Abstract
There is a push to fully document the biodiversity of the world within 25 years. However, the magnitude of this challenge, particularly in marine environments, is not well known. In this study, we apply DNA barcoding to explore the biodiversity of gonodactylid stomatopods (mantis shrimp) in both the Coral Triangle and the Red Sea. Comparison of sequences from 189 unknown stomatopod larvae to 327 known adults representing 67 taxa in the superfamily Gonodactyloidea revealed 22 distinct larval operational taxonomic units (OTUs). In the Western Pacific, 10 larval OTUs were members of the Gonodactylidae and Protosquillidae where success of positive identification was expected to be 96.5%. However, only five OTUs could be identified to species and at least three OTUs represent new species unknown in their adult form. In the Red Sea where the identification rate was expected to be 75% in the Gonodactylidae, none of four larval OTUs could be identified to species; at least two represent new species unknown in their adult forms. Results indicate that the biodiversity in this well-studied group in the Coral Triangle and Red Sea may be underestimated by a minimum of 50% to more than 150%, suggesting a much greater challenge in lesser-studied groups. Although the DNA barcoding methodology was effective, its overall success was limited due to the newly discovered taxonomic limitations of the reference sequence database, highlighting the importance of synergy between molecular geneticists and taxonomists in understanding and documenting our world's biodiversity, both in marine and terrestrial environments.
Keywords: DNA barcoding, marine biodiversity, Indo-Pacific, stomatopod
1. Introduction
The world is facing a global biodiversity crisis (Novacek & Cleland 2001; Bellwood et al. 2004). The rapid loss of marine and terrestrial biodiversity has prompted efforts to catalogue this biodiversity, such as the Census of Marine Life (www.coml.org) and the All Species Foundation (www.all-species.org), the goal of the latter being to catalogue all the biodiversity on the Earth within the next 25 years (Wilson 2003). The challenges facing such efforts are great in that a small fraction of the existing biodiversity is presently described (May 1988; Wilson 2003). Compounding this problem is the well-documented decline in number of taxonomists (Buyck 1999; Hopkins & Freckleton 2002) and funding for taxonomy (House of Lords 2002) resulting in both insufficient funding and personnel to pursue this vast taxonomic undertaking (Wilson 2003).
At the same time that traditional taxonomy has been declining, the advent of PCR followed by the ever-decreasing cost of DNA sequencing has catapulted molecular genetics to near ubiquity throughout the biological sciences. With the growing taxonomic coverage of sequences contained in online DNA sequence databases (e.g. GenBank) and the ease with which these sequences can be searched, it was logical that DNA sequence data would be used to aid in organismal identifications (e.g. Pace 1997; Bucklin et al. 1999). Hebert et al. (2003a) formalized this process as ‘DNA barcoding’, the goal of which is using ‘large-scale screening of one or a few reference genes in order to: (i) assign unknown individuals to species and (ii) enhance discovery of new species’ (Moritz & Cicero 2004 and references therein).
DNA barcoding is a hotly contested issue. Controversies range from how we will define species (Sites & Marshall 2003; Blaxter 2004) to the performance of different molecular markers in various taxonomic groups (Zardoya & Meyer 1996; Vences et al. 2005), to how to incorporate intraspecific variation (Moritz & Cicero 2004). Compounding these issues are errors known to be common in existing genetic databases (Harris 2003; Vilgalys 2003). Despite these issues, DNA barcoding is quickly gaining acceptance among many in the scientific community (see vol. 360 of the Philosophical Transactions of the Royal Society). Therefore, it is important to determine what can be learned from the application of barcoding beyond the basic goal of species identification.
The coral reefs of Indo–West Pacific are the world's most diverse (Roberts et al. 2002) and most threatened (Burke et al. 2002). However, data from many taxonomic groups suggest that the biodiversity of this region is underestimated (e.g. Barber et al. 2002b; Paulay & Meyer 2002; Meyer 2003). Thorough documentation of marine biodiversity in this region is difficult because the size of this region and complexity of the habitat. Yet, documentation of this biodiversity is essential to understanding the origins of high marine biodiversity in this region (Mora et al. 2003; Barber & Bellwood 2005).
Reef-dwelling stomatopods in the Indo–West Pacific are a well-studied group with a taxonomy that is relatively stable (Erdmann & Manning 1998; Moosa 2000; Ahyong 2001). However, new species are discovered with regularity (e.g. Erdmann & Manning 1998). Thus, while it is clear that the stomatopod biodiversity in the Indo-Pacific is not fully described, the magnitude of the problem is unknown. This question is important because understanding the degree to which the diversity of a well-studied taxa is described can provide insight into how well described the diversity in lesser-studied groups may be, improving our understanding of the taxonomic challenges that lay ahead.
Stomatopods, like a large percentage of marine species, have a bipartite lifecycle where dispersal is achieved through a planktonic larval developmental stage. These larvae provide an alternative life stage with which to study biodiversity, yet larval stages are notoriously difficult to identify morphologically. Through efforts to better understand the systematics of Indo–West Pacific stomatopods in the family Gonodactylidae, we have developed a substantial cytochrome c oxidase subunit-1 (CO1) database of described and newly discovered species of coral reef dwelling stomatopods in this region. Within the superfamily Gonodactyloidea, reference sequences exist for 61 of 67 (91%) morphologically described species in the Indo–West Pacific region, including 29 of 30 (97%) species in the Gonodactylidae and 26 of 27 (96%) of the Protosquillidae. Within the Red Sea, reference sequences exist for six of 12 (50%) morphologically described species within the Gonodactyloidea. However, an additional three species (Mesacturoides brevisquamatus, Haptosquilla lenzi and Pseudosquilla megalophthalma) are the only species within their genus for which sequences are not available, increasing the expected identification rate to 75% through process of elimination. This large molecular database of morphologically identified adults combined with a larval stage that defies traditional morphological identification provides a system ideally suited to studying stomatopod biodiversity through a barcoding approach.
