Abstract
The MYH (MutY glycosylase homologue) increases replication fidelity by removing adenines or 2-hydroxyadenine misincorporated opposite GO (7,8-dihydro-8-oxo-guanine). The 9-1-1 complex (Rad9, Rad1 and Hus1 heterotrimer complex) has been suggested as a DNA damage sensor. Here, we report that hMYH (human MYH) interacts with hHus1 (human Hus1) and hRad1 (human Rad1), but not with hRad9. In addition, interactions between MYH and the 9-1-1 complex, from both the fission yeast Schizosaccharomyces pombe and human cells, are partially interchangeable. The major Hus1-binding site is localized to residues 295–350 of hMYH and to residues 245–293 of SpMYH (S. pombe MYH). Val315 of hMYH and Ile261 of SpMYH play important roles for their interactions with Hus1. hHus1 protein and the 9-1-1 complex of S. pombe can enhance the glycosylase activity of SpMYH. Moreover, the interaction of hMYH–hHus1 is enhanced following ionizing radiation. A significant fraction of the hMYH nuclear foci co-localizes with hRad9 foci in H2O2-treated cells. These results reveal that the 9-1-1 complex plays a direct role in base excision repair.
Keywords: base excision repair, DNA damage checkpoint, DNA glycosylase, fission yeast, Hus1, MutY glycosylase homologue (MYH)
Abbreviations: the 9-1-1 complex, Rad9, Rad1 and Hus1 heterotrimer complex; AP, apurinic/apyrimidinic; APE1, AP endonuclease 1; ATM, ataxia telangiectasia mutated; ATR, ATM- and Rad3-related protein; ATRIP, ATR-interacting protein; BRCA1, breast-cancer susceptibility gene 1; DAPI, 4′,6′-diamidino-2-phenylindole; FEN1, flap endonuclease 1; GO, 7,8-dihydro-8-oxo-guanine; GST, glutathione S-transferase; HA, haemagglutinin; hHus1, human Hus1; hRad1, human Rad1; MSH2, MutS homologue 2; MLH1, MutL homologue 1; MYH, MutY glycosylase homologue; hMYH, human MYH; mMYH, mouse MYH; PCNA, proliferating-cell nuclear antigen; hPCNA, human PCNA; RFC, replication factor C; rMYH, rat MYH; RPA, replication protein A; SpHus1, S. pombe Hus1; SpMYH, S. pombe MYH; XPA, xeroderma pigmentosum group A; XPF, xeroderma pigmentosum group F
INTRODUCTION
Oxidative damage to DNA can induce mutagenesis and lead to degenerative diseases. One of the most abundant and highly mutagenic type of oxidative damage to DNA is GO (7,8-dihydro-8-oxo-guanine). If not repaired, GO lesions in DNA can produce A/GO (adenine/GO) mismatches during DNA replication [1] and result in G:C to T:A transversions [2,3]. hMYH [human MYH (MutY glycosylase homologue)] reduces G:C to T:A mutations by removing adenines or 2-hydroxyadenines mispaired with guanines or GO that arise through DNA replication errors [4–6]. Germline mutations in the hMYH gene cause autosomal recessive colorectal adenomatous polyposis, which is characterized by multiple adenomas, some of which progress to cancer [7,8].
Cell-cycle checkpoints are surveillance mechanisms that monitor the cell's state, maintain telomere stability and preserve genomic integrity (reviewed in [9]). The signal transduction pathways triggered by DNA damage involve many components including sensors, transducers and effectors. Human ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related protein) are phosphoinositol phosphate 3-kinase-related kinases. After stress, ATM or ATR is activated and can transduce the DNA damage signal by phosphorylating many proteins in a Rad9-, Rad1-, Hus1- and Rad17-dependent manner. Rad9, Rad1 and Hus1 form a heterotrimer complex [referred as the 9-1-1 complex (Rad9, Rad1 and Hus1 heterotrimer complex)] that has predicted structural homology to PCNA (proliferating-cell nuclear antigen) sliding clamp [10,11]. Rad17 protein is a paralogue of the largest subunit of RFC (replication factor C), and it forms the alterative clamp loader with RFC2-5. The 9-1-1 complex is loaded on to DNA by Rad17–RFC [12,13]. hATM/hATR, the 9-1-1 complex and Rad17 are proposed to act at an early step of DNA damage response to sense the DNA damage and to lead to cell-cycle arrest or apoptosis (reviewed in [9]). It has been suggested that these checkpoint proteins may detect a common intermediate, such as single-stranded DNA coated by RPA (replication protein A), which is processed by various DNA repair pathways [14]. RPA has been shown to directly interact with the 9-1-1 complex [15]. Recently, several reports support a hypothesis that checkpoint proteins may require a series of ‘adaptors’ to recognize DNA damage [16–18]. Such adaptor proteins may be DNA damage recognition proteins involved in mismatch repair, nucleotide excision repair and double-strand break repair.
We have shown that MYH is directly associated with PCNA in both the fission yeast Schizosaccharomyces pombe and human cells [19,20]. It has been suggested that the coupling between the hMYH base excision repair pathway and DNA replication may provide a signal to target the MYH repair to the daughter DNA strands [20–22]. In addition, we have shown that the S. pombe 9-1-1 complex is associated with SpMYH (S. pombe MYH) and that the DNA-damage-induced SpHus1 (S. pombe Hus1) phosphorylation is dependent on SpMYH expression [23]. In the present study, we show that hMYH physically interacts with hHus1 (human Hus1) and hRad1 (human Rad1), but not with hRad9. Interactions between MYH and the 9-1-1 complex, from both S. pombe and human cells, are partially interchangeable. hHus1 interacts with hMYH at a region that is different from the PCNA-interacting motif. We demonstrate, for the first time, that Val315 of hMYH and Ile261 of SpMYH are important for Hus1 interaction. The DNA glycosylase activity of SpMYH is stimulated by hHus1 and the S. pombe 9-1-1 complex. Moreover, the interaction of hMYH–hHus1 is enhanced by ionizing radiation. A significant fraction of the hMYH nuclear foci co-localizes with hRad9 foci in H2O2-treated cells. Recently, the 9-1-1 complex has been shown to interact with and stimulate the enzymes involved in base excision repair, which include polymerase β [24], FEN1 (flap endonuclease 1) [25,26] and DNA ligase 1[27,28]. Thus the 9-1-1 complex serves both as a damage sensor and as a component of base excision repair.
MATERIALS AND METHODS
Human cell lines and extracts
Human HeLa cell line was purchased from American Type Cell Culture (Manassas, VA, U.S.A.). The HeLa cells were grown in Joklik's minimum Eagle's medium with 5% (v/v) fetal bovine serum, 2 g/l NaHCO3, 1% non-essential amino acids, 4 mM L-glutamine and 1% penicillin/streptomycin at 37 °C in 5% CO2. Cells in T25 flasks were grown to approx. 80% confluence, treated by 10 Gy irradiation on a PantakHF320 X-ray machine and then recovered at different times in serum-free media. HeLa cells were treated with 5 mM H2O2 for 40 min and then recovered in serum-free media for 6 h. Nuclear and cell extracts were prepared as described in [20,29]. The protein concentration was determined by the Bio-Rad protein assay (Bio-Rad).
GST (glutathione S-transferase) fusion protein constructs
The constructs of GST fusions of intact, ΔC1, ΔC2 and ΔN2 hMYH, as well as GST–SpMYH-(245–461), have been described [20,23]. The cDNA fragments containing residues 1–350 of hMYH (ΔC3), residues 351–535 of hMYH (ΔN5) and residues 294–461 of SpMYH fused to the GST gene were made by PCR method (see Supplementary material at http://www.BiochemJ.org/bj/400/bj4000053add.htm). The PCR products were digested with BamHI and XhoI and ligated into the BamHI–XhoI-digested pGEX-4T-2 vector (GE Health). The V315A mutant of the hMYH gene and the I261A mutant of the SpMYH gene were constructed by the PCR splicing overlap extension method [30] (see Supplementary material). The plasmids pGEX-3X-hHus1 (hHus1 coding region inserted at BamHI and XhoI sites) and pGEX-4T3-hRad9 (hRad9 coding region inserted at BamHI and EcoRI sites), which contained GST-tagged hHus1 and hRad9 respectively, were obtained from Dr A. E. Tomkinson (University of Maryland, Baltimore, MD, U.S.A.). The pGEX-3X-hRad1 plasmid (Rad1 coding region inserted at BamHI and EcoRI sites) was obtained from Dr E. Y. Lee (University of California, Irvine, CA, U.S.A.) through Dr A. E. Tomkinson.
