induced in both BJABK3 and BL41K3, chemokine receptors like CCR7 or the orphan chemokine receptor 1 (RDC1) induced in BL41K3, and CXCR4 induced in BJABK3 (see http://www.gsf.de/kmolbi/group3_frameset.htm). In concert with activation markers like CD21, CD83, and CD23, they might significantly modulate lymphocyte trafficking of EBNA-2-expressing cells. The potential contribution of these target genes to B-cell immortalization and EBV pathogenesis will need to be investigated specifically and will require detailed functional studies.
Additional studies were performed in order to define overlapping EBNA-2 and Notch target gene pools. The majority of published Notch target genes were not induced by EBNA-2 in B-cell lines used for this study. In contrast, Northern blot analysis of all six family members of the bHLH genes revealed that HES-1, HERP1, and HERP2 are induced. HERP3 expression could be detected by RT-PCR but was not activated by EBNA-2 (data not shown). Neither HES-5 nor HES-7 was detected in BL41K3 or BJABK3. It appears that a general assumption that Notch equals EBNA-2 functions is not justified. Tissue-specific restrictions fine-tune expression patterns and override CBF1 signaling. Some of the target genes described in this study might be interesting candidates for Notch signaling and thus warrant further investigations. These include the cdk5 activator p35 (CDK5R1), which is typically expressed in neuronal cells but is also expressed in monocytes and neutrophils (63, 64; for details, see 56 and references therein).
In summary, we have performed a comprehensive screen and identified a new collection of EBNA-2 target genes. This collection of EBNA-2 targets will help elucidate the details of cis requirements necessary for EBNA-2 to activate cellular target genes. We confirmed subsets of our results by RT-PCR or Northern blot analysis, proving that high-quality and reliable data sets have been obtained. The entire data set obtained in this study is presented as an open resource to the reader. We hope our results foster the exchange of complex data sets obtained by independent groups and contribute to the generation of specific expression profiles governed by the individual viral factors in EBV-related disorders.
Acknowledgments
We thank R. Kageyama (Kyoto) and M. Gessler (Würzburg) for plasmids, B. Frankenberger for help with real-time RT-PCR, Anja Mantik for excellent technical assistance, and the people in the lab for suggestions on the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB455), die Deutsche Krebshilfe (grant 10-1963-Ke-I), and Wilhelm-Sanderstiftung (grant 2003.143.1) to B. Kempkes. The Microarray and Bioinformatics Core Unit at the Institute of Medical Microbiology, Immunology, and Hygiene is supported by the Bundesministerium für Bildung und Forschung (NGFN Network Infection and Inflammation FKZ 01G0113, TP 37 to R. Lang, R. Hoffmann, and H. Wagner).
REFERENCES
- 1.Bashir, R., J. Luka, K. Cheloha, M. Chamberlain, and F. Hochberg. 1993. Expression of Epstein-Barr virus proteins in primary CNS lymphoma in AIDS patients. Neurology 43:2358-2362. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 3.Bessho, Y., G. Miyoshi, R. Sakata, and R. Kageyama. 2001. Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6:175-185. [DOI] [PubMed] [Google Scholar]
- 4.Birkenbach, M., K. Josefsen, R. Yalamanchili, G. Lenoir, and E. Kieff. 1993. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 67:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgstahler, R., B. Kempkes, K. Steube, and M. Lipp. 1995. Expression of the chemokine receptor BLR2/EBI1 is specifically transactivated by Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 215:737-743. [DOI] [PubMed] [Google Scholar]
- 6.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κ B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, K. L., E. Cahir-McFarland, and E. Kieff. 2002. Epstein-Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76:10427-10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., W. H. Fischer, and G. N. Gill. 1997. Regulation of the ERBB-2 promoter by RBPJκ and NOTCH. J. Biol. Chem. 272:14110-14114. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deftos, M. L., E. Huang, E. W. Ojala, K. A. Forbush, and M. J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirmeier, U., R. Hoffmann, E. Kilger, U. Schultheiss, C. Briseno, O. Gires, A. Kieser, D. Eick, B. Sugden, and W. Hammerschmidt. 2005. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 24:1711-1717. [DOI] [PubMed] [Google Scholar]
- 13.Dudziak, D., A. Kieser, U. Dirmeier, F. Nimmerjahn, S. Berchtold, A. Steinkasserer, G. Marschall, W. Hammerschmidt, G. Laux, and G. W. Bornkamm. 2003. Latent membrane protein 1 of Epstein-Barr virus induces CD83 by the NF-κB signaling pathway. J. Virol. 77:8290-8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordadze, A. V., R. Peng, J. Tan, G. Liu, R. Sutton, B. Kempkes, G. W. Bornkamm, and P. D. Ling. 2001. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J. Virol. 75:5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale, G., S. Slavin, J. M. Goldman, S. Mackinnon, S. Giralt, and H. Waldmann. 2002. Alemtuzumab (Campath-1H) for treatment of lymphoid malignancies in the age of nonmyeloablative conditioning? Bone Marrow Transplant. 30:797-804. [DOI] [PubMed] [Google Scholar]