This study applies DNA barcoding methodology to the question of stomatopod biodiversity in the Indo–West Pacific. The goals of the project were threefold: (i) to identify stomatopod larvae through CO1 sequence data, (ii) to estimate levels of stomatopod biodiversity in the Indo–West Pacific and (iii) to determine what knowledge beyond species identification could be gained from applying a genetic barcoding methodology to the identification of marine larvae.
2. Material and methods
Planktonic larvae were collected via larval light traps deployed just beneath the surface for approximately 1–2 h at night in close proximity to coral reefs. Collections were made in the western Pacific from two coral reefs in Kimbe Bay, Papua New Guinea (May's Reef and Walindi) as well as one reef in the Red Sea (Eliat, Israel) and were preserved in 95% ethanol. Stomatopod larvae become positively phototaxic when capable of recruiting to the reef, ensuring that larval stomatopod collections only included terminal larval phases. Stomatopod larvae were removed from the bulk collections for further morphological and genetic analysis. All larvae are held by the author.
Prior to genetic analyses all larvae were examined morphologically and sorted into morphotypes to determine whether future studies could limit sequencing effort and expense by only sequencing morphologically unique larvae. Because there are no taxon-specific morphological keys for stomatopod larvae, larvae were grossly sorted into distinct morphotypes based on differences in telson and uropod structure (e.g. general shape, degree of armature and presences of pits, tubercles and spinules), raptorial claw morphology (e.g. shape and spination of dactylus and propodus), and carapace characteristics (presence/absence, shape and spination of lateral, dorsal and rostral spines). Exemplars of distinct morphotypes were then photographed for future reference.
DNA was extracted from all stomatopod larvae using a 5–10% Chelex (Biorad, Hercules, CA) solution (Walsh et al. 1991). An approximately 700 base pair (bp) fragment of mitochondrial cytochrome c oxidase subunit-1 gene (CO1) was then amplified via PCR using primers H1490 and L2198 (Folmer et al. 1994) following published protocols (Barber & Erdmann 2000). Double stranded PCR products were prepared for sequencing by incubating 5 μl of PCR product and 0.5 units of shrimp alkaline phosphatase and five units of exonuclease at 37 °C for 30 min, then 80 °C for 15 min. Cleaned PCR fragments were sequenced using BigDye (Applied Biosystems, Foster City, CA) terminator chemistry then visualized on an ABI 377 DNA sequencer. Forward and reverse sequences were proofread and compiled in Sequencher 4.0 (GeneCode, Ann Arbor, MI) and subsequently aligned by eye. Only data with fully overlapping heavy and light strand sequences was used for the analyses. Protein translations were confirmed in MacClade v. 4.05 (Maddison & Maddison 2002).
Barcoding methods followed that of Hebert et al. (2004a). Briefly, larval DNA sequences were compared to known DNA sequences from 327 morphologically identified adult stomatopods. Adult samples represented 72 unique species, as well as many highly distinct phylogroups (e.g. clades in excess of 5% sequence divergence) that may represent cryptic species (Barber et al. 2002b); 67 of these represent the superfamily Gonodactyloidea. The neighbour-joining algorithm in PAUP* v. 4.0b10 (Swofford 2002) was used with Kimura 2-parameter distances (K2P) to build a phylogram in which unknown larval samples were grouped with sequences of known taxonomic identity.
Intraspecific genetic variation can complicate the identification of unknown samples (Moritz & Cicero 2004). Therefore, to standardize attribution of unknown samples to species, we adopted the following approach. First, levels of K2P sequence variation were calculated both within clades containing unknown larval samples as well as between these clades and their nearest sister group. These values were then compared to CO1 data obtained by Barber et al. (2002b) in which 393 individual Haptosquilla pulchella fell into three clades: K2P divergence within clades ranged from 0 to 3.1% while divergence among clades (presumably cryptic species) ranged from 5.7 to 9.7%. As such, an unknown larval sample was determined to have been successfully identified if it was: (i) part of a reciprocally monophyletic group that included a known reference sample and (ii) K2P sequence divergence within this clade did not exceed 3%, similar to the values of 3% in lepidopterans and birds (Hebert et al. 2003a, 2004b). Samples in excess of 5% sequence divergence from their nearest sister group were considered to be unambiguously different species than reference samples, a value further supported by data from alpheid shrimp in which most sister taxa are separated by greater than 5% sequence variation (Knowlton & Weigt 1998). Values between 3 and 5% were considered to have ambiguous taxonomic identities. Although such a standardized procedure was necessary for the analysis, it does not imply that species boundaries can or should be determined solely through mtDNA sequence variation.
3. Results
Heavy and light strands combined to yield 551 usable base pairs of mitochondrial CO1 sequence data from 189 stomatopod larvae, including 91 larvae from May's Reef, 40 larvae from Walindi and 58 larvae from Eliat, Israel. All sequences aligned easily and translated into protein without stop codons. Ambiguous base calls at the terminal ends of a few sequences resulted in an average level of ambiguity of less than 0.5% and had no effect on the results.
