Abstract
Complete or partial genome sequences have recently become available for several medically and evolutionarily important parasitic protozoa. Through the application of bioinformatics complete metabolic repertoires for these parasites can be predicted. For experimentally intractable parasites insight provided by metabolic maps generated in silico has been startling. At its more extreme end, such bioinformatics reckoning facilitated the discovery in some parasites of mitochondria remodelled beyond previous recognition, and the identification of a non-photosynthetic chloroplast relic in malarial parasites. However, for experimentally tractable parasites, mapping of the general metabolic terrain is only a first step in understanding how the parasite modulates its streamlined, yet still often puzzlingly complex, metabolism in order to complete life cycles within host, vector, or environment. This review provides a comparative overview and discussion of metabolic strategies used by several different parasitic protozoa in order to subvert and survive host defences, and illustrates how genomic data contribute to the elucidation of parasite metabolism.
Keywords: parasitic protozoa, host–pathogen interaction, comparative genomics, Trypanosoma, Leishmania, Apicomplexa
1. Introduction
A fascinating adaptive evolution shapes the metabolism of parasites. In a classic paradigm this results in the evolution of novel pathways that are integral for parasite subversion of host defences. Yet the availability within the host of an assortment of metabolites—small organic molecules that provide substrates for energy generation or building blocks for assembling macromolecular structures—also means that parasites often dispense completely with various core pathways of metabolism that are present in many other organisms. Not unsurprisingly, the abandonment of some pathways is a driving force in the evolution of obligate, as opposed to opportunistic, parasitism (e.g. Borza et al. 2005). While the study of such comparative biochemistry is interesting in its own right, there is another important aspect to the study of parasite metabolism. A glance at recent statistics released by the World Health Organization reveals that infectious diseases caused by parasites remain major causes of mortality. Sadly, much of the disease burden occurs in the developing world, within countries that are perhaps the least well equipped to bring new medicines to the clinic. Parasitic diseases are also relevant to veterinary medicine. Problems associated with existing drugs, including efficacy, delivery, toxicity and resistance, mean that there is often an urgent need to develop new chemotherapeutic or prophylactic treatments for diseases caused by parasites.
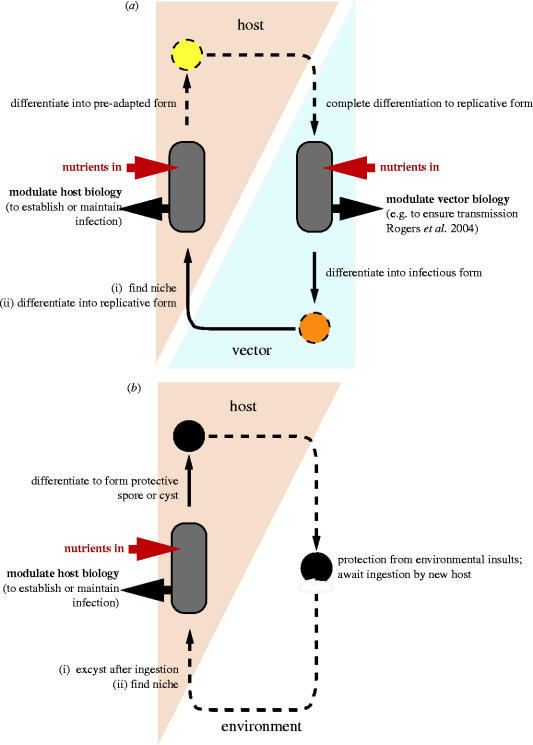
The aim in this review is to provide a comparative overview, which illustrates how the metabolic strategies employed by a range of different parasitic protozoa, including several human pathogens (table 1), is influenced by the environmental niche (or niches) occupied by each parasite. Many of the parasites included in the discussion replicate within more than one niche; others—the enteric parasites and Toxoplasma gondii—survive outside of the animal host in a stress-resistant encysted form (figure 1). The digenetic parasites considered here include a number which are transmitted between mammalian hosts by blood-feeding insect vectors. Each of these parasites undergoes a complex developmental cycle within its vector. The parasites are as follows. (i) The malarial parasite Plasmodium falciparum, which is transmitted between hosts by its mosquito vector. (ii) Trypanosoma brucei, which causes sleeping sickness in man, N'gana in cattle, and is transmitted between hosts by tsetse flies. (iii) T. cruzi, which is able to replicate within the cytoplasm of a number of host cells, most notably from the standpoint of pathology cardiac muscle cells. It is transmitted between hosts by a triatomine insect vector (commonly Rhodnius prolixus). (iv) Pathogenic Leishmania species, which replicate within host macrophages and are transmitted between hosts by female sand flies.
Table 1.
Statistics relating to important human diseases caused by some parasitic protozoa.
| parasite | disease | geographical distribution | disease incidence and mortality in 2002a |
|---|---|---|---|
| trypanosomatids | |||
| Trypanosoma brucei | African sleeping sickness | East and West Africa | 60 million at risk, 48 000 deaths |
| Trypanosoma cruzi | Chagas' disease | South and Central America | 100 million at risk, 18 million cases, 14 000 deaths |
| Leishmania species | Leishmaniasis | tropics and sub-tropics | 350 million at risk, 12 million cases, 51 000 deaths |
| apicomplexans | |||
| Plasmodium falciparum | malaria (including cerebral malaria) | sub-Saharan Africa, southeast Asia and the Americas | 2400 million at risk, 1.27 million deaths |
| Toxoplasma gondii | toxoplasmosis | global | see belowb |
| other parasitic protozoa | |||
| Giardia | giardiasis (includes severe diarrhoea) | global (prevalent in hot countries) | total deaths from all diarrhoeal diseases: 1.8 million (many in tropical countries) |
| Entamoeba histolytica | amoebic dysentery | global (prevalent in hot countries) | |
| comparisons with other pathogens | |||
| HIV | AIDS | global | 2.78 million deaths |
| Mycobacterium tuberculosis | tuberculosis | global | 1.57 million deaths |
Figures taken from World Heath Organization statistics for 2002.
Rarely causes disease in healthy individuals, although approximately one-third of humanity has been exposed. However, infection does cause severe neurological disorders in congenitally infected children and severe pathology in immunodeficient individuals.
Figure 1.
Complex interactions can occur between protozoan parasites and (a) host and vector or (b) host and environment. aRogers et al. (2004).
Adaptability to different niches requires an ability to sense environmental change as a cue for differentiation into a new morphological form, and, although not considered further here, metabolism plays its part here too, because it is often the threshold concentration of a low molecular weight metabolite that contributes toward a commitment to differentiate (Lujan et al. 1996; Billker et al. 1998; Engstler & Boshart 2004). In order to set the scene for the comparative discussion of niche-specific metabolism that is provided in §4, §2 is used to state some of the problems associated with studying metabolism in parasitic protozoa; and in §3, the unusual metabolic compartmentalization exhibited by some parasitic protozoa, which can arise as a consequence of metabolic streamlining within the niche environment, is summarized.
2. Interrogating the biology of experimentally refractory parasites
One limitation to the interrogation of any parasite's metabolism is an ability to replicate experimentally the environmental niches in which that parasite has evolved. Unfortunately, for many parasitic protozoa, this is not possible using experimentally tractable cultured parasites. Even with the advent of stable DNA transfection and the availability of reverse and forward genetics approaches to study gene function in some parasitic protozoa, our understanding of their metabolism is often limited by a necessity to maintain cultured parasites in complex semi-defined media containing a foetal calf serum supplement.
More significantly, some parasitic protozoa replicate inside host cells (e.g. Leishmania amastigotes or intraerythrocytic P. falciparum). Here, host cell metabolism can lead to ambiguities in the interpretation of labelling patterns arising from the use of isotopically enriched metabolites to study the chemical reactions in a given metabolic pathway. The presence of a host cell can also provide additional complications for purification or fractionation of an intracellular parasite. Yet the arrival of several complete genome sequences has recently unlocked the safe in which the metabolic secrets of many parasitic protozoa were previously hidden (e.g. Katinka et al. 2001; Gardner et al. 2002; Abrahamsen et al. 2004; Xu et al. 2004). In part, new insight into parasite metabolism comes from using comparative genomics approaches to produce hypothetical metabolic maps, but even a limited amount of (impure) parasite biomass can often be sufficient for proteomic analysis using modern mass spectrometry approaches. In this way, the temporal expression of many proteins during a parasite life cycle can be determined. A prime example of the metabolic output from this type of systems biology-based approach is the identification of distinct metabolic strategies that are used by malaria parasites in the mammalian host and mosquito vector, respectively (Hall et al. 2005; discussed further in §4).
3. Refining and redefining the cellular organization of a streamlined metabolism
A defining characteristic of eukaryotes, distinguishing them from bacteria and Archaea, is the compartmentalization of the chromosomes (DNA) within a nucleus. Eukaryotes also compartmentalize their metabolism, dividing catabolic (energy-generating) and anabolic (biosynthetic) pathways between the cytoplasm and multiple membrane-enclosed organelles. In consuming fuels, such as glucose, cells generate ATP—the main cellular energetic currency—through substrate-level phosphorylation using various catabolic intermediates. Glucose catabolism (glycolysis) in most cells is a cytoplasmic pathway that generates a small net gain of ATP, but in aerobic organisms mitochondria are the cellular powerhouses that oxidize carbon substrates completely to CO2, using the electrons acquired from such catabolic reactions to generate much larger amounts of energy in a process called oxidative phosphorylation. In plants and algae, photosynthetic fixation of CO2 into carbohydrates occurs in chloroplasts.
Large evolutionary distances separate free-living and parasitic protozoa from fungal, metazoan and plant lineages, concealing an intriguing evolutionary trail of gene acquisition and gene loss, which results in many extant protozoa, presenting an unusual organellar organization to their metabolism (table 2).
Table 2.
Organization of organellar metabolism in parasites discussed in this review. (+ denotes the presence of an organelle, − denotes the absence.)
| organism | aerobic mitochondrion | hydrogenosomes | mitosomes | peroxisomes | plastid relic |
|---|---|---|---|---|---|
| trypanosomatids | |||||
| T. brucei | + | − | − | + | − |
| T. cruzi | + | − | − | + | − |
| Leishmania | + | − | − | + | − |
| apicomplexans | |||||
| P. falciparum | + | − | − | − | + |
| T. gondii | + | − | − | − | + |
| Eimeria tenella | + | − | − | − | + |
| Cryptosporidium | − | − | + | − | − |
| other parasites | |||||
| Giardia | − | − | + | − | − |
| E. histolytica | − | − | + | − | − |
| Trichomonas | − | + | − | − | − |
(a) Remodelling mitochondrial function
There is now a prevailing consensus that all extant eukaryotes probably contain an organelle(s) of mitochondrial ancestry (Mai et al. 1999; Tovar et al. 1999, 2003; Dyall et al. 2000; Katinka et al. 2001; van der Giezen et al. 2002, 2003; Williams et al. 2002; Hrdy et al. 2004; Putignani et al. 2004). In Plasmodium, Toxoplasma, many other apicomplexan parasites and most trypanosomatids aerobic mitochondria with a capacity for oxidative phosphorylation are present. In the microaerophile Trichomonas vaginalis, capacity for oxidative phosphorylation is not present (Cerkasov et al. 1978; Muller & Lindmark 1978). However, as with some ciliates and some fungi, there are mitochondrial-related organelles in Trichomonas that function anaerobically, generating hydrogen gas and, through substrate-level phosphorylation, energy (Lindmark & Muller 1973; Cerkasov et al. 1978; Muller & Lindmark 1978). These organelles are called hydrogenosomes; the morphology of some ciliate hydrogenosomes was shown some years ago to resemble that of aerobic mitochondria (Finlay & Fenchel 1989). Subsequent molecular investigations provided confirmation of the mitochondrial ancestry of hydrogenosomes (Embley et al. 2003; Hrdy et al. 2004). The complete or extensive genome coverage that is available for Cryptosporidium, Giardia, the amoebozoan Entamoeba histolytica and the microsporidian parasite Encephalitozoon cuniculi indicates that a capacity for organellar energy generation has probably been lost completely (Katinka et al. 2001; Abrahamsen et al. 2004; Xu et al. 2004; Loftus et al. 2005a). Here, a mitochondrial relic remains for the purpose of Fe–S cluster assembly (Tovar et al. 2003), an ancient and ubiquitous biosynthetic pathway that occurs universally in mitochondria (Lill & Kispal 2000) and is also found in trichomonad hydrogenosomes (Sutak et al. 2004).