- 16.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 17.Hayward, S. D. 2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14:387-396. [DOI] [PubMed] [Google Scholar]
- 18.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubmann, R., J. D. Schwarzmeier, M. Shehata, M. Hilgarth, M. Duechler, M. Dettke, and R. Berger. 2002. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood 99:3742-3747. [DOI] [PubMed] [Google Scholar]
- 20.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 21.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 22.Jochner, N., D. Eick, U. Zimber-Strobl, M. Pawlita, G. W. Bornkamm, and B. Kempkes. 1996. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin mu gene: implications for the expression of the translocated c-myc gene in Burkitt's lymphoma cells. EMBO J. 15:375-382. [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen, L. M., C. D. Deppmann, K. D. Erickson, W. F. Coffin III, T. M. Thornton, S. E. Humphrey, J. M. Martin, and E. J. Taparowsky. 2003. EBNA2 and activated Notch induce expression of BATF. J. Virol. 77:6029-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, G., A. Bell, and A. Rickinson. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 8:1098-1104. [DOI] [PubMed] [Google Scholar]
- 26.Kelly, G. L., A. E. Milner, R. J. Tierney, D. S. Croom-Carter, M. Altmann, W. Hammerschmidt, A. I. Bell, and A. B. Rickinson. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt's lymphoma cells and with increased resistance to apoptosis. J. Virol. 79:10709-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kempkes, B., M. Pawlita, U. Zimber-Strobl, G. Eissner, G. Laux, and G. W. Bornkamm. 1995. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-Jκ in a conditional fashion. Virology 214:675-679. [DOI] [PubMed] [Google Scholar]
- 28.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempkes, B., U. Zimber-Strobl, G. Eissner, M. Pawlita, M. Falk, W. Hammerschmidt, and G. W. Bornkamm. 1996. Epstein-Barr virus nuclear antigen 2 (EBNA2)-oestrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J. Gen. Virol. 77:227-237. [DOI] [PubMed] [Google Scholar]
- 30.Kiss, C., J. Nishikawa, K. Takada, P. Trivedi, G. Klein, and L. Szekely. 2003. T cell leukemia I oncogene expression depends on the presence of Epstein-Barr virus in the virus-carrying Burkitt lymphoma lines. Proc. Natl. Acad. Sci. USA 100:4813-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein, G., T. Lindahl, M. Jondal, W. Leibold, J. Menezes, K. Nilsson, and C. Sundstrom. 1974. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc. Natl. Acad. Sci. USA 71:3283-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutson, J. C. 1990. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J. Virol. 64:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krebs, L. T., M. L. Deftos, M. J. Bevan, and T. Gridley. 2001. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 238:110-119. [DOI] [PubMed] [Google Scholar]
- 34.Krebs, L. T., N. Iwai, S. Nonaka, I. C. Welsh, Y. Lan, R. Jiang, Y. Saijoh, T. P. O'Brien, H. Hamada, and T. Gridley. 2003. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 17:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuppers, R. 2003. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat. Rev. Immunol. 3:801-812. [DOI] [PubMed] [Google Scholar]
- 36.Kurth, J., M. L. Hansmann, K. Rajewsky, and R. Kuppers. 2003. Epstein-Barr virus-infected B cells expanding in germinal centers of infectious mononucleosis patients do not participate in the germinal center reaction. Proc. Natl. Acad. Sci. USA 100:4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, E. C. 2002. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 3:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenoir, G. M., M. Vuillaume, and C. Bonnardel. 1985. The use of lymphomatous and lymphoblastoid cell lines in the study of Burkitt's lymphoma, p. 309-318. In G. Lenoir, G. O'Conor, and C. Olweny (ed.), Burkitt's lymphoma: a human cancer model. IARC Publications, Lyon, France. [PubMed]
- 39.Maier, M. M., and M. Gessler. 2000. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275:652-660. [DOI] [PubMed] [Google Scholar]
- 40.Maier, S., M. Santak, A. Mantik, K. Grabusic, E. Kremmer, W. Hammerschmidt, and B. Kempkes. 2005. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J. Virol. 79:8784-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohan, J., J. Dement-Brown, S. Maier, T. Ise, B. Kempkes, and M. Tolnay. 2006. Epstein-Barr virus nuclear antigen 2 induces FcRH5 expression through CBF1. Blood 107:4433-4439. [DOI] [PubMed] [Google Scholar]
- 42.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa, O., D. G. McFadden, M. Nakagawa, H. Yanagisawa, T. Hu, D. Srivastava, and E. N. Olson. 2000. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc. Natl. Acad. Sci. USA 97:13655-13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura, M., F. Isaka, M. Ishibashi, K. Tomita, H. Tsuda, S. Nakanishi, and R. Kageyama. 1998. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49:69-75. [DOI] [PubMed] [Google Scholar]
