Abstract
Gene expression profiling of diffuse large B-cell lymphoma (DLBCL) has revealed prognostically important subgroups: germinal center B-cell-like (GCB) DLBCL, activated B cell-like (ABC) DLBCL, and primary mediastinal large B-cell lymphoma. The t(14;18)(q32;q21) has been reported previously to define a unique subset within the GCB-DLBCL. We evaluated for the translocation in 141 cases of DLBCL that were successfully gene expression profiled. Using a dual-probe fluorescence in situ hybridization assay, we detected the t(14;18) in 17% of DLBCLs and in 34% of the GCB subgroup which contained the vast majority of positive cases. In addition, 12 t(14;18)-positive cases detected by polymerase chain reaction assays on additional samples were added to the fluorescence in situ hybridization-positive cases for subsequent analysis. Immunohistochemical data indicated that BCL2, BCL6, and CD10 protein were preferentially expressed in the t(14;18)-positive cases as compared to t(14;18)-negative cases. Within the GCB subgroup, the expression of BCL2 and CD10, but not BCL6, differed significantly between cases with or without the t(14;18): 88% versus 24% for BCL2 and 72% versus 32% for CD10, respectively. In the GCB-DLBCL subgroup, a heterogeneous group of genes is overexpressed in the t(14;18)-positive subset, among which BCL2 is a significant discriminator. Interestingly, the t(14;18)-negative subset is dominated by overexpression of cell cycle-associated genes, indicating that these tumors are significantly more proliferative, suggesting distinctive pathogenetic mechanisms. However, despite this higher proliferative activity, there was no significant difference in overall or failure-free survival between the t(14;18)-positive and -negative subsets within the GCB subgroup.
Diffuse large B-cell lymphoma (DLBCL) is an aggressive malignancy of mature B cells with an annual incidence of ∼25,000 cases in the United States. DLBCL is a heterogeneous entity both clinically and morphologically. We have recently shown by gene expression profiling that DLBCL can be classified into two major subgroups.1 The germinal center B-cell-like (GCB) subgroup expresses genes characteristic of normal GC B cells and is associated with a good outcome after multiagent chemotherapy, whereas the activated B-cell-like (ABC) subgroup expresses genes characteristic of activated blood B cells and is associated with a poor clinical outcome. Nonetheless, considerable molecular heterogeneity exists within each subgroup. A small number of DLBCL cases are unclassifiable and do not express the GCB or ABC signature genes at a high level.2 More recently, primary mediastinal large B-cell lymphoma (PMBL) has been identified as a distinct subgroup of DLBCL that can be distinguished by gene expression profiling from GCB- and ABC-DLBCL.3,4
The t(14;18)(q32;q21) is a characteristic feature of follicular lymphoma and is considered to be the initiating event in lymphomagenesis. The t(14;18) is the result of an error during the process of VDJ recombination, leading to deregulation of the expression of the anti-apoptotic gene BCL2 by bringing it into proximity of the immunoglobulin heavy chain (IgH) gene enhancer.5–8 In our initial study of 35 cases of DLBCL, we correlated BCL2 translocation data with gene expression profiles and showed that the t(14;18) defines a unique subset of DLBCL within the GCB subgroup.9 This observation suggested that important genetic lesions are associated with a unique, identifiable gene expression profile. To substantiate and further extend this finding, we have examined 141 new cases that were part of 240 cases of DLBCL studied by cDNA microarray for molecular predictors of survival after chemotherapy.2 These 141 cases of DLBCL with gene expression profiles, clinical data, and genetic data for BCL2 translocation were studied to determine: 1) the distribution of the t(14;18) among the subgroups of DLBCL identified by gene expression profiling; 2) whether t(14;18)-positive cases have a unique gene expression profile; and 3) whether there are differences in the tumor characteristics and clinical behavior between cases with and without the t(14;18).
Materials and Methods
Patient Information
We studied 240 previously analyzed cases of DLBCL with clinical data and gene expression profiles determined by complementary DNA (cDNA) microarray technology.2 A panel of hemopathologists including E Campo, ES Jaffe, G Ott, HK Müller-Hermelink, J Delabie, R Gascoyne, T Grogan, DD Weisenburger, and WC Chan confirmed the diagnosis of DLBCL and excluded the presence of follicular lymphoma in all patients. Informed consent was obtained and the Institutional Review Board of the University of Nebraska approved this study.
