Abstract
Bacteriophage T7 DNA primase recognizes 5′-GTC-3′ in single-stranded DNA. The primase contains a single Cys4 zinc-binding motif that is essential for recognition. Biochemical and mutagenic analyses suggest that the Cys4 motif contacts cytosine of 5′-GTC-3′ and may also contribute to thymine recognition. Residues His33 and Asp31 are critical for these interactions. Biochemical analysis also reveals that T7 primase selectively binds CTP in the absence of DNA. We propose that bound CTP selects the remaining base G, of 5′-GTC-3′, by base pairing. Our deduced mechanism for recognition of ssDNA by Cys4 motifs bears little resemblance to the recognition of trinucleotides of double-stranded DNA by Cys2His2 zinc fingers.
Single Cys4 zinc-binding motifs participate in protein–nucleic acid interactions. Predominantly, they are found in enzymes involved in DNA replication/transcription. For example, bacteriophage T4 and T7 primases each contain a single Cys4 motif and each recognize specific trinucleotides of single-stranded DNA (ssDNA) (1, 2), whereas peptide fragments containing the Cys4 motif of transcription factor TFIIS interact with ssDNA (3, 4), double-stranded DNA and RNA (5). These motifs occur in other primases (2, 6, 7), transcription factor TFIIE (8), and in subunits of RNA polymerase II and bacterial DNA repair proteins (9). Structural analysis of the single Cys4 zinc-binding motifs found in transcription factors TFIIS and TFIIB suggest that these “zinc ribbons” have little in common with other zinc-binding motifs: their secondary structure is almost entirely β-sheet (3, 9, 10).
T7 DNA primase has been characterized extensively and hence offers advantages for studying the Cys4 motif. Primase is the product of T7 gene 4, a gene that encodes two colinear proteins, a 63-kDa helicase/primase (referred to simply as primase) and a 56-kDa helicase (11). Helicase is translated from an internal AUG codon and lacks the Cys4 motif. It binds to ssDNA as a hexamer (12) and translocates 5′ to 3′ along the DNA by hydrolyzing dTTP (13, 14). On encountering a duplex region, the protein continues to translocate and thus unwinds the DNA.
The 63-kDa gene 4 primase/helicase possesses all of the above activities, but additionally catalyzes the template-directed synthesis of oligoribonucleotides on ssDNA. These are used as primers by T7 DNA polymerase (15–17). The functional primers pppACCC, pppACCA, and pppACAC are synthesized at the recognition sites 5′-GGGTC-3′, 5′-TGGTC-3′, and 5′-CTGTC-3′ (13). However, the minimal primase recognition site, 5′-GTC-3′, is sufficient for recognition and supports the synthesis of pppAC dimers (2). The 3′-cytidine is cryptic: it is required for recognition but is not copied into the primer.
There is homology between the 245 N-terminal amino acids of T7 primase and prokaryotic primases, whereas the C-terminal amino acids (residues 277–566) show homology to prokaryotic helicases (7). Recently, DNA sequences encoding these two domains have been cloned individually. The C-terminal domain has helicase activity (18) and contains the dTTPase site essential for binding to ssDNA and for translocation (19, 20) and the sites for hexamer formation and interaction with T7 DNA polymerase (21, 22). The N-terminal domain has only primase activity (23).
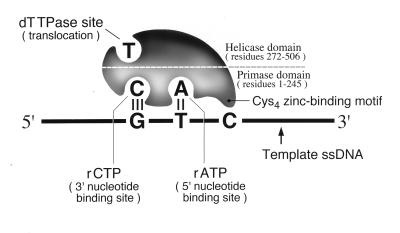
A cartoon depicting the 63-kDa gene 4 protein illustrates the dTTP binding site in the helicase domain and proposes the presence of two NTP binding sites in the primase domain, one for ATP and one for CTP (Fig. 1). Removal or disruption of the Cys4 motif destroys recognition of 5′-GTC-3′ (6, 24). In contrast, a chimeric T7 primase containing the Cys4 motif of bacteriophage T3 primase recognizes the same 5′-GTC-3′ site as native T7 primase. In this protein, the Cys4 loop region is conserved, whereas surrounding sequences are highly substituted (25). However, chimeric T7 primases that contain only the Cys4 loop region of either phage T4 primase or the Escherichia coli primase each recognize, albeit poorly, a novel trinucleotide sequence (26). Clearly, the Cys4 motif is not the sole determinant of sequence specificity.
Figure 1.
Model of T7 primase/helicase. The helicase domain comprises residues 272–566 (7) and contains the dTTP binding site (19, 20). The primase domain, including the Cys4 motif, comprises residues 1–245 (7) and recognizes the sequence 5′-GTC-3′ in ssDNA. Here it synthesizes the dinucleotide 5′-pppApC-3′ from ATP and CTP (23). We denote the ATP and CTP binding sites as the 5′ and 3′ nucleotide binding sites, respectively.
