Abstract
The mating pathway in Saccharomyces cerevisiae is one of the best understood signal transduction pathways in eukaryotes. It transmits the mating signal from plasma membrane into the nucleus through the G-protein coupled receptor and the mitogen-activated protein kinase (MAPK) cascade. According to current understanding of the mating pathway, we construct a system of ordinary differential equations to describe the process. Our model is consistent with a wide range of experiments, indicating that it captures some main characteristics of the signal transduction along the pathway. Investigation with the model reveals that the shuttling of the scaffold protein and the dephosphorylation of kinases involved in the MAPK cascade cooperate to regulate the response upon pheromone induction and to help preserve the fidelity of the mating signaling. We explored factors affecting the dose-response curves of this pathway and found that both negative feedback and concentrations of the proteins involved in the MAPK cascade play crucial roles. Contrary to some other MAPK systems where signaling sensitivity is being amplified successively along the cascade, here the mating signal is transmitted through the cascade in an almost linear fashion.
INTRODUCTION
Cells have to respond to changes in the environment and/or to the external stimuli. This is accomplished by signal transduction pathways which sense the signal, transduce it, and induce necessary changes in the cell, such as in gene expression. One of the best understood signaling pathways in eukaryotes is the mating pathway in the budding yeast Saccharomyces cerevisiae (1,2). Extensive studies of the mating pheromone response have contributed much to the understanding of the mechanisms of several conservative biological modules (3), such as the G protein cycle (2,4) and the mitogen-activated protein kinase (MAPK) cascade (2,5). Genetic, biochemical, and molecular analyses of the response have combined to establish basic principles of the signaling and regulation. Many important discoveries are made in the study of this pathway, such as the concept of a kinase-scaffold protein (6) and the role of regulator of G-protein signaling (RGS) proteins in the pathway (7).
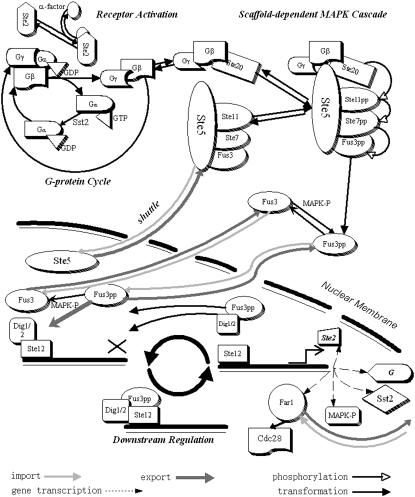
The budding yeast can exist in either of the two types, MATa or MATα. These two types of cells will mate when each one receives the mating signal, a peptide pheromone, secreted by the opposite type (a-factor by MATa and α-factor by MATα). Once the pheromone binds to the seven-transmembrane-segment receptor in the plasma membrane (Ste2 in MATa and Ste3 in MATα), the receptor is activated, which then activates the heterotrimeric G protein that couples to it (Fig. 1). The activated G protein transmits a signal to multiple effectors, resulting in the beginning of the MAPK cascade, which is embedded in a scaffold protein Ste5. This cascade consists of three kinases: Ste11 (MAPKKK), Ste7 (MAPKK), and Fus3 (MAPK). The activation of the cascade finally leads to the phosphorylation of Fus3. The phosphorylated MAPK then travels into the nucleus, and transmits the signal to downstream effectors, leading to preparation for mating, including the cell cycle arrest in G1 phase to assure synchronism of the mating partners, the induction of new gene expression necessary for mating, and the polarized growth in the direction of the pheromone source.
FIGURE 1.
Spatial structure of the mating pathway.
Much qualitative and quantitative information in this pathway has been documented. With the increasing amount of experimental data and information, it is now possible to study this pathway quantitatively at a system level. Several mathematical models have been employed to study this (8–12) and some other related systems (13–16), showing that mathematical modeling and simulation can be a powerful method in the analysis of functional and structural characteristics of biological pathways.
We set up an ordinary differential equation (ODE) model to describe the mating pathway in budding yeast. Although several models have been employed to illustrate mechanisms in the pathway, there has not been one that integrates all the known essential features with a comprehensive analysis of its dynamic properties. Some models were constrained to a single step (8,12), while others oversimplified the regulations and functions of the scaffold (10). In our model, biochemical interactions, induced gene expressions which feed back to the pathway, and translocations of key components such as the scaffold protein Ste5, are all considered. Results from our model are consistent with a wide range of experimental data. We then tested the current understanding of regulations of cellular responses and further explored the intrinsic mechanisms in the pathway, with special interest in the role of the scaffold protein Ste5. We find that the shuttling of the scaffold and dephosphorylation of the MAP kinases cooperate to regulate the responses upon pheromone induction, and to help keeping the fidelity of the mating pathway. We further explored the mechanisms of the dose-response curves of this pathway, and elucidated the role of enzyme concentration. We found that instead of an ultrasensitive response as in some other MAPK cascade (17), the mating signal here is transmitted through the cascade in an almost linear manner due to negative feedback.
THE MODEL
We choose a mutant (TMY101), a MATa type of Saccharomyces cerevisiae, as the main modeling subject. In this type of cell, the gene BAR1 is deleted. In a wild-type MATa cell, the product of BAR1 can be excreted from the cells and cleave the α-factor. To simulate a continuous and constant α-factor treatment, we use this mutant in our model.
The mating response can be divided into three modules in a temporal order: the activation of G protein cycle, the scaffold-dependent MAPK cascade, and the downstream effects of activated MAPK (See Fig. 1). Viewing the response as a series of modules arranged in the temporal order can help us to better understand the signaling process. In our model, couplings and feedback between these modules are also taken into account.
The activation of the G protein cycle
The α-factor secreted by MATα binds to, and hence activates, the seven-transmembrane-segment receptor (Ste2) on the plasma membrane surface of MATa. Pheromone binding enhances mono-ubiquitination of the receptor, and the ubiquitination in this case serves as a signal for endocytosis and delivery to the vacuole (18). This comprises a negative feedback loop (at short timescales). In our model, this process is treated as a process of accelerated degradation for simplicity. The synthesis of receptor Ste2 is included; the downstream effector Ste12 is responsible for the gene expression of Ste2. Thus, it comprises a positive feedback (at long timescales).
The interaction between the activated receptor and Gα leads to some conformational changes, which enable Gα to release GDP and to bind GTP (19). Gα·GTP cannot interact with Gβγ, resulting in a release of Gβγ from the receptor. The Gγ unit fixes the heterodimer on the plasma membrane surface, while the Gβ unit can interact with several effectors to transmit the signal. In this sense, Gα unit is a negative regulator of the pathway; it plays a role in an adaptational response to pheromone through preventing the availability of Gβγ when there is no signal (20). The Gα·GTP can be hydrolyzed into Gα·GDP, which can reassociate with Gβγ into a heterotrimer. The cycle of G protein is thus closed. Regulators of G protein signaling (RGS) proteins can accelerate the hydrolyzation of Gα·GTP (21). In this pathway, the most important RGS protein is Sst2, which is considered in our model. The gene expression of Sst2 is also regulated by Ste12 in the downstream. Therefore, Sst2 is part of a negative feedback loop that leads to the adaptation (8,22). Since there is experimental evidence that the amount of Gα increases significantly when the cells are treated with pheromone (8), we add G protein synthesis in our model. It is commonly accepted that Ste12 transcripts Gα genes. According to the above description, we formulate the reactions in the G protein cycle as
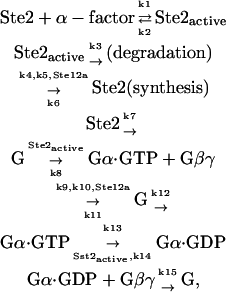 |
where the k-values are the kinetic parameters, and the protein above or below the arrow is the enzyme or transcription factor of the reaction
The scaffold-dependent MAPK pathway
The released Gβγ has several effectors (23). One effector for mating is Ste20, the first p21-activated protein kinase to be identified in any eukaryote (24). Ste20 is also activated by Cdc42, which is regulated by Cdc24. However, this process is not included in our model because the Cdc42 binding domain of Ste20 has been shown to be dispensable for pheromone signaling in yeast (25,26), and there should be enough active Cdc24GEF and Cdc42 constitutively at the membrane to activate the amount of Ste20 required for initial signaling. Besides, mutants in Cdc24 do not have much influence on the pathway (27,28).
Another effector of Gβγ is the scaffold protein Ste5. The correlation between the disruption of the Ste4(Gβ)-Ste5 interaction and sterility confirms the importance of this interaction in signal transduction (29). Gβγ can bind to Ste5 on the LIM domain of Ste5, which is required for Ste11 (MAPKKK) activation (30), probably through inducing a conformational change that enhances Ste20-dependent activation of Ste11. Also it interacts with Ste5 in the RING-H2 domain, which is essential for Ste5 oligomerization (31).