Preparation of Tissue Microarrays (TMAs) and Immunohistochemical Studies
TMAs were prepared from cases with adequate archival paraffin-embedded tissue. Hematoxylin and eosin-stained sections from each paraffin-embedded, formalin-fixed block were examined to define diagnostic areas and two to five (average, four) representative 0.6-mm cores were obtained from each case and inserted in a grid pattern into a recipient paraffin block using a tissue arrayer (Beecher Instruments, Silver Spring, MD). Five μm sections were then cut from each TMA and stained with antibodies to BCL2, BCL6, and CD10 as described.10 CD20 stains were performed to evaluate each core for involvement by tumor, and each case was evaluated independently by two pathologists (CPH, DDW) for the percentage of tumor cells stained and recorded in 10% increments. Disagreements were resolved by joint review on a multihead microscope. For each case, the core with the highest percentage of tumor cells stained was used for analysis. Cases were considered positive if 30% or more of the tumor cells were stained with an antibody.10
Detection of the t(14;18)(q32;q21) by Fluorescence in Situ Hybridization (FISH)
Among the 240 cases studied by gene expression profiling, 129 cases were studied by interphase FISH for the presence of the t(14;18)(q32;q21). To perform FISH studies, 4-μm sections were cut from the TMA paraffin blocks and mounted on positively charged slides. The sections were dewaxed in three changes of HEM-D (Scientific Safety Solvents, Keller, TX) followed by dehydration in 95% ethanol. They were then treated with 0.2 N HCl for 15 minutes, rinsed in distilled-deionized water, and incubated in a sodium thiocyanate solution at 80°C for 15 minutes. After rinsing in phosphate-buffered saline (PBS), the sections were digested with a protease solution at 37°C for 10 minutes; postfixed in 0.95% formaldehyde solution/PBS with 0.45% MgCl2 for 5 minutes at room temperature; rinsed in PBS; and sequentially dehydrated in 70%, 80%, and 95% ethanol.
For FISH, the dual-color LSI IgH Spectrum Green/LSI BCL2 Spectrum Orange Dual-Fusion Translocation Probe (Vysis, Downers Grove, IL) was used to detect the t(14;18), and the CEP 18 Spectrum Aqua probe (Vysis) was used simultaneously to evaluate the chromosome 18 copy number. The probe mixture (10 μl) was placed on the tissue sections, coverslipped, and sealed. Hybridization was performed overnight at 37°C using an automated hybridization chamber (HYBrite, Vysis) after denaturation at 75°C for 5 minutes. The slides were washed in 2× standard saline citrate/0.1% Nonidet P-40 for 2 minutes at 72°C and then at room temperature for 2 minutes. Nuclei were counterstained with 4,6-diamidino-2-phenylindole at a concentration of 125 ng/ml in anti-fade solution and the slides were visualized using an Olympus BX52 fluorescence microscope. Images were captured and archived using Cytovision software (Applied Imaging, Santa Clara, CA). To analyze the hybridization, a total of 50 to 100 nuclei were scored per case for the presence of the t(14;18). In normal cells, an interphase nucleus will exhibit individual red (BCL2) and green (IgH) signals. When the t(14;18) occurs, the red and green signals form two yellow fusion signals in the interphase cell. It has been established by the University of Nebraska Medical Center Human Genetics Laboratories that reactive lymphoid tissues show <2% positive cells at 2.5 SD for this FISH assay.
Detection of BCL2 Gene Rearrangement by Polymerase Chain Reaction
Extracted DNA from all 240 cases was tested for the t(14;18) by the polymerase chain reaction (PCR). Amplification of the BCL2/JH translocation at the major breakpoint region (mbr) and minor cluster region (mcr) was performed as described previously.11 Positive controls consisted of DNA extracted from the human B-cell lymphoma cell lines RL-7 for the mbr and DHL-16 for the mcr. Negative controls consisted of sterile water instead of DNA, and DNA from normal peripheral blood mononuclear cells obtained from healthy donors. Standard precautions were taken to guard against PCR contamination. The PCR-positive cases were added to the FISH-positive cases to form a final set of 34 cases with the t(14;18). The FISH-negative cases comprised the t(14;18)-negative group.
Analysis of Differential Gene Expression
We used the recently published Bayesian classification system to define the GCB, ABC, and unclassifiable subgroups of DLBCL.12 We also separated cases with the PMBL gene expression profile3 into a new subgroup, and examined the t(14;18) within each of these subgroups. The presence or absence of the BCL2 translocation was used to supervise the discovery of differentially expressed genes between the t(14;18)-positive and -negative cases in the GCB subgroup. The two-tailed Student’s t-test was used to compare the differences in gene expression levels. Genes differentially expressed between the two subsets with a P value of <0.01 were selected for further analysis using the Significance Analysis of Microarrays (SAM) approach, as described previously.13 In addition to the P value, we also calculated the t-statistics between the two groups and only genes that differed by a magnitude of >2.6 were selected for further analysis.