We have used mutational analysis and biochemical characterization of both native and altered primases to examine the role of the Cys4 motif. The results suggest contacts between the Cys4 motif and its trinucleotide recognition sequence. Our data imply that the 5′ and 3′ nucleotide binding sites of T7 primase contribute to sequence specificity.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
E. coli DH5α and BL21(DE3) and plasmid pGP4-6G64S10 have been described (27, 28). Plasmid pET4-6G64S10 was constructed by cloning the NdeI-BglII fragment of pGP4-6G64S10 into the NdeI-BamHI sites of pET11b (Novagen). Each plasmid encodes a T7 primase in which Met64 is substituted with Gly. Primase M64G has all the catalytic activities of T7 primase (28), and hereafter is referred to as wild type. Bacteriophage T7Δ4–1 was provided by S. Tabor (Harvard Medical School) (29).
Enzymes and DNAs.
All primases were purified from E. coli BL21(DE3) containing the expression vector pET4-6G64S10 (21). Enzymes were from Amersham Pharmacia or New England Biolabs. Sequences of oligonucleotide templates were as follows.
For analysis of the 3′ cryptic nucleotide: GTN30:15 (where N is alternatively A, C, G, and T): 5′-GGATTAGCGGAGATTTGAGAGGATATGCGAGGGTNTTTTTTTTTTTTTTT-3′ and GTCx5:10 (where Cx is alternatively C and 4HC): 5′-GCTGATGGTCxAGTGGTATCG-3′. For analysis of T analogs within 5′-GTC-3′: GTxC5:10, (where Tx is alternatively T, dU, C, and 5mC): 5′-CTATCGGGTxCATCCCACAGG-3′.
Construction of Expression Vectors.
Gene 4 mutants used solely for in vivo analyses of gene 4 were constructed from the vector pGP4-6G64S10 by the “2-primer” method (Strategene). Mutants used in both in vivo and in vitro analyses of gene 4 function were constructed in the plasmid pET4–6G64S10 by PCR. Full protocols for the construction of these mutants may be found in the supplemental material on the PNAS web site (www.pnas.org).
Enzyme Assays.
Nucleotide hydrolysis assays were carried out as described (26) with 50 nM M13 DNA and 150 nm primase (monomer). Radioactivity was quantified by using a BAS100 Fuji Bio-Imaging analyzer. Oligonucleotide synthesis assays (30-min incubations) were carried out as described (30) with 50 nM primase (hexamer), 100 μM NTPs as indicated and 100 nM synthetic oligonucleotide template or 20 nM M13 DNA. In template-independent assays, the DNA was omitted, 10 mM MnCl2 replaced the MgCl2, and reactions were treated with calf intestinal alkaline phosphatase before electrophoresis (2).
RESULTS
Polymerization of NTPs by Primase.
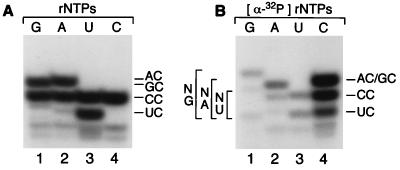
To synthesize the diribonucleotide pppAC, T7 primase must first bind ATP and CTP. Extension of a synthetic pppAC dinucleotide by primase suggested that the dinucleotide is bound before sequence recognition (30). Might the precursors, ATP and CTP, similarly be bound in the active site before template recognition? If so, the nucleotides might themselves play a role in ssDNA recognition. We examined nucleotide binding and polymerization in the absence of a ssDNA template. The 63-kDa primase catalyzes diribonucleotide synthesis, albeit at low levels, in the absence of a template (6), a reaction enhanced by manganese (24).
To examine the first phosphodiester bond formation, synthesis of dinucleotides was monitored in the absence of DNA with each of the four NTPs and MnCl2. Because cytosine is preferentially incorporated into the second position in template-directed dinucleotide synthesis (pppAC), we monitored template-independent dinucleotide synthesis with [α-32P]CTP. In the reaction, the 5′ triphosphate is removed by alkaline phosphatase, so that radiolabel remains only in the phosphodiester linkage. Hence, only dinucleotides that contain cytosine as the second base are detected. In the absence of template DNA, primase catalyzes the synthesis of AC (Fig. 2A, lane 1), CC (all lanes), GC (lane 3), and UC (lane 4) equally well. We conclude that the 5′ nucleotide binding site, to which ATP binds in the template-directed reaction (Fig. 1), is nonspecific.
Figure 2.
Template-independent diribonucleotide synthesis by T7 primase. (A) Variation of the 5′ nucleotide. Oligonucleotide synthesis reactions contained Mn2+ and were performed in the absence of template DNA, with the NTPs indicated and [α-32P]CTP. After synthesis, triphosphate groups were removed by treatment with calf intestinal alkaline phosphatase. Products were separated on a 25% denaturing PAGE gel and autoradiographed. Diribonucleotide species, AC, CC, GC, and UC are identified. (B) Variation of the 3′ nucleotide. Oligonucleotide synthesis reactions were performed and analyzed as above, except that reactions contained the indicated [α-32P]NTP and the three other NTPs. Diribonucleotide species NA, NC, NG, and NU are identified.
Recognition of Guanine in 5′-GTC-3′.
Because any nucleotide may be incorporated in the 5′ position of the dinucleotide, we repeated the reaction in the presence of individual [α-32P]NTPs, the other three NTPs being unlabeled. After phosphatase treatment, the relative incorporation of each nucleotide at the 3′ position can be determined. Relative to NC (100%), the synthesis of NA was 15.2%, NG: 11.6%, and NU: 5.8% (Fig. 2B). In contrast with the lack of selectivity for the 5′ nucleotide, primase preferentially incorporates cytosine at the 3′ end of the dinucleotide.