(b) Peroxisome loss in parasitic protozoa
While refinement and modification of mitochondrial function appears to be commonplace among microbial eukaryotes, microbodies of the peroxisome class provide an example of organelles that have actually been lost from microsporidian parasites, and probably numerous protozoan lineages too. The evidence for this comes not only from cell cytology, but also from the absence of a conserved cohort of peroxisome biogenesis proteins from the complete, or almost complete, genome sequences of apicomplexan parasites and the microsporidian E. cuniculi too (Ding et al. 2000; Katinka et al. 2001; Gardner et al. 2002; Abrahamsen et al. 2004; Xu et al. 2004). Microbodies of the peroxisome class come in several guises. In addition to canonical peroxisomes, there are also glyoxysomes and glycosomes; all appear to share a common role in lipid metabolism. β-Oxidation of fatty acids is a ubiquitous peroxisomal pathway, and in mammals and trypanosomatids, which incorporate ether phospholipids into their membranes, some steps of ether lipid biosynthesis are peroxisomal too (Michels et al. 2000; Zufferey et al. 2003). Specialized peroxisomes called glyoxysomes are the intracellular site for the glyoxylate shunt, a pathway used by some plants and micro-organisms when the only available carbon sources are acetyl-CoA (a C2 compound) precursors. In bypassing the decarboxylation steps of the Krebs cycle, acetyl-CoA can be used for the net synthesis of malate without draining the Krebs cycle of its key intermediates. The malate then provides a carbon precursor for anabolic pathways such as gluconeogenesis that become essential within an environment of nutritional deprivation. The glyoxylate shunt is not found in mammals.
Trypanosomatid parasites possess peroxisomes, albeit with unique biochemical properties (discussed further in §4). Thus, assuming that the phylum Euglenozoa, within which trypanosomatids are grouped, retains a deep-branching position in eukaryotic phylogeny (Baldauf 2003), then peroxisomes arose early in evolution. It is perhaps harder to determine whether Giardia and Trichomonas ever had peroxisomes because it is difficult to be certain of where, within the deeper branches of eukaryotic evolution, Giardia and Trichomonas diverged. However, in the examples of apicomplexan and microsporidian parasites and E. histolytica we can be confident that loss has occurred. Not only did these organisms appear later in evolution, after the divergence of the Euglenozoa, but in the example of the microsporidians these parasites share a close phylogenetic relationship with fungi; they either evolved from the fungi or are a sister group to the fungi (Keeling & Fast 2002). Fungi contain peroxisomes. Thus, the parsimonious explanation is that in streamlining their biology microsporidian parasites lost peroxisomes. Similarly, apicomplexans belong to the Alveolata (Baldauf 2003), where peroxisomes are retained by free-living members of the group (e.g. ciliates). If we consider why peroxisomes were lost from Entamoeba, the Apicomplexa, the Microsporidia, and probably Giardia and Trichomonas too, then a common feature of all these parasites' biology is a limited capability for lipid metabolism. Such limitations include an inability to use fatty acids as energy sources and little capacity to synthesize lipids or modify host-derived lipids (Katinka et al. 2001; Das et al. 2002), and none of the fore-mentioned parasites possess a classical glyoxylate cycle.
(c) Plastid relics
None of the pathogenic parasitic protozoa are photosynthetic, but the similarity of a 35 kb circular DNA element from Plasmodium and Toxoplasma to chloroplast genome sequences, and subsequent localization of the Toxoplasma element to an organelle surrounded by four membranes (Kohler et al. 1997) indicated the progenitor of the Apicomplexa acquired a photosynthetic plastid following endosymbiosis with an algal cell (Wilson et al. 1996). After millions of years, the only structural evidence from this encounter is a plastid-derived organelle surrounded by its own double membrane, a third membrane corresponding to the endosymbiont's plasma membrane and an outermost membrane derived from the food vacuole, in which the endosymbiont resided (Waller et al. 2000).
The presence in various Apicomplexa of a structure containing multiple membranes had been documented previously, but the revelation that the organelle's secret identity was a plastid relic, complete with its own genome (Kohler et al. 1997), heralded a determined effort to define function for an organelle that was subsequently shown to be essential for parasite development (Fichera & Roos 1997; McConkey et al. 1997).
The annotation of the plastid genome provided no direct clues to the metabolic function of the organelle, now known as the apicoplast. However, using the metabolic properties of photosynthetic plastids functioning in the dark as a template it was possible to identify in the P. falciparum genome the presence of nuclear genes encoding apicoplast-targeted enzymes, including components of the apicoplast-localized fatty acid and isoprenoid biosynthetic pathways (Waller et al. 1998; Jomaa et al. 1999). Subsequently, knowledge gathered on how apicoplast-targeted proteins reach their destination meant that, rather than look for apicoplast-targeted malaria proteins by comparison with a chloroplast functioning without illumination, it was realistic to mine the Plasmodium genome for proteins with a probable apicoplast targeting sequence (Foth et al. 2003). In this way, a picture has emerged of how the apicoplast functions as an anabolic organelle, probably autonomous in its ability to synthesize fatty acids and the isopentenyl diphosphate and dimethylallyl diphosphate intermediates of isoprenoid biosynthesis, following the import of appropriate carbon precursors (Ralph et al. 2004).
Prior to the initial characterization of apicoplast-targeted acyl carrier protein (ACP), β-ketoacyl-ACP synthase and β-hydroxyacyl-ACP dehydratase (Waller et al. 1998), Plasmodium had been considered to be a fatty acid auxotroph. Certainly, during asexual development in the infected erythrocyte Plasmodium does indeed acquire from the host significant amounts of phospholipid precursors (fatty acids and polar head groups) for incorporation into its own membranes and the membrane system it elaborates within the cytoplasm of the infected erythrocyte (Vial et al. 1990). In truth, the identity of the essential product(s) of the apicoplast type II fatty acid biosynthetic pathway has not yet been resolved. On the other hand, the expeditious identification of 2-deoxy-d-xylulose-5-phosphate (DXP) synthase and DXP reductoisomerase in P. falciparum (Jomaa et al. 1999) was only possible because of genetic dissection in Escherichia coli and plants of a biosynthetic isoprenoid pathway first documented in a series of milestone publications during the mid-1990s (Rohmer et al. 1993, 1996; Schwender et al. 1996). Thus, the example of the apicoplast provides an exquisite example and a paradigm for how genome sequencing provides a powerful tool for ascribing function to organelles that are otherwise refractory to experimental study. A similar in silico study of a mitochondrial relic provided the first indication of the metabolic functions that remain when the capacity for energy generation is lost from mitochondria (Katinka et al. 2001). The presence of the DXP pathway also highlights the dependence of Plasmodium on its apicoplast, because the acquisition of the pathway, along with the algal endosymbiont, probably facilitated loss of a cytoplasmic mevalonate pathway for isopentenyl diphosphate biosynthesis that operates in most eukaryotes (Smit & Mushegian 2000).
Finally, although there is no plastid relic in the trypanosomatids, this parasite family retains numerous plant-like traits of uncertain origin (Hannaert et al. 2003; Martin & Borst 2003). Just as was the case with the study of the evolution of the Apicomplexa, the continued characterization of such possible traits (Wilkinson et al. 2002; Allen et al. 2004; Vickers et al. 2004) continues to point towards a possibility that the unusual organellar metabolism of extant trypanosomatids has been shaped through contact and lateral gene transfer with a photosynthetic or secondarily non-photosynthetic microbe.
4. Comparative metabolism: survival within a niche
(a) Survival in a microaerobic environment
Section 3 began to illustrate how a parasite's organellar composition (e.g. classification of its mitochondrial-derived organelle, presence or absence of peroxisomes) can reflect external conditions within its niche environment, or a specific requirement for essential anabolic pathways (e.g. apicoplast). Yet, this is only part of the story. For parasitic protozoa in the gut or reproductive tract (Giardia, Cryptosporidium, E. histolytica, Trichomonas), reduced oxygen availability means that anaerobic fermentation of glucose and amino acid carbon sources, and not oxidative phosphorylation, is the strategy used for energy generation (Lockwood & Coombs 1991; Edwards et al. 1992; Schofield et al. 1992; Coombs & Muller 1995; North & Lockwood 1995; Zuo et al. 1995; Yarlett et al. 1996). Even here, however, specialized biochemical twists maximize the ATP yield from what could be considered an inefficient catabolism of fuel sources. For instance, in glycolysis, which provides the major energy generating pathway, the phosphofructokinase from several enteric (intestinal) parasites is pyrophosphate (PPi) rather than ATP dependent (Denton et al. 1996; Muller et al. 2001). In Giardia, E. histolytica, and probably Cryptosporidium too, the glycolytic end-product, pyruvate, can be metabolized to acetyl-CoA, which is then the substrate for acetyl-CoA synthetase. This enzyme belongs to the same super family as succinyl-CoA synthetase from the Krebs cycle and, just like succinyl-CoA synthetase, generates one additional ATP molecule in breaking the thioester bond of the acyl-CoA (acetyl-CoA in Giardia and E. histolytica) to liberate a free organic acid (acetate in the cases of Giardia and E. histolytica; Sanchez et al. 2000). Finally, the genome sequence of phagotrophic E. histolytica reveals this parasite's metabolism has been sculpted by lateral gene transfer (Loftus et al. 2005a). Through the acquisition of various bacterial enzymes, this amoebal parasite has expanded the range of carbohydrates and amino acids that can be used as carbon sources for anaerobic fermentation.
(b) A comparison between malarial parasites and trypanosomes
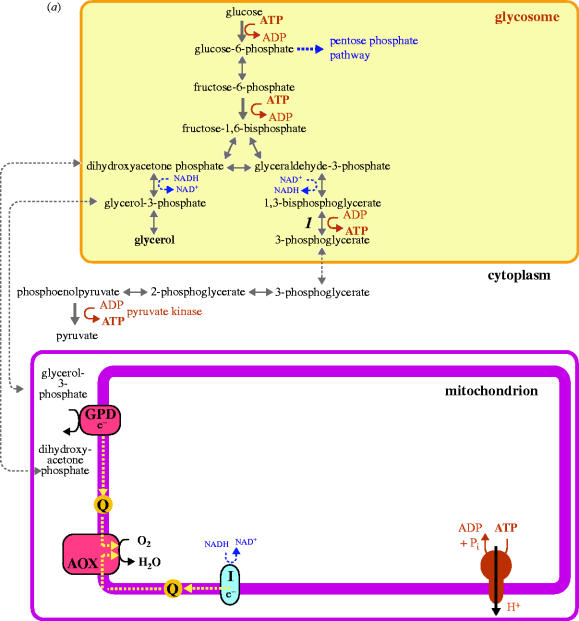
In contrast to intestinal parasites and Trichomonas, the African trypanosome T. brucei and malarial parasites reside in the bloodstream within an aerobic environment, but still use a strategy of ‘glycolysis only’ (i.e. without oxidative phosphorylation) as the sole (T. brucei) or predominant pathway (malarial parasites) for energy generation. In bloodstream trypanosomes glucose catabolism is aerobic, requiring an oxygen-dependent alternative terminal oxidase for regeneration of the NAD+ reduced during glycolysis, and pyruvate is the end-product of metabolism (figure 2a; Moyersoen et al. 2004). In P. falciparum, the formation of lactate as the end-product of anaerobic glycolysis maintains NAD+/NADH balance (Vander Jagt et al. 1990). Yet in both parasites, the strategy of glucose fermentation is fine because these parasites replicate in an environment where glucose availability is normally not limiting. There is, however, a unique twist to trypanosomatid glycolysis, because these parasites compartmentalize many of their glycolytic enzymes behind the peroxisomal membrane, giving rise to the classification of these organelles as glycosomes (Moyersoen et al. 2004).
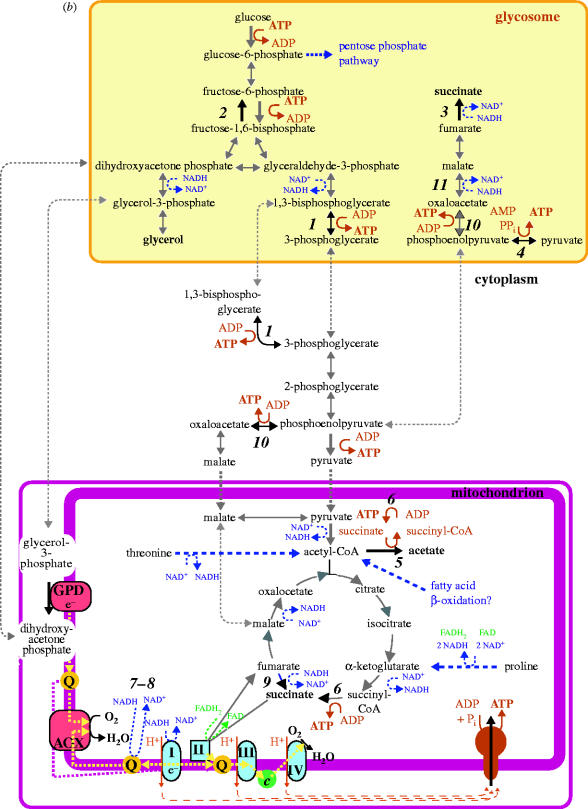
Figure 2.