As previously described, of the three different BCL2 clones (232714, 342181, 1336385) on the Lymphochip microarray, only clone 232714 with a more 5′ sequence could detect message with a truncated 3′ end because of translocation at the mbr. Overexpression of BCL2 as measured by clone 232714 was highly correlated with the BCL2 translocation. However, clone 232714 is located far from the 3′ end of the transcript and, therefore, may fail to measure much of the cDNA transcribed by reverse transcription reactions starting from the poly-A tail of the normal BCL2 transcript. We used the mean values measured by the BCL2 clones that are located close to the 3′ end (342181, 1336385) of the transcript to represent the gene expression level in cases in which this mean value was greater than the measurement by clone 232714.
Survival Analysis
The Kaplan-Meier method was used to estimate the overall and event-free survival of the patients, and the log-rank test was used to compare the survival experiences between the t(14;18)-positive and -negative cases. Overall survival was defined as the time from diagnosis to death from any cause or, for patients remaining alive, the time from diagnosis to last contact. Event-free survival was defined as the time from diagnosis to the first occurrence of relapse or death from any cause or, for patients remaining alive and relapse-free, the time from diagnosis to last contact. SAS software (SAS Institute Inc., Cary, NC) was used for the data analysis.
Results
Occurrence of the t(14;18) in DLBCL
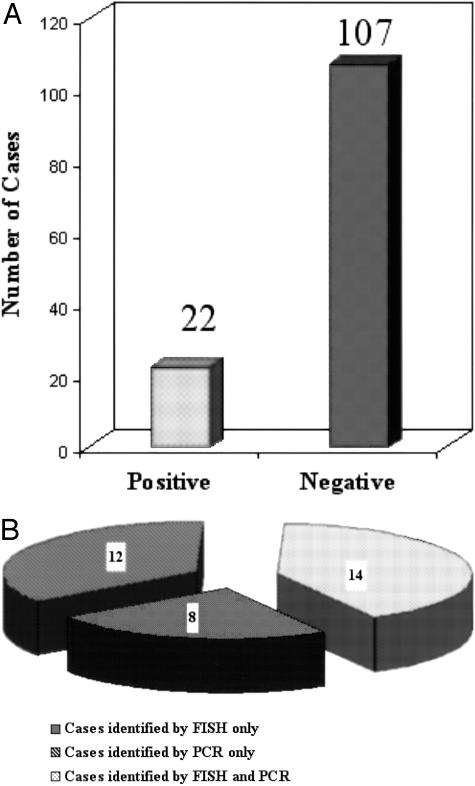
Based on the results of our recent study of PMBL,3 12 GCB cases were reclassified as PMBL. Therefore, we classified the DLBCL cases into GCB, ABC, PMBL, and unclassifiable subgroups and examined the distribution of the t(14;18). Interphase FISH was applied to the 129 TMA cases and detected 22 cases that were positive for the t(14;18). Based on FISH analysis, t(14;18) was detected in 17% (22 of 129) of the cases of DLBCL which represented 34% (19 of 55) of the cases in the GCB subgroup. We also performed PCR assays on 240 cases studied by cDNA microarray, 26 cases were found to be positive, including 14 of the cases previously found to be positive by FISH (Figure 1). The PCR-negative cases that were not tested by FISH were excluded from further analysis because of the recognized high false-negative rate of the PCR assay. Thus, PCR analysis revealed an additional 12 positive cases and in this cohort of 141 cases, 34 DLBCL cases carried the t(14;18) translocation, and 107 lacked this abnormality (Figure 1).
Figure 1.
Frequency of the t(14;18) in DLBCL. A: Among the 129 cases studied by interphase FISH for the presence of t(14;18), 22 cases were positive whereas 107 cases were negative. B: PCR analysis detected 26 positive cases, of which 14 were also shown to be positive by FISH. Therefore, an additional 12 positive cases detected by PCR were included in this study. Thus, of the 141 cases, 34 cases were BCL2 translocation-positive and 107 were negative.
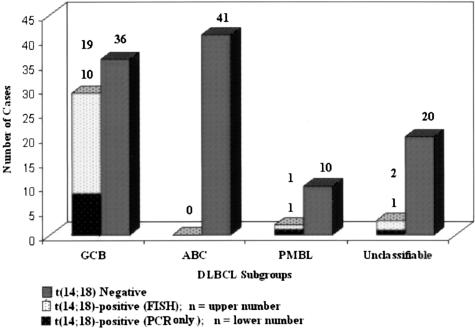
Review of the frequency of this anomaly in relation to the DLBCL subgroups (ie, GCB, ABC, PMBL, and unclassifiable) showed that the great majority (29 of 34) of the t(14;18)-positive cases occurred in the GCB subgroup. In contrast, only 3 cases occurred among the 23 cases in the unclassifiable subgroup, and none among the 41 ABC cases. Interestingly the t(14;18) translocation was found in 2 of the12 cases categorized by gene expression profiling as PMBL3 (Figure 2).
Figure 2.
Distribution of the t(14;18) among the DLBCL subgroups. Twenty-nine of 34 positive cases occurred in the GCB subgroup, whereas three cases occurred in the unclassifiable subgroup and none in the ABC subgroup. Among 12 PMBL cases identified using the PMBL predictor, 2 cases had the t(14;18).