We conclude that the 3′ nucleotide binding site (Fig. 1) preferentially binds CTP. CTP specificity should enable the site to bind CTP before interaction with the template. Instead of the ssDNA template “selecting” the incoming nucleotide, we propose that the bound nucleotide “selects” the template. In short, we propose that CTP, already bound by T7 primase, base pairs with guanine in the primase recognition site 5′-GTC-3′ and so contributes (although not solely) to ssDNA sequence recognition.
Recognition of Thymine in 5′-GTC-3′.
In the absence of ssDNA, the 5′ nucleotide binding site is nonspecific and binds any nucleotide. However, in the presence of ssDNA, oligonucleotide synthesis is initiated by primase almost exclusively from ATP (13, 17, 31). This preference indicates that in the presence of ssDNA, either the 5′ nucleotide binding site gains ATP specificity or primase recognizes thymine in the template 5′-GTC-3′, which in turn specifies ATP by base pairing.
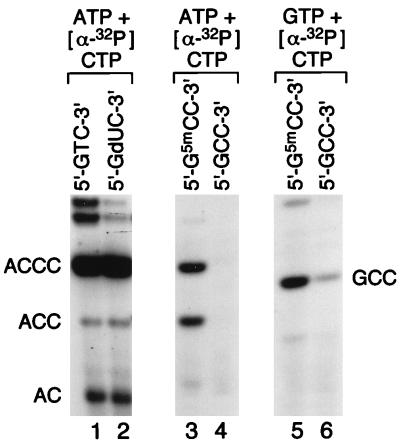
To examine these possibilities, oligonucleotide synthesis reactions were performed on templates containing the nucleotide analogs dU, 5mC, and C in place of thymine in 5′-GTC-3′ (Fig. 3). Primase initiates oligonucleotide synthesis from ATP at both T- and dU-containing sites (lanes 1 and 2) and may also initiate primer synthesis with ATP at a site containing 5-methylcytosine (lane 3) but not cytosine itself (lane 4). Because oligonucleotide synthesis may also be initiated with GTP albeit at a low rate (26), oligonucleotide synthesis reactions were examined with GTP instead of ATP on the templates containing cytosine. Whereas some primer synthesis can be initiated with GTP at the 5′-GCC-3′ site, synthesis is far more efficient at the 5′-G5mCC-3′ site (lanes 5 and 6).
Figure 3.
Oligonucleotide synthesis on templates containing base substitutions for thymine in the primase recognition site 5′-GTC-3′. Oligonucleotide synthesis assays were performed on synthetic oligonucleotide templates with ATP and [α-32P]CTP (lanes 1–4) or GTP and [α-32P]CTP (lanes 5 and 6). The thymine in the recognition site 5′-GTC-3′ was substituted with deoxyuridine (dU), 5-methylcytosine (5mC), or cytosine (C), as indicated (32). After synthesis, products were separated on a 25% denaturing PAGE gel and autoradiographed. Oligonucleotide products are identified.
The reactions illustrated in Fig. 3 allow the roles in specificity of base pairing, ATP selectivity (in the presence of ssDNA), and thymine recognition by primase to be evaluated individually. The slight but measurable incorporation of GTP opposite cytosine in the template 5′-GCC-3′ (lane 6) demonstrates that base pairing in isolation is sufficient to support oligonucleotide synthesis, albeit weakly. In contrast, neither ATP selectivity nor thymine recognition is sufficient individually to support primer synthesis. Thus ATP is not incorporated opposite cytosine in the template 5′-GCC-3′ (lane 4) and no incorporation of GTP is seen opposite thymine in the native template 5′-GTC-3′ (data not shown).
Lanes 3 and 4 in Fig. 3 show the results of two reactions designed to examine the relative contributions of ATP selectivity and thymine recognition toward sequence specificity. There can be no significant base pairing between ATP and 5-methylcytosine, yet primase can incorporate ATP at 5mC-containing sites (lane 3). However, ATP is not incorporated at sites that contain unmodified cytosine (lane 4). Thus the methyl group on C5 of 5-methyl cytosine must be recognized by primase. Extrapolation of this result suggests that primase recognizes the methyl group on C5 of thymine. Similarly, preferential incorporation of GTP at 5-methylcytosine-containing sites over sites containing unmodified cytosine (lanes 5 and 6) implicates the methyl moiety in sequence-specific recognition by primase, from which it can be deduced that the methyl group on C5 of thymine is recognized.
However, methyl group recognition makes only a modest contribution to sequence specificity, which is unmasked solely under suboptimal conditions. Thus, there is little difference between oligonucleotide synthesis initiated with ATP on thymine and uracil-containing templates (Fig. 3, lanes 1 and 2). Therefore, in addition to the major and minor contributions made by base pairing and methyl group recognition, respectively, a further mechanism, as yet undefined, is required to generate the ATP/5′-GTC-3′ specificity of primase that is normally observed. This specificity might involve additional contacts between primase and thymine in the template and/or ATP selectivity gained by primase in the presence of ssDNA.
