Abstract
JNKs are attractive targets for treatment of obesity and type-2 diabetes. A sustained increase in JNK activity was observed in dietary and genetic models of obesity in mice, whereas JNK deficiency prevented obesity-induced insulin resistance. A similar insulin-sensitizing effect was seen upon treatment of obese mice with JNK inhibitors. We now demonstrate that treatment with the saturated fatty acid palmitic acid results in sustained JNK activation and insulin resistance in primary mouse hepatocytes and pancreatic β-cells. In the latter, palmitic acid treatment inhibits glucose-induced insulin gene transcription, in part, by interfering with autocrine insulin signaling through phosphorylation of insulin-receptor substrates 1 and 2 at sites that interfere with binding to activated insulin receptors. This mechanism may account for the induction of central insulin resistance by free fatty acids.
Keywords: diabetes, insulin gene expression, lipotoxicity, obesity
Type-2 diabetes mellitus is a metabolic disease of alarming prevalence, affecting >170 million individuals worldwide in 2001, a number predicted to increase by almost 50% by 2010 (1). Recent studies focused attention on JNK family members, especially JNK1, as promising drug targets in the treatment of obesity and type-2 diabetes (2–4). Sustained JNK activation was observed in both dietary and genetic models of obesity (2), whereas mice with a targeted mutation at the Jnk1 locus are resistant to obesity-induced insulin resistance (2). Although JNK1 isoforms are the major contributors to insulin resistance, a recent study has shown that, upon reduction of the JNK1 dose, JNK2 isoforms also contribute to glucose intolerance (3). The proposed mechanism by which JNK activation leads to insulin resistance is centered on the phosphorylation of insulin-receptor substrate (IRS)1 at the inhibitory site Ser-307 (2, 5). Normally, insulin receptor (InsR) activation induces IRS1 Tyr phosphorylation, thereby triggering downstream signaling pathways, such as the phosphatidylinositol 3′-kinase (PI3K)-AKT cascade (6). However, Ser-307 phosphorylation inhibits recruitment of IRS1 to the activated receptor by interfering with the function of its phospho-Tyr-binding (PTB) motif, thereby preventing activation of PI3K-AKT signaling and other effector pathways (5, 7). Ablation of the Jnk1 locus or treatment with a peptide inhibitor of JNK (D-JNKi) prevents IRS1 Ser-307 phosphorylation and potentiates InsR signaling (2, 4). In addition to improved InsR signaling in peripheral insulin-responsive tissues (liver, muscle, and adipose tissue), treatment with D-JNKi increases insulin mRNA and protein content in pancreatic β-cells of obese mice (4). Furthermore, expression of catalytically inactive JNK in cultured β-cells prevents inhibition of insulin gene transcription in response to oxidative stress (8). The inhibitory effect of oxidative stress was suggested to be due to inhibition of AKT activation, which eventually results in cytoplasmic accumulation of PDX-1, a transcription factor that controls insulin gene transcription (9).
Although IRS1 phosphorylation at Ser-307 correlates with, and may account for, inhibition of AKT activation (2, 4), complete absence of IRS1 results in only a mild diabetogenic phenotype (10). Thus, it is likely that the absence of IRS1 is compensated for by the related adaptor molecule IRS2 (11, 12), suggesting that JNK activation may also lead to inhibition of IRS2 functions. In fact, the relative contributions of IRS1 and IRS2 to InsR signaling in liver was recently investigated by adenovirus-mediated gene transfer of small interfering hairpin RNAs (shRNA). That study demonstrated that silencing of either IRS1 or IRS2 alone did not affect PI3K and AKT activation, whereas delivery of shIRS1 plus shIRS2 adenoviruses decreased activation of both effector functions (13).
Here, we propose that lipotoxicity is a major trigger of JNK activation during obesity and show that long-chain saturated fatty acids, such as palmitic acid (PA), can trigger insulin resistance in both primary hepatocytes and pancreatic β-cells in a JNK-dependent manner. We also demonstrate that PA-mediated inhibition of insulin gene induction by either glucose or insulin is JNK-dependent and likely to be mediated through IRS1 and IRS2 phosphorylation. Indeed, we have identified a JNK phosphorylation site on IRS2 that may be functionally equivalent to Ser-307 of IRS1.
Results
Obesity and Saturated Fatty Acids, but Not High Glucose, Lead to JNK Activation.