Most scaffolds are contained in the nucleus during vegetative growth. Upon pheromone induction they undergo enhanced exportation from the nucleus and localize at the “shmoo” tip (1). Although the detailed controlling mechanism of exportation of Ste5 is not clear, it is plausible that mating pheromone increases the rate of Ste5 export (57). Here, we utilize an active control mechanism where the import rate is kept constant, while the export rate is dependent on the total concentration of the released Gβγ. When there is no signal, the export rate is very low, keeping most scaffolds in the nucleus. When the mating signal opens the G-protein cycle, released Gβγ enhances the export rate, driving scaffolds to the shmoo tip. In this way, the localization of the scaffolds can be regulated by G protein cycle.
The mating pathway is highly dependent on the scaffold protein Ste5. First, Ste5 functions as an adaptor protein. It recruits Ste11 to the plasma membrane, where Ste20 is also tethered, to facilitate Ste11's activation (28), triggering the MAPK cascade. Another function for Ste5 is scaffolding. Ste5 tethers Ste11 (MAPKKK), Ste7 (MAPKK), and Fus3 (MAPK) to form a complex (32), keeping the kinases and their substrates in proximity, as well as preventing the influence of phosphatases. This function is supposed to be important in enzyme regulation and in preventing cross talk (33,34).
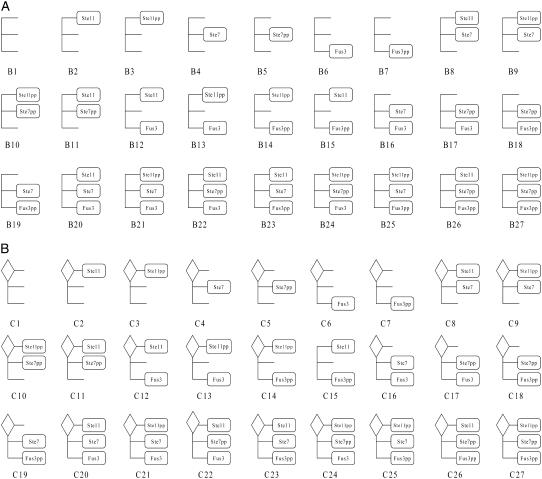
When Ste5 is in the cytosol, it can form scaffold-kinase complexes with Ste11, Ste7, and Fus3. Every kinase binding site on the scaffold is in one of the three possible states: without a kinase; with an unphosphorylated kinase; or with a dual-phosphorylated kinase. So for scaffold-kinase complexes in solution, there are 3 × 3 × 3 = 27 states: B1, B2, …B27 (See Fig. 2 A). Gβγ can bind to Ste20 and Bi (i = 1, 2…27). Because Ste20 is already on the plasmid membrane through the interaction with Cdc42 before signaling, and the scaffold must shuttle out from the nucleus to bind to Gβγ, we assume that Gβγ first binds to Ste20, then binds to Bi. Once Bi binds to GβγSte20 complex, it is fixed at the plasmid membrane and the whole complex is denoted Ci (see Fig. 2 B). Ci and Bi are the same in the interaction with MAPK kinases, except that Ci can phosphorylate Ste11 while Bi cannot. Note that
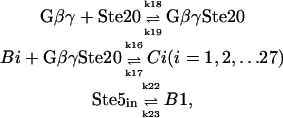 |
where the expression Ste5in denotes Ste5 in the nucleus, B1 denotes Ste5 outside of the nucleus, and k22 is dependent on the total concentration of released Gβγ:
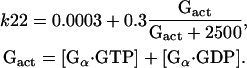 |
FIGURE 2.
(A) Twenty-seven solution-located scaffold complexes. (B) Twenty-seven membrane-located scaffold complexes. The diamond symbol at the upper-left corner indicates the GβγSte20 complex.
Ste11 is the MAPKKK of the yeast pheromone pathway, which consists of an N-terminal regulatory domain and a C-terminal kinase region (1). The interaction of these two domains keeps Ste11 in an anti-self state. CBD domain in N-terminal contains serine and threonine residues that can be phosphorylated by Ste20. Ste20-mediated phosphorylation of these residues activates Ste11 (35). In addition to recruiting Ste11 to a pool of its activator Ste20, Ste5 also binds to Ste11 in its N-terminal, making the CBD domain in N-terminal more accessible for Ste20. Ste50 also helps to make the CBD domain more accessible to Ste20 by a direct interaction between the SAM domain of itself and the SAM domain of Ste11 (36), but is less essential than Ste5. Cells lacking Ste50 are not truly sterile, thus we do not include Ste50 in our model. Ste11pp phosphorylates the target residues in the activation loops of Ste7 (MAPKK), and activates it (37). The activated Ste7pp then phosphorylates, and activates its targets, the MAPKs Fus3 and Kss1 (38). In the mating pathway, Fus3 plays a much more important role, while Kss1 is the main MAPK in the filamentation-invasion pathway in nitrogen-starved cells (5). Thus we do not consider Kss1 in our model, although analysis about cross talk will be given in the Discussion. While Ste11 and Ste7 are predominantly cytoplasmic proteins, Fus3 can shuttle between the nucleus and the cytoplasm. It concentrates in the nucleus after activation, thus bringing the signal to the nucleus (7,35,39). There are also several feedback loops: Ste11 (MAPKKK) undergoes ubiquitination and MAPK-dependent degradation (40); Ste7 (MAPKK) is assumed to undergo enhanced degradation after phosphorylation (41). In addition, Ste7pp in the scaffold is assumed to be hyperphosphorylated by activated Fus3pp, which reduces the binding efficiency between Ste7pp and the scaffold sharply (42).
All the kinases can be categorized into two pools: on the scaffold and in the solution. We assume that phosphorylation on the scaffold employs a processive mechanism, while phosphorylation in solution is distributive(11). “Processive mechanism” means that the active kinase collides with and binds to a substrate, phosphorylates it once, then it may slide to align the second phosphorylation site of the substrate with the active site of the kinase, and phosphorylates the substrate a second time before it finally dissociates. “Distributive mechanism” means that the active kinase collides with and binds to a substrate, phosphorylates it once and releases the monophosphorylated product, which then collides with a second molecule of the active kinase, and is phosphorylated a second time (14). In our model, we assume that a kinase is activated when and only when it is dual-phosphorylated, while a partial phosphorylated kinase possesses no activity. Dual phosphorylation in a distributive manner could lead to a sharp, sigmoidal stimulus-response curve (14,17), leading an all-or-none cell fate (14). However, the scaffold might diminish this property if phosphorylations on the scaffold occur in a processive manner (11). The dephosphorylations in the solution employ the distributive mechanism while dephosphorylations in scaffolds are precluded in our model due to sterical obstruction of the phosphatase groups. The proteins responsible for dephosphorylation of Ste11pp and Ste7pp are not clear. In the model, we add two proteins with constant concentration to dephosphorylate Ste11pp and Ste7pp, respectively. There are several phosphatases for Fus3pp: the dual-specificity phosphatase Msg5 (equally distributed in nucleus and cytoplasm), and the tyrosine phosphatases Ptp3 (cytoplasm) and Ptp2 (nucleus), all of which can result in the inactivation of Fus3pp (7,43,44). The basal level of Fus3 phosphorylation is controlled mainly by Ptp3, the amount of which is constant during the stimulation (44). Pheromone treatment induces the expression of Msg5 through the effects of Ste12 (43), which then act together with Ptp3 to inactivate Fus3pp. In our model, we use MAPK-P with an initial concentration and with a synthesis rate regulated by Ste12 to represent these three phosphatases. A recent experiment shows that different inputs by Ste5 and Msg5 phosphatase lead the MAPK cascade to multiple outcomes (45), indicating that MAPK-P is a key regulator in the network. Reactions of MAPK cascade in cytosol are formulated as
 |
where p indicates single phosphorylation, and pp indicates dual phosphorylation.
As for the scaffolds, we made the following assumptions in the model:
Inactive kinase can bind to Bi and Ci. On the scaffold, this inactive kinase can either dissociate from the scaffold without phosphorylation or undergo processive phosphorylation before getting off the scaffold if its upstream kinase happens to be on the same scaffold and in the active state.
Dephosphorylations on scaffolds are precluded due to sterical obstruction.