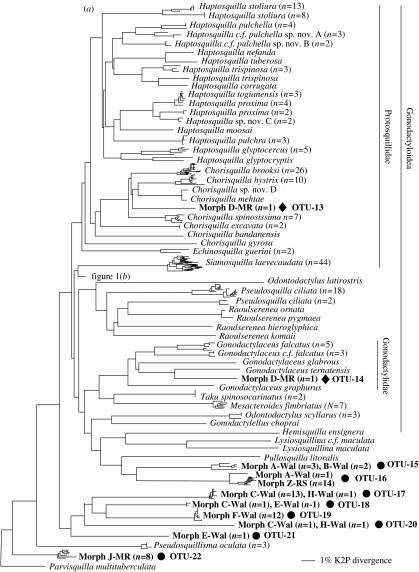
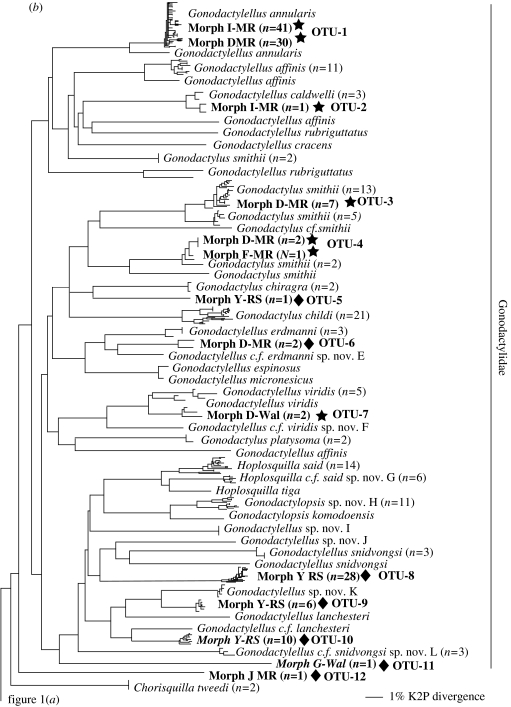
Larvae sorted into 12 distinct morphotypes (see electronic supplementary material) based on characters from the telson, raptorial claw and carapace. However, few morphotypes formed clades (figure 1). The superfamily Gonodactyloidea was characterized by morphotypes D, G, I and Y (figure 1b). Morphotype D and I were found to characterize the same operational taxonomic unit (OTU), yet individuals of both morphotype D (7 OTUs) and I (2 OTUs) were found in multiple positions on the neighbour-joining tree. Morphotype Y was only found in the Red Sea, but represented four distinct OTUs. Morphotype G was represented by one larva only. Morphotypes F and J were found both within the superfamily Gonodactyloidea as well as within clades of non-gonodactyloid stomatopods (figure 1a).
Figure 1.
Neighbour-joining phylogeny showing the phylogenetic placement of larval haplotypes for: (a) all stomatopods except most of the Gonodactylidae and (b) most of the Gonodactylidae. All larvae from May's Reef (MR), Walindi (Wal) and the Red Sea (RS), including sample sizes, are highlighted in bold and referenced by OTU numbers corresponding to table 1. Taxa listed as c.f. indicate uncertain taxonomic identity, but affinity to a specific species. Stars indicate successful barcoding identifications, diamonds indicate gonodactylid (from superfamily Gonodactyloidea) larvae that could not be identified and circles indicate non-gonodactylid larvae that could not be identified. Branch length scale is in Kimura 2-Parameter distance.
Morphotypes A, B, C, E, H and Z all fell outside the superfamily Gonodactyloidea (figure 1a). Morphotypes A and B formed a single clade. However, one larva of morphotype A grouped with a sister clade comprised only of larvae of morphotype Z. Most individuals with morphotype C formed a single clade. However, two individuals of morphotype C were part of a larger clade containing morphotypes C, E, F and H. Within this larger clade, morphotype F formed a monophyletic group, but morphotypes E and H were found in multiple positions.
The neighbour-joining tree indicated a minimum of 22 putative larval OTUs (GenBank accession numbers DQ440591–DQ440612) that likely represent distinct species based on depth of genetic divergence (table 1). A total of 14 OTUs were from the superfamily Gonodactyloidea, where sampling of known taxa was high. However, only 5 of these 14 OTUs (36%) grouped with known reference species. These five OTUs contained 85 larvae, a 45% overall rate of larval identification to species. However, this rate is inflated by 71 larvae representing Gonodactylellus annularis. Excluding these larvae, only 14 of 118 (11.9%) larvae were identified to species. Examining only Gonodactylids, 85 of 133 (63.9%) larvae were identified to species; excluding the 71 G. annularis larvae, only 14 of 62 (22.6%) were successfully identified.
Table 1.