GST pull-down assay
The GST pull-down assay was similar to the described procedures [20]. The GST-tagged proteins were expressed in Escherichia coli (BL21-Star/DE3) cells (Stratagene), which harbour the expression plasmids, and immobilized on glutathione–Sepharose 4B. The beads, containing approx. 300 ng of proteins, were incubated with 0.1 μg of target proteins, 0.4 mg of nuclear extracts or 0.3 μg of partially purified S. pombe 9-1-1 complex overnight at 4 °C. A control was run concurrently with immobilized GST alone. After centrifugation at 1000 g for 2 min, the pellets were washed four times. The pellets were analysed on a 10% polyacrylamide gel containing SDS and transferred to a nitrocellulose membrane. Western-blot analyses were performed (see below).
Co-immunoprecipitation
HeLa whole cell extracts (0.8 mg) were precleared by adding 30 μl of Protein G–agarose (Invitrogen) for 1–4 h at 4 °C. After centrifugation at 1000 g for 2 min, the supernatant was incubated with 1 μg of anti-hMYH peptide 516 (α516 against residues 516–534) antibodies [31] overnight at 4 °C. Protein G–agarose (30 μl) was added and incubated for 4–12 h at 4 °C. After centrifugation at 1000 g for 2 min, the supernatant was saved and the pellet was washed. Both the supernatant (∼10% of total volume) and pellet fractions were resolved by SDS/10% PAGE and Western-blot analyses for PCNA and hHus1 were performed (see below).
Western blotting
Proteins were fractionated by SDS/PAGE and transferred to a nitrocellulose membrane. The membranes were allowed to react with antibodies against an hMYH peptide (α516) [31], SpMYH [32], hPCNA (human PCNA) (Calbiochem–Novabiochem), hHus1 (sc-8323; Santa Cruz Biotechnology), HA (haemagglutinin) probe (sc-7392; Santa Cruz Biotechnology), N-terminal histidine-probe (sc-8036; Santa Cruz Biotechnology), C-terminal histidine-probe (Invitrogen), S-tag (sc-802; Santa Cruz Biotechnology) and actin (sc-1616; Santa Cruz Biotechnology). Western blotting was detected by the ECL® (enhanced chemiluminescence) analysis system (GE Health) according to the manufacturer's protocol.
Expression and purification of the hHus1 protein
The E. coli BL21-Star cells (Stratagene) that harbour the expression plasmid of hHus1–His protein (see Supplementary material) were grown and induced such as for GST–hMYH. The hHus1–His protein was purified by Ni-NTA (Ni2+-nitrilotriacetate) resin (Qiagen) under native conditions according to the manufacturer's protocol. The hHus1–His protein was dialysed twice with TEG buffer (50 mM Tris/HCl, pH 7.4, 0.1 mM EDTA, 50 mM KCl, 10% glycerol, 0.5 mM dithiothreitol and 0.1 mM PMSF) and further purified by a 1 ml heparin column (GE Health) equilibrated with TEG buffer. Upon washing with 5 ml of equilibration buffer, the column was eluted with a 30 ml linear gradient of KCl (0.05–0.8 M) in TEG buffer. The fractions that contain most of the hHus1–His protein eluted at 0.5 M KCl. This was confirmed by SDS/PAGE and Western blotting with histidine antibody (Santa Cruz Biotechnology). The fractions were then divided into small aliquots and stored at −80 °C. The protein concentration was determined by the Bradford method [33].
Expression and purification of the recombinant SpRad9–Rad1–Hus1 complex
E. coli BL21-Star cells (Stratagene), harbouring both pET21a-SpHus1 and pACYCD-SpRad1-SpRad9 (see Supplementary material), were grown and induced such as for GST–hMYH. The 9-1-1 complex was purified from 14 g of cell paste by 45% ammonium sulfate precipitation, heparin and phosphocellulose chromatographies similar to the described procedures [34]. Fraction V (the pool from phosphocellulose column) was precipitated by 45% ammonium sulfate and dialysed against buffer A containing 200 mM KCl to obtain FVAd. The sample was then divided into small aliquots and stored at −80 °C. The FVAd (0.2 ml, 31 μg of protein) was layered on top of 5.0 ml 15–35% (v/v) glycerol gradient. The gradients were spun for 22 h at 45000 rev./min in an AH650 rotor (Sovall) at 4 °C. The fractions of two drops were collected from the bottom of the tube and aliquots were analysed by SDS/PAGE and Western blotting. The elution positions of the marker proteins (bovine thyroglobin, apoferritin, β-amylase and BSA) were determined by Coomassie Blue staining.
Other proteins used
The recombinant SpMYH expressed in E. coli was purified as described in [35]. HA-tagged SpHus1 in S. pombe extracts has been described [23]. Histidine-tagged SpHus1 in E. coli extracts was prepared from Rosetta cells (EMD Biosciences) that expressed pET21a-SpHus1 (see Supplementary material). hMYH, with streptococcal Protein G (GB1 domain) at its N-terminus and a 6-Histidine tag at its C-terminus, was partially purified as described in [36].
Assays of SpMYH binding and glycosylase activities
The binding and glycosylase assays for purified recombinant SpMYH with an A/GO-containing DNA were described previously [35]. The DNA substrate was a 20-mer duplex DNA containing an A/GO mismatch (see Supplementary Table S1 in Supplementary material at http://www.BiochemJ.org/bj/400/bj4000053add.htm) that was labelled at the 5′-end of the mismatched A-containing strand as described in [34].
Immunofluorescent staining
Human HeLa cells were grown in Lab-Tek chamber slides (Nunc) overnight, treated with 5 mM H2O2 for 40 min and then recovered in serum-free media for 6 h. The cells were fixed with 4% formaldehyde for 15 min and permeabilized at room temperature (23 °C) in PBS+0.1% Triton for 10 min. After being blocked in PBS containing 15% fetal bovine serum for 15 min at 37 °C, the cells were reacted with hMYH polyclonal antibody and hRad9 monoclonal antibody (Imgenex) at 37 °C for 30 min. Next, the cells were washed three times for 15 min each in PBS and incubated with Alexa Fluor® 594 goat anti-rabbit and Alexa Fluor® 488 goat anti-mouse IgG antibodies (Invitrogen) at a 1:250 dilution in PBS for 30 min at 37 °C. The cells were then washed three times in PBS. Nuclear DNA was counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories). Images were captured with a Nikon E400 fluorescent microscope with an attached CCD (charge-coupled-device) camera.
RESULTS
Human MYH physically interacts with hHus1 and hRad1, but not with hRad9
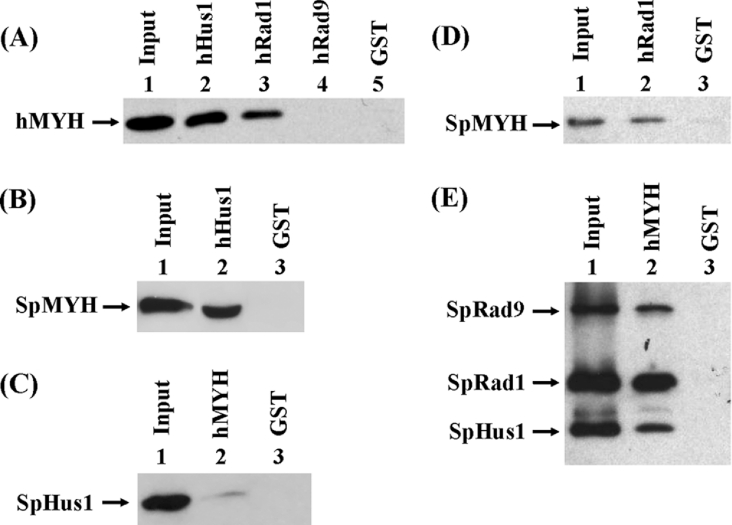
Because we have shown that the S. pombe PCNA-like 9-1-1 complex is associated with SpMYH [23], we tested whether hMYH has any interaction with hHus1, hRad1 and hRad9. The GST-tagged hHus1, hRad1 and hRad9 proteins were immobilized on beads and used to pull down partially purified hMYH. As shown in Figure 1(A), hMYH bound strongly to GST–hHus1 (lane 2), bound weakly to GST–hRad1 (lane 3), but not to GST–Rad9 (lane 4). As a negative control, hMYH did not bind to GST alone (lane 5). Thus hMYH binds to the 9-1-1 complex asymmetrically. The individual proteins used in Figure 1(A) were expressed in E. coli; thus hMYH can interact with hHus1 and hRad1 when they are not in a complex.