There is no binding of partially activated kinases to the scaffold proteins. For free fully activated kinases, only Ste11pp can bind to the scaffold. Experimental evidence indicates that active Ste5 can also accept Ste11pp activated by other pathways and channel those signals to Fus3 (46), and that a greater amount of scaffold proteins interact with Ste11 rather than the other two kinases. As for Ste7, it undergoes hyperphosphorylation by activated Fus3pp, which accelerates its dissociation from the scaffold (42). Fus3pp dissociates rapidly from the scaffold after phosphorylation (47) and travels into the nucleus. Thus, the reassociation of Ste7pp and Fus3pp to scaffold seems to be unlikely.
Scaffold molecules do possess some catalytic properties (33), so that the reaction rates within a scaffold complex are greater than in the solution. Moreover, Ste7 and Fus3 can bind firmly (23), and the residues for Ste7 binding in Fus3 are the same as the residues for Ste5 binding (48), so it is reasonable to assume that Ste7 competes with Ste5 for binding to Fus3. Fig. 3 illustrates the scaffold-dependent reactions of Ste11.
FIGURE 3.
Scaffold-dependent reactions.
Downstream effects
After activation, Fus3pp dissociates rapidly from the scaffold, while the scaffold remains tethered to the plasma membrane and partly in the solution (47,49), acting as a platform for activation of many molecules of Fus3 and leading to the propagation of the signal. The activated Fus3pp transmits the signal into the nucleus, resulting in the activation of transcription and the induction of cell cycle arrest. Fus3pp is assumed to mediate the pheromone-induced transcription of PRE-containing genes through phosphorylation and activation of at least three nuclear proteins: Dig1, Dig2, and Ste12 (1,2). In unstimulated cells, Dig1 and Dig2 bind to and thus repress Ste12 (50). Fus3pp phosphorylates Dig1, Dig2, and Ste12, and induces the release of Ste12 from the complex (50,51). The free Ste12 then interacts with other proteins of the transcription machinery and thereby activates transcription of many different genes. Among the products of these genes are proteins that activate (e.g., Fus3, receptor) or inhibit (e.g., Msg5, Sst2) the pathway (2). Therefore, the transcription affords several feedback loops in the pathway. Another important substrate of Fus3pp is Far1. Activated Ste12 increases the transcription of Far1, and Fus3pp is able to phosphorylate Far1 and thus to stabilize it (52). Far1 is a bifunctional scaffold protein. In the cytoplasm, Far1 is involved in polarized growth; in the nucleus, it has a key function in controlling cell cycle (53,54). Far1 inhibits Cln-Cdc28 complex, the master regulator of the yeast cell cycle in the G1 phase. In our model, the binding of Far1 to Cln-Cdc28 is treated as a symbol for cell cycle arrest. Ste12 is also the transcription factor of Bar1, which is then excreted from the cell and inactivates α-factor (53). It is deleted in our model because most experiment results that we use were from bar1Δ mutants. However, bar1Δ could cause >100-fold sensitivity increase in downstream transcription response (12). Reactions in the downstream are
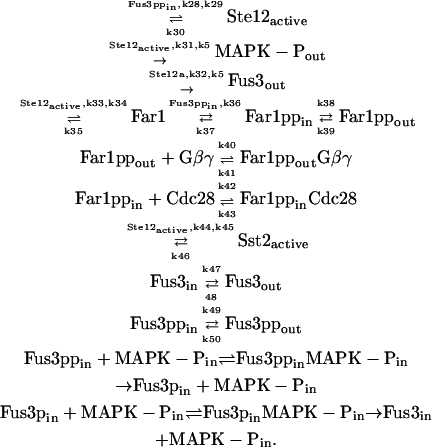 |
Spatial location
We consider two compartments in the cell: the nucleus and the shmoo tip (a projection toward the direction of pheromone formed as a result of polarized growth). The nucleus is where downstream effects take place. The shmoo tip is where the many signaling proteins are concentrated (55) and the main place for the upstream reactions, including the G-protein cycle and Ste5-related reactions. Thus in our model we neglected the rest part of the cytosol. In other words, we restricted the cytosol to the shmoo tip. The scaffold protein Ste5, the MAPK Fus3 (both activated and inactivated), and Far1 are shuttling between the nucleus and the shmoo tip. Ste5 is mostly sequestered in the nucleus in the absence of pheromone while pheromone enhances nuclear exportation of Ste5 (56). Nuclear localization of Fus3 is slightly enhanced by the pheromone treatment (7,39,57,58).
The mathematical model
We employ a set of ordinary differential equations (ODEs) to describe the changes in the concentration of proteins involved in the mating pathway. Generally, in a system of l biochemical species with the concentration ci (i = 1,2,···,l) and m biochemical reactions with the rates vj (j = 1,2,···,m), the following series of equations can be utilized to describe the biochemical mechanism in the system:
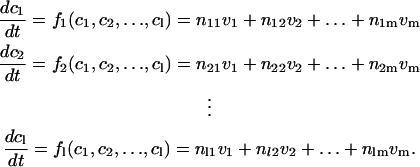 |
The quantity nij denotes the stoichiometric coefficient. The rate of a reaction is a function of the concentrations of substrates, products, and probable effectors (10). If we treat the gene expression as a special kind of reaction which can be described with Hill functions, the equations listed above can be employed to describe the dynamics of our system. In our model, all the unbound substances in various phosphorylation states and complexes formed by them are viewed as individual species. All complex formations, dissociations, degradations, phosphorylations, and dephosphorylations are treated as reactions. The parameters and initial concentrations in the model are derived from experiments whenever possible. For the remaining parameters, some are determined by fitting the results of the model to indirect experiments; others are estimated according to the mechanisms and similar reactions in other organisms. The list of the model parameters as well as the detailed ODEs are presented in Supplementary Material. For simulation, we use MatLab, version 6.5 (The MathWorks, Natick, MA).
RESULTS
Temporal characteristics
G-protein cycle
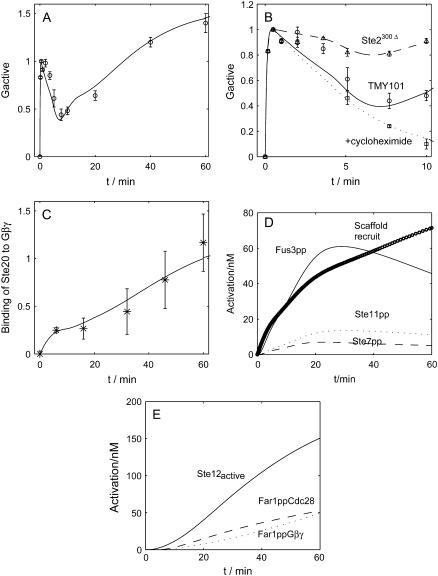
Fig. 4 summarizes the dynamics of the G-protein cycle. Upon saturated pheromone induction (1 μM α-factor), the level of the activated G-protein climbs up rapidly, reaches its peak at ∼30 s, and then gradually declines to a bottom at ∼7.5 min before it gradually increases again, as shown in Fig. 4 A. The simulation result (solid line) fits quite well with the experiment data (12) (circles with error bars). One crucial factor that might contribute to enhancing the closure of G-protein cycle is the endocytosis of activated receptor Ste2. This hypothesis is supported by experiment with mutant Ste2300Δ (the C-terminal tail of the α-factor receptor gene STE2 is removed to impair its endocytosis) (12). We slowed down the degradation rate of the active Ste2 to simulate the Ste2300Δ mutant. Consistent with the experiment data, the closure of G-protein cycle is apparently impaired and the amount of the activated G-protein levels off after reaching its peak, as shown in Fig. 4 B. The behavior of Ste2300Δ cells (dashed line for simulation, and up-triangle with error bars for experimental data) indicates that endocytosis is a key factor that causes the G protein cycle to close up. Fig. 4 A shows that after 10 min, the activated G-protein continues to rise steadily. We attribute it to protein synthesis, because that is the timescale for gene expression. To test this hypothesis, we delete protein synthesis of all proteins considered in our model, and find that the level of the activated G-protein does not rise in the simulation. The behavior of cycloheximide-treated cells, as shown in Fig. 4 B (dotted line for simulation and square with error bars for experiment data (12)), supports this hypothesis. For comparison, the time-course for TMY101 cells is also shown in the figure (solid line for simulation, and circles with error bars for experiment data).
FIGURE 4.
Time-course of the G protein cycle activation. (A) G protein activation. The values are normalized to the maximum concentration at ∼30 s. Result from simulation is shown as a solid line, and experiment data (12) are plotted in circles with error bars. (B) G protein activation in Ste2300Δ cells, the wild-types and cycloheximide-treated cells (experiment data are from (12)). (C) The time-course of binding Ste20 to Gβγ. The values are normalized to the maximum concentration (experiment data are from (24)). (D) The recruitment of scaffold protein Ste5 (circles) which is the sum of scaffolds in the solution and scaffolds at the membrane, the activation of MAPK cascade components Ste11pp (MAPKKKK) (dotted line), Ste7pp (MAPKK) (dashed line), and Fus3pp (MAPK) (solid line). (E) Downstream responses to α-factor induction: activated Ste12 (solid line), Far1pp•Cdc28 (dashed line), and Far1pp•Gβγ.