Kimura 2-parameter distances within and among clades. (OTUs correspond to distinct larval groups identified on figure 1. The maximum K2P pairwise sequence divergence is reported within each of these larval OTUs and the minimum K2P pairwise sequence divergence is reported between each larval OTU and its nearest sister group. Asterisks indicate larval OTUs that were successfully identified to species.)
| OTU | maximum divergence within clades (%) | minimum divergence among clades (%) | closest sister group |
|---|---|---|---|
| 1* | 1.7 | 4.5 | G. annularis |
| 2* | 2.2 | 14.6 | G. affinis |
| 3* | 2.6 | 15.00 | G. smithii |
| 4* | 2.2 | 8.5 | G. smithii |
| 5 | n.a. | 11.4 | G. chiragra |
| 6 | 1.9 | 6.6 | G. c.f. erdmanni sp. nov. E |
| 7* | 2.0 | 4.1 | G. viridis |
| 8 | 3.2 | 14.8 | G. svidongi |
| 9 | 0.6 | 5.6 | G. sp. nov. K |
| 10 | 1.5 | 6.6 | G. lanchesteri |
| 11 | n.a. | 19.4 | E. guerini |
| 12 | n.a. | 18.9 | C. tweedi |
| 13 | n.a. | 8.6 | C. spinosissima |
| 14 | n.a. | 14.2 | G. glabrous |
| 16 | 3.6 | 14.9 | OTU15 |
| 15 | 3.5 | 14.9 | OTU16 |
| 17 | 0.7 | 15.0 | OTU18 |
| 18 | 0.4 | 12.9 | OTU19 |
| 19 | 0.9 | 12.9 | OTU18 |
| 20 | 0.0 | 19.0 | OTU19 |
| 21 | n.a. | 15.0 | OTU19 |
| 22 | 2.2 | 8.1 | P. multituberculata |
The 71 larvae in OTU-1 allied with Gonodactylellus annularis. Maximum K2P divergence within this group was 1.7% and its closest sister group was a more divergent G. annularis sequence 4.5% different. These sequences were 13.9% divergent from Gonodactylellus affinis. OTU-2 included one larva that grouped with Gonodactylellus caldwelli. Maximum K2P variation within this group was 2.2% while being 14.6% divergent from G. affinis. OTU-3 and OTU-4 contained seven and three larvae, respectively, as well as known Gonodactylus smithii; variation between these two smithii clades, however, exceeded 13.3% K2P distance. Levels of variation within OTU-3 was 2.6% while this group was 3.4% divergent from another clade of G. smithii; together they were 15.0% divergent from G. c.f. smithii. Variation within OTU-4 was 2.2 and 8.5% divergent from another highly distinct G. smithii sequence. Lastly, OTU-7 included two larvae that grouped with Gonodactylellus viridis. Variation within this group was 2.0% while being 4.1% distinct from another grouping of G. viridis; these two groups were in turn 9.0% divergent from G. c.f. viridis sp. nov. F.
Of the unidentified larvae in the superfamily Gonodactyloidea, most were in one of four clades of unknown stomatopods from the Red Sea (45 larvae of morphotype Y). All four of these OTUs fell within the genera Gonodactylus and Gonodactylellus even though the database contained all but three described species in these two genera from the Red Sea. Similarly, an additional five unidentified gonodactylid larval OTUs were comparatively rare larvae from Papua New Guinea. All five of these allied with the Gonodactylidae and Protosquillidae, where reference sequences exist for all but two described species from these families in this region.
Outside of the Gonodactyloidea, there were eight OTUs of non-gonodactylid larvae that could not be identified. These included two clades allied with Pullosquilla comprised mostly of morphotypes A, B and Z, five clades of unknown taxonomic affinity comprised of larvae of morphotypes C, E, F and H and a final clade of unknown taxonomic affinity at the base of the phylogeny comprised of morphotype J.
4. Discussion
(a) Biodiversity implications
The DNA barcoding approach applied in this study was effective in identifying larvae from only 5 of 14 (36%) gonodactylid OTUs to the species-level and only 22.6% of all gonodactylid larvae. Despite having reference sequences for 91% of described Indo–West Pacific Gonodactylid stomatopods, only 5 of 10 (50%) larval OTUs were successfully identified to species. Furthermore, all five unidentified OTUs grouped with the Gonodactylidae and Protosquillidae, where all but two species in this region are represented in the database. Thus, our results indicate a minimum of three to five additional species within this region that are not yet known in their adult forms. However, this number could be much greater.
Given that the larvae were collected in light traps, they are likely to represent a random sample. Discovering a minimum of three new species in a random sample of 10 larval OTUs in the Gonodactylidae and Protosquillidae suggests that biodiversity of stomatopods in the Indo-Pacific region may be underestimated by a minimum of 43%; this value would increase to at least 100% if all five of these larval OTUs represent undescribed species. Thus, although this region is a known biodiversity hotspot (Hughes et al. 2002; Roberts et al. 2002), results of this study indicate that biodiversity in this region is much higher.
Similar patterns were seen in the Red Sea. Despite the potential to identify 75% of the Gonodactyloidea in this region, none of the larvae from this region could be identified to species based on DNA barcodes. One unidentified larva allied with Gonodactylus chiragra and may represent Gonodactylus botti, a similar species of Gonodactylus from the Red Sea. The remaining three are clearly Gonodactylellus, yet there is only one described species of Gonodactylellus from the Red Sea that is missing from the database. This result suggests a significant amount of stomatopod biodiversity in the Red Sea has yet to be discovered and described. All of the larvae from this region fell within the Gonodactylidae in which identification through DNA barcodes should be possible for four of seven species. Given the discovery of at least two unknown taxa, stomatopod diversity in this region is underestimated by at least 29%, but may exceed 133%.
The above values of underestimated biodiversity take into account cryptic lineages in H. pulchella, Haptosquilla glyptocercus, G. viridis and G. erdmanni in which genetic data and preliminary morphological analyses suggest the presence of cryptic species (Barber et al. in press). However, these estimates do not take into account the many polyphyletic lineages of Gonodactylellus rubriguttatus, Gonodactylellus affinis, Gonodactylellus lanchesteri, Haptosquilla trispinosa and Pseudosquilla ciliata or the deep divergences in Gonodactylus smithii, Haptosquilla said, Gonodactylellus svidongi, Haptosquilla stoliura, P. ciliata and Gonodactylus falcatus that indicate the potential for an additional 17 cryptic species. Thus, the act of creating the barcoding database resulted in the discovery of an additional 25% potentially new species. Combined with the results above, stomatopod biodiversity in the Indo-Pacific is underestimated by at least 68–125% and 54–158% in the Red Sea.