Mutagenic Studies of the Cys4 Zinc Finger of T7 Primase.
Because the Cys4 motif is critical for primase activity, we examined its role in direct protein–DNA interactions. Previous studies involving a T3/T7 chimeric primase suggested that the Cys4 loop is involved in sequence-specific recognition of ssDNA (25). Consequently, we altered individual amino acids within the Cys4 loop by in vitro mutagenesis and examined their effects on T7 growth and catalytic activities. With the exception of glycine residues, we replaced each of the Cys4 loop residues conserved between T3 and T7 primases with alanine. In some instances, we also made conservative substitutions. The gene 4 mutants were screened for ability to support the growth of T7Δ4–1, a bacteriophage lacking gene 4. T7Δ4–1 is unable to grow in E. coli unless complemented with gene 4 protein expressed from a plasmid (29, 33). The locations and identities of the substitutions and their effects on T7Δ4–1 growth are presented in Fig. 4.
Figure 4.
The Cys4 motif of T7 primase, illustrating position, identity and effect on plating efficiency of single amino acid substitutions in T7 primase. Homology of residues between T3 and T7 primases (11, 34) and identities of individual substitutions are indicated. Numbers represent the efficiency of plating (EOP) of bacteriophage T7Δ4–1, a bacteriophage in which gene 4 is deleted, on E. coli cells containing vectors expressing the indicated gene 4. Efficiencies of plating were calculated relative to those obtained for T7Δ4–1 on an E. coli strain in which wild-type gene 4 was expressed (EOP = 1.0). Mutations encoding alanine substitutions were generated in the vector pGP4-6G64S10. Mutations encoding other substitutions were generated in the vector pET4-6G64S10.
All of the altered primases that contained alanine substitutions were able to support the growth of T7Δ4–1, as evidenced by plating efficiencies greater than 0.1 (Fig. 4). In fact, the only substitution to have a major effect on the growth of T7Δ4–1 was the replacement of serine 27 with aspartate. However, because replacement of this same residue with alanine has no effect on T7 viability, we do not believe that serine 27 plays a crucial role in sequence recognition.
Neither altered sequence recognition by primase nor a lowered overall primase activity would be expected to eliminate primer synthesis altogether. Therefore, provided the altered primases are marginally functional, plating efficiency should not be affected. Consequently, we purified several of the altered T7 primases and characterized them biochemically. T7 primase catalyzes the ssDNA-dependent hydrolysis of dTTP, which fuels unidirectional translocation (14). Hence, dTTPase activity provides a quantitative assay of helicase activity. To ensure that none of our alterations affected primase simply by disrupting helicase activity, we analyzed the dTTPase activity of each of the altered primases. All had relatively normal levels of dTTPase activity. (The results are presented in the supplemental material on the PNAS web site, Table 1, column 2).
Each of the purified mutated primases was next examined for its ability to catalyze the synthesis of oligonucleotides. Because sequence specificity might have been changed by individual amino acid substitutions, oligonucleotide synthesis reactions were performed on M13 ssDNA with both ATP + CTP and GTP + CTP. Little or no synthesis was observed from primases S27D, D31A and D31L. All other primases catalyzed oligoribonucleotide synthesis from ATP + CTP. The products of these reactions are illustrated in Fig. 7, which is published as supplemental material on the PNAS web site.
Recognition of the 3′ Cryptic Base.
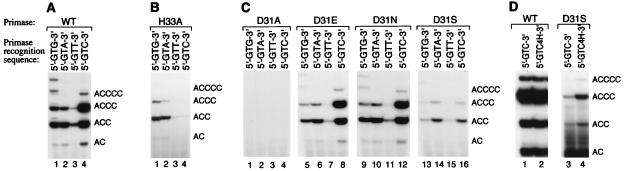
The cryptic base, cytosine, of 5′-GTC-3′ is not involved in base pairing with the primer but is required for initiation of oligonucleotide synthesis (2). This base must therefore be recognized directly by primase. Accordingly, we examined the effect of amino acid substitutions on the recognition of the cryptic cytosine. As a control, oligonucleotide synthesis reactions were carried out with wild-type primase by using synthetic templates that contained the sequences 5′-GTA-3′, 5′-GTC-3′, 5′-GTG-3′, and 5′-GTT-3′ (Fig. 5A). In agreement with previous studies, these data demonstrated that the recognition site 5′-GTT-3′ is used minimally (3.1%). The significant use of 5′-GTA-3′ and 5′-GTG-3′ (12.8% and 15.7%, respectively) was surprising, although RNA-primed DNA synthesis assays used in early studies (13) likely were insufficiently sensitive to detect use of these primase recognition sites.
Figure 5.
Oligonucleotide synthesis by wild-type primase and primases genetically altered at positions 31 and 33 on templates containing base substitutions for the cryptic cytosine. Oligonucleotide synthesis assays were performed with ATP and [α-32P]CTP on synthetic ssDNA oligonucleotide templates containing primase recognition sites of sequence 5′-GCN-3′, where N is A, C, G, or T, as indicated (Fig. 5 A–C) or 5′-GCCx-3′, where Cx is either C or 4HC, as indicated (Fig. 5D). After synthesis, products were separated on a 25% denaturing PAGE gel and autoradiographed. Oligonucleotide products are identified. (A) Wild-type primase. (B) Primase H33A. (C) Primases D31A, D31E, D31N and D31S. (D) Wild-type and D31S primases (for comparison with D31S, the products of wild-type primase were diluted 10-fold before electrophoresis).