Figure 1. Interactions between MYH and the Hus1/Rad1/Rad9 checkpoint proteins.
(A) Pull-down of hMYH by GST–hHus1, GST–Rad1 and GST–Rad9. GST–hHus1 (lane 2), GST–Rad1 (lane 3), GST–Rad9 (lane 4) and GST alone (lane 5) were immobilized to glutathione–Sepharose and incubated with partially purified hMYH, which was tagged with both GB1 and histidine as described in the Materials and methods section. The pellets were fractionated by SDS/10% PAGE followed by Western-blot analysis with the hMYH antibody. Lane 1 contains 10 ng (10% of the total input) of partially purified hMYH. (B) Pull-down of purified SpMYH by GST–hHus1 bound to glutathione–Sepharose. Lane 1 contains 10 ng (10% of the total input) of purified SpMYH. Lanes 2 and 3 are pull-down pellets from GST–hHus1 and GST alone respectively. The Western blot was detected with the antibody against SpMYH. (C) Interaction of hMYH with HA-tagged SpHus1. Cell extracts (0.4 mg) of S. pombe expressing HA-tagged SpHus1 were added to GST–hMYH bound to glutathione–Sepharose. Lane 1 contains 40 μg (10% of total input) of cell extracts. Lanes 2 and 3 are pulled down pellets from GST–hMYH and GST alone respectively. The Western blot was detected by the antibody against HA. (D) Interaction of SpMYH with hRad1. Purified SpMYH can be pull-down by GST–hRad1 bound to glutathione–Sepharose. Lane 1 contains 2.5 ng (2.5% of total input) of purified SpMYH. Lanes 2 and 3 are pull-down pellets from GST–hRad1 and GST alone respectively. The Western blot was detected with the antibody against SpMYH. (E) Interaction of hMYH with the S. pombe 9-1-1 complex. The partially purified S. pombe 9-1-1 complex (fraction FVAd) was added to GST–hMYH bound to glutathione–Sepharose. The SpHus1, SpRad1 and SpRad9 proteins were tagged with a C-terminal histidine, N-terminal histidine and C-terminal S-tag respectively. Lane 1 contains 30 ng (10% of total input) of the S. pombe 9-1-1 complex. Lanes 2 and 3 are pull-down pellets from GST–hMYH and GST alone respectively. The Western blot was detected by a mixture of against S-tag, N-terminal histidine probe and C-terminal histidine probe.
The interactions between MYH and the 9-1-1 complex from human and S. pombe cells are interchangeable
Because we have shown that MYH–PCNA interactions are interchangeable between human and S. pombe cells [19], we tested whether this interchangeable property extends to MYH and the 9-1-1 complex. In binding experiments using GST–hHus1 fusion proteins with purified SpMYH, SpMYH was detected in the GST–hHus1 pellets (Figure 1B, lane 2), but not in the GST beads (Figure 1B, lane 3). Conversely, HA-tagged SpHus1 expressed in yeast cells was detected in the pellets of GST–hMYH (Figure 1C, lane 2), but not in the GST beads (Figure 1C, lane 3). However, the interaction between hMYH and SpHus1 is weak. We also tested whether SpMYH can bind to hRad1. As shown in Figure 1(D), SpMYH was detected in the GST–hRad1 pellets, but not in the GST beads. Also, the partially purified S. pombe 9-1-1 complex (see Supplementary material) could be pulled-down by GST–hMYH (Figure 1E, lane 2). Thus the interactions between MYH and the 9-1-1 complex, from S. pombe and human cells, are partially interchangeable.
Mapping the Hus1 interacting domain within MYH
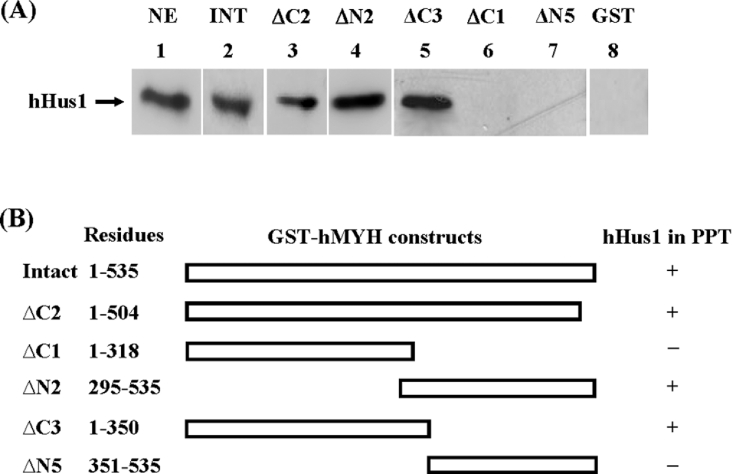
We have shown that the hPCNA-binding site is located at the C-terminus of hMYH containing residues 505–527 [20]. To test whether hPCNA and hHus1 bind to the same region of hMYH, we analysed the hHus1 binding to a truncated hMYH (ΔC2), which contains residues 1–504 with the hPCNA-binding motif deleted. The result (Figure 2A, lane 3) shows that ΔC2 could associate with hHus1 in HeLa nuclear extracts. Thus hHus1 and hPCNA bind to different regions of hMYH.
Figure 2. Determination of regions within hMYH involved in hHus1 binding.
(A) The GST pull-down assay was employed using various GST–hMYH constructs to determine the binding regions within hMYH for interaction with hHus1 in HeLa nuclear extracts (0.4 mg). Lane 1 contains 15% of input (60 μg) of nuclear extracts (NE). In lanes 2–7, GST fusion proteins, indicated on top of each lane, were used in the pull-down experiments. INT stands for intact hMYH. Western-blot analyses of the pellets were performed with the antibody against hHus1. A control was run concurrently with immobilized GST alone (lanes 8). The results represent four separate experiments in which 15% of total input is used as a standard and GST alone is used as a negative control for each pull-down analysis. (B) Graphic depiction of GST–hMYH constructs and the hHus1 binding to the hMYH fusion proteins. The amino acid residues of hMYH in the GST constructs are indicated. The ‘+’ and ‘–’ listed on the right of each construct indicate the presence and absence of hHus1 in the pellets (PPT) of GST beads respectively.
By using constructs containing different portions of hMYH fused to GST, we determined the regions of hMYH engaged in the physical interaction with the 9-1-1 complex in HeLa nuclear extracts. These hMYH deletion mutants were constructed based on the domain structures of prokaryotic MutY [37] and hMYH molecular model. The results are shown in Figure 2(A) and summarized in Figure 2(B). Constructs ΔN2 and ΔC3 retained interaction (Figure 2A, lanes 4 and 5); however, construct ΔC1 exhibited limited interaction (Figure 2A, lane 6) and ΔN5 had no binding (Figure 2A, lane 7) with hHus1. Thus the hHus1-interacting domain is localized to residues 295–350 of hMYH.