The possibility that a great deal of biodiversity in the oceans may be unknown has been noted previously (Knowlton 1993; Reaka-Kudla 1997). However, the DNA barcoding approach applied here provides a mechanism to quantify this unknown biodiversity. Even discounting complexes of likely sibling species (e.g. G. viridis, G. smithii, H. pulchella, H. glyptocercus and H. said) and potentially cryptic taxa (e.g. polyphyletic lineages of G. affinis, G. rubriguttatus and G. smithii) it is remarkable that the biodiversity in such a well-studied fauna is likely to have been underestimated by at least half. Having been the subject of intensive study for decades (e.g. Manning 1978; Erdmann 1997; Moosa 2000; Ahyong 2001), there are probably few groups as well collected and described as stomatopods, particularly within the Indo–West Pacific. As such, levels of unknown biodiversity in lesser-known groups will surely exceed that seen in stomatopods, suggesting that Reaka-Kudla's (1997) estimate that diversity on coral reefs is underestimated by 90% may be correct. Similar arguments follow for terrestrial taxa.
(b) Evolutionary and ecological implications
Identifying unique DNA sequences in the absence of morphological identification is problematic in that a species definition cannot be assigned to the sequence. It has been suggested that this problem be approached through ‘reverse taxonomy’ in which discovery of new OTUs guides the collection and identification of the novel sequences (Markmann & Tautz 2005). Although such an approach may be possible for the benthic meiofauna, where large environmental samples can be carefully sifted and sorted, the challenges of locating a particular unknown stomatopod on the reef is extremely daunting, a result that may be characteristic of many marine and terrestrial systems. An alternate approach is to rear larval stages to adulthood as Janzen et al. (2005) did for lepidopterans unknown as adults. However, unlike butterflies, it can take years for stomatopods to achieve their adult morphology, making this approach largely impractical for stomatopods and many other reef invertebrates.
Ascribing a species name to a sequence may not always be possible, yet knowing that this biodiversity exists and knowing the DNA sequence is still valuable. Understanding the magnitude and distribution of biodiversity is important in terms of examining regional patterns of biodiversity and biodiversity gradients (Connolly et al. 2003; Mora et al. 2003) and defining regional conservation units (Green & Mous 2004). Accuracy of phylogenetic inference (see Bergsten (2005) for a recent review) and/or biogeographic reconstruction (Barber & Bellwood 2005) depends on completeness of taxonomic sampling, particularly when examining regional patterns of lineage diversification (Barber & Bellwood 2005) or calibrating molecular clocks among putative geminate species (e.g. Bermingham et al. 1997; Marko 2002). Thus, the inclusion of these undescribed OTUs in phylogenetic or biogeographic analysis may facilitate a more accurate reconstruction of relationships among those taxa that are known.
Understanding the nature of marine larval dispersal has been a key issue in marine biology over the past decade because it is key to understanding the evolution and dynamics of marine populations and for developing effective management strategies (see the 2003 special issue of Ecological Applications). However, while traditional approaches can examine dynamics of recruitment over time, such studies often cannot describe these patterns down to the species-level (e.g. Reyns & Sponaugle 1999). Our results indicate gonodactylid larvae were dominant in May's reef while rare in Walindi and that G. annularis represented 78% of this sample. Moreover, 18 of these latter larvae (25%) had identical mtDNA sequences. Even larger percentages of identical haplotypes are seen in the clades comprised of morphotype C (5 of 15, 33%), morphotype F (5 of 12, 42%) and morphotype Y clade, where 14 of 28 (50%) of larvae have identical CO1 sequences. Although such high levels of genetic similarity can be observed in stomatopods (Barber et al. 2002b), few of the taxa in figure 1 show high levels of genetic similarity. Thus, either these four taxa have lower genetic diversity than other stomatopods or the larval cohort includes a large number of sibs, suggesting schooling during larval dispersal. While such conclusions will require verification through studies of adult populations, it is clear that applying barcoding to studies of larval recruitment could greatly improve our understanding of the dynamics of larval dispersal as previous molecular ecology approaches have in the past (e.g. Medeiros-Bergen et al. 1995).
(c) Methodological concerns
For this study, identifications of larval OTUs were confirmed only if they fell into a monophyletic group with less than 3% K2P sequence divergence. Although this criterion was satisfied in all cases, three of these groupings (OTU 1/G. annularis, OTU 3/G. smithii and OTU 7/G. viridis) were associated with other putative conspecific sequences greater than 3%, a pattern seen within other OTUs as well (table 1). Whether these intermediately divergent sequences are conspecific or heterospecific is a question that will require subsequent morphological study and/or the examination of nuclear genes. However, this pattern raises two important issues. First, levels of intraspecific variation in this study were found to be much greater than reported in Hebert et al. (2003b, 2004b). Although the 3% cut-off was not a problem in this study in terms of the successful identification of five larval OTUs, the high levels of variation among putative conspecific indicates the potential for assignment difficulties. Large effective population sizes and geographic ranges in many marine taxa may result in levels of intraspecific variation that are problematic for barcoding efforts. Second, this deeper variation was only observed through sampling multiple geographic locations, highlighting the importance of regional intraspecific genetic structure for successful DNA barcoding (Moritz & Cicero 2004; DeSalle et al. 2005). Similar conclusions are drawn from the remaining two OTUs in which morphologically identified taxa (e.g. G. smithii and G. affinis) from different geographic locations have sequence divergences suggestive of cryptic species. Combined, these results indicate that regional sampling will be essential to DNA barcoding so that the full range of intraspecific sequence variation is represented within databases. It will also be essential to work closely with taxonomists to determine whether groups with high levels of intraspecific variation are simply highly diverse or may represent cryptic species diversity.