A comparison of energy metabolism in (a) bloodstream form T. brucei and (b) procyclic (insect form) T. brucei. (a) Bloodstream T. brucei, although an obligate aerobe, produces a net gain in ATP only through the cytoplasmic substrate-level phosphorylation catalysed by pyruvate kinase. The figure shows the compartmentalization of glycolysis between glycosomes and cytoplasm; the mitochondrion does not contribute directly to ATP production. ATP synthase operates in the direction of ATP hydrolysis to generate a proton motive force. A partial respiratory chain complex I enzyme (NADH : ubiquinone oxidoreductase, denoted I in the figure) oxidizes NADH generated in the mitochondrial matrix, but does not contribute to proton-pumping across the inner mitochondrial membrane. There is no classical respiratory chain; the terminal oxidase is alternative oxidase (AOX). Intraglycosomal redox balance is maintained using a glycerol-3-phosphate dehydrogenase (GPD) shuttle. The additional enzyme in this scheme is 1, phosphoglycerate kinase. Other abbreviation: Q, ubiquinone. (b) In procyclic trypanosomes, the organization and compartmentalization of energy metabolism is more complex. Enzymes discussed in the text and additional to those listed in (a) are as follows: 2, fructose-1,6-bisphosphatase; 3, glycosomal fumarate reductase; 4, pyruvate phosphate dikinase; 5, acetate : succinate CoA transferase; 6, succinyl-CoA synthetase; 7–8, mitochondrial NADH dehydrogenases distinct from rotenone-sensitive complex I; 9, mitochondrial fumarate reductase; 10, phosphoenolpyruvate carboxykinase; 11, glycosomal malate dehydrogenase. Other abbreviations: II, succinate dehydrogenase (complex II); III, cytochrome c reductase (complex III); IV, cytochrome c oxidase (complex IV); c, cytochrome c. It is not known whether mitochondrial oxidation of fatty acids (by far the most efficient substrate in terms of amount of ATP that can be generated per carbon molecule oxidized to CO2) contributes to energy generation in African trypanosomes. End-products of energy metabolism are highlighted in bold.
In bloodstream T. brucei, approximately 90% of the peroxisomal, or glycosomal, matrix proteins are glycolytic enzymes. It is not known how or why glycolytic enzymes came to be compartmentalized behind peroxisomal membranes in T. brucei and the other trypanosomatids, but it is a phenomenon that is not likely to be readily reversed. Such compartmentalization is not required to achieve a high rate of glycolysis (Moyersoen et al. 2004). Rather, it reflects an absence of regulatory control on the two ATP-consuming steps at the head of the glycolytic pathway, catalysed by hexokinase and phosphofructokinase, respectively. Originally, computer modelling of trypanosome glycolysis predicted that redistribution of hexokinase and phosphofructokinase to the cytoplasm would cause a rapid, irreversible depletion of cytosolic ATP and concomitant build-up of phosphorylated sugars (Bakker et al. 2000). Subsequently, this prediction was supported experimentally when it was shown that relocation of glycolytic enzymes to the cytoplasm, by preventing the expression of components of the glycosomal protein import machinery, is lethal (Furuya et al. 2002; Guerra-Giraldez et al. 2002). In procyclic (insect form) T. brucei, which can use multiple carbon sources for energy production (see below), this lethal phenotype is ameliorated if the parasite is grown in the absence of external glucose (Furuya et al. 2002). Death occurs because the activities of hexokinase and phosphofructokinase are not allosterically regulated by a build-up of either products from the reactions catalysed by these enzymes or other glycolytic intermediates.
Comparisons between small molecule permeability and the transporter properties of peroxisomal membranes in other organisms suggest that the glycosomal matrix is probably a closed compartment with respect to ATP/ADP exchange with the cytoplasm. Compartmentalization of the ATP-consuming steps of glycolysis and an ATP-regenerating step, catalysed by phosphoglycerate kinase (Blattner et al. 1998; figure 2a), therefore, provides bloodstream T. brucei with a mechanism to not only maintain ATP/ADP homeostasis, facilitating a continuous glycolytic flux, but also prevents the ‘uncontrollable’ hexokinase and phosphofructokinase enzymes consuming ATP required for other cellular processes.
In trypanosomes and Plasmodium, is the reliance on glycolysis a stage-specific metabolic strategy that reflects the availability of nutrients in the external environment? The answer is probably yes. Although not particularly amenable to large-scale biochemical studies, all the major developmental forms of Plasmodium berghei, which causes malaria in rodents, can be obtained in culture or from infected mosquitoes in quantities sufficient for proteomic analysis. A recent systems biology-based comparison of the P. berghei proteome at different points in the life cycle revealed that ookinetes, which traverse across the mid gut epithelium of the mosquito, exhibit increased expression of proteins required for a mitochondrial respiratory chain. This expression profile is supported by ultra-structural changes to mitochondrial morphology that occur during the malarial parasite life cycle—ookinetes, but not asexual blood stages, have well-developed cristate mitochondria (Sinden 1978). Moreover, it suggests a metabolic switch to oxidative phosphorylation during development in the mosquito (Hall et al. 2005), because either alternative carbon sources are available or the parasite needs to have a more efficient metabolism at this point in its life cycle.
In contrast to malarial parasites, the replicative forms of T. brucei that are found in the mid gut of its tsetse fly vector can be readily grown in culture. This means biochemical studies are possible. Procyclic (tsetse form) T. brucei relies on mitochondrial energy generation for its viability (Bochud-Allemann & Schneider 2002; van Weelden et al. 2003). For this parasite, the steady release over the last few years of sequence information from the T. brucei genome project has accelerated progress in our understanding of its metabolism, but a complex, perhaps murky picture (figure 2b) is emerging.
In addition to an apparently characteristic aerobic metabolism, procyclic trypanosomes express a cohort of unusual enzymes, some of which are more characteristic of anaerobic metabolism (e.g. Riviere et al. 2004; Coustou et al. 2005; figure 2b and discussed below). Glycolysis can be used for energy generation by cultured parasites, but in contrast to bloodstream trypanosomes there is a cytoplasmic phosphoglycerate kinase (Misset & Opperdoes 1987), which means the parasite needs multiple pathways in order to maintain a balance of ATP and ADP concentrations inside the glycosomes. The glycerol-3-phosphate shuttle, which is essential in bloodstream T. brucei (figure 2a), is again operative, but there is a second route for maintaining glycosomal NAD+/NADH homeostasis (Besteiro et al. 2002). This pathway utilizes NADH-dependent fumarate reductase to generate succinate from the phosphoenolpyruvate intermediate of glycolysis (figure 2b). Intriguingly, this fumarate reductase activity is present on a modular multifunctional protein, which also has NADH-dependent cytochrome c reductase activity and an N-terminal ApbE protein-like domain, but the functional significance of these additional domains is not known (Besteiro et al. 2002). There is also pyruvate phosphate dikinase, a glycosomal protein, for which the functional role is not known (Bringaud et al. 1998; Coustou et al. 2003). It catalyses reversible formation of pyruvate from phosphoenolpyruvate, and requires one molecule of AMP and two of PPi to generate ATP when operating in the direction of pyruvate formation.
Although glucose is a staple constituent of the medium normally used to cultivate procyclic trypanosomes and is the energy substrate of choice for these cultured parasites (Coustou et al. 2003; Lamour et al. 2005), sugars are not thought to be readily available in the digestive tract of the tsetse fly. The glucose content of the blood meal is depleted rapidly following ingestion, and tsetse flies, unlike many other invertebrate vectors, do not acquire nutrients by feeding on sugar-rich plant sap. The discovery that procyclic trypanosomes express a high-affinity glucose transporter, rather than the low-affinity, high-capacity transporter expressed within the glucose-rich environment of the mammalian bloodstream, is consistent with a view that glucose availability is low within the tsetse environment (Tetaud et al. 1997; Vedrenne et al. 2000). The shift towards mitochondrial energy generation in procyclic T. brucei can, therefore, be considered to reflect a predominance of peptides and amino acids, liberated by digestion of the blood meal, as the primary carbon sources available for ATP production.
Proline, which is also the fuel oxidized by the tsetse in order to generate the energy needed for flight, and threonine are two of the amino acids most readily catabolized by procyclic T. brucei (Coustou et al. 2003). In the presence of glucose, the end-products of proline catabolism are equimolar amounts of CO2 and succinate (van Weelden et al. 2003, 2005), and the enzymes required for this catabolism result in the generation of ATP, through substrate-level phosphorylation, and NADH and FADH2, which both provide electrons for the respiratory chain (van Weelden et al. 2005; figure 2b). Threonine, on the other hand, is metabolized via the intermediacy of acetyl-CoA to acetate (Linstead et al. 1977; van Weelden et al. 2005). Acetate production requires the transfer of CoA to succinate, and is catalysed by acetate : succinate CoA-transferase (ASCT; Van Hellemond et al. 1998a). The succinyl-CoA is recycled to succinate by succinyl-CoA synthetase, generating one ATP for every molecule of acetyl-CoA metabolized by ASCT (Bochud-Allemann & Schneider 2002). Although the trypanosome ASCT exhibits a high degree of sequence identity and similarity to mammalian succinyl-CoA : 3-ketoacid-CoA transferases (Riviere et al. 2004), the ASCT reaction has only previously been found in the anaerobic mitochondria of some marine invertebrates and parasitic helminths and in hydrogenosomes (van Hellemond et al. 2003). The occurrence of an ASCT activity within the solely aerobic mitochondria of T. brucei, and other trypanosomatids too, is therefore somewhat surprising. What is perhaps more surprising however is that, at least in procyclic T. brucei, acetate is an end-product of energy metabolism because, despite the presence of a complete cohort of Krebs cycle enzymes (with either predicted or experimentally confirmed mitochondrial locations), no culture condition has been found in which acetyl-CoA enters the Krebs cycle to be completely oxidized to CO2 (van Weelden et al. 2003, 2005). This observation does not of course preclude a role for the Krebs cycle in T. brucei energy generation at some point during the transmission cycle through tsetse (van Weelden et al. 2003), but it highlights a conundrum for which an experimental explanation is required—what is the true functional role(s) of the Krebs cycle enzymes in T. brucei?
In the absence of glucose, procyclic trypanosomes are, not surprisingly, critically dependent upon oxidative phosphorylation for energy generation (Lamour et al. 2005). The trypanosome electron transport chain is branched at its terminal point; alternative oxidase and cytochrome c oxidase can be used to reduce O2, and electrons from mitochondrial NADH can enter into the respiratory chain by multiple routes. A rotenone-sensitive NADH : ubiquinone oxidoreductase (complex I) and two alternative rotenone-insensitive NADH dehydrogenases, which do not combine reduction of ubiquinone with proton-pumping across the inner mitochondrial membrane, are present (Fang et al. 2001; Fang & Beattie 2002, 2003). One of these alternative enzymes is orthologous to the NADH dehydrogenases found in Saccharomyces cerevisiae, which lacks rotenone-sensitive complex I, and certain other fungi. The individual contributions that complex I and the alternative NADH dehydrogenases provide to the mitochondrial metabolism of T. brucei is unclear. Unexpectedly, a soluble mitochondrial fumarate reductase has recently been identified as a further enzyme able to oxidize NADH (Coustou et al. 2005). However, unlike the parasitic helminths and marine invertebrates that use fumarate instead of O2 as a terminal electron acceptor, reduction of mitochondrial fumarate in trypanosomes is not linked to the respiratory chain. In helminths and marine invertebrates, fumarate reductase is a membrane-bound enzyme, considered to be evolutionarily related to the Krebs cycle enzyme succinate dehydrogenase (van Hellemond et al. 2003). The NADH-dependent trypanosome enzyme, by contrast, is soluble and exhibits extensive structural and sequence homology with the glycosomal fumarate reductase (Coustou et al. 2005), indicating that the enzymes from both sub-cellular compartments share an evolutionary ancestry. Thus, it appears that fumarate reduction in anaerobic mitochondria and trypanosomes is an example of convergent evolution. However, cultured procyclic trypanosomes (van Weelden et al. 2003), and indeed other trypanosomatids, lack the capacity for anaerobic growth, yet the ability to use fumarate as a mitochondrial electron sink is fundamentally an anaerobic trait. Thus, although trypanosomes can only multiply under aerobic conditions one can speculate that mechanisms are retained for maintaining redox balance during putative transient hypoxia during tsetse transmission.