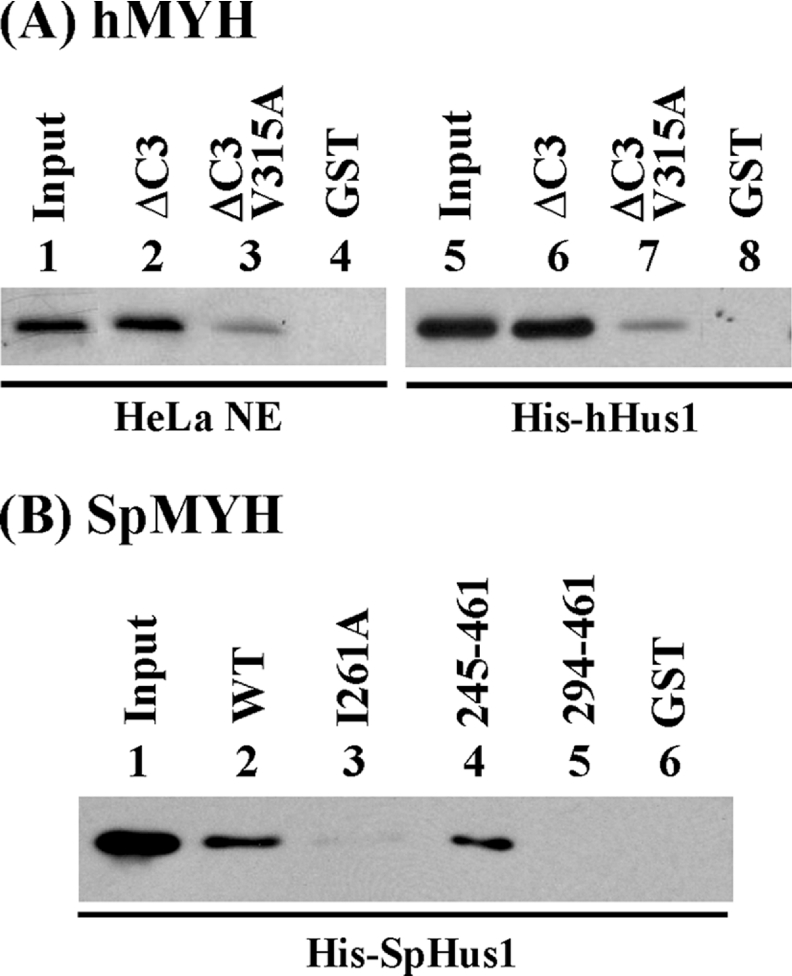
Computational analysis of residues 295–350 of hMYH reveals that several amino acids are conserved among other eukaryotic MutY family members, including mMYH (mouse MYH) [38], rMYH (rat MYH) [39] and SpMYH [35] (Figure 3). Based on known clamp binding signatures [40,41], we expect that the MYH–Hus1 interaction may involve conserved hydrophobic residues. The analysis revealed that Val315 of hMYH and Ile261 of SpMYH (marked with a star in Figure 3) are the best candidates for mediating the binding to Hus1. As shown in Figure 4(A), the interaction between the V315A mutant of hMYH and Hus1 was reduced by approx. 3.5-fold using either HeLa nuclear extracts (Figure 4A, lanes 2–4) or recombinant hHus1 protein expressed in E. coli (Figure 4A, lanes 6–8). Previous results indicate that the C-terminal half of SpMYH (residues 245–461) contains the major SpHus1-binding site [23]. Consistent with the data of hMYH–hHus1 binding, SpMYH-(294–461) showed no binding with SpHus1 (Figure 4B, lane 5). The interaction of SpMYH(I261A) with SpHus1 (Figure 4B, lane 3) was reduced 5-fold as compared with that of wild-type SpMYH (Figure 4B, lane 2). Therefore both Val315 of hMYH and Ile261 of SpMYH play important roles in hHus1 binding.
Figure 3. Alignment of the Hus1-binding motifs of MutY homologues.
Sequences are: Homo sapiens MYH (hMYH; accession no. U63329), Mus musculus MYH (mMYH; accession no. AY007717), Rattus norvegicus MYH (rMYH; accession no. NP579850) and SpMYH (accession no. Z69240). Identical amino acid residues are shaded in black and conserved residues are boxed in grey. Val315 of hMYH and Ile261 of SpMYH are marked by a star.
Figure 4. Val315 of hMYH and Ile261 of SpMYH are important for hHus1 binding.
The GST pull-down assay was employed similarly to those described in Figure 2. (A) GST–ΔC3hMYH (lanes 2 and 6), GST–ΔC3hMYH(V315A) (lanes 3 and 7) and GST alone (lanes 4 and 8) were immobilized on beads and then incubated with HeLa nuclear extracts (0.4 mg) (lanes 2–4) or 100 ng of His–hHus1 (lanes 6–8). Lanes 1 and 5 contain 80 μg (20% of total input) of nuclear extracts and 10 ng (10% of total input) of purified His–hHus1 respectively. Western-blot analysis was performed with antibody against hHus1 (lanes 1–4) or N-terminal histidine probe (lanes 5–8). (B) E. coli extracts containing His–SpHus1 (48 μg) were added to different constructs of GST–hMYH bound to glutathione–Sepharose. Lane 1 contains 10% of input of cell extracts. Lanes 2–6 are pull-down pellets from GST–SpMYH (wild-type), GST–SpMYH(I261A), GST–SpMYH(245–461), GST–SpMYH(294–461) and GST alone respectively. Western-blot analysis was performed with antibody against histidine probe.
Human Hus1 and S. pombe 9-1-1 complex can enhance SpMYH activity
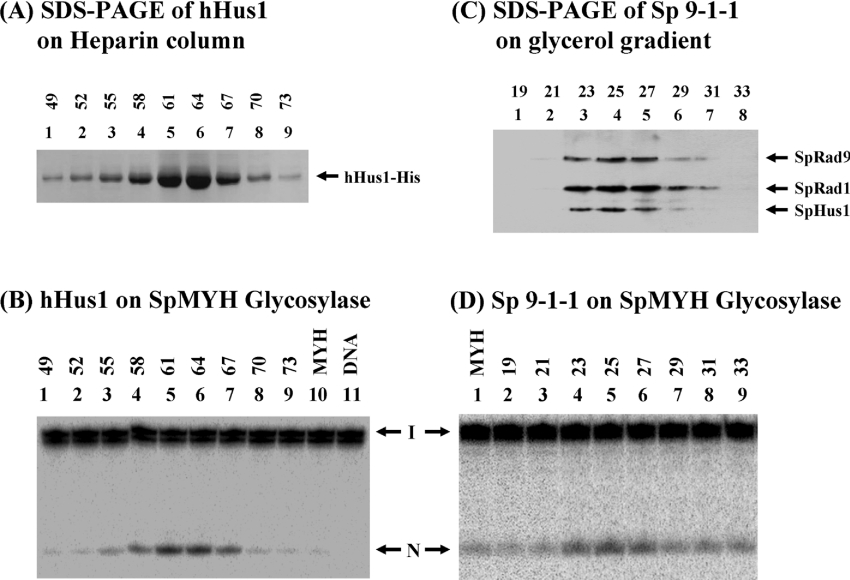
We further tested whether MYH activity can be affected by the 9-1-1 complex. Because it is difficult to express hMYH in E. coli [4,6] and SpMYH is similar to hMYH in Hus1 binding (Figures 1B and 1C), we used SpMYH for the activity assays. The hHus1 protein was purified to approx. 95% homogeneity (Figure 5A) and the S. pombe 9-1-1 complex was partially purified (see Supplementary Figure S1 in Supplementary material at http://www.BiochemJ.org/bj/400/bj4000053add.htm). The heparin column fractions of hHus1 were added to the SpMYH glycosylase reactions with the A/GO-containing DNA substrate. As shown in Figure 5, the stimulatory activity of the SpMYH glycosylase (Figure 5B) exactly matches the peak of hHus1 protein as shown by SDS/PAGE (Figure 5A). The S. pombe 9-1-1 complex was purified by a 15–35% glycerol gradient in the final step. By comparison with the size markers, the S. pombe 9-1-1 complex sedimented at the position indicating a molecular mass of approx. 110 kDa, which is in line with the theoretical value (120 kDa) of a trimeric complex. By silver staining (Supplementary Figure S1 in Supplementary material) and Coomassie Blue staining (results not shown), the complex appears to have equal molar amounts of SpHus1, SpRad1 and SpRad9. The enhancement of SpMYH glycosylase activity (Figure 5D) is in agreement with the peak of the S. pombe 9-1-1 complex as detected by Western blotting (Figure 5C). Therefore hHus1 alone and the S. pombe 9-1-1 complex can stimulate the SpMYH glycosylase activity.
Figure 5. The stimulatory activity on SpMYH glycosylase is correlated to the hHus1 and S. pombe 9-1-1 complex peaks during purification.