Another potential concern is the possibility that mitochondrial introgression or incomplete lineage sorting may blur taxonomic boundaries. In all of our previous work (Barber & Erdmann 2000; Barber et al. 2000, 2002a,b) as well as the present study we have yet to find evidence for shared mitochondrial haplotypes among highly distinct morphospecies. However, such species can have indistinct genetic boundaries, such as Haptosquilla proxima and Haptosquilla togianensis, where incomplete lineage sorting may result in the inability to accurately identify species based solely on CO1 sequence data. Similarly, species boundaries can be subtle such as with C. mehtae and C. sp. nov. D and G. micronesicus and G. espinosus, where pairwise K2P distances are both 0.92% suggesting that a 3% cut-off for species definition may not always be appropriate, a result similar to that of Hebert et al. (2004a) that indicates that a 3% cut-off for species boundaries may occasionally be too high. Although shallow or incomplete genetic boundaries may limit the ability for specific identifications for some species, it affects a minority of the taxa and does not affect the results of this study. The larger challenge is identifying and describing the many potentially cryptic species suggested by deeply divergent sequences among putative conspecifics. However, the genetic identification of these cryptic taxa should provide a mechanism to facilitate traditional taxonomic descriptions (Knowlton 1993, 2000). Whether too much genetic diversity or too little, the issue of species boundaries (Sites & Marshall 2003; Blaxter 2004) will likely remain a challenge in DNA barcoding.
Ideally, barcoding efforts will sequence all samples collected during a biological survey. However, given the present expense of DNA sequencing, a more likely approach is that only subsamples will be analysed. These will either have to be random samples or samples selected based on morphological distinctiveness. Despite our knowledge of decapod taxonomy, single species included multiple larval morphotypes and single larval morphotypes contained multiple species. The former probably results from the presence of supernumary instars that are common in stomatopods. More troubling is the presence of multiple OTUs within a single morphotype, a pattern that may be a function of a lack of a key to stomatopod larvae and/or the lack of larval characters to reliably distinguish among species. Either way, it will be difficult to rely on morphological sorting of samples to choose salient units for barcoding analysis. The alternative strategy is random sampling of specimens for sequencing. However, this approach would have missed a large portion of the diversity in the sample as 64% (9 of 14) of the Gonodactylid larval OTUs were represented by less than three individuals, and thus could be easily missed by random sampling. As such, complete documentation of the biodiversity contained within a sample will likely require sequencing of all specimens. This strategy may be possible for some systems, but will likely represent a costly challenge to others.
The neighbour-joining algorithm employed following Hebert et al. (2004a) generated distinct OTUs and indicated taxonomic affinities of larvae. Given the very low sequence divergence within clades of successfully identified larvae, these identifications are unlikely to be affected by phylogenetic reconstruction method; indeed, a maximum parsimony analysis of the data resulted in identical number, composition and identification of the 22 larval OTUs (analyses not shown). However, due to the modest length of sequences and the generally poor performance of neighbour-joining compared to other methods of phylogenetic inference (Huelsenbeck et al. 1996), the higher level interspecific relationships should be viewed with scepticism.
5. Conclusions
The DNA barcoding approach applied in this study was successful in unambiguously assigning only 22.6% gonodactylid larval OTUs to species. This pattern resulted not from a failure of the barcoding methodology, but instead from stomatopod diversity being much higher than previously believed—diversity discovered through the application of DNA barcoding. This result demonstrates both the utility of DNA barcoding and its Achilles heel. The foundation of successful barcoding efforts must be robust and complete taxonomy. Given the decline in funding for traditional taxonomy with the concomitant decline in number of taxonomists, it is unclear that sufficient taxonomic resources are available to support present DNA barcoding initiatives.
Barcoding sequencing efforts in both marine and terrestrial environments will undoubtedly reveal novel biodiversity. However, without taxonomic descriptions of this diversity, these sequences lose much of their value. The iterative process of revealing this diversity through barcoding followed by describing this diversity through taxonomic study should result in increasingly reliable barcoding up to the inherent limits of the technique (e.g. species boundaries). Such a synergy between molecular geneticists and taxonomists will greatly advance our understanding, description and cataloguing of our planet's biodiversity, moving us closer to the goal of documenting the entirety of the world's biodiversity in both marine and terrestrial environments. However, this synergy will only be possible if funding is directed both towards barcoding efforts as well as traditional morphological-based taxonomy that successful barcoding efforts will require.
Acknowledgments
This paper was stimulated by N. Knowlton, B. Bermingham and the Coral Reef Barcoding and Environmental Sampling Workshop at the Smithsonian Tropical Research Institute. Boston University and the National Science Foundation (OCE-0349177) provided funding. M. V. Erdmann and S. Ahyong contributed samples and taxonomic expertise. N. Shashar provided larvae from the Red Sea. N. Lindquist supported field collections of larvae in Papua New Guinea. M. E. Jones contributed to adult sequence collection. The manuscript was improved by comments from B. Chick, E. Crandall and M. E. Jones and three anonymous reviewers.