Trypanosoma brucei also retain an ability to synthesize carbohydrates, as evidenced by (i) the presence in the genome of the key gluconeogenic enzyme fructose-1,6-bisphosphatase (Morris et al. 2002; Hannaert et al. 2003) and (ii) labelling studies using 14C-labelled proline, which indicated that in the absence of glucose procyclic T. brucei is likely to metabolize proline-derived succinate to produce the gluconeogenic precursor phosphenolpyruvate (van Weelden et al. 2005). Although oxidative phosphorylation is likely to be sufficient to satisfy the cellular energy demand, glucose and other sugars are required for nucleic acid and glycoconjugate synthesis and for the glycosylation of proteins exported to the procyclic cell surface. Thus, the available genomic and biochemical evidence are consistent with a widely held view that glucose is not likely to be available in significant quantities in the tsetse mid gut. This assertion suggests that within the context of the natural life cycle glycolytic enzymes, and potentially the other enzymes of carbohydrate metabolism too, are required by procyclic T. brucei for purposes other than energy generation. The presence of fructose-1,6-bisphosphatase in T. brucei also provides a contrast with malarial parasites, since the latter lack this key gluconeogenic enzyme (Gardner et al. 2002).
There is uncertainty with regard to the identity of the carbon sources available to malarial ookinetes and sporulating oocysts in the mosquito (Lang-Unnasch & Murphy 1998). Glucose is considered to be present only in low quantities, and there is evidence that glucose availability is likely to be an important regulator of gene expression in P. falciparum (Fang et al. 2004a; Fang et al. 2004b). However, the absence of fructose-1, 6-bisphosphatase from malarial parasites is informative, because it indicates that carbohydrates are available in the mosquito and, moreover, that this availability is sufficient for Plasmodium to complete its life cycle. This is a conclusion that is also consistent with an old observation that sugar concentrations are lower in the haemolymph of Plasmodium-infected mosquitoes than in uninfected mosquitoes (Mack et al. 1979).
(c) Wider comparisons between metabolic strategies in evolutionarily related parasites
Although it is possible to draw parallels between the metabolic strategies employed by African trypanosomes and malarial parasites, these parasites do not share a recent evolutionary ancestry. Both, however, have parasitic relations.
(i) Comparative metabolism in the Trypanosomatidae
The trypanosomatid family provides an excellent vehicle for comparative studies of parasite biochemistry. This is not least because they tend to be experimentally tractable, but also because there is the availability of genome sequences for several different trypanosomatids. In addition, the parsimonious view is that the ancestor of extant trypanosomatids was itself a parasitic organism, probably a parasite of insects. While many trypanosomatids are still parasites only of insects, digenetic parasitism has arisen multiple times during trypanosomatid evolution (Fernandes et al. 1993). No free-living trypanosomatid species has ever been described, although this is not surprising given that the reacquisition of traits which would facilitate free-living growth and replication in the environment is always unlikely in the face of extensive metabolic streamlining and remodelling that can occur during an adaptation to parasitism.
It is known that many of the enzymes found in T. brucei, including components of the branched respiratory chain, ASCT, NADH-dependent fumarate reductases and pyruvate phosphate dikinase, are expressed by other trypanosomatids too (Bringaud et al. 1998; Van Hellemond et al. 1998a,b; Besteiro et al. 2002). However, as with the examples of bloodstream and procyclic T. brucei, the available evidence indicates that differences in the expression of these enzymes, either between different trypanosomatids or different life cycle stages of a single trypanosomatid species, are always likely to reflect the environment in which that cell type lives. Here, I will provide examples of such metabolic adaptation in three further trypanosomatids: the plant pathogen Phytomonas, and the human pathogens Leishmania and Trypanosoma cruzi.
There is no nuclear genome sequencing project for Phytomonas, but the absence from its mitochondrial genome of genes encoding universally conserved components of the cytochrome c reductase and cytochrome c oxidase complexes (Maslov et al. 1999; Nawathean & Maslov 2000) supports the biochemical evidence that this parasite is incapable of cytochrome-dependent respiration (Chaumont et al. 1994; Maslov et al. 1999). Like bloodstream T. brucei, Phytomonas gets by on inefficient (at least in terms of ATP yield) aerobic glucose fermentation because of the availability of a sugar-rich meal ticket. Here, sugars come from breakdown of plant carbohydrates, either from the phloem of the plant host or the digestive tract of the sap-feeding invertebrate vector. The parasite adapts to its niche by secreting enzymes—amylase, amylomaltase, invertase, carboxymethylcellulase and oligo-galactosiduronate lyase—that break down the cellulose and pectin available from the host (Sanchez-Moreno et al. 1992). A high glycolytic capacity is sustained by a large number of glycosomes (Sanchez-Moreno et al. 1992). Looking in further detail at the biochemistry of Phytomonas glycosomes, it appears that another specific adaptation is an expansion of the glycosomal malate dehydrogenase gene family (Uttaro et al. 2000). In T. brucei, glycosomal malate dehydrogenase is required for glycosomal formation of succinate (figure 2b), and the enzyme is encoded by a single copy gene. In Phytomonas, the orthologous single gene copy has expanded through tandem duplication to yield a gene family of approximately 28 diverged copies. It is possible that some of these additional copies are pseudogenes—genes that do not encode functional proteins—although other members of this gene family encode catalytically active enzymes. At least one of these dehydrogenases has no activity with an oxaloacetate substrate, but instead is able to reduce a broad range of 2-oxoacids. Such alterations in substrate specificity have presumably arisen through the accumulation of mutational changes within a duplicated malate dehydrogenase gene (Uttaro et al. 2000). The functional significance of divergent substrate specificity within the 2-oxoacid dehydrogenase family is not clear. Yet for a parasite that requires glycosomal sugar metabolism for energy generation, but still retains an ability to deaminate a range of amino acids, thus providing substrates for hydroxyacid dehydrogenase, it means there are many options for regenerating glycosomal NAD+ (Uttaro et al. 2000).
The physiological switch from development at near neutral pH to development at an acidic pH during the life cycle of Leishmania provides another interesting example of niche adaptation (Mukkada et al. 1985; Burchmore & Barrett 2001). The biochemistry of cultured Leishmania promastigotes appears to be compatible with the likely nutrient availability in the digestive tract of the sandfly vector, where, following the digestion of the blood meal, amino acid and peptides carbon sources are replaced by carbohydrates ingested from a subsequent meal of plant sap. At low densities, cultured promastigotes consume amino acids and glucose for both catabolic and anabolic processes (Cazzulo et al. 1985; Ginger et al. 1999; Burchmore et al. 2003), switching to the consumption of glucose during the later stages of growth (Cazzulo et al. 1985). The identification of a sucrase secreted by Leishmania donovani provided an early indication of adaptation towards the availability of plant-derived nutrients (Blum & Opperdoes 1994). However, the completed genome sequence for Leishmania major recently revealed further adaptations that would enable Leishmania parasites to consume nutrients available within plant sap: the presence of genes encoding several sugar kinases and an ability to metabolize disaccharides (Berriman et al. 2005). By contrast, genes encoding similar enzymes are absent from T. brucei (Berriman et al. 2005), which unlike Leishmania is transmitted by an obligate blood-feeding invertebrate vector.
The metabolic strategies used by Leishmania amastigotes are, by comparison with Leishmania promastigotes, much less well understood. Despite this gap in our knowledge, an intriguing comparison of amastigote metabolism with the metabolic strategies employed by three other microbes within macrophage environments can be made.
Macrophages are phagocytes, and constitute an important part of the innate immune system by virtue of their ability to engulf and destroy microbial invaders and activate an immune response to such intrusions. Following phagocytosis, lysosomes fuse with the phagosomal compartment in which microbial cells are enclosed, creating the acidic phagolysosome where microbes are killed by a combination of low pH and an arsenal of antimicrobial molecules. A few pathogens, however, either choose the macrophage as home, or are able to resist its lethal defences. Three examples illustrate these macrophage–microbe interactions: (i) following phagocytosis the yeast S. cerevisiae is killed within the phagolysosome; (ii) the bacterial pathogen Mycobacterium tuberculosis (causal agent of tuberculosis) thrives within the macrophage environment because it arrests the maturation of the phagolysosome; and (iii) the fungal pathogen Candida albicans responds brutally to entry into the macrophage by switching to a hyphal form of growth, and breaks out of the macrophage, lysing the phagocyte from inside out. The significant point for this discussion is that the microbes from these examples use the glyoxylate cycle within the intracellular niche provided by the macrophage.
I pointed out earlier that under conditions of nutritional deprivation the glyoxylate shunt allows plants and certain microbes to use the C2 metabolite acetyl-CoA for anabolic pathways such as carbohydrate biosynthesis. Although it is destined to die within a phagolysosome, S. cerevisiae still senses that it has entered a nutrient-poor environment and responds with a starvation response, turning on the expression of genes required for the glyoxylate cycle and the key gluconeogenic enzyme fructose-1,6-bisphosphatase (Lorenz & Fink 2001). A similar transcriptional response is seen in C. albicans, together with increased expression of genes required for fatty acid oxidation (Lorenz & Fink 2001; Lorenz et al. 2004). Cells that are deficient in a key glyoxylate cycle enzyme, isocitrate lyase, are unable to escape from the macrophage and are, as a result, avirulent (Lorenz & Fink 2001). Mycobacterium tuberculosis contains an abundance of genes encoding enzymes used for fatty acid oxidation (Cole et al. 1998), and mutants which lack glyoxylate cycle activity are unable to survive or proliferate within macrophages (Boshoff & Barry 2005; Munoz-Elias & McKinney 2005). Thus, phagocytosis by macrophages places microbes within a nutritionally challenging environment and they adapt by drawing on the availability of fatty acids, either from internal reserves or from the vacuolar membrane, to produce acetyl-CoA for both energy generation and anabolic pathways. The completion, or near completion, of two Leishmania genome sequences perhaps challenges this paradigm.
Leishmania prevents lysosomal fusion during its differentiation from an invasive metacyclic promastigote form into a replicative intracellular amastigote, but then replicates in its amastigote form within an acidic phagolysosome (Duclos & Desjardins 2000). Amino acids liberated through proteolysis may provide carbon sources for metabolism, but like the other macrophage dwellers, Leishmania amastigotes have a high rate of fatty acid oxidation (Coombs et al. 1982; Hart & Coombs 1982). In that regard it appears that fatty acids represent a ubiquitous source of carbon inside macrophages. Yet, Leishmania amastigotes lacking an ability to transport glucose in from the environment are unable to replicate within macrophages (Burchmore et al. 2003), which suggests other carbon sources are, or can be, made available to this macrophage dweller within its niche environment. The assertion that Leishmania amastigotes can gain access to host cell nutrients from beyond the confines of the parasitophorous vacuole is supported by the labelling of infected macrophages with Lucifer yellow and fluoresecent dextran (Schaible et al. 1999). Interestingly, the complete and almost complete genomes available for two Leishmania species indicate that a classical glyoxylate cycle is absent from Leishmania, and also the trypanosomatids T. brucei and T. cruzi too (Berriman et al. 2005). Biochemical evidence for the presence or absence of a Leishmania glyoxylate cycle is equivocal: one group has documented evidence for the occurrence of the glyoxylate cycle in Leishmania (Mukkada 1977; Simon et al. 1978), but these results have not been replicated elsewhere (Mottram & Coombs 1985). Incorporation of radioactivity from 14C-labelled acetate and lauric acid into the carbohydrate polymer mannan is also consistent with the operation of the glyoxylate cycle in Leishmania (Keegan & Blum 1993), but it does not constitute proof because alternative routes for the incorporation of radioactivity into mannan are possible. For instance, following condensation with oxaloacetate to form citrate the two carbons of acetyl-CoA are not lost as CO2 in a first turn of the Krebs cycle; this means that radio-labelled Krebs cycle intermediates could be substrates for conversion into cytoplasmic phosphoenolpyruvate (see figure 2b for how this pathway would operate). Thus, the jury should remain out regarding a decision on whether the glyoxylate cycle is present or absent in Leishmania. In the absence of further evidence, one hypothesis perhaps worthy of further consideration is that in the absence of a classical glyoxylate cycle, changes in the substrate specificity of enzymes that catalysed mechanistically similar reactions to isocitrate lyase and malate synthase have resulted in the convergent evolution of an unconventional glyoxylate cycle. The example of the 2-hydroxyacid dehydrogenase in Phytomonas (Uttaro et al. 2000) possibly provides an example of how such convergent evolution could arise, as does the phylogenetic evidence that lactose dehydrogenases in T. vaginalis and Cryptosporidium arose following duplication and divergence of malate dehydrogenase genes (Wu et al. 1999; Madern et al. 2004) and that fumarate reductase in anaerobic helminths evolved from succinate dehydrogenase (van Hellemond et al. 2003).