Comparison with Immunohistochemical Data from the TMA
We reviewed the immunohistochemical reactions of 109 TMA samples to correlate the presence and absence of the t(14;18) translocation with the expression of three relevant proteins, namely BCL-2 (encoded by the gene deregulated by the translocation) and two markers of normal germinal center B cells (BCL6 and CD10).
The protein expression of BCL2 was similar in the GCB (28 of 50, 56%) and ABC (19 of 31, 61%) subgroups, but was less frequent in the unclassifiable (6 of 19, 31%) or the PMBL subgroups (2 of 9, 22%). The difference did not reach statistical significance (P = 0.054) (Table 1). Within the GCB subgroup there was a significant difference (P < 0.0001) in BCL2 protein expression between thet(14;18)-positive cases (88%) and t(14;18)-negative cases (24%) (Table 2) indicating that the t(14;18) is highly associated with BCL2 protein expression in this subgroup. In other subgroups BCL2 was generally up-regulated by mechanisms other than the t(14;18), as frequently seen in the ABC cases.
Table 1.
Expression of BCL2, BCL6, and CD10 in DLBCL Subgroups
| Protein expression | DLBCL subgroups
|
P value | |||
|---|---|---|---|---|---|
| GCB n = 50 | ABC n = 31 | Unclassifiable n = 19 | PMBL n = 9 | ||
| BCL2 | 28 (56%) | 19 (61%) | 6 (31%) | 2 (22%) | 0.054 |
| BCL6 | 42 (84%) | 11 (35%) | 6 (31%) | 4 (44%) | <0.0001 |
| CD10 | 26 (52%) | 1 (3%) | 3 (16%) | 1 (11%) | <0.0001 |
Table 2.
Expression of BCL2, BCL6, and CD10 with Regard to BCL2 Translocation
| Protein expression | BCL2 translocation
|
P value | |
|---|---|---|---|
| Positive n = 28 | Negative n = 81 | ||
| BCL2 | 25 (89%) | 30 (37%) | <0.0001 |
| BCL6 | 21 (75%) | 42 (52%) | 0.033 |
| CD10 | 19 (67%) | 12 (15%) | <0.0001 |
BCL6 protein was detected in 58% (63 of 109) of the cases of DLBCL and it was very frequently expressed in the GCB subgroup (42 of 50, 84%; P < 0.0001), but within this category there was no correlation with the t(14;18) translocation (Table 3). CD10 protein expression was detected overall less commonly than BCL6 (28%, 31 of 109), and the great majority of these cases (26 of 31) were in the GCB subgroup (Table 1). This accounted for the fact that CD10 expression was much commoner overall in t(14;18)-positive cases than in t(14;18)-negative cases (67% versus 15%; P < 0.0001) (Table 2).
Table 3.
Expression of BCL2, BCL6, and CD10 in GCB DLBCL Cases According to BCL2 Translocation Status
| Protein expression | BCL2 translocation in GCB DLBCL
|
P value | |
|---|---|---|---|
| Positive n = 25 | Negative n = 25 | ||
| BCL2 | 22 (88%) | 6 (24%) | <0.0001 |
| BCL6 | 21 (84%) | 21 (84%) | 1.0 |
| CD10 | 18 (72%) | 8 (32%) | 0.0046 |
Differential Gene Expression between t(14;18)-Positive and t(14;18)-Negative Cases in GCB-DLBCL
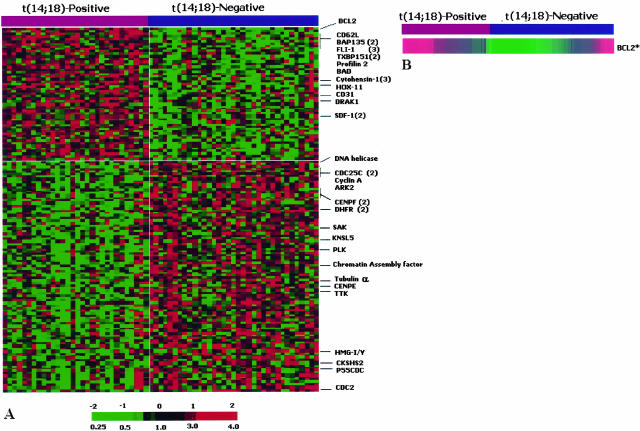
To search for genes that are differentially expressed between the t(14;18)-positive and t(14;18)-negative cases in the GCB subgroup, we examined all informative genes among 7399 known or uncharacterized genes on the Lymphochip.2 Clones with a P value of <0.01 were selected for SAM analysis.13 Among the 146 clones selected, 53 clones were overexpressed in the t(14;18)-positive group and 93 clones were overexpressed in the t(14;18)-negative group. The genes that were overexpressed in the t(14;18)-positive group represented a heterogeneous set including genes involved in apoptosis (BCL2, BAD, DRAK1, and TXBP151), a number of transcription factors (FLI1, HOX11, and BAP135) and genes associated with cell adhesion/migration (CD62L, cytohesin-1, profilin 2, SDF1, and CD31). Interestingly, a large number of genes overexpressed in the t(14;18)-negative group are associated with cell cycle progression and regulation (Figure 3A). These include genes that control several events in mitosis (PLK, KNSL5, TTK, P55CDC, ARK2, CENE, and CENF), genes involved in cell cycle progression (CYCLIN-A, CDC2, CDC25C, and SAK) and DNA replication (HMG-I/Y, DNA helicase, and DHFR).