Binding of Ste20 to Gβγ
After activation, Gβγ activates two effectors: Ste20 (MAPKKKK) and Bi (scaffold in the solution), hence transmitting the signal downwards. In our model, the time-course for the pheromone-induced binding of Ste20 to Gβγ fits well with experimental data (24) (Fig. 4 C). It shows that Ste20 binds quickly to Gβγ during the first 5 min. The binding slows down afterwards and then speeds up. This time-course seems to follow the activation of G-protein cycle upstream (Fig. 4 A), consistent with the presumption that Gβγ-dependent activation rather than Cdc42-dependent activation of Ste20 is critical in the mating pathway.
Activation of MAPK pathway
After the recruitment of the scaffold protein Ste5 to the membrane, the signal passes down through the MAPK cascade (Fig. 4 D). Note that except for the gradual recruitment of Ste5, the signal transduction is very fast.
Downstream effects
Fig. 4 E shows the activation of Ste12, Far1ppGβγ, and Far1ppCdc28 to illustrate the downstream effects of Fus3pp (MAPK). The formation of complex Far1pp-Cln-Cdc28 is responsible for the cell cycle arrest, and Far1pp-Gβγ causes the polarized growth and the formation of the shmoo tip. Since Gβγ is a part of the upstream complex involving Ste5 to provide a scaffold for the MAPK pathway, excess Gβγ is not available until the pathway is attenuated to some extent. Thus, the curve for Far1-Gβγ begins to rise at 20 min after pheromone treatment, relatively late compared to other downstream effectors.
Features of the pathway
Scaffold shuttling and dephosphorylation cooperate to regulate the MAPK cascade quantitatively and to keep its fidelity to the mating signal
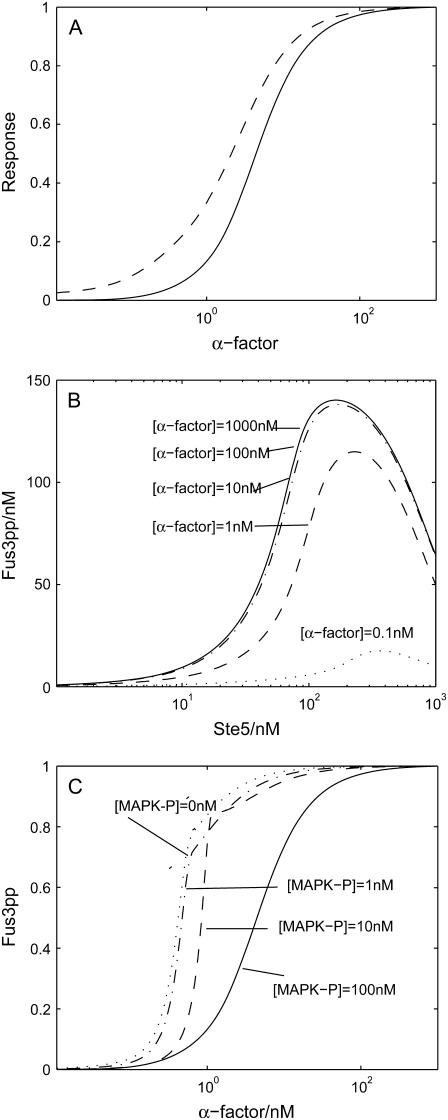
One of the most distinctive features of the mating pathway is its dependence on the scaffold. Interestingly, we found that the amount of Ste5 localizing out of nuclear upon pheromone induction coincides with activated Fus3pp (MAPK) in the dose-response curve, as shown in Fig. 5 A. Thus we speculate that the mating pathway is tightly controlled by scaffold protein Ste5 and its shuttling. It is highly possible that with different concentrations of scaffolds out of the nucleus, the efficiency of the MAPK cascade varies. When scaffold concentration is relatively low, the pathway efficiency should increase with scaffold concentration because of the catalytical function and the spatial protection function of the scaffold. When scaffold concentration is too high, it may reduce the mating efficiency because an effective complex suitable for signal transduction is hardly found (9). To investigate this possibility, we shut down the shuttling of the scaffold protein Ste5 and varied the concentration of total Ste5 at the shmoo tip from 1 nM to 1000 nM. For the levels of pheromone induction, we varied the concentration from 0.1 nM to 1000 nM. Consistent with our expectation, the intensity of the output of MAPK cascade, indicated by the amount of the activated Fus3pp, first increases with scaffold concentration, then decreases, with the optimal scaffold concentration at ∼100 nM, as shown in Fig. 5 B. Thus, with our choice of Ste5 concentration in the model (∼100 nM), monotonic increases with scaffold available in the shmoo tip, and it is reasonable to suggest that scaffold proteins have the ability to quantitatively control the strength of signal transmission through the MAPK cascade. This function of Ste5 could be achieved by facilitating Ste11's activation by Ste20 through binding to Gβγ, concentrating MAPK cascade components, and sequestering them from inhibition by phosphatases (see below).
FIGURE 5.
(A) Predicted dose-response curves of recruitment of scaffold protein Ste5 (dashed line) and activation of Fus3 (MAPK) (solid line). (B) The dependence of Fus3pp on the concentration of scaffold proteins Ste5, with different concentration of α-factor. (C) Predicted dose-response curves for mutants in which MAPK-P is underexpressed. All the values are normalized. In all these simulations, cells are treated as indicated with α-factor for 20 min.
Dephosphorylation is the other mechanism we speculate that might contribute to controlling the activation of the mating pathway. To test this hypothesis, we varied MAPK-P's concentration (both in the nucleus and at the shmoo tip) from 100 nM down to 0 nM. As expected, Fus3pp (MAPK) shows super-sensitivity upon pheromone induction (see Fig. 5 C). How does dephosphorylation control activation of MAPK cascade? Since the kinases are exposed to phosphatase only when they are in the cytosol and the scaffold could help to prevent the influence of the phosphatase on the kinases bound to it, we suggest that certain levels of phosphatase concentration can keep the kinase phosphorylation in the cytosol at a very low level, and thus constrain the signal transduction on the scaffold. Hence, when the phosphatases are attenuated, a large amount of activated Ste11pp, Ste7pp, and Fus3pp could be accumulated in the cytosol even at a lower level of scaffold protein recruited to the shmoo tip, bypassing the control of scaffold protein Ste5. In short, the shuttling of the scaffold and the dephosphorylation of the MAP kinases cooperate to control the activation of MAPK cascade quantitatively.
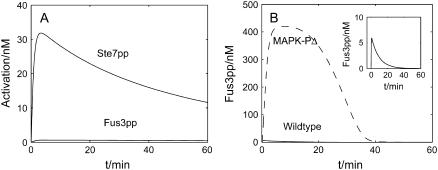
The cooperation between the scaffold shuttling and the MAPK dephosphorylation is also crucial to the specificity of the pathway. There are at least five MAPK signal transduction pathways in budding yeast (5), some of which share the same proteins, such as Ste11 and Ste7. A big puzzle is how specificity is achieved. To investigate the mating pathway's ability to isolate inappropriate signals leaking in from other pathways such as the filamentation-invasion pathway, we tested the behavior of some mutants. We set all Ste11 molecules in the dual phosphorylated state at t = 0 and shut off MAPKKK-P to simulate the constitutive activation of Ste11pp in invasive growth in the absence of mating pheromone. From the time-course curve of Fig. 6 A, we observed that although Ste7pp (MAPKK) is activated to a relatively low extent, little Fus3pp (MAPK) is stimulated. We then set all Ste7 molecules in the dual phosphorylated state at t = 0 and shut off MAPKK-P to simulate constitutive Ste7 activation, and found that Fus3pp (MAPK) could only be activated transiently in cytosol; the activation dropped down immediately (within 10 s) (inset in Fig. 6 B). This result is consistent with the experiments, revealing that persistent activation by constitutive Ste7pp fails to support Fus3-dependent mating in the yeast (42). Further simulation by removing MAPK-P indicates that the later deactivation of Fus3pp is caused by dephosphorylation (Fig. 6 B). Note that only the kinases in the solution are exposed to phosphatases, and the scaffold could help to shield from the influence of phosphatases. Thus, phosphatases constrain the signal on the scaffold. While on the scaffold, Ste7pp prefers to phosphorylate Fus3 instead of Kss1; the signal constrained on the scaffold will lead to the activation of Fus3, rather than Kss1, the main MAPK in the filamentation-invasion pathway in nitrogen-starved cells. That means that the activation of Fus3 strictly relies on the scaffold protein. When there is no pheromone induction, no scaffold protein is recruited to the shmoo tip, and the phosphatases inhibit the basal activation of Fus3. With pheromone treatment, the active scaffold proteins (the activation means recruitment to the plasma membrane in our model) help to assemble MAP kinases cascade components and accelerate the activation of Fus3pp, the main MAPK in the mating pathway. When activation exceeds dephosphorylation, the signal will be transmitted into the nucleus.