(A) SDS/polyacrylamide gel of corresponding hHus1–His fractions from heparin chromatography; 20 μl of each fraction was loaded on to each lane and the gel was stained with Coomassie Blue. (B) SpMYH glycosylase activity is enhanced by hHus1. Lane 11 has DNA substrates containing A/GO. The DNA substrate (0.18 nM) was incubated with SpMYH (0.2 nM) (lane 10). Fractions of hHus1–His protein (1 μl of 1:10 dilution) from heparin chromatography were added to the SpMYH glycosylase reactions as in lane 10 (lanes 1–9). Reactions were carried out at 37 °C for 30 min and the products were separated on a 14% DNA sequencing gel. Arrows mark the intact DNA substrate (I) and the nicked product (N). (C) SDS/polyacrylamide gel of corresponding S. pombe 9-1-1 complex fractions from glycerol gradient; 20 μl of each fraction was loaded on to each lane and SpRad9, SpRad1 and SpHus1 were analysed by Western blotting with a mixture of antibodies against S-tag, N-terminal histidine probe and C-terminal histidine probe. (D) SpMYH glycosylase activity is enhanced by the S. pombe 9-1-1 complex. Fractions of S. pombe 9-1-1 complex (1 μl) from a 15–35% (v/v) glycerol gradient were added to the SpMYH glycosylase reactions as described in (B). DNA substrate containing A/GO (0.18 nM) was incubated with SpMYH (0.2 nM) (lane 1).
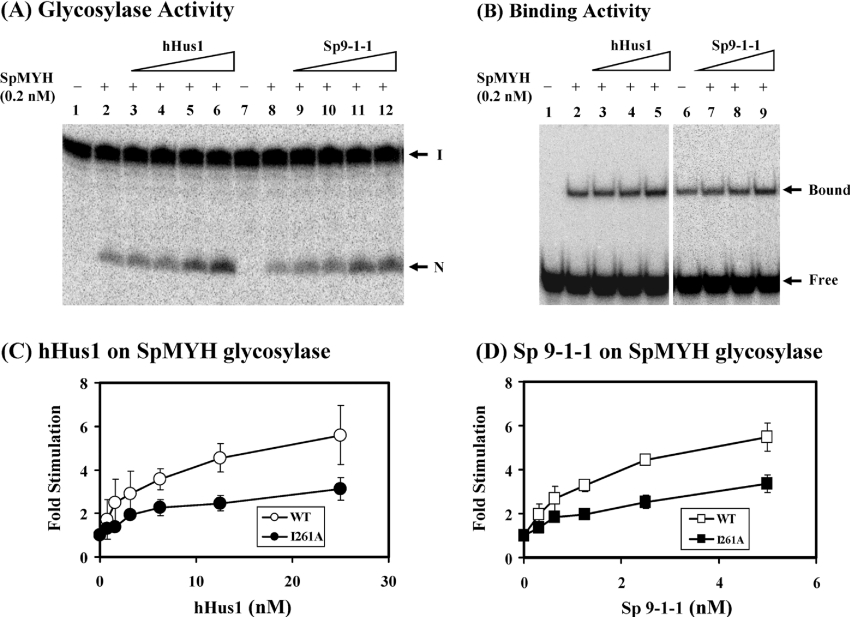
Next, we added increasing amounts of hHus1 and the S. pombe 9-1-1 complex to the SpMYH glycosylase reactions. As shown in Figure 6(A) (lanes 3–6), the SpMYH glycosylase activity was enhanced significantly by hHus1–His protein. The difference between SpMYH (0.2 nM) alone and SpMYH with 15 nM of hHus1 was 5-fold (Figure 6C, open circles). Similar stimulation of the SpMYH glycosylase activity was observed with the partially purified S. pombe 9-1-1 complex (Figure 6A, lanes 9–12). The difference between SpMYH (0.2 nM) alone and SpMYH with 5 nM of S. pombe 9-1-1 complex was 5.5-fold (Figure 6D, open squares). Interestingly, the concentrations of human hHus1 and the S. pombe 9-1-1 complex required to produce a given stimulatory effect on SpMYH glycosylase are similar to one another (e.g. 15 nM Hus1 is equivalent to 5 nM of the 9-1-1 complex). hHus1 and the S. pombe 9-1-1 complex did not have any adenine glycosylase activity at the highest tested concentrations (results not shown). In addition, E. coli histidine-tagged β-clamp and hPCNA could not stimulate the SpMYH glycosylase activity (results not shown). The SpMYH binding to A/GO mismatch was only slightly enhanced by hHus1 protein and the S. pombe 9-1-1 complex (Figure 6B).
Figure 6. hHus1 and the S. pombe 9-1-1 complex stimulate SpMYH activity.
(A) SpMYH glycosylase activity with A/GO-containing DNA was stimulated by hHus1 and S. pombe 9-1-1 complex. Lanes 1 and 7 include DNA substrates containing A/GO. In lanes 2 and 8, the DNA substrates (0.18 nM) were incubated with SpMYH (0.2 nM). In lanes 3–6, the DNA substrates (0.18 nM) were incubated with 0.2 nM SpMYH and 3.13, 6.25, 12.5 and 25 nM of His-hHus1 (fraction 61 of heparin chromatography). In lanes 9–12, the DNA substrates (0.18 nM) were incubated with 0.2 nM SpMYH and 0.63, 1.25, 2.5 and 5 nM of S. pombe 9-1-1 complex. The reactions were carried out at 37 °C for 30 min. The products were separated on a 14% DNA sequencing gel. Arrows mark the intact DNA substrate (I) and the nicked product (N). (B) SpMYH binding to A/GO-containing DNA was slightly stimulated by hHus1 and S. pombe 9-1-1 complex. Lane 1 has DNA substrates containing A/GO. In lanes 2 and 6, the DNA substrates (0.18 nM) were incubated with 0.2 nM SpMYH. In lanes 3–5, the DNA substrates (0.18 nM) were incubated with 0.2 nM SpMYH and 6.25, 12.5 and 25 nM of His–hHus1 (fraction 61 of heparin chromatography). In lanes 7–9, the DNA substrates (0.18 nM) were incubated with 0.2 nM SpMYH and 1.25, 2.5 and 5 nM of S. pombe 9-1-1 complex (the pooled fraction from phosphocellulose chromatography, FVAd). The products were fractionated on a 6% non-denaturing gel. Arrows indicate the positions of the SpMYH–DNA complex (Bound) and free DNA substrate (Free). (C, D) Quantitative analyses of fold stimulation of hHus1 and S. pombe 9-1-1 complex respectively, on wild-type (open circles and open squares) and I261A mutant (closed circles and closed squares) SpMYH glycosylase activities from three experiments. The error bars reported are the standard deviations of the averages.
The purified SpMYH(I261A) mutant protein was shown to exhibit similar glycosylase activity as the wild-type enzyme (results not shown). Consistent with its reduced physical interaction with SpHus1, SpMYH(I261A) required greater amounts of hHus1 (Figure 6C, closed circles) and the S. pombe 9-1-1 complex (Figure 6D, closed squares) for stimulation. Human Hus1 (15 nM) and the S. pombe 9-1-1 complex (5 nM) stimulated the glycosylase activity of SpMYH(I261A) by approx. 3-fold.
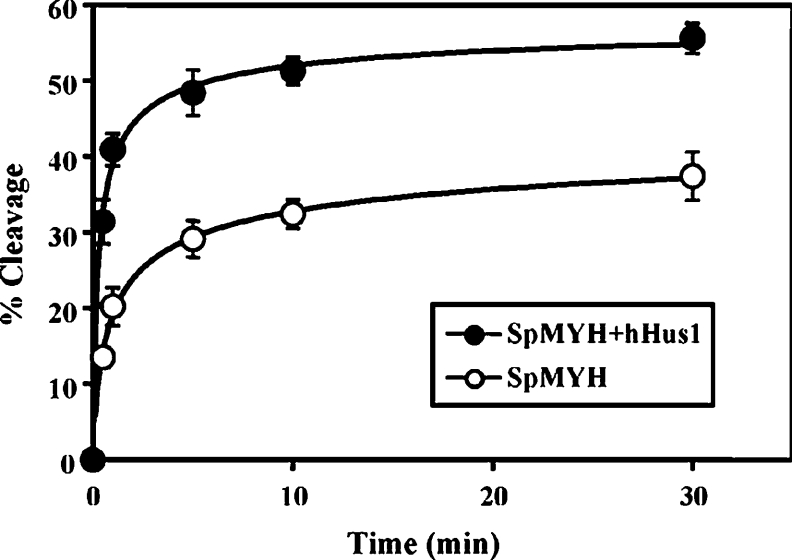
Like many DNA glycosylases, MYH exhibits slow enzymatic turnover due to strong binding to its AP (apurinic/apyrimidinic)/GO product [38]. The glycosylase assays carried out in Figure 6 measured the steady-state kinetics and could not determine which step of the DNA glycosylase reaction is affected by Hus1 and the 9-1-1 complex. Using single turnover kinetics and saturating enzyme conditions (Figure 7), SpMYH removed A from A/GO mispairs with a rate of kobs=0.80±0.17 min−1 at room temperature. In the presence of 10-fold excess of hHus1, the rate of kobs was increased to 1.98±0.34 min−1. Thus the rate of the MYH glycosylase activity is increased by 2.5-fold by Hus1.