Figure 3.
Differential gene expression in the two cytogenetic subsets of GCB-DLBCL. A: Differential expression of genes in cases with or without t(14;18) in the GCB subgroup. Differentially expressed genes are illustrated with red indicating increased expression and green decreased expression. Each column represents a single GCB DLBCL case and each row represents a single gene. Among the 146 clones selected, 53 clones were overexpressed in the t(14;18)-positive group and 93 clones were overexpressed in the t(14;18)-negative group. Genes discussed in the text are listed on the right with the number of clones selected (when >1) indicated in the bracket. The full gene list is available in the supplemental table at http://ajp.amjpathol.org. B: BCL2 gene expression in cases with and without the t(14;18) in the GCB subgroup. BCL2 gene expression was calculated as detailed in the text to more accurately reflect the true expression levels. These new values avoided the false-negative measurements because of 3′ truncation of the transcript resulting from BCL2 translocation at the mbr and the falsely low values on many normal transcripts, when measured by the clone (232714). Using these values, BCL2 is significantly overexpressed in the t(14;18)-positive subset (P = 0.0015 and t-statistic value = 3.35).
Some of the cDNAs were represented by multiple clones on the array and these clones were selected independently by the computer algorithm, providing confidence in the reproducibility of the experimental data and the analytical approach. BCL2 gene expression by the different cDNA clones had been adjusted to obtain a more accurate measurement as described in the Materials and Methods section. It was found to be a highly significant discriminator between the two cytogenetic groups (Figure 3B).
Correlation between the t(14;18) and Survival in the GCB Subgroup
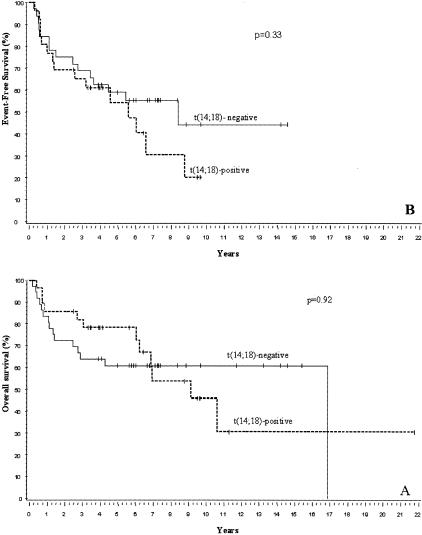
The GCB subgroup (excluding PBML) consisted of 39 men and 26 women with a median age of 61 years (range, 24 to 88 years). The median follow-up of the surviving patients was 7.3 years (range, 0.8 to 21.8 years) in this group. Figure 4, A and B, shows Kaplan-Meier estimates of overall and event-free survival experiences, respectively, of the t(14;18)-positive and negative subsets of the patients in the GCB subgroup as defined by the Bayesian classifier. P values are based on a log-rank comparison of the outcome of the two subsets. There is no significant difference between the two cytogenetic subsets.
Figure 4.
Survival of t(14;18)-positive versus t(14;18)-negative patients in the GCB DLBCL subgroup. A: Overall survival; B: event-free survival. There is no significant difference 0in survival between the two subsets of patients.