FIGURE 6.
Dephosphorylation prevents improper signal to leak in. (A) All Ste11 is dual-phosphorylated at t = 0, and MAPKKK-P is shut off. This simulates the condition in which signal in invasive growth pathway is “on”. (B) All Ste11 is dual-phosphorylated at t = 0, and MAPKK-P is shut off. The activation of Fus3 is still repressed downwards except for the small pulse at the very beginning (Inset graph). Dashed line indicates the activation of Fus3 when MAPK-P is eliminated.
To conclude, when there is no pheromone induction, phosphatases repress the activation of the mating pathway and prevent inappropriate signals from leaking in. When pheromone exists, scaffolds are recruited to the shmoo tip by the activated G protein, gathering MAPK cascade components and sequestering them from phosphatases so that the mating signal can be transmitted downwards. Thus, the mating pathway is highly dependent on scaffolds. This conclusion is consistent with experiments (45). Although there are other factors that contribute to suppress the cross talk between the two pathways (34), the mechanism outlined above could also play an important role.
Desensitization to pheromone induction
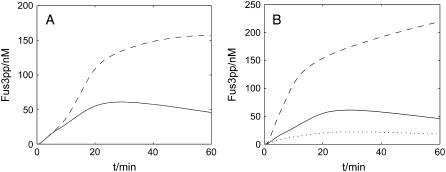
The amount of activated Fus3pp (MAPK) decreases with time even when the cells are exposed to prolonged α-factor induction, as shown in Fig. 7 A, our wild-type cell simulation (solid line). This indicates a desensitization effect. Desensitization is a key feature of the pathway, which enables cells to reenter the cell cycle to resume vegetative growth. We investigated the possible factors that might contribute to this desensitization, and found that multiple negative feedback loops—such as the degradation of Ste11 (MAPKKK) and Ste7pp (MAPKK), the synthesis of Msg5 (MAPK phosphatase) and Sst2—should be the major cause. As shown in Fig. 7 A, a wild-type cell with all the negative feedback shows desensitization, but a mutant without these negative feedback does not (dashed line): the activation of Fus3pp does not decrease even after 1-h treatment with saturating pheromone.
FIGURE 7.
Desensitization. (A) Time-course of activation of Fus3 (MAPK) in a wild-type cell (solid line) and in a mutant (dashed line). (B) Effect of feedback hyperphosphorylation of Ste7 in the scaffold. Predicted time-course activation of Fus3 for wild-type (solid line) and for mutants (dashed line and dotted line).
Another important cause is the negative regulation of Ste7's (MAPKK) binding ability to scaffold by Fus3pp (MAPK). In the scaffold, Ste7pp, which undergoes feedback phosphorylation by activated Fus3pp, dissociates more quickly from the scaffold (42), hence exposing itself to ubiquitination and degradation (41,59). This feedback can also accelerate the disassembling of scaffold complexes. In our model, we assume that the dissociation rate for Ste7pp on scaffolds with both Ste7pp and Fus3pp is larger than that for Ste7pp on scaffolds without Fus3pp. If we change the former parameter to be the same as the latter one, activation of Fus3pp continues to rise after prolonged stimulation, as shown in Fig. 7 B (dashed line). On the other hand, the dissociation rate for Ste7pp from the scaffold without Fus3pp has to be relatively slow to keep the intensity of the signal transduction. A mutant in which the dissociation rate for the normal phosphorylated Ste7pp (with no Fus3pp on the scaffold) is enhanced (dotted line in Fig. 7 B) results in a very low intensity of the signal transduction. Thus our model shows that differentiated binding abilities of Ste7 to the scaffold ensure the correct behavior of the MAPK cascade.
Sensitivity to pheromone
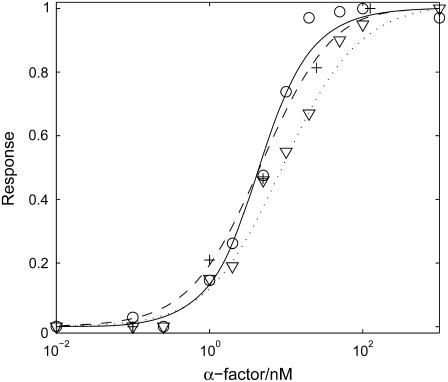
Aside from the temporal characteristics, the sensitivity to different levels of pheromone induction is another key feature of the signal transduction pathway. We compared the dose-response curves predicted by our model to those observed in experiments (12,45), and there is a quantitative agreement (Fig. 8). While our model specifically simulates the ΔBar1 strain, our results are also consistent with experiments of the wild-type, taking into account the 100-fold sensitivity shift (8,60). In our study, cells are assumed to be treated with indicated concentration (0.001 nM–1000 nM) of α-factor for 20 min. The response of every component to a certain concentration of α-factor is represented by the maximum amount of that component. For the activated Ste2, the peak is obtained within seconds; for G protein activation (measured by the sum of Gα-GTP and Gα-GDP), the maximum amount is achieved at ∼30 s; for Ste11pp, Ste7pp, and Fus3pp, the peak value appears at ∼20 min. These different timescales are consistent with experimental observations and illustrate the characteristics of the activation of different components. All these curves are normalized. We fit the dose-response curves of Ste2a, G-protein activation, scaffold recruitment, Ste11pp, Ste7pp, and Fus3pp with the Hill Function,
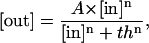 |
where n is the Hill coefficient and th the threshold where the response reaches half its maximum (Table 1). Note that the sensitivity to α-factor is well conserved throughout the whole pathway, from receptor Ste2 at the very beginning throughout the MAPK cascade.
FIGURE 8.
Dose-response curves for key components in the mating pathway in TMY101 cells: activated Ste2 (dotted line for simulation, down-triangles for experiment data from (12)); G protein activation, i.e., the sum of Gα-GTP and Gα-GDP (dashed line for simulation, circles for experiment data from (12)); and Fus3pp (MAPK) (solid line for simulation and crosses for experiment data from (45)).
TABLE 1.
Coefficients in Hill function
| Coefficient | Ste2a | G activation | Ste11pp | Ste7pp | Fus3pp |
|---|---|---|---|---|---|
| n | 0.9 | 1.0 | 1.2 | 1.2 | 1.2 |
| th (nM) | 9.1 | 4.8 | 4.0 | 4.8 | 4.8 |
Simulation results with the whole model.
As stated before, our model separates the whole mating pathway into different modules. It is interesting to explore the dose-response curve of each module. The first module is ligand binding. The reason why the dose-response curve of the activated Ste2 is a Hill function with n ≈ 1 is the reaction it takes. Consider the reaction 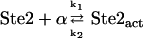 , with the input concentration of α (on the left-hand side) fixed at [α] and the total concentration of Ste2 fixed at [Ste2]0. At steady state,
, with the input concentration of α (on the left-hand side) fixed at [α] and the total concentration of Ste2 fixed at [Ste2]0. At steady state,
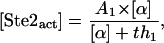 |
(1) |
with A1 = [Ste2]0 and th1 = k2/k1, which takes the value th1 = 5.0 with our choice of parameters k1 and k2. The coefficients derived from our simulation with our whole model are n = 0.9, th = 9.1. The differences come from protein synthesis, degradation and signal dependent feedback. When these effects are deleted from the whole model, the simulation results agree perfectly with the analysis.
We further explored the dose-response curve for G protein activation. Again, for simplicity, we do not take into account in the calculation the signal-dependent production, degradation and the feedback of the RGS protein (we treat the amount of Sst2 as a constant) and look for the steady-state solution. The result of the calculation is
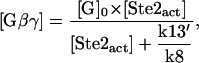 |
(2) |
where k13′ = k13 + k14[Sst2] . To get this result, we made the assumption that the hydrolysis is a relatively slow process, so that 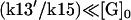 . If we substitute Eq. 1 into Eq. 2, we get
. If we substitute Eq. 1 into Eq. 2, we get
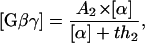 |
(3) |
where 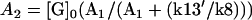 and
and 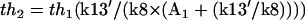 . Equation 3 is a Hill function with n = 1.0 and th = 2.9. The simulation result of our whole model is n = 1.0 and th = 4.8. Again, if the protein synthesis, degradation, and feedbacks are deleted, the simulation results of n = 1.0 and th = 2.9 agree well with the analysis.