The American trypanosome T. cruzi is the aetiologic agent of Chagas' disease (table 1), and although it is experimental tractable, in comparison to microsporidian or many apicomplexan parasites it is perhaps less amenable to experimental study or genetic manipulation than other trypanosomatids. Thus, we had less insight into its metabolism than for its distantly related African counterpart T. brucei or Leishmania. The arrival of its complete genome sequence (El-Sayed et al. 2005) allows the comparison between its metabolic repertoire and that of other trypanosomatids to be made (Berriman et al. 2005), and a recent proteomic survey of the T. cruzi (Atwood et al. 2005) provided a way around some of the technical difficulties associated with studying the biology of T. cruzi. Three examples of niche-specific metabolic adaptations highlight the significant new insights that comparative genomic and proteomic analyses can provide. (i) Trypanosoma cruzi, but not L. major or T. brucei is capable of metabolizing histidine to glutamate, which can then be converted to the Krebs cycle precursor α-ketoglutarate (Atwood et al. 2005). The enzymes required for this metabolism of histidine are upregulated in parasite forms found in the insect vector. Importantly, this result correlates with the occurrence of histidine as the predominant free amino acid in the excreta and haemolymph of the T. cruzi vector R. prolixus (Atwood et al. 2005). (ii) Following cell invasion and encapsulation in a host cell phagolysosome, but before escape into the host cytoplasm, T. cruzi switches to fatty acid metabolism as a strategy for energy generation (Atwood et al. 2005). This is reminiscent of the increased oxidation rate of fatty acids by Leishmania amastigotes compared to Leishmania promastigotes. (iii) The presence in the T. cruzi genome of hexose phosphate transporters, not present in T. brucei or L. major, and sugar kinases, also present in L. major but not T. brucei, can be linked to possible carbohydrate availability for amastigotes in the cytoplasm of the host cell (Berriman et al. 2005).
(ii) Comparative metabolism in the Apicomplexa
The trypanosomatids constitute a family of parasites, but apicomplexans constitute a phylum of parasites. Over 5000 species are included in the phylum Apicomplexa; all are obligate intracellular parasites. Their intracellular life style means they are difficult to work with, but with genome sequences available for representative pathogens from different sub-groups of the phylum, comparative genomics is feasible (Li et al. 2003; Templeton et al. 2004). Malarial parasites are part of the Haemosporidia; another group, the Coccidia, can be divided into tissue-dwellers (including T. gondii) and invaders of intestinal cells (the enteric Coccidia, which includes the chicken parasite Eimeria tenella). In comparison to Plasmodium, the genome sequences of Toxoplasma and E. tenella indicate that coccidian parasites retain the ability to synthesize carbohydrates, as evidenced by the presence of fructose-1,6-bisphosphatase (M. L. Ginger, unpublished observations 2005). What, therefore, is the key biological difference that results in these two coccidian parasites retaining a gluconeogenic pathway?
A general theme of apicomplexan life cycles is invasion of a host cell followed by growth, escape, and then re-invasion of another host cell. Transmission between hosts requires an obligatory sexual cycle and results in production of an oocyst. One difference between Plasmodium and the Coccidia is that in the latter, sporulation of the oocyst occurs in the external environment, whereas maturation of Plasmodium oocysts occurs in a mosquito. At least in the case of Eimeria, sporulation also utilizes an internal fuel supply, the complex carbohydrate mannitol (Liberator et al. 1998; Allocco et al. 1999). Perhaps the presence of a gluconeogenic pathway ensures that the carbohydrate reserve is sufficient in parasites that find themselves in the external environment prior to sporulation for life cycle completion. In contrast to other intestinal parasites considered in this review, Eimeria also retains an ability to use its mitochondrion for oxidative phosphorylation. One possible reason for this is that the maximum energetic potential of mannitol metabolism in an (aerobic) external environment can be realized by the use of an oxidative metabolism.
Cryptosporidium is another apicomplexan that can be included in a comparative survey of metabolic strategies. This water- and food-borne parasite resides on the surface of the intestinal epithelium, just beneath the plasma membrane of its infected host cell. Infection can result in severe and potentially serious diarrhoea, particularly in immunodeficient individuals, and the oocysts which are shed in the stool are extremely resistant to environmental insults. Thus far, Cryptosporidium has also proved extremely difficult to work with; it cannot be genetically manipulated, and virtually all that has been learnt about its metabolism has come from the recent sequencing and annotation of its compact genome (Abrahamsen et al. 2004; Xu et al. 2004). From the genome sequence, it has been possible to confirm the suspicion that this parasite presents an extremely streamlined metabolism, relying on the host cell for virtually all the precursors necessary to assemble macromolecular structures (proteins, membranes, DNA), although with the genome sequence in hand the extent of this streamlining is still nonetheless surprising (e.g. Striepen et al. 2004 and see below). Like many other enteric parasites, Cryptosporidium has dispensed with the capacity for its mitochondrial relic organelle to undertake oxidative phosphorylation, although the parasite does retain an ability to metabolize carbon from a complex carbohydrate store (Abrahamsen et al. 2004; Xu et al. 2004). Energy is generated by anaerobic glucose fermentation. A more interesting example of this parasite's metabolic streamlining is, however, the disposal of its apicoplast.
Loss of the apicoplast might imply that an intimate interface between Cryptosporidium and the cytoplasm of the host cell limits the need for Cryptosporidium to retain much capacity for anabolic biochemistry. However, the presence of genes that encode enzymes which probably catalyse (i) the condensation of the isopentenyl diphosphate and dimethylallyl diphosphate intermediates normally synthesized in the apicoplast, probably facilitating the endogenous formation of dolichols, and (ii) the covalent modification of proteins by farnesylation or geranylgeranylation (M. L. Ginger, unpublished observations) indicates that like other organisms Cryptosporidium needs isoprenoids. Moreover, the presence of such enzymes indicates that the isoprenoids required by Cryptosporidium cannot be acquired directly from the host cell.
If, as seems likely, the acquisition during apicomplexan evolution of a plastid facilitated the loss of a cytoplasmic mevalonate pathway for isopentenyl diphosphate synthesis (Smit & Mushegian 2000), then in disregarding its apicoplast, did Cryptosporidium lose the ability to synthesize the isopentenyl diphosphate and dimethylallyl diphosphate precursors, from which all isoprenoid metabolites are derived? The failure to detect in the Cryptosporidium genome any gene encoding an enzyme known to be involved in isopentenyl diphosphate production could suggest that Cryptosporidium has evolved a new biochemical strategy for synthesizing isoprenoids de novo; a similar hypothesis has been suggested for the gut pathogen E. histolytica, which also retains enzymes for the metabolism of isopentenyl diphosphate and dimethylallyl diphosphate, but lacks any known capacity for synthesis of these compounds (Loftus et al. 2005a). The precedent for parasites re-engineering the active sites of an enzyme in order to carry out mechanistically similar reactions with different substrates has been established (discussed earlier), but an alternative explanation is that both parasites are (perhaps the first) examples of organisms that can acquire the essential precursors for isoprenoid biosynthesis, probably required only in low amounts from the host environment. The identification of 3-hydroxy-3-methylglutaryl-CoA reductase (Boucher & Doolittle 2000) and mevalonate diphosphate decarboxylase (Accession number EAA39903) from the incomplete genome sequence of Giardia, however, indicates that an inability to synthesize isoprenoid precursors is not common to all anaerobic enteric parasitic protozoa and amoebae. Rather, the putative acquisition of the key isoprenoid precursors could reflect the nature of extracytoplasmic interface that forms between Cryptosporidium and its host cell (discussed in Mead 2002) and, in E. histolytica, phagotrophy.
(d) Nutrient acquisition: gene families shaped by the constraints of genome architecture
Thus far, the discussion has mainly focused on the need to generate energy, but there are obviously other crucial facets to any parasite's metabolic repertoire. The most obvious is taking organic molecules—carbohydrates, amino acids, lipids and nitrogenous purine and pyrimidine bases—as the precursors for assembling proteins, cellular membranes and DNA. Another important aspect of survival in microaerophilic as well as aerobic environments is limiting the accumulation of reactive oxygen species, arising from oxygen metabolism, which readily and irreversibly damage the cell's macromolecular structures (Coombs et al. 2004; Loftus et al. 2005a). However, if an aim here is to illustrate how a metabolic repertoire uncovered by genome sequencing reflects the niche in which the parasite resides, then the requirement for trypanosomatids to acquire nucleotide precursors provides an elegant comparative example of how, at least in trypanosomatids, constraints on genome architecture drive the evolution of gene families.
Rather than synthesize amino acids, lipids or DNA precursors de novo, many parasites acquire ready-made precursors from the host and assemble macromolecular structures in much the same way as people assemble flat-packed furniture. This is not metabolic laziness; the reliance on macromolecular precursors arises as a consequence of a highly adapted biology. Cryptosporidium and E. histolytica provide examples of acquisitive extremes (Abrahamsen et al. 2004; Xu et al. 2004; Loftus et al. 2005a), but all obligate parasitic protozoa are united in their complete reliance on the salvage of preformed purines from the host. While many mammalian cells readily assemble these key intermediates of nucleic acid biosynthesis, parasitic protozoa rely on an intricate network of transporters and salvage pathways to first bring purines in from the external environment—normally in the form of nucleosides and nucleobases—and then transform these scavenged metabolites into either adenosine monophosphate or guanosine monophosphate (Landfear et al. 2004). Insight into the substrate specificity and physiological function of nucleoside/nucleotide delivery systems, and potentially other parasite transporters, is significant because such permeases often provide the delivery routes for anti-parasitic drugs; changes in substrate specificity or expression levels of these transporters can result in drug resistance.
In T. brucei, the complexity of nucleoside/nucleobase transport systems is reflected by the expression of a variety of different transporters, including a family of six clustered genes, TbNT2–TbNT7 (Sanchez et al. 2002). The individual transporters encoded by this gene cohort are very closely related, reflecting their emergence following a series of gene duplication events, but they differ in their ability to transport different substrates. While all are expressed in bloodstream form T. brucei, only two isoforms are expressed by procyclic trypanosomes. Why, though, do such complexities with regard to substrate affinities and stage-specific expression need to be encoded by a gene family? In many eukaryotes, including some microbial parasites (e.g. Muhia et al. 2003; Loftus et al. 2005b), cohorts of highly related proteins with subtly different biochemical properties can be generated either by alternatively splicing different exons from a single gene, or initiating the transcription of a single gene at alternative start sites. Trypanosomatid parasites are unique, however, in that they fail to regulate gene expression at the level of transcript initiation (Clayton 2002)—there are no individual transcription promoters for protein-coding genes transcribed by RNA polymerase II—and the coding regions of individual genes are, with only four possible exceptions, not interrupted by intervening non-coding sequences (introns). A general consequence of such divergent gene regulation is that even for closely related gene products trypanosomatids can only generate diversity by retaining a separate gene for each isoform that is required. Among the trypanosomatids there are several examples of gene families, such as the glucose transporter families (Tetaud et al. 1997; Vedrenne et al. 2000), that encode distinct enzymes or transporters which nonetheless share a high degree of primary sequence identity; such gene families have evolved following tandem duplication of an ancestral single-copy gene.
5. The future
We now have in our possession metabolic maps for some of the medically most significant parasitic protozoa. Thus far, however, the general terrain has only been mapped in its broadest sense: different strategies for generating energy can be matched with environments where they are likely to be the most profitable, and we have a perspective on the biosynthetic precursors that many parasitic protozoa require—or, depending upon your point of view—acquire from the host. Yet, there is still much to do if we are to understand fully at a molecular level how these parasites modulate and regulate their specialized metabolism in order to complete life cycles within host, vector, or environment (figure 1). Specialized metabolism also extends to manipulation of the host environment, as illustrated by the discovery of a lipoxygenase secreted by T. gondii. This enzyme uses host arichidonic acid to initiate eicosanoid production, perhaps influencing the host immune system to suit the needs of the Toxoplasma parasite (Bannenberg et al. 2004). We have, however, only just begun to gain metabolic insight into how such host manipulation by parasitic protozoa can occur.