Discussion
DLBCL is the most common type of lymphoma, comprising 30 to 40% of all non-Hodgkin’s lymphoma.14 The recent development of DNA microarray technology provides an opportunity to take a genome-wide approach to the study of DLBCL. We have previously identified two molecularly distinct subtypes of DLBCL indicative of different functional stages of B-cell differentiation.1,2 One type expressed genes characteristic of GC B cells (GCB-DLBCL), whereas the other type expressed genes normally seen during in vitro activation of peripheral blood B cells (ABC-DLBCL). In a previous study,9 we found that the t(14;18)-positive cases of DLBCL were all in the GCB group but, because of the limited number of cases, a number of interesting questions could not be addressed. In this study, we have examined the presence of the t(14;18) in a much larger series of DLBCL and have correlated this genetic abnormality with the gene expression profiles, tumor characteristics, and survival of the patients. The FISH assay detected the t(14;18) in 17% (22 of 129) of the cases and in 34% (19 of 55) of the GCB cases, similar to our previous findings.9 By PCR analysis, we detected the translocation in an additional 12 cases outside of the cohort studied by FISH. We observed that the vast majority (29 of 34) of the t(14;18)-positive cases occurred in the GCB subgroup. These cases had a gene expression profile resembling normal GCB cells, suggesting that this translocation plays an important role in the pathogenesis of this subset of DLBCL, just as it does in follicular lymphoma. The complete absence of t(14;18) in the ABC-DLBCL confirmed our previous observation and supported the conclusion that GCB and ABC DLBCL are distinct pathogenetic and biological entities. Three t(14;18)-positive cases were found in the unclassifiable subgroup. However the unclassifiable cases are not well characterized and are heterogeneous, likely including some GCB cases with atypical profiles. Recently, Rosenwald and colleagues3 and Savage and colleagues4 have defined a unique gene expression profile for PMBL and this subgroup can now be distinguished from the GCB-DLBCL. It is interesting to note that a low percentage (22%) of PMBL cases had the t(14;18), suggesting that there may be further heterogeneity within this subgroup.
We also analyzed the immunohistochemical findings available in the cases with t(14;18) data. The vast majority of patients with this translocation had increased BCL2 protein expression. However, BCL2 protein was also frequently present in cases without the translocation, especially in the ABC subgroup, which characteristically had high BCL2 mRNA levels. This indicates that up-regulation of BCL2 expression may occur by other mechanisms such as activation of the nuclear factor-kB pathway or an increased copy number of the BCL2 gene. It is interesting to note that, in the GCB subgroup, BCL2 protein expression was markedly different in cases with and without the t(14;18) (89% versus 37%, respectively). BCL6 protein was more frequently expressed in the GCB versus the ABC and other subgroups, but was not specifically correlated with the t(14;18) within the GCB subgroup. On the other hand, CD10 expression was highly specific for the GCB subgroup and was most frequently expressed in cases with the t(14;18). In the PMBL subgroup, the majority have the immunohistochemical profile of the t(14;18)-negative cases, as expected.
We also attempted to correlate the t(14;18) status with gene expression profiles within the GCB subgroup to discover genes that characterize the two cytogenetic subsets and might provide insight into their pathobiology. SAM analysis was used to find genes that were differentially expressed between the two subsets after an initial data reduction based on the Student’s t-test (P < 0.01). SAM analysis identified 53 up-regulated genes in the t(14;18)-positive cases and 93 up-regulated genes for t(14;18)-negative cases. BCL2 gene expression is an important discriminator between the two cytogenetic subsets. BCL2 was overexpressed in most cases of GCB-DLBCL with the t(14;18). However, there were a few cases in which BCL2 expression was low, indicating that overexpression of this gene may no longer be necessary in some cases of established DLBCL. The microarray analysis was further substantiated by BCL2 immunohistochemistry, which revealed that a high percentage (88%) of t(14;18)-positive GCB-DLBCL cases had BCL2 protein expression, whereas BCL2 was expressed in only 24% of the t(14;18)-negative cases. In addition to BCL2, several genes that may be involved in apoptosis were also overexpressed in t(14;18)-positive cases including BAD, a proapoptotic gene in the BCL2 family. DRAK1, a positive mediator of apoptosis, is related to Death-associated Protein Kinase 1 (DAP1).15 TXBP151 is a novel anti-apoptotic gene that has been shown to mediate the activity of A20, which is induced by CD40/CD40L interaction,16,17 and its proteolysis is associated with FAS-mediated apoptosis. IL15 was also overexpressed in the BCL2 translocated cases and it has been implicated in protecting B cells from apoptosis.18 The precise functional alterations in the apoptotic pathway are difficult to predict from the gene expression data and it is possible that the apoptotic pathway is dominated by BCL2 overexpression in the vast majority of t(14;18)-positive cases. In addition to these genes, there is also overexpression of a heterogeneous group of