. Equation 3 is a Hill function with n = 1.0 and th = 2.9. The simulation result of our whole model is n = 1.0 and th = 4.8. Again, if the protein synthesis, degradation, and feedbacks are deleted, the simulation results of n = 1.0 and th = 2.9 agree well with the analysis.
We see that the curve of [Gβγ] ∼ [α-factor] employs the same type of function as the curve of [Ste2act] ∼ [α-factor], with the same Hill coefficient. The only difference is that there is a shift in threshold.
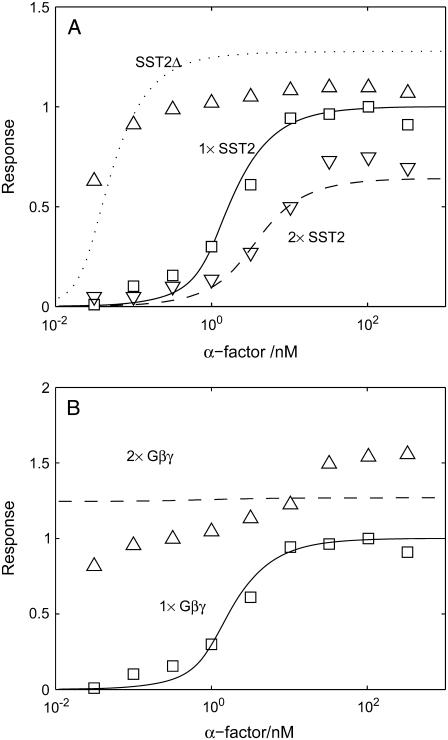
The above analysis suggests that the amount of Sst2 can affect the threshold of the dose-response curve. Fig. 9 A indicates that Sst2, which can accelerate the closure of the G protein cycle, is indeed a key regulator of the mating pathway's sensitivity. Comparison of the dose-response curves for mutants SST2Δ (dotted line for simulation, up-triangle with error bars for experiment data), wild-type cells (solid line for simulation, squares for experiment data (8)), and 2 × SST2 (dashed line for simulation, down-triangle for experiment data) clearly shows that the system is sensitive to the amount of Sst2, which is consistent with the theoretical analysis above. Another mutant we studied in this module is one with excess Gβγ copies. Fig. 9 B compares dose-response curves for TMY101 cells (solid line for simulation and squares for experiment data) and cells with 2 × Gβγ (dashed line for simulation, and up-triangles for experiment data (8)), indicating that Gβγ alone is sufficient to switch on the downstream signal transduction.
FIGURE 9.
Response curve of Ste12 to α-factor for (A) different Sst2 expression level; and (B) different Gβγ expression level. To simulate the 2 × Gβγ cells, we separately add another 1000 nM Gβγ at t = 0 min to double the total concentration of Gβγ. Cells are treated with α-factor for 60 min. Note that the experiment data are shifted left by >100-fold because Bar1 is deleted in TMY101 cells. Experiment data are from Hao et al. (8).
The next module in the pathway is the scaffold-dependent phosphorylation cascade. Again, we do not take into account the protein synthesis and degradation or all the feedback. Here, Gβγ is the input, while the concentration of Ste11pp, Ste7pp, and Fus3pp is chosen as the output. Gβγ first binds to Ste20, then binds to scaffold in the solution to generate C (we use C to indicate the ensemble of C1, C2,…C27), which then initiates MAPK cascade. We calculate the Hill coefficients of Ste11pp, Ste7pp, and Fus3pp using Gβγ as input, and find that each of them shows ultrasensitivity (numerical results: nste11pp = 2.3, nste7pp = 2.2, and nfus3pp = 1.8). Since Gβγ experiences two simple reversible reactions to generate C, the relation between C and Gβγ must be a Hill function with n = 1. Therefore, the ultrasensitivity in this module must arise from C to Ste11pp, Ste7pp, and Fus3pp. Let us first consider the phosphorylation and dephosphorylation cycle of Ste11. Ste11 can be dual-phosphorylated on the scaffold in a processive mechanism and dual-dephosphorylated in the cytosol in a distributive mechanism:
 |
where we assume that Ste11pp leaves the scaffold so fast that CSte11pp can be neglected in the process. Introduce Michaelis constant 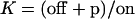 ,
, 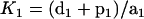 , and
, and 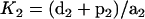 ; define
; define 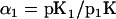 ,
, 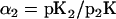 ,
, 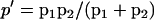 , and
, and 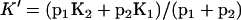 ; and we get
; and we get
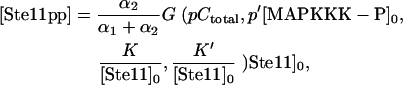 |
(4) |
where
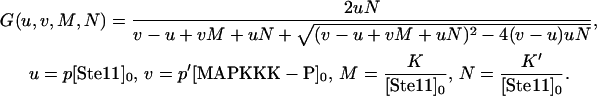 |
The above G function can be fitted to a sigmoidal curve (Hill function) (61), with the Hill coefficient and the threshold value to be
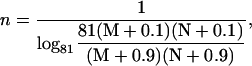 |
(5) |
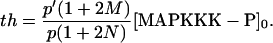 |
(6) |
According to the parameters in the Ste11 cycle, M = K/[Ste11]0 ≈ 0.125 and N = K′/[Ste11]0 = 0.255. So Eq. 7 approximates a Hill function with n ≈ 2.6 and th ≈ 21 nM. Therefore, it is the 0th-order ultrasensitivity that leads to the ultrasensitive response of Ste11pp. Similarly, in the Ste7 cycle, M ≈ 0.183 and N = 0.324; in the Fus3 cycle, M ≈ 0.135 and N = 0.181. Therefore, Ste7 and Fus3 are also located in the 0th-order region, making them ultrasensitive.
The analysis above together with Eq. 6 indicates that the relation between Ste11pp, Ste7pp, Fus3pp, and α should exhibit ultrasensitivity (numerical result: nste11pp = 2.1, nste7pp = 1.8, and nfus3pp = 1.9). This appears to contradict the results in Table 1, where all the dose-response curves overlap with Hill coefficient n ≈ 1. The reason for this contradiction is that in the analysis we cut off all the feedback in the original model. There are nine feedbacks altogether in the entire model, six of which are negative feedbacks: the transcription of Sst2; MAPK-P; the degradation of activated Ste2, Ste11, Ste7pp; and the hyperphosphorylation of Ste7pp. When [α] is low, all of these negative feedbacks are kept low, leaving the output nearly unaffected. When [α] is high, the negative feedbacks will also be strong, significantly reducing the output. Therefore, negative feedback can make the dose-response curve less steep. It may be the counterbalance between the 0th-order ultrasensitivity and the negative feedback that keeps the Hill coefficients of Ste11pp, Ste7pp, and Fus3pp as 1. To test this view, we add each of the six negative feedbacks into the simplified model where all feedbacks are cut off (Table 2). Numerical result shows that each feedback can reduce the Hill coefficients, and if all the negative feedbacks are added together, the Hill coefficients can decrease to nearly 1, indicating that negative feedback indeed can reduce the Hill coefficients.
TABLE 2.
Hill coefficient n when negative feedback are added
| Added feedback | Ste11pp | Ste7pp | Fus3pp |
|---|---|---|---|
| None | 2.1 | 1.8 | 1.9 |
| Transcription of Sst2 | 1.8 | 1.5 | 1.7 |
| Transcription of MAPK-P | 2.0 | 1.7 | 1.8 |
| Degradation of Ste2act | 1.8 | 1.4 | 1.8 |
| Degradation of Ste11 | 1.3 | 1.2 | 1.8 |
| Degradation of Ste7pp | 2.1 | 1.7 | 1.7 |
| Hyperphosphorylation of Ste7pp | 1.8 | 1.6 | 1.4 |
| All six feedbacks | 1.2 | 1.2 | 1.2 |
We further study how the Hill coefficients of Ste11pp, Ste7pp, and Fus3pp depend on the scaffold, the substrate, and the phosphatase concentrations (Table 3). First, we increase Ste11, Ste7, or Fus3 concentration by 10 times, and find that the Hill coefficients only change a little. The probable reason is that the original kinase concentration is already much larger than the scaffold concentration, so when Ste11, Ste7 or Fus3 concentration increases, the added part could not get to the scaffold to be phosphorylated, and thus does not contribute to response. Then, we increase the kinase concentration and scaffold concentration together, and find that Hill coefficients have a substantial increase. This is because when scaffold and kinase concentration increase together, effective substrate concentration increases. Therefore, the 0th-order ultrasensitivity becomes more significant, while feedback, which is mainly dependent on the downstream regulation, does not increase as fast. Table 3 also shows that when each phosphatase concentration is decreased by 10 times, Hill coefficients rise as well. This is because low phosphatase concentration allow more substrate to be phosphorylated in the cytosol in a distributive mechanism, which is regarded as another mechanism to generate ultrasensitivity aside from the 0th-order ultrasensitivity (17). Thus, the scaffold, the substrate, and the phosphatase concentration play an important role in determining the Hill coefficients of the MAPK pathway.