Finally, the key translational application resulting from the sequencing of parasite genomes and the study of parasite metabolism—the arrival of new drugs that can cure the devastating tropical diseases that are caused by some parasites—was stated in the introduction to this review. Inefficacy, toxicity and resistance associated with many drugs presently used to treat these diseases means there is an urgent need for new drugs. Yet the development of new drugs is challenging, not least because market forces are insufficient to support discovery and development pipelines in many of the countries where important diseases are endemic. However, with support from public–private partnerships and philanthropic funding progress is being made. A recent review (Pink et al. 2005) provides perspectives on both the opportunities and challenges that are at the forefront of antiparasitic drug discovery. The admission of fosmidomycin, an inhibitor of isoprenoid biosynthesis through the DXP pathway, into human clinical trials for malaria (Borrmann et al. 2004) provides one example of how the mining of genome sequences can result in discoveries that lend themselves toward crucial practical application.
Acknowledgments
M.L.G. is a Royal Society University Research Fellow, and would like to gratefully acknowledge Keith Gull (Oxford) and Graham Coombs (Glasgow) for helpful discussions, and Bill Wickstead (Oxford) for constructive criticism of the manuscript.
References
- Abrahamsen M.S, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. doi:10.1126/science.1094786 [DOI] [PubMed] [Google Scholar]
- Allen J.W, Ginger M.L, Ferguson S.J. Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway. Biochem. J. 2004;383:537–542. doi: 10.1042/BJ20040832. doi:10.1042/BJ20040832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocco J.J, Profous-Juchelka H, Myers R.W, Nare B, Schmatz D.M. Biosynthesis and catabolism of mannitol is developmentally regulated in the protozoan parasite Eimeria tenella. J. Parasitol. 1999;85:167–173. [PubMed] [Google Scholar]
- Atwood J.A, III, Weatherly D.B, Minning T.A, Bundy B, Cavola C, Opperdoes F.R, Orlando R, Tarleton R.L. The Trypanosoma cruzi proteome. Science. 2005;309:473–476. doi: 10.1126/science.1110289. doi:10.1126/science.1110289 [DOI] [PubMed] [Google Scholar]
- Bakker B.M, Mensonides F.I, Teusink B, van Hoek P, Michels P.A, Westerhoff H.V. Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc. Natl Acad. Sci. USA. 2000;97:2087–2092. doi: 10.1073/pnas.030539197. doi:10.1073/pnas.030539197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. doi:10.1126/science.1085544 [DOI] [PubMed] [Google Scholar]
- Bannenberg G.L, Aliberti J, Hong S, Sher A, Serhan C. Exogenous pathogen and plant 15-lipoxygenase initiate endogenous lipoxin A4 biosynthesis. J. Exp. Med. 2004;199:515–523. doi: 10.1084/jem.20031325. doi:10.1084/jem.20031325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. doi:10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- Besteiro S, Biran M, Biteau N, Coustou V, Baltz T, Canioni P, Bringaud F. Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J. Biol. Chem. 2002;277:38 001–38 012. doi: 10.1074/jbc.M201759200. doi:10.1074/jbc.M201759200 [DOI] [PubMed] [Google Scholar]
- Billker O, Lindo V, Panico M, Etienne A.E, Paxton T, Dell A, Rogers M, Sinden R.E, Morris H.R. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature. 1998;392:289–292. doi: 10.1038/32667. doi:10.1038/32667 [DOI] [PubMed] [Google Scholar]
- Blattner J, Helfert S, Michels P, Clayton C. Compartmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc. Natl Acad. Sci. USA. 1998;95:11 596–11 600. doi: 10.1073/pnas.95.20.11596. doi:10.1073/pnas.95.20.11596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum J.J, Opperdoes F.R. Secretion of sucrase by Leishmania donovani. J. Eukaryot. Microbiol. 1994;41:228–231. doi: 10.1111/j.1550-7408.1994.tb01502.x. [DOI] [PubMed] [Google Scholar]
- Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 2002;277:32 849–32 854. doi: 10.1074/jbc.M205776200. doi:10.1074/jbc.M205776200 [DOI] [PubMed] [Google Scholar]
- Borrmann S, et al. Fosmidomycin-clindamycin for the treatment of Plasmodium falciparum malaria. J. Infect. Dis. 2004;190:1534–1540. doi: 10.1086/424603. doi:10.1086/424603 [DOI] [PubMed] [Google Scholar]
- Borza T, Popescu C.E, Lee R.W. Multiple metabolic roles for the nonphotosynthetic plastid of the green alga Prototheca wickerhamii. Eukaryot. Cell. 2005;4:253–261. doi: 10.1128/EC.4.2.253-261.2005. doi:10.1128/EC.4.2.253-261.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff H.I, Barry C.E. A low-carb diet for a high-octane pathogen. Nat. Med. 2005;11:599–600. doi: 10.1038/nm0605-599. doi:10.1038/nm0605-599 [DOI] [PubMed] [Google Scholar]
- Boucher Y, Doolittle W.F. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol. Microbiol. 2000;37:703–716. doi: 10.1046/j.1365-2958.2000.02004.x. doi:10.1046/j.1365-2958.2000.02004.x [DOI] [PubMed] [Google Scholar]
- Bringaud F, Baltz D, Baltz T. Functional and molecular characterization of a glycosomal PPi-dependent enzyme in trypanosomatids: pyruvate, phosphate dikinase. Proc. Natl Acad. Sci. USA. 1998;95:7963–7968. doi: 10.1073/pnas.95.14.7963. doi:10.1073/pnas.95.14.7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchmore R.J, Barrett M.P. Life in vacuoles–nutrient acquisition by Leishmania amastigotes. Int. J. Parasitol. 2001;31:1311–1320. doi: 10.1016/s0020-7519(01)00259-4. doi:10.1016/S0020-7519(01)00259-4 [DOI] [PubMed] [Google Scholar]
- Burchmore R.J, Rodriguez-Contreras D, McBride K, Merkel P, Barrett M.P, Modi G, Sacks D, Landfear S.M. Genetic characterization of glucose transporter function in Leishmania mexicana. Proc. Natl Acad. Sci. USA. 2003;100:3901–3906. doi: 10.1073/pnas.0630165100. doi:10.1073/pnas.0630165100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzulo J.J, Franke de Cazzulo B.M, Engel J.C, Cannata J.J. End products and enzyme levels of aerobic glucose fermentation in trypanosomatids. Mol. Biochem. Parasitol. 1985;16:329–343. doi: 10.1016/0166-6851(85)90074-x. doi:10.1016/0166-6851(85)90074-X [DOI] [PubMed] [Google Scholar]
- Cerkasov J, Cerkasovova A, Kulda J, Vilhelmova D. Respiration of hydrogenosomes of Tritrichomonas foetus. I. ADP-dependent oxidation of malate and pyruvate. J. Biol. Chem. 1978;253:1207–1214. [PubMed] [Google Scholar]
- Chaumont F, Schanck A.N, Blum J.J, Opperdoes F.R. Aerobic and anaerobic glucose metabolism of Phytomonas sp. isolated from Euphorbia characias. Mol. Biochem. Parasitol. 1994;67:321–331. doi: 10.1016/0166-6851(94)00141-3. doi:10.1016/0166-6851(94)00141-3 [DOI] [PubMed] [Google Scholar]
- Clayton C.E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. doi:10.1093/emboj/21.8.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.T, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. doi:10.1038/31159 [DOI] [PubMed] [Google Scholar]
- Coombs G.H, Muller M. Energy metabolism in anaerobic protozoa. In: Marr J.J, Muller M, editors. Biochemistry and molecular biology of parasites. Academic Press; London: 1995. pp. 33–49. [Google Scholar]
- Coombs G.H, Craft J.A, Hart D.T. A comparative study of Leishmania mexicana amastigotes and promastigotes. Enzyme activities and subcellular locations. Mol. Biochem. Parasitol. 1982;5:199–211. doi: 10.1016/0166-6851(82)90021-4. doi:10.1016/0166-6851(82)90021-4 [DOI] [PubMed] [Google Scholar]
- Coombs G.H, Westrop G.D, Suchan P, Puzova G, Hirt R.P, Embley T.M, Mottram J.C, Muller S. The amitochondriate eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system. J. Biol. Chem. 2004;279:5249–5256. doi: 10.1074/jbc.M304359200. doi:10.1074/jbc.M304359200 [DOI] [PubMed] [Google Scholar]
- Coustou V, et al. ATP generation in the Trypanosoma brucei procyclic form: cytosolic substrate level is essential, but not oxidative phosphorylation. J. Biol. Chem. 2003;278:49 625–49 635. doi: 10.1074/jbc.M307872200. doi:10.1074/jbc.M307872200 [DOI] [PubMed] [Google Scholar]
- Coustou V, Besteiro S, Riviere L, Biran M, Biteau N, Franconi J.M, Boshart M, Baltz T, Bringaud F. A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J. Biol. Chem. 2005;280:16 559–16 570. doi: 10.1074/jbc.M500343200. doi:10.1074/jbc.M500343200 [DOI] [PubMed] [Google Scholar]
- Das S, Stevens T, Castillo C, Villasenor A, Arredondo H, Reddy K. Lipid metabolism in mucous-dwelling amitochondriate protozoa. Int. J. Parasitol. 2002;32:655–675. doi: 10.1016/s0020-7519(02)00006-1. doi:10.1016/S0020-7519(02)00006-1 [DOI] [PubMed] [Google Scholar]
- Denton H, Roberts C.W, Alexander J, Thong K.W, Coombs G.H. Enzymes of energy metabolism in the bradyzoites and tachyzoites of Toxoplasma gondii. FEMS Microbiol. Lett. 1996;137:103–108. doi: 10.1111/j.1574-6968.1996.tb08090.x. doi:10.1016/0378-1097(96)00047-X [DOI] [PubMed] [Google Scholar]
- Ding M, Clayton C, Soldati D. Toxoplasma gondii catalase: are there peroxisomes in toxoplasma? J. Cell Sci. 2000;113:2409–2419. doi: 10.1242/jcs.113.13.2409. [DOI] [PubMed] [Google Scholar]
- Duclos S, Desjardins M. Subversion of a young phagosome: the survival strategies of intracellular pathogens. Cell. Microbiol. 2000;2:365–377. doi: 10.1046/j.1462-5822.2000.00066.x. doi:10.1046/j.1462-5822.2000.00066.x [DOI] [PubMed] [Google Scholar]
- Dyall S.D, Koehler C.M, Delgadillo-Correa M.G, Bradley P.J, Plumper E, Leuenberger D, Turck C.W, Johnson P.J. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell. Biol. 2000;20:2488–2497. doi: 10.1128/mcb.20.7.2488-2497.2000. doi:10.1128/MCB.20.7.2488-2497.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.R, Schofield P.J, O'Sullivan W.J, Costello M. Arginine metabolism during culture of Giardia intestinalis. Mol. Biochem. Parasitol. 1992;53:97–103. doi: 10.1016/0166-6851(92)90011-8. doi:10.1016/0166-6851(92)90011-8 [DOI] [PubMed] [Google Scholar]
- El-Sayed N.M, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. doi:10.1126/science.1112631 [DOI] [PubMed] [Google Scholar]
- Embley T.M, van der Giezen M, Horne D.S, Dyal P.L, Foster P. Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Phil. Trans. R. Soc. B. 2003;358:191–201. doi: 10.1098/rstb.2002.1190. doi:10.1098/rstb.2002.1190 discussion 201–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. doi:10.1101/gad.323404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Beattie D.S. Novel FMN-containing rotenone-insensitive NADH dehydrogenase from Trypanosoma brucei mitochondria: isolation and characterization. Biochemistry. 2002;41:3065–3072. doi: 10.1021/bi015989w. doi:10.1021/bi015989w [DOI] [PubMed] [Google Scholar]
- Fang J, Beattie D.S. Identification of a gene encoding a 54 kDa alternative NADH dehydrogenase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;127:73–77. doi: 10.1016/s0166-6851(02)00305-5. doi:10.1016/S0166-6851(02)00305-5 [DOI] [PubMed] [Google Scholar]
- Fang J, Wang Y, Beattie D.S. Isolation and characterization of complex I, rotenone-sensitive NADH:ubiquinone oxidoreductase, from the procyclic forms of Trypanosoma brucei. Eur. J. Biochem. 2001;268:3075–3082. doi: 10.1046/j.1432-1327.2001.02205.x. doi:10.1046/j.1432-1327.2001.02205.x [DOI] [PubMed] [Google Scholar]