Footnotes
Present address: Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA.
Supplementary Material
Photographic examples of the 12 unique stomatopod morphotypes identified in this study.
References
- Ahyong S.T. Revision of the Australian stomatopod Crustacea. Rec. Aust. Mus. 2001;26(Suppl.):1–326. [Google Scholar]
- Barber P.H, Bellwood D.R. Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Mol. Phylogenet. Evol. 2005;35:235–253. doi: 10.1016/j.ympev.2004.10.004. 10.1016/j.ympev.2004.10.004 [DOI] [PubMed] [Google Scholar]
- Barber P.H, Erdmann M.V. Molecular systematics of the Gonodactylidae (Stomatopoda) using mitochondrial cytochrome oxidase C (subunit 1) DNA sequence data. J. Crustacean Biol. 2000;20:20–36. [Google Scholar]
- Barber P.H, Palumbi S.R, Erdmann M.V, Moosa M.K. A marine Wallace's line? Nature. 2000;406:692–693. doi: 10.1038/35021135. 10.1038/35021135 [DOI] [PubMed] [Google Scholar]
- Barber P.H, Moosa M.K, Palumbi S.R. Rapid recovery of genetic diversity of stomatopod populations on Krakatau: temporal and spatial scales of marine larval dispersal. Proc. R. Soc. B. 2002a;269:1591–1597. doi: 10.1098/rspb.2002.2026. 10.1098/rspb.2002.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber P.H, Palumbi S.R, Erdmann M.V, Moosa M.K. Sharp genetic breaks among populations of a benthic marine crustacean indicate limited oceanic larval transport: patterns, causes, and consequences. Mol. Ecol. 2002b;11:659–674. doi: 10.1046/j.1365-294x.2002.01468.x. 10.1046/j.1365-294X.2002.01468.x [DOI] [PubMed] [Google Scholar]
- Barber P.H, Erdmann M.V, Palumbi S.R. Comparative phylogeography of three co-distributed stomapods: origins and timing of regional lineage diversification in the Coral Triangle. Evolution. In press [PubMed] [Google Scholar]
- Bellwood D.R, Hughes T.P, Folke C, Nystrom M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. 10.1038/nature02691 [DOI] [PubMed] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. 10.1111/j.1096-0031.2005.00059.x [DOI] [PubMed] [Google Scholar]
- Bermingham E, McCafferty S.S, Martin A.P. Fish biogeography and molecular clocks: perspectives from the Panamanian Isthmus. In: Kocher T.D, Stepien C.A, editors. Molecular systematics of fishes. Academic Press; San Diego, CA: 1997. pp. 113–128. [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. 10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucklin A, Guarnieri M, Hill R.S, Bentley A.M, Kaartvedt S. Taxonomic and systematic assessment of planktonic copepods using mitochondrial COI sequence variation and competitive, species-specific PCR. Hydrobiologia. 1999;401:239–254. 10.1023/A:1003790411424 [Google Scholar]
- Burke L, Selig E, Spalding M. UNEP-WCMC; Cambridge, UK: 2002. Reefs at risk in Southeast Asia. [Google Scholar]
- Buyck B. Taxonomists are an endangered species in Europe. Nature. 1999;401:321. doi: 10.1038/43762. 10.1038/43762 [DOI] [PubMed] [Google Scholar]
- Connolly S.R, Bellwood D.R, Hughes T.P. Geographic ranges and species richness gradients: a re-evaluation of coral reef biogeography. Ecology. 2003;84:2178–2190. [Google Scholar]
- DeSalle R, Egan M.G, Siddall M. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1905–1916. doi: 10.1098/rstb.2005.1722. 10.1098/rstb.2005.1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, M. V. N. 1997 The Ecology, distribution, and bioindicator potential of Indonesian coral reef stomatopod communities. Ph.D. thesis, University of California, Berkeley, p. 290.