Figure 7. Time course studies of SpMYH glycosylase activity in the presence of hHus1.
The DNA substrates containing A/GO (0.18 nM) were incubated with 1.8 nM SpMYH in the absence (open circles) or presence (closed circles) of 18 nM His–hHus1 (fraction 61 of heparin chromatography). Reactions were carried out at 23 °C for various time intervals. The percentages of DNA cleaved were plotted as a function of time. Data were obtained from phosphoimager quantitative analyses of gel images over three experiments. The error bars reported are the standard deviations of the averages. The rate constants were determined by fitting of the data to a single exponential equation: P=Pf(1–e−kt) where P is the percentage of cleavage, Pf is the final value P, t is the time and k is the rate constant.
DNA damage stimulates hMYH–hHus1 interaction
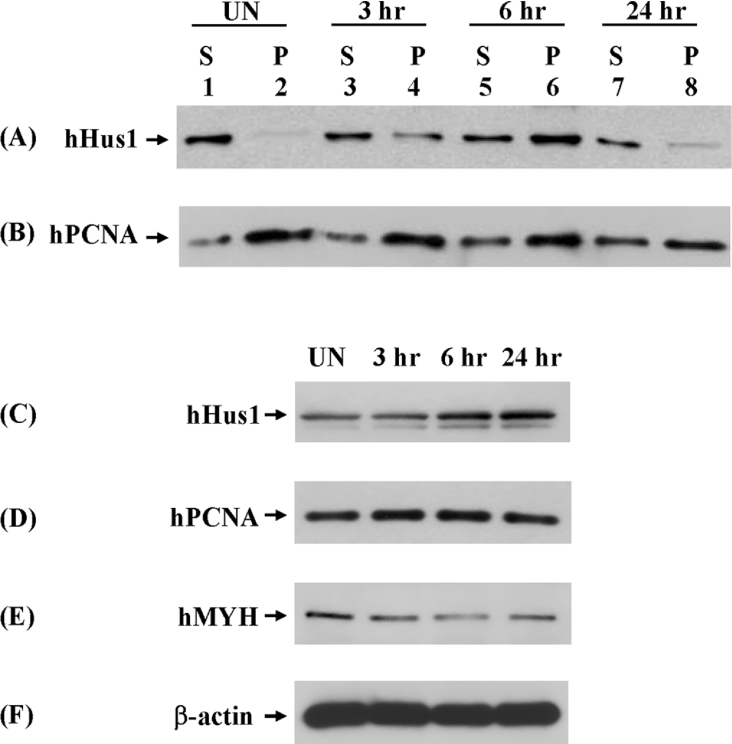
To investigate any alterations of hMYH–hHus1 interaction following DNA damage, we performed co-immunoprecipitation experiments with extracts from ionizing-radiation-treated HeLa cells. In these experiments, hHus1 could be immunoprecipitated by hMYH antibodies from HeLa cell extracts (Figure 8A, lanes 2, 4, 6 and 8), further confirming the physical interaction of hHus1 and hMYH. Interestingly, the interaction between hMYH and hHus1 was enhanced after ionizing radiation treatment. In untreated cells, the hMYH–hHus1 interaction was weak (Figure 8A, lane 2). Exposure to 10 Gy ionizing radiation resulted in an increased hMYH–hHus1 interaction, which reached a peak at 6 h after ionizing radiation treatment (Figure 8A, lane 6). At 24 h after ionizing radiation treatment, the hMYH–hHus1 interaction was then reduced to a level similar to that found in the untreated extracts (Figure 8A, lanes 2 and 8). In contrast, the hMYH–hPCNA interaction had minimal changes after ionizing radiation treatment (Figure 8B). The hMYH–hPCNA interaction was strong in untreated cells (Figure 8B, lane 2) and was slightly weaker at 24 h after ionizing radiation treatment (Figure 8B, lane 8). We also determined the total protein levels in cell extracts by direct Western blotting. The protein level of hHus1 was increased, while hPCNA and hMYH levels remained almost the same after ionizing radiation treatment (Figures 8C–8F). At 24 h after ionizing radiation treatment, while the hHus1 protein level was still elevated (Figure 8C, lane 4), the hHus1–hMYH interaction was decreased (Figure 8A, lane 8). The results of Figure 8 indicate that hMYH–hHus1 interaction is enhanced following ionizing radiation treatment, which probably induces oxidative DNA damage.
Figure 8. The interaction of hHus1 and hMYH is enhanced upon ionizing radiation.
HeLa cells were exposed to 10 Gy X-rays or left unirradiated (UN) and recovered for various times as indicated. Total cell extracts were prepared and analysed by immunoprecipitation with antibody against hMYH. S represents the supernatant and P represents the pellet. Western blotting was performed to detect hHus1 (A) and hPCNA (B). (C–F) Protein expression in irradiated HeLa cells. Extracts were subjected to direct Western blotting to detect hHus1 (C), hPCNA (D), hMYH (E) and β-actin (F).
Co-localization of hMYH with hRad9 in nuclear foci after oxidative stress
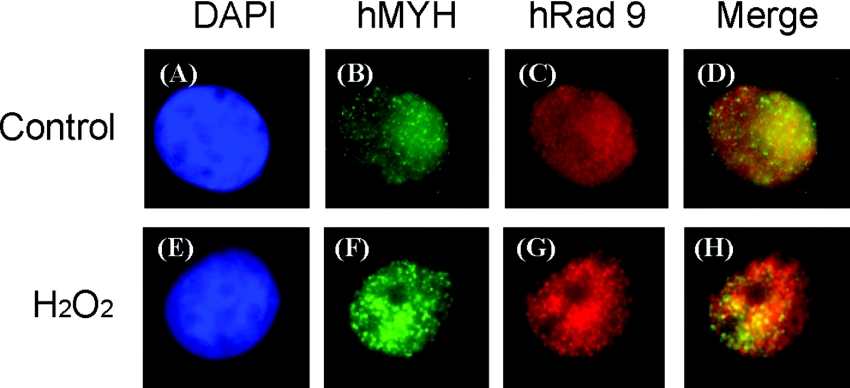
Based on the above results, we determined whether hMYH and hRad9 move to the same nuclear foci following H2O2 treatment. By immunofluorescent staining analyses, both hMYH and hRad9 appeared granulated in faint spots throughout the nucleus of untreated HeLa cells (Figures 9B and 9C). Some hMYH protein molecules were localized to cytoplasm (Figure 9B). In H2O2-treated cells, hMYH and hRad9 form discrete nuclear foci (Figures 9F and 9G). A significant fraction of the hMYH nuclear foci were found to co-localize with hRad9 foci in H2O2-treated cells (Figure 9H). Because oxidative stress generates many types of oxidized DNA base lesions, hRad9 may form foci with other protein partners. Together with the co-immunoprecipitation results, our results indicate that hMYH and the 9-1-1 complex translocate to some lesion sites following DNA damage.
Figure 9. Co-localization of hMYH with hRad9 following oxidative stress.
HeLa cells were not treated with H2O2, as in (A–D), or treated with 5 mM H2O2 for 40 min and then recovered for 6 h as in (E–H). The cells were stained with antibody against hMYH (green, B, F) and anti-hRad9 antibody (red, C, G). (A, E) DAPI-stained nuclei. (D) is the merged images of (B) and (C). (H) is the merged images of (F) and (G). Co-localization of hMYH (green) and hRad9 (red) is visualized as yellow.