- Fang J, Sullivan M, McCutchan T.F. The effects of glucose concentration on the reciprocal regulation of rRNA promoters in Plasmodium falciparum. J. Biol. Chem. 2004a;279:720–725. doi: 10.1074/jbc.M308284200. doi:10.1074/jbc.M308284200 [DOI] [PubMed] [Google Scholar]
- Fang J, Zhou H, Rathore D, Sullivan M, Su X.Z, McCutchan T.F. Ambient glucose concentration and gene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 2004b;133:125–129. doi: 10.1016/j.molbiopara.2003.09.004. doi:10.1016/j.molbiopara.2003.09.004 [DOI] [PubMed] [Google Scholar]
- Fernandes A.P, Nelson K, Beverley S.M. Evolution of nuclear ribosomal RNAs in kinetoplastid protozoa: perspectives on the age and origins of parasitism. Proc. Natl Acad. Sci. USA. 1993;90:11 608–11 612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichera M.E, Roos D.S. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. doi:10.1038/37132 [DOI] [PubMed] [Google Scholar]
- Finlay B.J, Fenchel T. Hydrogenosomes in some anaerobic protozoa resemble mitochondria. FEMS Microbiol. Lett. 1989;65:311–314. doi:10.1016/0378-1097(89)90236-X [Google Scholar]
- Foth B.J, Ralph S.A, Tonkin C.J, Struck N.S, Fraunholz M, Roos D.S, Cowman A.F, McFadden G.I. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. doi:10.1126/science.1078599 [DOI] [PubMed] [Google Scholar]
- Furuya T, Kessler P, Jardim A, Schnaufer A, Crudder C, Parsons M. Glucose is toxic to glycosome-deficient trypanosomes. Proc. Natl Acad. Sci. USA. 2002;99:14 177–14 182. doi: 10.1073/pnas.222454899. doi:10.1073/pnas.222454899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.J, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. doi:10.1038/nature01097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger M.L, Chance M.L, Goad L.J. Elucidation of carbon sources used for the biosynthesis of fatty acids and sterols in the trypanosomatid Leishmania mexicana. Biochem. J. 1999;342:397–405. doi:10.1042/0264-6021:3420397 [PMC free article] [PubMed] [Google Scholar]
- Guerra-Giraldez C, Quijada L, Clayton C.E. Compartmentation of enzymes in a microbody, the glycosome, is essential in Trypanosoma brucei. J. Cell Sci. 2002;115:2651–2658. doi: 10.1242/jcs.115.13.2651. [DOI] [PubMed] [Google Scholar]
- Hall N, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. doi:10.1126/science.1103717 [DOI] [PubMed] [Google Scholar]
- Hannaert V, Saavedra E, Duffieux F, Szikora J.P, Rigden D.J, Michels P.A, Opperdoes F.R. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl Acad. Sci. USA. 2003;100:1067–1071. doi: 10.1073/pnas.0335769100. doi:10.1073/pnas.0335769100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart D.T, Coombs G.H. Leishmania mexicana: energy metabolism of amastigotes and promastigotes. Exp. Parasitol. 1982;54:397–409. doi: 10.1016/0014-4894(82)90049-2. doi:10.1016/0014-4894(82)90049-2 [DOI] [PubMed] [Google Scholar]
- Hrdy I, Hirt R.P, Dolezal P, Bardonova L, Foster P.G, Tachezy J, Embley T.M. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. doi:10.1038/nature03149 [DOI] [PubMed] [Google Scholar]
- Jomaa H, et al. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. doi:10.1126/science.285.5433.1573 [DOI] [PubMed] [Google Scholar]
- Katinka M.D, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. doi:10.1038/35106579 [DOI] [PubMed] [Google Scholar]
- Keegan F.P, Blum J.J. Incorporation of label from acetate and laurate into the mannan of Leishmania donovani via the glyoxylate cycle. J. Eukaryot. Microbiol. 1993;40:730–732. doi: 10.1111/j.1550-7408.1993.tb04467.x. [DOI] [PubMed] [Google Scholar]
- Keeling P.J, Fast N.M. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. doi:10.1146/annurev.micro.56.012302.160854 [DOI] [PubMed] [Google Scholar]
- Kohler S, Delwiche C.F, Denny P.W, Tilney L.G, Webster P, Wilson R.J, Palmer J.D, Roos D.S. A plastid of probable green algal origin in Apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. doi:10.1126/science.275.5305.1485 [DOI] [PubMed] [Google Scholar]
- Lamour N, Riviere L, Coustou V, Coombs G.H, Barrett M.P, Bringaud F. Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 2005;280:11 902–11 910. doi: 10.1074/jbc.M414274200. doi:10.1074/jbc.M414274200 [DOI] [PubMed] [Google Scholar]
- Landfear S.M, Ullman B, Carter N.S, Sanchez M.A. Nucleoside and nucleobase transporters in parasitic protozoa. Eukaryot. Cell. 2004;3:245–254. doi: 10.1128/EC.3.2.245-254.2004. doi:10.1128/EC.3.2.245-254.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Unnasch N, Murphy A.D. Metabolic changes of the malaria parasite during the transition from the human to the mosquito host. Annu. Rev. Microbiol. 1998;52:561–590. doi: 10.1146/annurev.micro.52.1.561. doi:10.1146/annurev.micro.52.1.561 [DOI] [PubMed] [Google Scholar]
- Li L, et al. Gene discovery in the apicomplexa as revealed by EST sequencing and assembly of a comparative gene database. Genome Res. 2003;13:443–454. doi: 10.1101/gr.693203. doi:10.1101/gr.693203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberator P, Anderson J, Feiglin M, Sardana M, Griffin P, Schmatz D, Myers R.W. Molecular cloning and functional expression of mannitol-1-phosphatase from the apicomplexan parasite Eimeria tenella. J. Biol. Chem. 1998;273:4237–4244. doi: 10.1074/jbc.273.7.4237. doi:10.1074/jbc.273.7.4237 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G. Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem. Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. doi:10.1016/S0968-0004(00)01589-9 [DOI] [PubMed] [Google Scholar]
- Lindmark D.G, Muller M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 1973;248:7724–7728. [PubMed] [Google Scholar]
- Linstead D.J, Klein R.A, Cross G.A. Threonine catabolism in Trypanosoma brucei. J. Gen. Microbiol. 1977;101:243–251. doi: 10.1099/00221287-101-2-243. [DOI] [PubMed] [Google Scholar]
- Lockwood B.C, Coombs G.H. Amino acids catabolism in anaerobic protists. In: Coombs G.H, North M.J, editors. Biochemical protozoology. Taylor & Francis; London: 1991. pp. 113–122. [Google Scholar]
- Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005a;433:865–868. doi: 10.1038/nature03291. doi:10.1038/nature03291 [DOI] [PubMed] [Google Scholar]
- Loftus B.J, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005b;307:1321–1324. doi: 10.1126/science.1103773. doi:10.1126/science.1103773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.C, Fink G.R. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. doi: 10.1038/35083594. doi:10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- Lorenz M.C, Bender J.A, Fink G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. doi:10.1128/EC.3.5.1076-1087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan H.D, Mowatt M.R, Byrd L.G, Nash T.E. Cholesterol starvation induces differentiation of the intestinal parasite Giardia lamblia. Proc. Natl Acad. Sci. USA. 1996;93:7628–7633. doi: 10.1073/pnas.93.15.7628. doi:10.1073/pnas.93.15.7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack S.R, Samuels S, Vanderberg J.P. Hemolymph of Anopheles stephensi from noninfected and Plasmodium berghei-infected mosquitoes. 3. Carbohydrates. J. Parasitol. 1979;65:217–221. [PubMed] [Google Scholar]
- Madern D, Cai X, Abrahamsen M.S, Zhu G. Evolution of Cryptosporidium parvum lactate dehydrogenase from malate dehydrogenase by a very recent event of gene duplication. Mol. Biol. Evol. 2004;21:489–497. doi: 10.1093/molbev/msh042. doi:10.1093/molbev/msh042 [DOI] [PubMed] [Google Scholar]
- Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol. Cell. Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Borst P. Secondary loss of chloroplasts in trypanosomes. Proc. Natl Acad. Sci. USA. 2003;100:765–767. doi: 10.1073/pnas.0437776100. doi:10.1073/pnas.0437776100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov D.A, Nawathean P, Scheel J. Partial kinetoplast-mitochondrial gene organization and expression in the respiratory deficient plant trypanosomatid Phytomonas serpens. Mol. Biochem. Parasitol. 1999;99:207–221. doi: 10.1016/s0166-6851(99)00028-6. doi:10.1016/S0166-6851(99)00028-6 [DOI] [PubMed] [Google Scholar]
- McConkey G.A, Rogers M.J, McCutchan T.F. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J. Biol. Chem. 1997;272:2046–2049. doi: 10.1074/jbc.272.4.2046. doi:10.1074/jbc.272.4.2046 [DOI] [PubMed] [Google Scholar]
- Mead J.R. Cryptosporidiosis and the challenges of chemotherapy. Drug Resist. Updat. 2002;5:47–57. doi: 10.1016/s1368-7646(02)00011-0. doi:10.1016/S1368-7646(02)00011-0 [DOI] [PubMed] [Google Scholar]
- Michels P.A, Hannaert V, Bringaud F. Metabolic aspects of glycosomes in trypanosomatidae—new data and views. Parasitol. Today. 2000;16:482–489. doi: 10.1016/s0169-4758(00)01810-x. doi:10.1016/S0169-4758(00)01810-X [DOI] [PubMed] [Google Scholar]
- Misset O, Opperdoes F.R. The phosphoglycerate kinases from Trypanosoma brucei. A comparison of the glycosomal and the cytosolic isoenzymes and their sensitivity towards suramin. Eur. J. Biochem. 1987;162:493–500. doi: 10.1111/j.1432-1033.1987.tb10667.x. doi:10.1111/j.1432-1033.1987.tb10667.x [DOI] [PubMed] [Google Scholar]
- Morris J.C, Wang Z, Drew M.E, Englund P.T. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 2002;21:4429–4438. doi: 10.1093/emboj/cdf474. doi:10.1093/emboj/cdf474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram J.C, Coombs G.H. Leishmania mexicana: enzyme activities of amastigotes and promastigotes and their inhibition by antimonials and arsenicals. Exp. Parasitol. 1985;59:151–160. doi: 10.1016/0014-4894(85)90067-0. doi:10.1016/0014-4894(85)90067-0 [DOI] [PubMed] [Google Scholar]
- Moyersoen J, Choe J, Fan E, Hol W.G, Michels P.A. Biogenesis of peroxisomes and glycosomes: trypanosomatid glycosome assembly is a promising new drug target. FEMS Microbiol. Rev. 2004;28:603–643. doi: 10.1016/j.femsre.2004.06.004. doi:10.1016/j.femsre.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Muhia D.K, Swales C.A, Eckstein-Ludwig U, Saran S, Polley S.D, Kelly J.M, Schaap P, Krishna S, Baker D.A. Multiple splice variants encode a novel adenylyl cyclase of possible plastid origin expressed in the sexual stage of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 2003;278:22 014–22 022. doi: 10.1074/jbc.M301639200. doi:10.1074/jbc.M301639200 [DOI] [PubMed] [Google Scholar]
- Mukkada A.J. Tricarboxylic acid and glyoxylate cycles in the Leishmaniae. Acta Trop. 1977;34:167–175. [PubMed] [Google Scholar]
- Mukkada A.J, Meade J.C, Glaser T.A, Bonventre P.F. Enhanced metabolism of Leishmania donovani amastigotes at acid pH: an adaptation for intracellular growth. Science. 1985;229:1099–1101. doi: 10.1126/science.4035350. [DOI] [PubMed] [Google Scholar]
- Muller M, Lindmark D.G. Respiration of hydrogenosomes of Tritrichomonas foetus. II. Effect of CoA on pyruvate oxidation. J. Biol. Chem. 1978;253:1215–1218. [PubMed] [Google Scholar]