- 45.Oakley, F., J. Mann, R. G. Ruddell, J. Pickford, G. Weinmaster, and D. A. Mann. 2003. Basal expression of IκBα is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J. Biol. Chem. 278:24359-24370. [DOI] [PubMed] [Google Scholar]
- 46.Oka, C., T. Nakano, A. Wakeham, J. L. de la Pompa, C. Mori, T. Sakai, S. Okazaki, M. Kawaichi, K. Shiota, T. W. Mak, and T. Honjo. 1995. Disruption of the mouse RBP-Jκ gene results in early embryonic death. Development 121:3291-3301. [DOI] [PubMed] [Google Scholar]
- 47.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: MarvelouslY complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 48.Oswald, F., S. Liptay, G. Adler, and R. M. Schmid. 1998. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol. Cell. Biol. 18:2077-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pages, F., J. Galon, G. Karaschuk, D. Dudziak, M. Camus, V. Lazar, S. Camilleri-Broet, C. Lagorce-Pages, S. Lebel-Binay, G. Laux, W. H. Fridman, and B. Henglein. 2004. Epstein-Barr virus nuclear antigen 2 induces interleukin-18 receptor expression in B cells. Blood 105:1632-1639. [DOI] [PubMed] [Google Scholar]
- 50.Pajic, A., M. S. Staege, D. Dudziak, M. Schuhmacher, D. Spitkovsky, G. Eissner, M. Brielmeier, A. Polack, and G. W. Bornkamm. 2001. Antagonistic effects of c-myc and Epstein-Barr virus latent genes on the phenotype of human B cells. Int. J. Cancer 93:810-816. [DOI] [PubMed] [Google Scholar]
- 51.Radtke, F., A. Wilson, S. J. Mancini, and H. R. MacDonald. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5:247-253. [DOI] [PubMed] [Google Scholar]
- 52.Rangarajan, A., C. Talora, R. Okuyama, M. Nicolas, C. Mammucari, H. Oh, J. C. Aster, S. Krishna, D. Metzger, P. Chambon, L. Miele, M. Aguet, F. Radtke, and G. P. Dotto. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reizis, B., and P. Leder. 2002. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 16:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizmann, and S. E. Strauss (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 55.Ronchini, C., and A. J. Capobianco. 2001. Induction of cyclin D1 transcription and CDK2 activity by Notchic: implication for cell cycle disruption in transformation by Notchic. Mol. Cell. Biol. 21:5925-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosales, J. L., J. D. Ernst, J. Hallows, and K. Y. Lee. 2004. GTP-dependent secretion from neutrophils is regulated by Cdk5. J. Biol. Chem. 279:53932-53936. [DOI] [PubMed] [Google Scholar]
- 57.Schlee, M., T. Krug, O. Gires, R. Zeidler, W. Hammerschmidt, R. Mailhammer, G. Laux, G. Sauer, J. Lovric, and G. W. Bornkamm. 2004. Identification of Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) target proteins by proteome analysis: activation of EBNA2 in conditionally immortalized B cells reflects early events after infection of primary B cells by EBV. J. Virol. 78:3941-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroeder, T., H. Kohlhof, N. Rieber, and U. Just. 2003. Notch signaling induces multilineage myeloid differentiation and up-regulates PU. 1 expression. J. Immunol. 170:5538-5548. [DOI] [PubMed] [Google Scholar]
- 59.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spender, L. C., G. H. Cornish, B. Rowland, B. Kempkes, and P. J. Farrell. 2001. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75:3537-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spender, L. C., G. H. Cornish, A. Sullivan, and P. J. Farrell. 2002. Expression of transcription factor AML-2 (RUNX3, CBFα-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J. Virol. 76:4919-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strobl, L. J., H. Hofelmayr, G. Marschall, M. Brielmeier, G. W. Bornkamm, and U. Zimber-Strobl. 2000. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J. Virol. 74:1727-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Studzinski, G. P., and J. S. Harrison. 2003. The neuronal cyclin-dependent kinase 5 activator p35Nck5a and Cdk5 activity in monocytic cells. Leuk. Lymphoma 44:235-240. [DOI] [PubMed] [Google Scholar]
- 64.Tsai, L. H., I. Delalle, V. S. Caviness, Jr., T. Chae, and E. Harlow. 1994. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature 371:419-423. [DOI] [PubMed] [Google Scholar]
- 65.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, F., C. D. Gregory, M. Rowe, A. B. Rickinson, D. Wang, M. Birkenbach, H. Kikutani, T. Kishimoto, and E. Kieff. 1987. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc. Natl. Acad. Sci. USA 84:3452-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yalamanchili, R., X. Tong, S. Grossman, E. Johannsen, G. Mosialos, and E. Kieff. 1994. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204:634-641. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, B., S. Maruo, A. Cooper, M. R. Chase, E. Johannsen, E. Kieff, and E. Cahir-McFarland. 2006. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. USA 103:1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zweidler-McKay, P. A., Y. He, L. Xu, C. G. Rodriguez, F. G. Karnell, A. C. Carpenter, J. C. Aster, D. Allman, and W. S. Pear. 2005. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood 106:3898-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]