The oligonucleotide synthesis reactions were repeated with the mutationally altered primases. Alterations to positions Asp24 and Asn26 yielded primases that deviated from wild-type primase activity only in that recognition of 5′-GTA/G-3′ was diminished (data not shown). As might be predicted from earlier data, no synthesis was detected from primase S27D. However, alterations to two positions, His33 and Asp31, significantly affected the sequence specificity of primase.
As shown in Fig. 5B, substitution of alanine at position 33 had a striking effect on sequence recognition. Primase H33A is unable to synthesize oligonucleotides on a template that contains the normal recognition site 5′-GTC-3′. Surprisingly, however, this altered primase does have activity on the templates 5′-GTA/G-3′. Thus primase H33A can recognize templates in which the cryptic nucleotide is a purine but is incapable of oligonucleotide synthesis on templates in which the cryptic base is cytosine. We conclude that His33 plays an essential role in the recognition of the cryptic cytosine. Underlying cryptic purine recognition is unmasked only when the normal recognition of cytosine is prevented.
The Role of Asp31 in Primer Synthesis.
The second interesting residue identified in the Cys4 motif is Asp31. Substitution of Asp31 with alanine yields a primase that is inactive on all synthetic templates (Fig. 5C, lanes 1–4). Similarly, substitution of Asp31 with leucine yields a primase with undetectable activity on these templates (data not shown). In contrast, substitution of Asp31 with another acidic residue, glutamate, or an amide, asparagine (Fig. 5C, lanes 5–12), yields primases that synthesize primers in proportions similar to wild-type primase (Fig. 5A). The inactivities of primases D31A and D31L on all templates suggest that Asp31 plays an essential role in primer synthesis. In conjunction, the activity of primase D31N (Fig. 5C, lanes 9–12) negates a requirement for negative charge and suggests the importance of a hydrogen bond acceptor at position 31.
Interestingly, unlike the other alterations at position 31, the substitution of serine for aspartate affects the relative use of the synthetic templates. Comparison of the data in Fig. 5 A and C, lanes 13–16, shows that use of the template 5′-GTC-3′ by primase D31S is significantly reduced. Because cytosine recognition is disrupted by the replacement of Asp (a hydrogen bond acceptor) with Ser (which can act either as a hydrogen bond donor or acceptor), we examined cytosine itself for moieties that might hydrogen bond with Asp in the wild-type enzyme. Cytosine contains only one such moiety: the amino group on C4. To investigate the role of this amino group, we carried out oligonucleotide synthesis reactions with both wild-type and the D31S primases on a template containing the sequence 5′-GT4HC-3′. In agreement with earlier results (32), removal of the amino group had little effect on wild-type primase activity (Fig. 5D). However, removal of this amino group stimulated primer synthesis 2.6-fold by the substituted primase D31S (Fig. 5D).
We conclude that the amino group on C4 of cytosine inhibits oligonucleotide synthesis by primase D31S. Inhibition most likely results from unfavorable interaction(s) involving the amino group of cytosine. Most probably, substitution of Ser for Asp31 leads to a conformational change within primase that in turn causes a steric conflict between primase and the amino group on C4 of cytosine. There is, however, an alternative explanation. In the wild-type primase, Asp31 might also form a hydrogen bond with the amino group of cytosine. This electrostatic attraction could be converted to repulsion by the substitution of Ser for Asp31, but might be maintained by the substitutions D31E and D31N.
DISCUSSION
We have examined the role of the Cys4 zinc motif, or “zinc ribbon,” in sequence-specific recognition of ssDNA. We now summarize our findings and consider their relevance to other single Cys4 motif-containing proteins.
Sequence-Specific Recognition of ssDNA by T7 Primase.
At least three independent regions of T7 primase contribute to the sequence-specific recognition of 5′-GTC-3′. We propose that the primase 3′ nucleotide binding site (Fig. 1) initially binds CTP, which in turn, by base pairing, recognizes G of 5′-GTC-3′. Meanwhile, the Cys4 motif, using residue His33, contributes to the recognition of C in 5′-GTC-3′. Our results also suggest that primase recognizes the methyl group on C5 of the central nucleotide, T, of 5′-GTC-3′ by an unknown mechanism. Because we have demonstrated that ATP specificity of the 5′ nucleotide binding site is gained only in the presence of ssDNA, we suggest that this specificity results from base pairing between ATP and T in 5′-GTC-3′. However, Asp31 may also contribute to ATP recognition, perhaps by hydrogen bonding with ATP in the 5′ nucleotide binding site (see below).
The Mechanism of Oligonucleotide Synthesis by T7 Primase.