transcription factors including FLI1 that regulates expression of the TGF-β type II receptor,19 HOX11 that belongs to a family of DNA-binding transactivators,20 and BAP135. BAP135 exists as a complex with Bruton Tyrosine Kinase (BTK) before B-cell antigen receptor engagement and is transiently tyrosine-phosphorylated in response to B-cell receptor cross-linking.21 BAP135 is identical to the putative transcription factor TFII-I and there is evidence that tyrosine phosphorylation may enhance its activity, thereby linking receptor activation with changes in gene expression. A group of genes that have diverse functions were also overexpressed including a number of genes that are involved in cell adhesion, migration, and cytoskeletal function: CD62L, cytohesin-1, profilin 2, SDF1, and CD31. CD62L is involved in cell adhesion and migration.22 Cytohesin-1 is a guanine nucleotide exchange factor for ARF GTPases and is a regulator of integrin-mediated cell adhesion.23 The function of integrins is regulated through cytoplasmic signaling, and profilin 224 may form an important link between integrin cell adhesion signals and F-actin polymerization, as has been postulated in T lymphocytes.25 SDF1 is a highly potent lymphocyte chemoattractant and is also involved in intracellular actin polymerization in lymphocytes, a prerequisite for cell motility.26 CD31 has widespread distribution and in addition to cell adhesion, may have a role in regulating cell signaling and angiogenesis.27,28
The gene expression profile associated with t(14;18)-negative cases was dominated by genes involved in cell cycle progression and regulation. These included genes involved in regulating mitotic spindle functions, activation of anaphase promoting complex (APC) and events essential for cytokinesis (eg, PLK, mitotic kinesin-like protein-1, or KNSL5, P55CDC, and CKS1).29–32 In addition to these genes, many important regulators of cell cycle-related events including centrosome separation/segregation (eg, ARK2),33 kinetochore association (eg, CENF),34 functional components of mitotic check points (eg, CENPE and TTK orMps1),35,36 chromatin association and DNA replication (eg, HMG-I/Y, chromatin assembly factor-I (p150), DNA helicase and DHFR).37–39 A group of genes that promotes G2/M transit or are expressed in the M phase of the cell cycle, including CYCLIN-A, CDC2, CDC25C, and SAK,40–42 were also up-regulated. These results suggest that t(14;18)-negative GCB-DLBCL is significantly more mitotically active than the t(14;18)-positive cases, and indicates that the two cytogenetic subsets have distinctive biological characteristics. In normal cells, BCL2 has been reported to retard G1/S phase progression43 and the higher expression of BCL2 in the translocated cases may have the same effects on cell cycle progression as has been observed in developing B lymphocytes.44 However, further studies are needed to determine whether other mechanisms are involved in the higher expression of genes associated with cell cycle regulation and progression in the nontranslocated subset.
Overall and failure-free survival analysis did not show any significant differences between t(14;18)-positive and -negative cases within the GCB subgroup. However, there seems to be a continued occurrence of late deaths in the t(14;18)-positive subset but this finding needs to be confirmed in a larger series of patients.
In conclusion, we found that t(14;18) was present in a large subset of cases in the GCB-DLBCL subgroup. This specific translocation was primarily confined to GCB-DLBCL, without any cases seen in ABC-DLBCL, thus supporting the view that DLBCL can be divided into pathogenetically distinct subgroups with unique gene expression profiles. Cases in the ABC subgroups also frequently overexpressed BCL2, but through mechanisms other than t(14;18). The t(14;18)-positive subset within the GCB subgroup showed a clear decreased expression of proliferation signature genes when compared with the t(14;18)-negative subset. This could be partly explained by the anti-proliferation effect of BCL2, but there may be other underlying biological mechanisms. These two subsets did not show any significant differences on survival under conventional therapy. However, the increase in BCL2 expression in one subset and the higher proliferative activity in the other suggest biological differences that could be exploited in future clinical trials.
Supplementary Material
Acknowledgments
We thank Lynette M. Smith for statistical analysis and Martin Bast for data management.
Footnotes
Address reprint requests to Wing C. Chan. M.D., Department of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198-3135. E-mail: jchan@unmc.edu.
Supported in part by United States Public Health Service grants CA36727 and CA84967 awarded by the National Cancer Institute, Department of Health and Human Services.
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Campo E, Montserrat E, Lopez-Guillermo A, Ott G, Muller-Hermelink HK, Connors JM, Braziel R, Grogan TM, Fisher RI, Miller TP, LeBlanc M, Chiorazzi M, Zhao H, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Staudt LM. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, Aguiar RC, Li S, Salles G, Berger F, Jing W, Pinkus GS, Habermann T, Dalla-Favera R, Harris NL, Aster JC, Golub TR, Shipp MA. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin’s lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- Bakhshi A, Jensen JP, Goldman P, Wright JJ, McBride OW, Epstein AL, Korsmeyer SJ. Cloning the chromosomal breakpoint of t(14;18) human lymphomas: clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41:899–906. doi: 10.1016/s0092-8674(85)80070-2. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- Magrath I. Molecular basis of lymphomagenesis. Cancer Res. 1992;52:5529s–5540s. [PubMed] [Google Scholar]