DISCUSSION
DNA damage checkpoints are surveillance mechanisms that ensure a proper cellular response to stress and maintain genome stability (reviewed in [9]). The checkpoint proteins Rad9, Rad1 and Hus1 are proposed to function as damage sensor proteins. However, the mechanisms involved in the sensing step of these proteins are still poorly understood. In the present study, we show that Hus1 and the 9-1-1 complex interact with and stimulate MYH DNA glycosylase. Human MYH physically interacts with Hus1 and Rad1, but not with Rad9. hMYH can interact with hHus1 even in the absence of hRad1 and hRad9. Also, we have observed similar asymmetrical interactions between the S. pombe 9-1-1 complex and SpMYH [23]. Because E. coli β-clamp and hPCNA cannot stimulate the SpMYH glycosylase activity, the 9-1-1 complex exhibits a damage-specific enhancement on MYH base excision repair. The 9-1-1 complex may replace PCNA when the active PCNA concentration is low, such as when PCNA is inactivated by p21 during cell-cycle arrest in response to DNA damage [42].
The major Hus1-interacting region is localized to residues 295–350 of hMYH and residues 245–293 of SpMYH (Figure 3), which are different from their PCNA-binding sites [19,20]. A consensus PCNA-binding motif [QXX(L/V)XXF(F/Y)] is found in many proteins involved in DNA replication, DNA repair, DNA methylation and chromatin assembly [43,44]. Recently, the list of proteins that interact with 9-1-1 has been expanding, yet the interacting regions within these proteins have not been mapped to specific motifs. Thus the consensus Hus1-, Rad1- and Rad9-binding motifs and how they differ from the PCNA-binding signature are still to be determined. Here, we show that Val315 of hMYH and Ile261 of SpMYH play important roles for their interactions with Hus1. Although PCNA and the 9-1-1 complex interact at different regions of MYH, their points of contact with MYH appear to involve hydrophobic interactions. The region containing residues 295–350 of hMYH is conserved among eukaryotic MutY family members (Figure 3). The molecular model of hMYH, based on the prokaryotic MutY structure [37], indicates that this region is part of the N-terminal domain and is close to the junction with the C-terminal domain (Supplementary Figure S2 in Supplementary material at http://www.BiochemJ.org/bj/400/bj4000053add.htm). The region represents a long insertion compared with prokaryotic MutY proteins. The results from both secondary structure [45] and disorder [46] prediction suggest that this insertion is likely unstructured in solution when hMYH is in unbound form. Our model suggests that the 9-1-1 complex may interact with MYH and then promote the catalytic activity of MYH. Wang et al. [28] have suggested that the stimulation mechanism of human DNA ligase 1 by the 9-1-1 complex does not involve a structural change in the DNA ligase nor requires encirclement of the DNA. Thus a direct protein–protein interaction may mediate the role of the 9-1-1 complex in base excision repair. It is interesting to note that the hHus1-binding domain of hMYH (residues 295–350) overlaps with the binding domain of APE1 (AP endonuclease 1) on hMYH (residues 295–318) [20]. Yang et al. [38] have reported that APE1 can stimulate MYH glycosylase activity and turnover from its AP/G (AP/guanine) products. We did not observe significant enhancement of SpMYH binding to A/GO mismatches by Hus1 protein (Figure 6B), nor the ternary complex of DNA–MYH–Hus1. It remains to be tested whether hHus1 enhances MYH release from AP/GO-DNA products.
Stimulation of MYH glycosylase activity requires substantial molar excess of Hus1 and the 9-1-1 complex over MYH protein. With 75-fold and 25-fold molar excess of hHus1 and S. pombe 9-1-1 complex respectively, SpMYH glycosylase activity was stimulated approx. 5-fold (Figures 6C and 6D). The results suggest that the 9-1-1 complex may function more efficiently than the Hus1 alone or that hHus1 may not fully substitute SpHus1 to stimulate SpMYH activity. Alternatively, Hus1 may form a homotrimer since Hus1 can interact with itself [47,48]. Because Hus1 by itself can stimulate MYH activity, it remains to be determined whether 9-1-1 clamp formation or clamp loading to DNA is necessary for the stimulation. The required molar excess is also observed in the stimulation by the 9-1-1 complex of FEN1 [25,26], polymerase β [24] and DNA ligase 1 [27,28].
The association of MYH with the 9-1-1 complex is similar in both human and S. pombe cells [23]. First, MYH physically interacts with the Rad9–Rad1–Hus1 complex mainly via the Hus1 subunit. Secondly, the physical and functional interactions of MYH–Hus1 and MYH–Rad1 from S. pombe and human cells are interchangeable. It is interesting to note that MYH–Hus1 interactions, just like MYH–PCNA interactions, are interchangeable despite the fact that human and S. pombe Hus1 proteins share only 28% identical residues compared with 50% identity between corresponding PCNA proteins. Thirdly, interactions between MYH and Hus1 proteins are stress-inducible. hMYH–hHus1 interaction reached a peak at 6 h and reduced to a low level at 24 h following ionizing radiation treatment. The interaction between SpHus1 and SpMYH reached a peak at 3 h and then decreased after 6 h following H2O2 treatment [23]. In contrast, the MYH–PCNA interaction had minimal changes after stress in both organisms. In addition, a significant fraction of the hMYH nuclear foci was found to co-localize with hRad9 foci in H2O2-treated human cells. Along with the similarities, we also observed two differences in the association of MYH with the 9-1-1 complex between human and S. pombe cells. First, we have reported that the increase in the SpHus1–SpMYH interaction correlates with the presence of SpHus1 phosphorylation following H2O2 treatment. However, we did not observe mobility change of hHus1 after ionizing radiation treatment. Thus the state of hHus1 phosphorylation is unclear. Secondly, the protein level of hHus1 increases up to 24 h following ionizing radiation treatment, while SpHus1 protein level remains unchanged after H2O2 treatment [23].
How the sensor checkpoint proteins detect different types of DNA lesions remains elusive. Zou and Elledge [14] have shown that RPA-coated single-stranded DNA may be the common intermediate of many DNA repair pathways to recruit ATR/ATRIP (ATR-interacting protein). Several reports suggest that the sensor proteins may need a series of adaptor proteins for their recruitment to the lesion sites. The nucleotide excision repair proteins Rad14 [hXPA (human xeroderma pigmentosum group A) homologue] and Rad1 [hXPF (human xeroderma pigmentosum group F) homologue] of S. cerevisiae, as well as hXPA, are required for the damage response [49,50]. Direct interactions between ScRad14 (hXPA homologue) and the checkpoint proteins ScDdc1 (hRad9 homologue) and ScMec3 (hHus1 homologue) have been demonstrated [16]. In addition, human mismatch repair enzyme MSH2 (MutS homologue 2) interacts with the ATR in response to alkylating agents [18]. Brown et al. [51] have shown that MSH2 interacts with CHK2 and that MLH1 (MutL homologue 1) associates with ATM. Recently, Pandita et al. have shown that hRad9 interacts with recombination protein Rad51 and telomeric protein TRF2 [52]. The Mre11–Rad50–Nbs1 complex has been shown to sense double-strand breaks in DNA and to activate cell-cycle checkpoint pathways after exposure to radiation (reviewed in [17]).
Our results from current and previous studies [23] with human and S. pombe cells support the model that MYH may be one of the adaptors for sensor checkpoint proteins following oxidative DNA damage. Particularly, DNA-damage-induced SpHus1 phosphorylation is dependent on SpMYH expression [23]. In this model, MYH first recognizes the lesions and then recruits Rad9–Rad1–Hus1. Alternatively, the 9-1-1 complex may be loaded to DNA by Rad17–RFC and then it interacts and stimulates MYH. The 9-1-1 complex at the lesion sites serves both as a damage sensor to activate checkpoint control and as a component of base excision repair to enhance MYH glycosylase (Figures 7 and 8) and downstream polymerase β, FEN1 and DNA ligase 1 activities [24–28,53]. Our findings provide new insight on the initiation of signal transduction mechanism following DNA damage. In addition, other DNA damage recognition proteins may serve to recruit checkpoint proteins to different lesion sites. It is also possible that the damage-recognition involves a large complex such as BASC [BRCA1 (breast-cancer susceptibility gene 1)-associated genome surveillance complex], which contains DNA repair and replication proteins, including BRCA1, MSH2, MSH6, MLH1, ATM, RAD50 and RFC [54]. DNA damage response may be co-ordinated with ongoing DNA repair.