E. coli RNA polymerase synthesizes RNA by a mixture of monotonous and “inchworm-like” movements (35). In monotonous movement, the entire RNA polymerase moves forward by one nucleotide at a time. During “inchworm-like” movement, the front end of the enzyme remains anchored while the catalytic site continues to polymerize nucleotides (35). E. coli RNA polymerase is thought to use the “inchworm” model of elongation during initiation of RNA synthesis, and it is possible that E. coli primase uses a similar mechanism. In each case, if the initiating nucleotide, ATP, is crosslinked to the enzyme, nucleotide extension can continue for four or five bases in the case of primase (36) or for eight or nine bases in the case of RNA polymerase (37). We believe that an analogous mechanism may operate in T7 primase, in which ATP is held in the 5′ nucleotide binding site, while elongation is performed by the 3′ nucleotide binding site.
In accordance with this model, our finding that the 3′ nucleotide binding site is CTP specific may account for hitherto unexplained results. During “pseudotemplating” an additional cytosine, not specified by the template, is added to the 3′ end of a primer (2). Furthermore, on long polyguanidine templates, long poly-C-containing oligonucleotides may be synthesized (38). Although T7 primase can synthesize long oligonucleotides (25, 38), the incorporation of cytidylate is favored greatly, and only those with high cytosine contents are extended by T7 DNA polymerase (13, 16, 17, 38). We propose that this bias also results from the high affinity of the 3′ nucleotide binding site: a primer containing cytosine at its 3′ end is held more tightly by T7 primase and is therefore passed more efficiently to T7 DNA polymerase.
The Cys4 Motif of T7 Primase: Interactions with ssDNA and ATP.
Our results demonstrate that two residues within the Cys4 motif, Asp31 and His33, play major roles in sequence recognition. We have suggested the importance of a hydrogen bond acceptor at position 31. As such, Asp31 might simply be involved in the structural integrity of primase. Alternatively, Asp31 might accept a hydrogen bond from thymine in the site 5′-GTC-3′ or from the incoming 5′ nucleotide, ATP. Of these, we consider a hydrogen bond between thymine and Asp31 unlikely because thymine has only one hydrogen bond donor, which is required for base pairing with ATP. In contrast, after base pairing ATP still has a hydrogen bond donor available, the second proton of the C6 amino group. Studies with E. coli primase have shown that the initiating nucleotide, ATP, can be crosslinked both to residues within the nucleotide binding site and to residues in its CysHisCys2 zinc finger (36). We therefore propose a hydrogen bond between the amino group on C6 of ATP and the carboxyl group of Asp31.
The role of the second important residue, His33, is more easily defined. We have demonstrated that His33 is essential for cryptic cytosine recognition. Substitution of alanine for His33 might alter the specificity of primase simply by making space for a larger purine base at this position in the template. More likely, the substitution may directly eliminate an interaction with cytosine. Inasmuch as earlier studies had shown that N3 of the cryptic cytosine is essential for recognition (32), we propose that this nitrogen hydrogen bonds with an imidazole proton of His33.
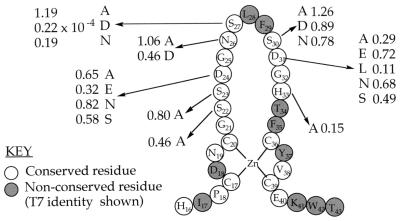
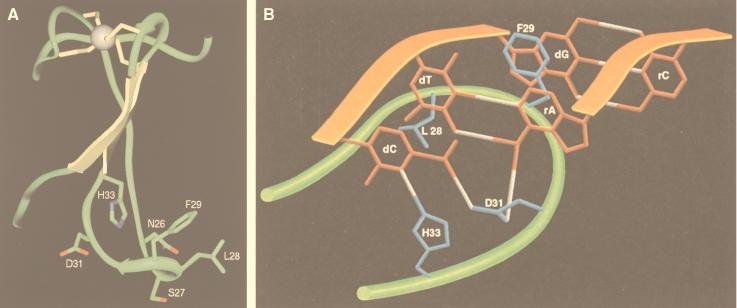
Because the Cys4 motifs of T7 primase and TFIIS share a common consensus sequence (9), a molecular model of the T7 primase Cys4 domain based on the Cys4 domain of TFIIS (PDB:1TFI) was constructed (Fig. 6A). Unfortunately, attempts to generate a similarly minimized segment of an RNA–DNA helix comprising 5′-GTC-3′ (DNA) and 5′-AC-3′ (RNA) were unsuccessful: energy minimization merely corrupted the base pairing between the two strands of nucleic acid. We therefore constructed a physical replica of residues Asp24-His33 of the energy-minimized Cys4 motif and a DNA/RNA helix of sequence 5′-GTC-3′/5′-AC-3′. We then docked the two models together by connecting Asp31 to the amino groups of ATP and dCMP and the imidazole proton of His33 to N3 of cytosine with hydrogen bonds. Rotation around the bonds between the α, β, and γ carbons of each side chain permitted parallel orientation of all bases in the RNA/DNA strands. The resulting model of the Cys4 motif/DNA/RNA complex is illustrated in cartoon format (Fig. 6B). Although highly speculative, the model is consistent with observed results. For example, in addition to the proposed hydrogen bonds, Ser27 is in sufficiently close proximity to form a hydrogen bond with the sugar-phosphate backbone between bases G and T of the trinucleotide. Consistent with such an interaction, we find that an acidic residue cannot be tolerated at position 27. If Ser27 does form a hydrogen bond with the sugar-phosphate backbone, then substitution S27D, which destroyed primase activity, would lead to a repulsion between the protein and ssDNA. Two hydrophobic interactions are also suggested by the model: a van der Waals interaction between the methyl group of thymine and Leu28 and an intercalation of Phe29 between bases T and G. These hydrophobic residues are functionally conserved in the Cys4 motif of bacteriophage T3 primase (34). The proposed contacts are all formed with the major groove of the RNA/DNA helix.