TABLE 3.
Hill Coefficient n when concentration changes
| Concentration changes | Ste11pp | Ste7pp | Fus3pp |
|---|---|---|---|
| [Ste11] × 10 | 1.2 | 1.2 | 1.3 |
| [Ste7] × 10 | 1.3 | 1.6 | 1.2 |
| [Fus3] × 10 | 1.2 | 1.2 | 1.2 |
| [Ste11] × 10, [Ste5] × 10 | 1.7 | 1.8 | 1.4 |
| [Ste7] × 10, [Ste5] × 10 | 1.8 | 2.5 | 2.8 |
| [Fus3] × 10, [Ste5] × 10 | 1.4 | 1.7 | 1.7 |
| [MAPKKK – P]/10 | 1.7 | 1.7 | 1.4 |
| [MAPKK – P]/10 | 1.2 | 1.7 | 1.2 |
| [MAPK – P]/10 | 1.2 | 1.3 | 2.1 |
Analysis of parameters
Due to the lack of experimental data to determine all the parameters, it is necessary to analyze the sensitivity of the system to changes of the parameters. To do this, we define a quantity
 |
where [Fus3pp]cal denotes the calculated output when a parameter is changed and [Fus3pp]obs denotes the original output, and the bar denotes the average of the relative variance of the output over an input (α-factor) range of 10−3 nM to 103 nM. We multiply and divide one parameter by 2 at each time, calculate D, and then take the average for the two situations of increasing and decreasing the parameter. The most sensitive parameters are listed in Table 4. Note that these parameters all have direct or indirect experimental support, which is reassuring. These parameters also give some clues about the mechanisms of the mating pathway. The parameters that influence the outcome of the pathway most are those involved in the receptor activation, which is consistent with our finding that the shape of dose-response curve is determined by the first step in the pathway—the receptor activation. The parameter in the production of receptor Ste2 is also essential to the outcome of the system. The other two influential parameters are the rates of the kinases to get off the scaffold, so the scaffold protein is also an important factor in this pathway.
TABLE 4.
Influential parameters
| Parameter | 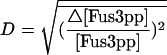 |
Related reaction |
|---|---|---|
| k1 | 0.519 | Ste2+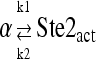
|
| k2 | 0.266 | |
| k6 | 0.167 | 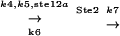 |
| **offK | 0.156 | Fus3pp gets off from the scaffold protein. |
| **off′KK | 0.150 | Ste7pp gets off from the scaffold protein. |
DISCUSSION
Our model describes the entire mating pathway comprising the G protein cycle, the scaffold-dependent MAPK cascade, and the downstream effects in the nucleus. We have investigated multiple features of the pathway, including its various characteristic timescales, desensitization, effect of the scaffold, specificity, sensitivity to different levels of pheromone induction, the role of feedback, and sensitivity amplification. Although many of the parameters in our model do not have solid experimental support and the detailed mechanisms of some steps are still not clear, the results given by our model are consistent with the current understanding of the pathway and with a wide range of experimental data.
The duration and sensitivity of the mating pathway have to be tightly regulated; an inappropriate activation of Fus3 will block the normal invasive growth. Our model shows that activation of the mating pathway attenuates with time even when the pathway is exposed to prolonged pheromone induction. This desensitization is attributed to several feedbacks, such as the enhanced degradation of Ste7 (MAPKK). These feedbacks enable the cells to recover from mating and continue their vegetative growth upon prolonged pheromone induction.
Evidence shows that oligomerization of the scaffold protein is required for its activation (31,57,62,63). However, for simplicity, oligomerization is not included in our model. Since we consider all possible complexes involving the scaffold protein, taking the oligomerization into account would make the model extremely complicated (with 27 × 27 = 729 possible complexes involving the scaffold protein). Moreover, there are few experimental data concerning the interactions between the scaffold protein and the kinases. Therefore, we have to make some simplifications. A clue for the simplification is the experimental evidence that nuclear export and shmoo tip recruitment of Ste5 are coordinated with oligomerization (57,61). Thus we use nuclear export as a controlling step. In our model, the recruitment of the scaffold protein to the shmoo tip implies its activation, including the effect of nuclear transportation and oligomerization. However, oligomerization may have effects on the pathway other than the activation of scaffold protein. Due to the lack of experimental data pertaining to this process, it is difficult to consider it in detail in our model. Experimental investigation into this process is much needed to further improve the model.
The scaffold protein undergoes continuous shuttling and enhanced exportation upon pheromone induction. The obvious question is why yeast cells take so much trouble to shuttle a huge protein through the nucleus when it functions predominantly in cytoplasm. Our model indicates that nuclear shuttling might be a key step controlling the availability of the scaffold protein to the pathway. Since the activation of the MAPK cascade in the mating pathway is dependent on the scaffold protein, whether and how many scaffold proteins are available determine whether and how efficient the MAPK pathway is stimulated. However, the mechanism of scaffold shuttling are still not clear. The complex Msn5p/Ste21 is suggested to be responsible for the export of Ste5p. Further work is required to establish the accurate and detailed mechanism of this controlling step. In our model, we employ an active control mechanism. That is, the scaffold export rate is dependent on the concentration of the separated Gβγ, which is released by the pheromone. The more pheromone, the more the activated Gβγ, and the higher the export rate. In other words, the G-protein cycle controls the shuttling of the scaffold and the concentration of the scaffold at the shmoo tip.
Another function of the scaffold is its role in keeping the pathway's fidelity to the signal. The capability of the scaffold protein to prevent kinases from dephosphorylation assures the mating pathway's dependence on the scaffold protein, and the availability of scaffold in the shmoo tip is further controlled by the G protein cycle. The specificity of different pathways in yeast is under intensive study (34). It is interesting to see that different cellular signals, which can be transmitted by the same components, result in distinct responses. Of special interest is that the haploid invasive growth pathway employs the same MAPK components (Ste11 as MAPKKK, Ste7 as MAPKK, Fus3 and Kss1 as MAPK) as the mating pathway, except that Fus3 is more active during mating while Kss1 is preferentially activated during invasive growth. Then how are the different outputs controlled? Our model suggests that dephosphorylation and scaffolds work in coordination to prevent improper signal from leaking in, and thus contributes to the mating pathway's fidelity to pheromone induction. Further work is needed to include the parallel pathway of the invasive growth, through which a more comprehensive understanding of specificity might be obtained.
MAPK cascade, which is conserved in all eukaryotic cells, is composed of both phosphorylation and dephosphorylation. Our model reveals that dephosphorylation has several roles in the mating pathway. It is obvious that it contributes to the desensitization of the pathway that enables the cell to reenter the cell cycle. Furthermore, it cooperates with the shuttling of the scaffold proteins, to realize other important features of the signaling pathway. First, it helps to preserve the consistence of the sensitivity from the G-protein cycle to the MAPK cascade. Second, the amount of MAPK-P, together with the scaffold protein, contributes to the pathway fidelity.
Notably, we suggest that negative feedback plays an important role in the experimentally observed preservation of sensitivity along the MAPK cascade. Whether or not the sensitivity is amplified as the MAPK cascade descends is also determined by the concentrations of the kinases and phosphatases involved. We call for new experiments to test this hypothesis.
Supplementary Material
Acknowledgments
We thank members of the Peking University Center for Theoretical Biology for their valuable suggestions.
This research is supported by the National Key Basic Research Project of China, the Chinese Natural Science Foundation, and the Chun-Tsung Endowment at Peking University. C.T. acknowledges support from the Sandler Family Supporting Foundation.
Danying Shao and Wen Zheng contributed equally to this work.