Online data
Acknowledgments
We thank Dr Evan Y. Lee and Alan E. Tomkinson for kindly providing the plasmids containing GST-tagged hHus1, hRad1 and hRad9. We are grateful to Dr David Beach, a Howard Hughes Medical Institute Investigator at Cold Spring Harbor Laboratory, for providing an S. pombe cDNA library. We acknowledge Cell Culture Center (Minneapolis, MN, U.S.A.) for growing the HeLa cells. We appreciate the critical reading and suggestions from Dr Alan E. Tomkinson and Ms Judy Chang. This work was supported by the National Institutes of Health grants CA/ES78391 and GM35132 to A. L. and in part by grants from Howard Hughes Medical Institute and the 6th European Community Framework Programme to Č. V.
References
- 1.Shibutani S., Takeshita M., Grollman A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 2.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C to T.A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood M. L., Dizdaroglu M., Gajewski E., Essigmann J. M. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 4.Gu Y., Lu A.-L. Differential DNA recognition and glycosylase activity of the native human MutY homolog (hMYH) and recombinant hMYH expressed in bacteria. Nucleic Acids Res. 2001;29:2666–2674. doi: 10.1093/nar/29.12.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohtsubo T., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slupska M. M., Luther W. M., Chiang J. H., Yang H., Miller J. H. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J. Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al Tassan N., Chmiel N. H., Maynard J., Fleming N., Livingston A. L., Williams G. T., Hodges A. K., Davies D. R., David S. S., Sampson J. R., et al. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 8.Sieber O. M., Lipton L., Crabtree M., Heinimann K., Fidalgo P., Phillips R. K., Bisgaard M. L., Orntoft T. F., Aaltonen L. A., Hodgson S. V., et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 9.Zhou B. B., Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 10.Shiomi Y., Shinozaki A., Nakada D., Sugimoto K., Usukura J., Obuse C., Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9–Rad1–Hus1 and Rad17–RFC. Genes Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 11.Venclovas C., Thelen M. P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bermudez V. P., Lindsey-Boltz L. A., Cesare A. J., Maniwa Y., Griffith J. D., Hurwitz J., Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17–replication factor C complex in vitro. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison V., Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 15.Wu X., Shell S. M., Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005;24:4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannattasio M., Lazzaro F., Longhese M. P., Plevani P., Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavin M. F. The Mre11 complex and ATM: a two-way functional interaction in recognizing and signalling DNA double strand breaks. DNA Repair (Amsterdam) 2004;3:1515–1520. doi: 10.1016/j.dnarep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang D. Y., Lu A.-L. Functional interaction of MutY homolog (MYH) with proliferating cell nuclear antigen (PCNA) in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2002;277:11853–11858. doi: 10.1074/jbc.M111739200. [DOI] [PubMed] [Google Scholar]
- 20.Parker A., Gu Y., Mahoney W., Lee S.-H., Singh K. K., Lu A.-L. Human homolog of the MutY protein (hMYH) physically interacts with protein involved in long-patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 21.Boldogh I., Milligan D., Lee M. S., Bassett H., Lloyd R. S., McCullough A. K. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi H., Tominaga Y., Hirano S., McKenna A. E., Nakabeppu Y., Matsumoto Y. Replication-associated repair of adenine:8-oxoguanine mispairs by MYH. Curr. Biol. 2002;12:335–339. doi: 10.1016/s0960-9822(02)00686-3. [DOI] [PubMed] [Google Scholar]
- 23.Chang D. Y., Lu A. L. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 24.Toueille M., El Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M. O., Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich-Heineken E., Toueille M., Tannler B., Burki C., Ferrari E., Hottiger M. O., Hubscher U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Wang W., Brandt P., Rossi M. L., Lindsey-Boltz L., Podust V., Fanning E., Sancar A., Bambara R. A. The human Rad9–Rad1–Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnova E., Toueille M., Markkanen E., Hubscher U. The human checkpoint sensor and alternative DNA clamp Rad9–Rad1–Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Lindsey-Boltz L. A., Sancar A., Bambara R. A. Mechanism of stimulation of human DNA ligase I by the Rad9–Rad1–Hus1 checkpoint complex. J. Biol. Chem. 2006;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- 29.Gu Y., Parker A., Wilson T. M., Bai H., Chang D. Y., Lu A. L. Human MutY homolog (hMYH), a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins hMSH2/hMSH6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 30.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 31.Parker A., Gu Y., Lu A. L. Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria. Nucleic Acids Res. 2000;28:3206–3215. doi: 10.1093/nar/28.17.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang D.-Y., Gu Y., Lu A.-L. Fission yeast (Schizosaccharomyces pombe) cells defective in the MutY-homologous glycosylase activity have a mutator phenotype and are sensitive to hydrogen peroxide. Mol. Genet. Genomics. 2001;266:336–342. doi: 10.1007/s004380100567. [DOI] [PubMed] [Google Scholar]
- 33.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Lu A. L. Repair of A/G and A/8-oxoG mismatches by MutY adenine DNA glycosylase. In: Vaughan P., editor. DNA Repair Protocols: Prokaryotic Systems. Totowa, NJ: Humana Press; 2000. pp. 3–16. [Google Scholar]
- 35.Lu A.-L., Fawcett W. P. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from Schizosacchromyces pombe. J. Biol. Chem. 1998;273:25098–25105. doi: 10.1074/jbc.273.39.25098. [DOI] [PubMed] [Google Scholar]
- 36.Bai H., Jones S., Guan X., Wilson T. M., Sampson J. R., Cheadle J. P., Lu A. L. Functional characterization of two human MutY homolog (hMYH) missense mutations (R227W and V232F) that lie within the putative hMSH6 binding domain and are associated with hMYH polyposis. Nucleic Acids Res. 2005;33:597–604. doi: 10.1093/nar/gki209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromme J. C., Banerjee A., Huang S. J., Verdine G. L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 38.Yang H., Clendenin W. M., Wong D., Demple B., Slupska M. M., Chiang J. H., Miller J. H. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29:743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H. M., Wang C., Hu Z., Greeley G. H., Makalowski W., Hellmich H. L., Englander E. W. Hypoxia induces mitochondrial DNA damage and stimulates expression of a DNA repair enzyme, the Escherichia coli MutY DNA glycosylase homolog (MYH), in vivo, in the rat brain. J. Neurochem. 2002;80:928–937. doi: 10.1046/j.0022-3042.2002.00774.x. [DOI] [PubMed] [Google Scholar]
- 40.Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 41.Venclovas C., Colvin M. E., Thelen M. P. Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 2002;11:2403–2416. doi: 10.1110/ps.0214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waga S., Hannon G. J., Beach D., Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 43.Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P., Mo J. Y., Perez A., Leon A., Liu L., Mazloum N., Xu H., Lee M. Y. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase δ. J. Biol. Chem. 1999;274:26647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 45.Jones D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 46.Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C. J., Dunker A. K. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(Suppl. 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 47.Hang H., Lieberman H. B. Physical interactions among human checkpoint control proteins HUS1p, RAD1p, and RAD9p, and implications for the regulation of cell cycle progression. Genomics. 2000;65:24–33. doi: 10.1006/geno.2000.6142. [DOI] [PubMed] [Google Scholar]
- 48.Hang H., Zhang Y., Dunbrack R. L., Jr, Wang C., Lieberman H. B. Identification and characterization of a paralog of human cell cycle checkpoint gene HUS1. Genomics. 2002;79:487–492. doi: 10.1006/geno.2002.6737. [DOI] [PubMed] [Google Scholar]
- 49.Neecke H., Lucchini G., Longhese M. P. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson W. G., Kastan M. B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol. Cell. Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown K. D., Rathi A., Kamath R., Beardsley D. I., Zhan Q., Mannino J. L., Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 52.Pandita R. K., Sharma G. G., Laszlo A., Hopkins K. M., Davey S., Chakhparonian M., Gupta A., Wellinger R. J., Zhang J., Powell S. N., et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helt C. E., Wang W., Keng P. C., Bambara R. A. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.