Figure 6.
Models of the Cys4 zinc-binding motif of T7 primase. (A) Model based on the zinc ribbon of TFIIS. Coordinates of the NMR ensemble of TFIIS (nucleic acid binding domain, Homo sapiens, Protein Data Bank ID code 1TFI) (9) were used to model the Cys4 motif of bacteriophage T7 primase. Initially, the first nine conformers of TFIIS were aligned and structurally conserved regions (SCRs) were identified by automatic structure alignment [insightII ver. 95.0 Homology module, default parameters (43)]. The amino acid sequence of TFIIS was sequentially aligned, and coordinates of the T7 primase were built from that of TFIIS by using coordinates previously determined by SCRs. The gaps were consequentially closed by using coordinates of one of the conformers of TFIIS. Energy minimization was then applied by using the Amber forcefield within the insightII ver. 95.0 Discover module (44). Fifty steps of steepest decent energy minimization were used, followed by 200 steps of conjugate gradient energy minimization. (B) Illustration of proposed interaction between residues Asp24–His33 of the T7 primase Cys4 motif, the ssDNA recognition sequence, 5′-GTC-3′, and the diribonucleotide AC. Peptide and phosphodiester backbones are represented as green and orange ribbons, respectively.
Is There a Conserved Role for the Single Cys4 Motif?
Many examples of Cys2His2 zinc fingers are known and the role of these “mini-domains,” first proposed by Miller et al. (39), has been analyzed from structures of protein–DNA complexes (e.g., 40). In essence, one zinc finger contacts three bases of double-stranded DNA. Protein–DNA contacts are made by residues in known positions within the motif and consecutive zinc fingers may bind successive trinucleotides of double-stranded DNA (40). In comparison, single Cys4 “zinc ribbons” are structurally dissimilar to any other type of DNA-binding domain (3). Although the Cys4 motif of T7 primase also mediates the recognition of three bases of DNA, it is ssDNA rather than double-stranded DNA. Moreover, existing characterizations of single Cys4 motifs suggest that the motif contributes to protein–DNA interactions, rather than making those interactions per se. For example, in TFIIS both the Cys4 motif and a contiguous region of the protein are required for protein–ssDNA interaction (4). Similarly, although the two subunits of TFIIE bind ssDNA in concert, the large subunit, which contains a Cys4 motif, facilitates binding to ssDNA by the small subunit, rather than interacting with the DNA itself (41). Again, adenovirus E1A protein is thought to activate transcription by interacting with sequence-specific transcription factors, an activity for which both its Cys4 motif and a contiguous region are essential (42).
Given that single TFIIS-like Cys4 motifs are found in functionally related proteins that bind ssDNA, we suggest that Cys4 “zinc ribbons” have a conserved role. Our results suggest that the Cys4 motif of T7 primase recognizes only one base of the trinucleotide 5′-GTC-3′ directly, and that it contributes to the recognition of a second base. However, the third base appears to be recognized by a different domain of T7 primase: the 3′ nucleotide binding site. We postulate that rather than recognizing a trinucleotide of ssDNA directly, the Cys4 motif instead recognizes one or two bases of ssDNA and so stabilizes DNA recognition by other domains of T7 primase. By extrapolation, we propose that the single Cys4 motif may act similarly in other proteins.
Supplementary Material
Acknowledgments
We are grateful to Benjamin B. Beauchamp, Joonsoo Lee, and Stanley Tabor for providing purified wild-type T7 gene 4 primase and bacteriophage T7Δ4–1. We thank Dr. Ian O. Sutherland (University of Liverpool, Liverpool, U.K.) and Dr. Thomas Ellenberger (Harvard Medical School) for helpful discussions and David Frick, Leo Guo, Ingrid Richardson, and Michael Sawaya for critical readings of the manuscript. This investigation was supported by grant number AI-06045 from the United States Public Health Service and grant number NP-1–28 from the American Cancer Society to C.C.R. and by a grant from the Pharmaceutical Institute, Aston University, Birmingham, U.K. to A.V.H..