References
- 1.Bardwell, L. 2004. A walk-through of the yeast mating pheromone response pathway. Peptides. 25:1465–1476. [DOI] [PubMed] [Google Scholar]
- 2.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703–754. [DOI] [PubMed] [Google Scholar]
- 3.Wang, Y., and H. G. Dohlman. 2004. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science. 306:1508–1509. [DOI] [PubMed] [Google Scholar]
- 4.Dohlman, H. G. 2002. G protein and pheromone signaling. Annu. Rev. Physiol. 64:129–152. [DOI] [PubMed] [Google Scholar]
- 5.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K., B. Satterberg, D. M. Lyons, and E. A. Eloin. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 78:499–512. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell, E., I. M. Halatek, H. N. Kim, A. T. Ellicott, A. A. Obukhov, and D. E. Stone. 2003. Effect of the pheromone-responsive Gα and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol. Cell. Biol. 23:1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao, N., N. Yildirim, Y. Wang, T. C. Elston, and H. G. Dohlman. 2003. Regulators of G protein signaling and transient activation of signaling. J. Biol. Chem. 278:46506–46515. [DOI] [PubMed] [Google Scholar]
- 9.Kholodenko, B. N. 2000. Negative feedback and ultrasensitivity can bring about oscillation in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 267:1583–1588. [DOI] [PubMed] [Google Scholar]
- 10.Kofahl, B., and E. Klipp. 2004. Modeling the dynamics of the yeast pheromone pathway. Yeast. 21:831–850. [DOI] [PubMed] [Google Scholar]
- 11.Levchenko, A., J. Bruck, and P. W. Sternberg. 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA. 97:5818–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi, T., H. Kitano, and M. Simon. 2003. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc. Natl. Acad. Sci. USA. 100:10764–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhalla, U. S., P. T. Ram, and R. Lyengar. 2002. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Sci. Rep. 297:1018–1023. [DOI] [PubMed] [Google Scholar]
- 14.Ferrell, J. E., and R. R. Bhatt. 1997. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 272:19008–19016. [DOI] [PubMed] [Google Scholar]
- 15.Ferrell, J. E., and E. M. Machleder. 1998. The biochemical basis of an all-or-none cell fate within Xenopus oocytes. Sci. Rep. 280:895–898. [DOI] [PubMed] [Google Scholar]
- 16.Xiong, W., and E. Ferrell. 2003. A positive-feedback-based bistable “memory module” that governs a cell fate decision. Nature. 426:460–465. [DOI] [PubMed] [Google Scholar]
- 17.Huang, C. F., and J. E. Ferrell. 1996. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA. 93:10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicke, L., and H. Reizman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 84:277–288. [DOI] [PubMed] [Google Scholar]
- 19.Conklin, B. R., and H. R. Bourne. 1993. Structural elements of Gα subunits that interact with Gβγ, receptors and effectors. Cell. 73:631–641. [DOI] [PubMed] [Google Scholar]
- 20.Cole, G. M., D. E. Stone, and S. I. Reed. 1990. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol. Cell. Biol. 10:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohlman, H. G., and J. Thorner. 1997. RGS proteins and signaling by heterotrimeric G protein. J. Biol. Chem. 272:3871–3874. [DOI] [PubMed] [Google Scholar]
- 22.Dohlman, H. G., D. Apaniesk, Y. Chen, J. Song, and D. Nusskern. 1995. Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3635–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardwell, L., J. G. Cook, E. C. Chang, B. R. Cairns, and J. Thorner. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16:3637–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 391:191–195. [DOI] [PubMed] [Google Scholar]
- 25.Leberer, D. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter, M., A. M. Neiman, H. O. Park, M. Van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 27.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 22:2939–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβ γ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12:2684–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowell, S. J., A. L. Bishop, S. L. Dyos, A. J. Brown, and M. S. Whiteway. 1998. Mapping of a yeast G βγ signaling interaction. Genetics. 150:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng, Y., L. Y. Song, E. Kincaid, S. K. Mahanty, and E. A. Elion. 1998. Functional binding between Gβ and LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 8:267–278. [DOI] [PubMed] [Google Scholar]
- 31.Sette, C., C. J. Inouye, S. L. Stroschein, P. J. Iaquinta, and J. Thorner. 2000. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell. 11:4033–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Printen, J. A., and G. F. Sprague. 1994. Protein-protein interaction in the yeast pheromone response pathway: Ste5 interacts with all members of the MAP kinase cascade. Genetics. 138:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, S., A. Zarrinpar, and W. A. Lim. 2003. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 299:1061–1064. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, M. A., and H. D. Madhani. 2004. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38:725–748. [DOI] [PubMed] [Google Scholar]
- 35.Van Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630–639. [DOI] [PubMed] [Google Scholar]
- 36.Kwan, J. J., N. Warner, T. Pawson, and L. W. Donaldson. 2004. The solution structure of the S. cerevisiae Ste11 MAPKKK SAM domain and its partnership with Ste50. J. Mol. Biol. 342:681–693. [DOI] [PubMed] [Google Scholar]
- 37.Neiman, A. M., and I. Herskowitz. 1994. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homolog STE7 by STE11. Proc. Natl. Acad. Sci. USA. 91:3398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Errede, B., A. Gartner, Z. Z. Hou, K. Nasmyth, and G. Ammerer. 1993. MAP kinase-related Fus3 from S. cerevisiae is activated by Ste7 in vitro. Nature. 362:261–264. [DOI] [PubMed] [Google Scholar]
- 39.Choi, K., J. E. Kranz, S. K. Mahanty, K. Park, and E. A. Elion. 1999. Characterization of Fus3 localization: active Fus3 localizes in complexes of varying size and specific activity. Mol. Biol. Cell. 10:1553–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esch, R. K., and B. Errede. 2002. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA. 99:9160–9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Y., and H. G. Dohlman. 2002. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 277:15766–15772. [DOI] [PubMed] [Google Scholar]
- 42.Maleri, S., Q. Ge, E. A. Hackett, Y. Wang, H. G. Dohlman, and B. Errede. 2004. Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast. Mol. Cell. Biol. 24:9221–9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doi, K., A. Gartner, G. Ammerer, B. Errede, H. Shinkawa, and K. Sugimoto. 1994. MSG5, a novel protein phosphatase, promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan, X. L., R. J. Deschenes, and K. L. Guan. 1997. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 11:1690–1702. [DOI] [PubMed] [Google Scholar]
- 45.Andersson, J., D. M. Simpson, M. Qi, Y. Wang, and E. A. Elion. 2004. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 23:2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flatauer, L. J., S. F. Zadeh, and L. Bardwell. 2005. Mitogen-activated protein kinase with distinct requirements for Ste5 scaffolding influence signaling specificity in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Drogen, F., and M. Peter. 2001. MAP kinase dynamics in yeast. Biol. Cell. 93:63–70. [DOI] [PubMed] [Google Scholar]
- 48.Kusari, A. B., D. M. Molina, W. Sabbagh, C. S. Lau, and L. Bardwell. 2004. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromone in budding yeast. Nat. Cell Biol. 3:1051–1059. [DOI] [PubMed] [Google Scholar]
- 50.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228–238. [DOI] [PubMed] [Google Scholar]
- 51.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831–2848. [DOI] [PubMed] [Google Scholar]
- 52.Gartner, A., A. Jovanovic, D. I. Jeoung, S. Bourlat, F. R. Cross, and G. Ammerer. 1998. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol. Cell. Biol. 18:3681–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballensiefen, W., and H. D. Schmitt. 1997. Periplasmic Bar1 protease of Saccharomyces cerevisiae is active before reaching its extracellular destination. Eur. J. Biochem. 247:142–147. [DOI] [PubMed] [Google Scholar]
- 54.Chang, F., and I. Herskowitz. 1990. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a Gly cyclin, CLN2. Cell. 63:999–1011. [DOI] [PubMed] [Google Scholar]
- 55.Bagnat, M., and K. Simon. 2002. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 99:14183–14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahanty, S. K., Y. Wang, F. W. Farley, and E. A. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 98:501–512. [DOI] [PubMed] [Google Scholar]
- 57.Elion, E. A. 2001. The Ste5p scaffold. J. Cell Sci. 114:3967–3978. [DOI] [PubMed] [Google Scholar]
- 58.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Cell Biol. 3:573–581. [DOI] [PubMed] [Google Scholar]
- 59.Wang, Y., Q. Ge, D. Houston, J. Thorner, B. Errede, and H. G. Dohlman. 2003. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1-Cullin-F-box complex. J. Biol. Chem. 278:22284–22289. [DOI] [PubMed] [Google Scholar]
- 60.Garrison, T. R., Y. Zhang, M. Pausch, D. Apanovitch, R. Aebersold, and H. G. Dohlman. 1999. Feedback phosphorylation of an RGS protein by MAP kinase in yeast. J. Biol. Chem. 274:36387–36391. [DOI] [PubMed] [Google Scholar]
- 61.Goldbeter, A., and D. E. Koshland. 1981. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. USA. 78:6840–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Y., and E. A. Elion. 2003. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol. Biol. Cell. 14:2543–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yablonski, D., I. Marbach, and A. Levitzki. 1996. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc. Natl. Acad. Sci. USA. 93:13864–13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.