- Muller M, Lee J.A, Gordon P, Gaasterland T, Sensen C.W. Presence of prokaryotic and eukaryotic species in all subgroups of the PP(i)-dependent group II phosphofructokinase protein family. J. Bacteriol. 2001;183:6714–6716. doi: 10.1128/JB.183.22.6714-6716.2001. doi:10.1128/JB.183.22.6714-6716.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias E.J, McKinney J.D. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005;11:638–644. doi: 10.1038/nm1252. doi:10.1038/nm1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P, Maslov D.A. The absence of genes for cytochrome c oxidase and reductase subunits in maxicircle kinetoplast DNA of the respiration-deficient plant trypanosomatid Phytomonas serpens. Curr. Genet. 2000;38:95–103. doi: 10.1007/s002940000135. doi:10.1007/s002940000135 [DOI] [PubMed] [Google Scholar]
- North M.J, Lockwood B.C. Amino acid and protein metabolism. In: Marr J.J, Muller M, editors. Biochemistry and molecular biology of parasites. Academic Press; London: 1995. pp. 67–89. [Google Scholar]
- Pink R, Hudson A, Mouries M.A, Bendig M. Opportunities and challenges in antiparasitic drug discovery. Nat. Rev. Drug Discov. 2005;4:727–740. doi: 10.1038/nrd1824. doi:10.1038/nrd1824 [DOI] [PubMed] [Google Scholar]
- Putignani L, Tait A, Smith H.V, Horner D, Tovar J, Tetley L, Wastling J.M. Characterization of a mitochondrion-like organelle in Cryptosporidium parvum. Parasitology. 2004;129:1–18. doi: 10.1017/s003118200400527x. doi:10.1017/S003118200400527X [DOI] [PubMed] [Google Scholar]
- Ralph S.A, Van Dooren G.G, Waller R.F, Crawford M.J, Fraunholz M.J, Foth B.J, Tonkin C.J, Roos D.S, McFadden G.I. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. doi:10.1038/nrmicro843 [DOI] [PubMed] [Google Scholar]
- Riviere L, et al. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J. Biol. Chem. 2004;279:45 337–45 346. doi: 10.1074/jbc.M407513200. doi:10.1074/jbc.M407513200 [DOI] [PubMed] [Google Scholar]
- Rogers M.E, Ilg T, Nikolaev A.V, Ferguson M.A, Bates P.A. Transmission of cutaneous leishmaniasis by sand flies is enhanced by regurgitation of fPPG. Nature. 2004;430:463–467. doi: 10.1038/nature02675. doi:10.1038/nature02675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M, Seemann M, Horbach S, BringerMeyer S, Sahm H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 1996;118:2564–2566. doi:10.1021/ja9538344 [Google Scholar]
- Sanchez L.B, Galperin M.Y, Muller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming) J. Biol. Chem. 2000;275:5794–5803. doi: 10.1074/jbc.275.8.5794. doi:10.1074/jbc.275.8.5794 [DOI] [PubMed] [Google Scholar]
- Sanchez M.A, Tryon R, Green J, Boor I, Landfear S.M. Six related nucleoside/nucleobase transporters from Trypanosoma brucei exhibit distinct biochemical functions. J. Biol. Chem. 2002;277:21 499–21 504. doi: 10.1074/jbc.M202319200. doi:10.1074/jbc.M202319200 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moreno M, Lasztity D, Coppens I, Opperdoes F.R. Characterization of carbohydrate metabolism and demonstration of glycosomes in a Phytomonas sp. isolated from Euphorbia characias. Mol. Biochem. Parasitol. 1992;54:185–199. doi: 10.1016/0166-6851(92)90111-v. doi:10.1016/0166-6851(92)90111-V [DOI] [PubMed] [Google Scholar]
- Schaible U.E, Schlesinger P.H, Steinberg T.H, Mangel W.F, Kobayashi T, Russell D.G. Parasitophorous vacuoles of Leishmania mexicana acquire macromolecules from the host cell cytosol via two independent routes. J. Cell Sci. 1999;112:681–693. doi: 10.1242/jcs.112.5.681. [DOI] [PubMed] [Google Scholar]
- Schofield P.J, Edwards M.R, Matthews J, Wilson J.R. The pathway of arginine catabolism in Giardia intestinalis. Mol. Biochem. Parasitol. 1992;51:29–36. doi: 10.1016/0166-6851(92)90197-r. doi:10.1016/0166-6851(92)90197-R [DOI] [PubMed] [Google Scholar]
- Schwender J, Seemann M, Lichtenthaler H.K, Rohmer M. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem. J. 1996;316:73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.W, Martin E, Mukkada A.J. Evidence for a functional glyoxylate cycle in the leishmaniae. J. Bacteriol. 1978;135:895–899. doi: 10.1128/jb.135.3.895-899.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R.E. Cell biology. In: Killick-Kendrick R, Peters W, editors. Rodent malaria. Academic Press; London: 1978. pp. 85–168. [Google Scholar]
- Smit A, Mushegian A. Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 2000;10:1468–1484. doi: 10.1101/gr.145600. doi:10.1101/gr.145600 [DOI] [PubMed] [Google Scholar]
- Striepen B, Pruijssers A.J, Huang J, Li C, Gubbels M.J, Umejiego N.N, Hedstrom L, Kissinger J.C. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc. Natl Acad. Sci. USA. 2004;101:3154–3159. doi: 10.1073/pnas.0304686101. doi:10.1073/pnas.0304686101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutak R, Dolezal P, Fiumera H.L, Hrdy I, Dancis A, Delgadillo-Correa M, Johnson P.J, Muller M, Tachezy J. Mitochondrial-type assembly of FeS centres in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl Acad. Sci. USA. 2004;101:10 368–10 373. doi: 10.1073/pnas.0401319101. doi:10.1073/pnas.0401319101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton T.J, Iyer L.M, Anantharaman V, Enomoto S, Abrahante J.E, Subramanian G.M, Hoffman S.L, Abrahamsen M.S, Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. doi:10.1101/gr.2615304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetaud E, Barrett M.P, Bringaud F, Baltz T. Kinetoplastid glucose transporters. Biochem. J. 1997;325:569–580. doi: 10.1042/bj3250569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar J, Fischer A, Clark C.G. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. doi:10.1046/j.1365-2958.1999.01414.x [DOI] [PubMed] [Google Scholar]
- Tovar J, Leon-Avila G, Sanchez L.B, Sutak R, Tachezy J, van der Giezen M, Hernandez M, Muller M, Lucocq J.M. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. doi:10.1038/nature01945 [DOI] [PubMed] [Google Scholar]
- Uttaro A.D, Altabe S.G, Rider M.H, Michels P.A, Opperdoes F.R. A family of highly conserved glycosomal 2-hydroxyacid dehydrogenases from Phytomonas sp. J. Biol. Chem. 2000;275:31 833–31 837. doi: 10.1074/jbc.M006080200. doi:10.1074/jbc.M006080200 [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Slotboom D.J, Horner D.S, Dyal P.L, Harding M, Xue G.P, Embley T.M, Kunji E.R. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 2002;21:572–579. doi: 10.1093/emboj/21.4.572. doi:10.1093/emboj/21.4.572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M, Birdsey G.M, Horner D.S, Lucocq J, Dyal P.L, Benchimol M, Danpure C.J, Embley T.M. Fungal hydrogenosomes contain mitochondrial heat-shock proteins. Mol. Biol. Evol. 2003;20:1051–1061. doi: 10.1093/molbev/msg103. doi:10.1093/molbev/msg103 [DOI] [PubMed] [Google Scholar]
- Vander Jagt D.L, Hunsaker L.A, Campos N.M, Baack B.R. d-Lactate production in erythrocytes infected with Plasmodium falciparum. Mol. Biochem. Parasitol. 1990;42:277–284. doi: 10.1016/0166-6851(90)90171-h. doi:10.1016/0166-6851(90)90171-H [DOI] [PubMed] [Google Scholar]
- Van Hellemond J.J, Opperdoes F.R, Tielens A.G. Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc. Natl Acad. Sci. USA. 1998a;95:3036–3041. doi: 10.1073/pnas.95.6.3036. doi:10.1073/pnas.95.6.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hellemond J.J, Simons B, Millenaar F.F, Tielens A.G. A gene encoding the plant-like alternative oxidase is present in Phytomonas but absent in Leishmania spp. J. Eukaryot. Microbiol. 1998b;45:426–430. doi: 10.1111/j.1550-7408.1998.tb05094.x. [DOI] [PubMed] [Google Scholar]
- van Hellemond J.J, van der Klei A, van Weelden S.W, Tielens A.G. Biochemical and evolutionary aspects of anaerobically functioning mitochondria. Phil. Trans. R. Soc. B. 2003;358:205–213. doi: 10.1098/rstb.2002.1182. doi:10.1098/rstb.2002.1182 discussion 213–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weelden S.W, Fast B, Vogt A, van der Meer P, Saas J, van Hellemond J.J, Tielens A.G, Boshart M. Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J. Biol. Chem. 2003;278:12 854–12 863. doi: 10.1074/jbc.M213190200. doi:10.1074/jbc.M213190200 [DOI] [PubMed] [Google Scholar]
- van Weelden S.W, van Hellemond J.J, Opperdoes F.R, Tielens A.G. New functions for parts of the krebs cycle in procyclic Trypanosoma brucei, a cycle not operating as a cycle. J. Biol. Chem. 2005;280:12 451–12 460. doi: 10.1074/jbc.M412447200. doi:10.1074/jbc.M412447200 [DOI] [PubMed] [Google Scholar]
- Vedrenne C, Bringaud F, Barrett M.P, Tetaud E, Baltz T. The structure–function relationship of functionally distinct but structurally similar hexose transporters from Trypanosoma congolense. Eur. J. Biochem. 2000;267:4850–4860. doi: 10.1046/j.1432-1327.2000.01543.x. doi:10.1046/j.1432-1327.2000.01543.x [DOI] [PubMed] [Google Scholar]
- Vial H.J, Ancelin M.L, Philippot J.R, Thuet M.J. Biosynthesis and dynamics of lipids in Plasmodium-infected mature mammalian erythrocytes. Blood Cells. 1990;16:531–555. discussion 556–561. [PubMed] [Google Scholar]
- Vickers T.J, Greig N, Fairlamb A.H. A trypanothione-dependent glyoxalase I with a prokaryotic ancestry in Leishmania major. Proc. Natl Acad. Sci. USA. 2004;101:13 186–13 191. doi: 10.1073/pnas.0402918101. doi:10.1073/pnas.0402918101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller R.F, et al. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 1998;95:12 352–12 357. doi: 10.1073/pnas.95.21.12352. doi:10.1073/pnas.95.21.12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller R.F, Reed M.B, Cowman A.F, McFadden G.I. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. doi:10.1093/emboj/19.8.1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S.R, Obado S.O, Mauricio I.L, Kelly J.M. Trypanosoma cruzi expresses a plant-like ascorbate-dependent hemoperoxidase localized to the endoplasmic reticulum. Proc. Natl Acad. Sci. USA. 2002;99:13 453–13 458. doi: 10.1073/pnas.202422899. doi:10.1073/pnas.202422899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.A, Hirt R.P, Lucocq J.M, Embley T.M. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. doi:10.1038/nature00949 [DOI] [PubMed] [Google Scholar]
- Wilson R.J, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. doi:10.1006/jmbi.1996.0449 [DOI] [PubMed] [Google Scholar]
- Wu G, Fiser A, ter Kuile B, Sali A, Muller M. Convergent evolution of Trichomonas vaginalis lactate dehydrogenase from malate dehydrogenase. Proc. Natl Acad. Sci. USA. 1999;96:6285–6290. doi: 10.1073/pnas.96.11.6285. doi:10.1073/pnas.96.11.6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, et al. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. doi:10.1038/nature02977 [DOI] [PubMed] [Google Scholar]
- Yarlett N, Martinez M.P, Moharrami M.A, Tachezy J. The contribution of the arginine dihydrolase pathway to energy metabolism by Trichomonas vaginalis. Mol. Biochem. Parasitol. 1996;78:117–125. doi: 10.1016/s0166-6851(96)02616-3. doi:10.1016/S0166-6851(96)02616-3 [DOI] [PubMed] [Google Scholar]
- Zufferey R, et al. Ether phospholipids and glycosylinositolphospholipids are not required for amastigote virulence or for inhibition of macrophage activation by Leishmania major. J. Biol. Chem. 2003;278:44 708–44 718. doi: 10.1074/jbc.M308063200. doi:10.1074/jbc.M308063200 [DOI] [PubMed] [Google Scholar]
- Zuo X, Lockwood B.C, Coombs G.H. Uptake of amino acids by the parasitic, flagellated protist Trichomonas vaginalis. Microbiology. 1995;141:2637–2642. doi: 10.1099/13500872-141-10-2637. [DOI] [PubMed] [Google Scholar]