- Erdmann M.V, Manning R.B. Preliminary descriptions of nine new stomatopod crustaceans from coral reef habitats in Indonesia and Australia Raf. Bull. Zool. 1998;46:615–626. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit 1 from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Green A, Mous P.J. In The nature conservancy expert workshop. South East Asia Center for Marine Protected Areas; Bali, Indonesia: 2004. Delineating the Coral Triangle, its ecoregions and functional seascapes; p. 24. [Google Scholar]
- Harris D.J. Can you bank on GenBank? Trends Ecol. Evol. 2003;18:317–319. 10.1016/S0169-5347(03)00150-2 [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, DeWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003b;270:S96–S99. doi: 10.1098/rsbl.2003.0025. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004;101:14 812–14 817. doi: 10.1073/pnas.0406166101. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PLoS. 2004b;2:1657–1663. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins G.W, Freckleton R.P. Declines in the numbers of amateur and professional taxonomists: implications for conservation. Anim. Conserv. 2002;5:245–249. 10.1017/S1367943002002299 [Google Scholar]
- House of Lords. H.M. Stationary Office Ltd; London, UK: 2002. What on Earth? The threat to the science underpinning conservation. [Google Scholar]
- Huelsenbeck J.P, Hillis D.M, Jones R. Parametric bootstrapping in molecular phylogenetics: applications and performance. In: Ferraris J.D, Palumbi S.R, editors. Molecular zoology: advances, strategies, and protocols. Wiley-Liss; New York, NY: 1996. pp. 19–45. [Google Scholar]
- Hughes T.P, Bellwood D.R, Connolly S.R. Biodiversity hotspots, centres of endemicity, and the conservation of coral reefs. Ecol. Lett. 2002;5:775–784. [Google Scholar]
- Janzen D.H, Hajibabaei M, Burns J.M, Hallwachs W, Remigio E, Hebert P.D.N. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1835–1845. doi: 10.1098/rstb.2005.1715. 10.1098/rstb.2005.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton N. Sibling species in the sea. Annu. Rev. Ecol. Syst. 1993;24:189–216. 10.1146/annurev.es.24.110193.001201 [Google Scholar]
- Knowlton N. Molecular genetic analyses of species boundaries in the sea. Hydrobiologia. 2000;420:73–90. 10.1023/A:1003933603879 [Google Scholar]
- Knowlton N, Weigt L.A. New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. B. 1998;265:2257–2263. 10.1098/rspb.1998.0568 [Google Scholar]
- Maddison W.P, Maddison D.R. Sinauer Associates; Sunderland, MA: 2002. MacClade. [Google Scholar]
- Manning R.B. New and rare stomatopod Crustacea from the Indo-West-Pacific region. Smithsonian Contrib. Zool. 1978;264:1–36. [Google Scholar]
- Markmann M, Tautz D. Reverse taxonomy: an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Proc. Natl Acad. Sci. USA. 2005;360:1917–1924. doi: 10.1098/rstb.2005.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko P.B. Fossil calibration of molecular clocks and the divergence times of geminate species pairs separated by the Isthmus of Panama. Mol. Biol. Evol. 2002;19:2005–2021. doi: 10.1093/oxfordjournals.molbev.a004024. [DOI] [PubMed] [Google Scholar]
- May R.M. How many species are there on Earth? Science. 1988;241:1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- Medeiros-Bergen D.E, Olson R.E, Conroy J.A, Kocher T.D. Disribution of Holothurian larvae determined with species-specific genetic probes. Limnol. Oceanogr. 1995;40:1225–1235. [Google Scholar]
- Meyer C.P. Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biol. J. Linn. Soc. 2003;79:401–459. 10.1046/j.1095-8312.2003.00197.x [Google Scholar]
- Moosa M.K. Marine biodiversity of the South China Sea: a checklist of stomatopod Crustacea. Raf. Bull. Zool. 2000;8:405–457. [Google Scholar]
- Mora C, Chittaro P.M, Sale P.F, Kritzer J.P, Ludsin S.A. Patterns and processes in reef fish diversity. Nature. 2003;421:933–936. doi: 10.1038/nature01393. 10.1038/nature01393 [DOI] [PubMed] [Google Scholar]
- Moritz C, Cicero C. DNA barcoding: promise and pitfalls. PLoS. 2004;2:1529–1531. doi: 10.1371/journal.pbio.0020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novacek M.J, Cleland E.E. The current biodiversity extinction event: scenarios for mitigation and recovery. Proc. Natl Acad. Sci. USA. 2001;98:5466–5470. doi: 10.1073/pnas.091093698. 10.1073/pnas.091093698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. 10.1126/science.276.5313.734 [DOI] [PubMed] [Google Scholar]
- Paulay G, Meyer C. Diversification in the tropical pacific: comparisons between marine and terrestrial systems and the importance of founder speciation Integr. Comp. Biol. 2002;42:922–934. doi: 10.1093/icb/42.5.922. [DOI] [PubMed] [Google Scholar]
- Reaka-Kudla M.L. The global biodiversity of coral reefs: a comparison with rain forests. In: Reaka-Kudla M.L, Wilson D.E, Wilson E.O, editors. Biodiversity II: understanding and protecting our biological resources. Joseph Henry Press; Washington, DC: 1997. pp. 83–108. [Google Scholar]
- Reyns N, Sponaugle S. Patterns and processes of brachyuran crab settlement to Caribbean coral reefs. Mar. Ecol. Prog. Ser. 1999;185:155–170. [Google Scholar]
- Roberts C.M, et al. Marine biodiversity hotspots and conservation priorities for tropical reefs. Science. 2002;295:1280–1284. doi: 10.1126/science.1067728. 10.1126/science.1067728 [DOI] [PubMed] [Google Scholar]
- Sites J.W, Marshall J.C. Delimiting species: a renaissance issue in systematic biology. Trends Ecol. Evol. 2003;18:462–470. 10.1016/S0169-5347(03)00184-8 [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. [Google Scholar]
- Vences M, Thomas M, van der Meijden A, Chiari Y, Vieites D.R. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Front. Zool. 2005;2:1–12. doi: 10.1186/1742-9994-2-5. 10.1186/1742-9994-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R. Taxonomic misidentification in public DNA databases. New Phytol. 2003;160:4–5. doi: 10.1046/j.1469-8137.2003.00894.x. 10.1046/j.1469-8137.2003.00894.x [DOI] [PubMed] [Google Scholar]
- Walsh P.S, Metzger D.A, Higuchi R. Chelex-100 as a medium for simple extraction of DNA for PCR based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Wilson E.O. The encyclopedia of life. Trends Ecol. Evol. 2003;18:77–80. 10.1016/S0169-5347(02)00040-X [Google Scholar]
- Zardoya R, Meyer A. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol. Biol. Evol. 1996;13:933–942. doi: 10.1093/oxfordjournals.molbev.a025661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Photographic examples of the 12 unique stomatopod morphotypes identified in this study.