- Yang E, Korsmeyer SJ. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- Huang JZ, Sanger WG, Greiner TC, Staudt LM, Weisenburger DD, Pickering DL, Lynch JC, Armitage JO, Warnke RA, Alizadeh AA, Lossos IS, Levy R, Chan WC. The t(14;18) defines a unique subset of diffuse large B-cell lymphoma with a germinal center B-cell gene expression profile. Blood. 2002;99:2285–2290. doi: 10.1182/blood.v99.7.2285. [DOI] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Sharp JG BM, Chan WC. Basel: Karger; Application of Malignant Cell Detection Techniques to Improve the Outcome of High-Dose Therapy and Transplantation for Lymphoma, Leukemia, Breast Cancer. 1997 [Google Scholar]
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A clinical evaluation of the international lymphoma group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- Sanjo H, Kawai T, Akira S. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem. 1998;273:29066–29071. doi: 10.1074/jbc.273.44.29066. [DOI] [PubMed] [Google Scholar]
- De Valck D, Jin DY, Heyninck K, Van de Craen M, Contreras R, Fiers W, Jeang KT, Beyaert R. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene. 1999;18:4182–4190. doi: 10.1038/sj.onc.1202787. [DOI] [PubMed] [Google Scholar]
- Sarma V, Lin Z, Clark L, Rust BM, Tewari M, Noelle RJ, Dixit VM. Activation of the B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apoptosis. J Biol Chem. 1995;270:12343–12346. doi: 10.1074/jbc.270.21.12343. [DOI] [PubMed] [Google Scholar]
- Bulfone-Paus S, Ungureanu D, Pohl T, Lindner G, Paus R, Ruckert R, Krause H, Kunzendorf U. Interleukin-15 protects from lethal apoptosis in vivo. Nat Med. 1997;3:1124–1128. doi: 10.1038/nm1097-1124. [DOI] [PubMed] [Google Scholar]
- Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG, Sorensen PH, Thiele CJ, Kim SJ. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet. 1999;23:222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- Dear TN, Sanchez-Garcia I, Rabbitts TH. The HOX11 gene encodes a DNA-binding nuclear transcription factor belonging to a distinct family of homeobox genes. Proc Natl Acad Sci USA. 1993;90:4431–4435. doi: 10.1073/pnas.90.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Desiderio S. BAP-135, a target for Bruton’s tyrosine kinase in response to B cell receptor engagement. Proc Natl Acad Sci USA. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenkovic I. The L-selectin adhesion system. Curr Opin Hematol. 1995;2:68–75. doi: 10.1097/00062752-199502010-00010. [DOI] [PubMed] [Google Scholar]
- Meacci E, Tsai SC, Adamik R, Moss J, Vaughan M. Cytohesin-1, a cytosolic guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. The three faces of profilin. Cell. 1993;75:835–838. doi: 10.1016/0092-8674(93)90527-w. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Minami Y, Mine S, Hirano H, Hu CD, Fujimoto H, Fujii K, Saito K, Tsukada J, van Kooyk Y, Figdor CG, Kataoka T, Eto S. H-Ras signals to cytoskeletal machinery in induction of integrin-mediated adhesion of T cells. J Immunol. 1999;163:6209–6216. [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DE. The unfolding tale of PECAM-1. FEBS Lett. 2003;540:7–14. doi: 10.1016/s0014-5793(03)00224-2. [DOI] [PubMed] [Google Scholar]
- Thompson RD, Noble KE, Larbi KY, Dewar A, Duncan GS, Mak TW, Nourshargh S. Platelet-endothelial cell adhesion molecule-1 (PECAM-1)-deficient mice demonstrate a transient and cytokine-specific role for PECAM-1 in leukocyte migration through the perivascular basement membrane. Blood. 2001;97:1854–1860. doi: 10.1182/blood.v97.6.1854. [DOI] [PubMed] [Google Scholar]
- Glover DM, Hagan IM, Tavares AA. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature. 2003;424:1009–1013. doi: 10.1038/nature01720. [DOI] [PubMed] [Google Scholar]
- Goepfert TM, Brinkley BR. The centrosome-associated Aurora/Ipl-like kinase family. Curr Top Dev Biol. 2000;49:331–342. doi: 10.1016/s0070-2153(99)49016-7. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrieu A, Kahana JA, Wood KW, Cleveland DW. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Wood LJ, Mukherjee M, Dolde CE, Xu Y, Maher JF, Bunton TE, Williams JB, Resar LM. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol. 2000;20:5490–5502. doi: 10.1128/mcb.20.15.5490-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A). J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Doree M, Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner? J Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- Hudson JW, Chen L, Fode C, Binkert C, Dennis JW. Sak kinase gene structure and transcriptional regulation. Gene. 2000;241:65–73. doi: 10.1016/s0378-1119(99)00467-9. [DOI] [PubMed] [Google Scholar]
- Deng X, Gao F, May WS., Jr Bcl2 retards G1/S cell cycle transition by regulating intracellular ROS. Blood. 2003;102:3179–3185. doi: 10.1182/blood-2003-04-1027. [DOI] [PubMed] [Google Scholar]
- O’Reilly LA, Harris AW, Tarlinton DM, Corcoran LM, Strasser A. Expression of a bcl-2 transgene reduces proliferation and slows turnover of developing B lymphocytes in vivo. J Immunol. 1997;159:2301–2311. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.