ABBREVIATION
- ssDNA
single-stranded DNA
References
- 1.Cha T A, Alberts B M. J Biol Chem. 1986;261:7001–7010. [PubMed] [Google Scholar]
- 2.Mendelman L V, Richardson C C. J Biol Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- 3.Qian X, Jeon C J, Yoon H S, Agarwal K, Weiss M A. Nature (London) 1993a;365:277–279. doi: 10.1038/365277a0. [DOI] [PubMed] [Google Scholar]
- 4.Qian X, Jeon C J, Yoon H S, Agarwal K, Weiss M A. Nature (London) 1995;376:279. [Google Scholar]
- 5.Agarwal K, Baek K H, Jeon C J, Miyamoto K, Ueno A, Yoon H S. Biochemistry. 1991;30:7842–7851. doi: 10.1021/bi00245a026. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein J A, Richardson C C. Proc Natl Acad Sci USA. 1988;85:396–400. doi: 10.1073/pnas.85.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilyina T V, Gorbalenya A E, Koonin E V. J Mol Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 8.Feaver W J, Henry N L, Bushnell D A, Sayre M H, Brickner J H, Gileadi O, Kornberg R D. J Biol Chem. 1994;269:27549–27553. [PubMed] [Google Scholar]
- 9.Qian X, Gozani S N, Yoon H S, Jeon C J, Agarwal K, Weiss M A. Biochemistry. 1993b;32:9944–9959. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Zeng Q, Colangelo C M, Lewis L M, Summers M F, Scott R A. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 11.Dunn J J, Studier F W. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 12.Hingorani M M, Patel S S. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- 13.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matson SW, Richardson C C. J Biol Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- 15.Scherzinger E, Lanka E, Hillenbrand G. Nucleic Acids Res. 1977;4:4151–4163. doi: 10.1093/nar/4.12.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romano L J, Richardson C C. J Biol Chem. 1979a;254:10483–10489. [PubMed] [Google Scholar]
- 17.Fujiyama A, Kohara Y, Okazaki T. Proc Natl Acad Sci USA. 1981;78:903–907. doi: 10.1073/pnas.78.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird L E, Hakansson K, Pan H, Wigley D B. Nucleic Acids Res. 1997;25:2620–2626. doi: 10.1093/nar/25.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notarnicola S M, Richardson C C. J Biol Chem. 1993;268:27198–27207. [PubMed] [Google Scholar]
- 20.Patel S S, Hngorani M M, Ng W M. Biochemistry. 1994;33:7857–7868. doi: 10.1021/bi00191a013. [DOI] [PubMed] [Google Scholar]
- 21.Notarnicola S M, Park K, Griffith J D, Richardson C C. J Biol Chem. 1995;270:20215–20224. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- 22.Notarnicola S M, Mulcahy H L, Lee J, Richardson C C. J Biol Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 23.Frick D N, Baradaran K, Richardson C C. Proc Natl Acad Sci USA. 1998;95:7957–7962. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendelman L V, Beauchamp B B, Richardson C C. EMBO J. 1994;13:3909–3916. doi: 10.1002/j.1460-2075.1994.tb06702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hine A V, Richardson C C. Proc Natl Acad Sci USA. 1994;91:12327–12331. doi: 10.1073/pnas.91.25.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusakabe T, Richardson C C. J Biol Chem. 1996;271:19563–19570. doi: 10.1074/jbc.271.32.19563. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, New York: Cold Spring Harbor Lab. Press; 1989. pp. A9–A12. [Google Scholar]
- 28.Mendelman L V, Notarnicola S M, Richardson C C. J Biol Chem. 1993;268:27208–27213. [PubMed] [Google Scholar]
- 29.Tabor S. Ph.D. thesis. Boston, MA: Harvard University Medical School; 1987. [Google Scholar]
- 30.Kusakabe T, Richardson C C. J Biol Chem. 1997a;272:12446–12453. doi: 10.1074/jbc.272.19.12446. [DOI] [PubMed] [Google Scholar]
- 31.Romano L J, Richardson C C. J Biol Chem. 1979b;254:10476–10482. [PubMed] [Google Scholar]
- 32.Mendelman L V, Kuimelis R G, McLaughlin L W, Richardson C C. Biochemistry. 1995;34:10187–10193. doi: 10.1021/bi00032a012. [DOI] [PubMed] [Google Scholar]
- 33.Mendelman L V, Notarnicola S M, Richardson C C. Proc Natl Acad Sci USA. 1992;89:10638–10642. doi: 10.1073/pnas.89.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck P J, Gonzalez S, Ward C L, Molineux I J. J Mol Biol. 1989;210:687–701. doi: 10.1016/0022-2836(89)90102-2. [DOI] [PubMed] [Google Scholar]
- 35.Nudler E, Goldfarb A, Kashlev M. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 36.Mustaev A A, Godson G N. J Biol Chem. 1995;270:15711–15718. doi: 10.1074/jbc.270.26.15711. [DOI] [PubMed] [Google Scholar]
- 37.Mustaev A, Kashlev M, Zaychikov E, Grachev M, Goldfarb A. J Biol Chem. 1993;268:19185–19187. [PubMed] [Google Scholar]
- 38.Kusakabe T, Richardson C C. J Biol Chem. 1997b;272:5943–5951. doi: 10.1074/jbc.272.9.5943. [DOI] [PubMed] [Google Scholar]
- 39.Miller J, McLachlan A D, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim C A, Berg J M. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 41.Kuldell N H, Buratowski S. Mol Cell Biol. 1997;17:5288–5298. doi: 10.1128/mcb.17.9.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisberg J V, Lee W S, Berk A J, Ricciardi R P. Proc Natl Acad Sci USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Homology User Guide (1995) (Biosym/Molecular Simulations Inc., San Diego, CA).
- 44.Discover User Guide (1995) (Biosym/Molecular Simulations, Inc., San Diego, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.