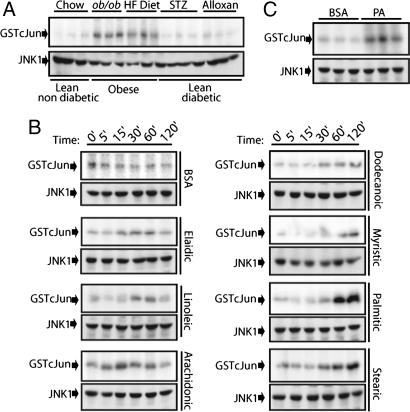
Obese mice exhibit sustained JNK activation (2). Two major obesity-induced complications are glucotoxicity and lipotoxicity (14). To test whether glucotoxicity can lead to JNK activation, we generated lean diabetic animals using a single injection of streptozotocin (STZ) or alloxan, two different toxic agents that induce insulin-dependent diabetes (15). STZ- or alloxan-injected mice developed severe hyperglycemia 3 days after injection that persisted for at least 10 days (Fig. 6, which is published as supporting information on the PNAS web site). Despite this profound effect, JNK activity in the liver was not elevated (Fig. 1A). By contrast, genetically obese (ob/ob) mice or mice kept on high fat diet exhibited a substantial elevation of JNK activity in their livers (Fig. 1A).
Fig. 1.
Free fatty acids and lipotoxic stress lead to sustained JNK activation. (A) Comparison of JNK activity using a solid-state kinase assay and GST-cJun (1–79) as a substrate in liver extracts from lean/nondiabetic, obese, and lean/diabetic mice. JNK1 levels were determined by immunoblotting. (B) JNK kinase activity in lysates of primary hepatocytes treated for the indicated times with delipidated BSA or BSA loaded with the indicated unsaturated (Left) or saturated (Right) fatty acids. (C) JNK activity in livers of mice that were perfused for 1 h with either delipidated or PA-loaded BSA.
If high blood glucose does not lead to JNK activation, perhaps this response is caused by blood lipids, such as free fatty acids (FFA), whose concentration is also elevated in genetic and dietary obesity (14). To examine this possibility we tested the capacity of different FFA to activate JNK in primary hepatocytes. The specific fatty acid to be examined (at 0.5 mM) was bound to fatty acid-free BSA (0.5%) as described in Materials and Methods. By and large, only modest effects on JNK activity were observed upon incubation with BSA loaded with unsaturated FFA, but incubation with BSA loaded with saturated FFA, such as PA, resulted in substantial JNK activation after 1–2 h (Fig. 1B; and see Fig. 7, which is published as supporting information on the PNAS web site). Liver perfusion with PA adsorbed to BSA also led to substantial JNK activation (Fig. 1C), suggesting that saturated FFA also cause JNK activation in vivo.
PA Induces Insulin Resistance in Primary Hepatocytes in a JNK-Dependent Manner.
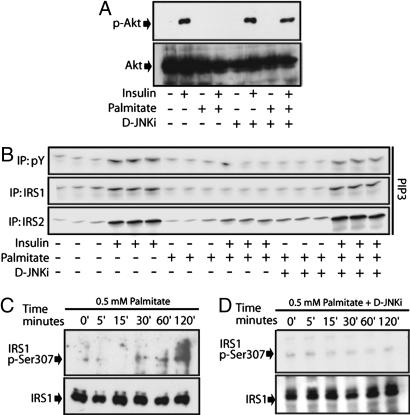
In addition to JNK activation, incubation of primary hepatocytes with PA for 2 h inhibited insulin-induced AKT activation (Fig. 2A) and led to IRS1 phosphorylation at the inhibitory site Ser-307 (Fig. 2C). Treatment with the JNK peptide inhibitor D-JNKi restored AKT activation (Fig. 2A) and prevented PA-induced IRS1 Ser-307 phosphorylation (Fig. 2D). Similar effects of PA were observed on insulin-induced PI3K activity immunoprecipitated with antibodies to phosphotyrosine (PY), IRS1, or IRS2 (Fig. 2B; and see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 2.
PA inhibits insulin signaling in primary mouse hepatocytes through a JNK-dependent mechanism. (A) Effect of JNK inhibition on PA-induced insulin resistance. Cells were incubated with or without PA (0.5 mM) for 2 h. When indicated, the peptide inhibitor of JNK, D-JNKi, was added during PA treatment before and 10 min after addition of insulin. AKT phosphorylation was assessed by immunoblotting. (B) PA inhibits IRS1–2-associated PI3K activity. Primary hepatocytes were treated as above, and PI3K activity was separately measured in PY, IRS1, and IRS2 immunoprecipitates. (C) Effects of PA on IRS1 Ser-307 phosphorylation. Cells were treated with PA (0.5 mM) for the indicated times before IRS1 Ser-307 phosphorylation was assessed by immunoblotting. (D) Effect of JNK inhibition on PA-induced IRS1 Ser-307 phosphorylation. Cells were treated as above, except that D-JNKi was present during the PA treatment.
As expected, hepatocytes from Jnk1−/− mice were protected from PA-induced IRS1 Ser-307 phosphorylation and PA-induced inhibition of either PY-associated PI3K activity or AKT activation by insulin (Fig. 9, which is published as supporting information on the PNAS web site). Reconstitution of Jnk1−/− hepatocytes with a lentivirus expressing JNK1 restored PA-induced IRS1 Ser-307 phosphorylation and inhibition of the PI3K–AKT cascade (Fig. 9).
PA Inhibits Glucose-Induced Insulin Gene Expression in a JNK-Dependent Manner.
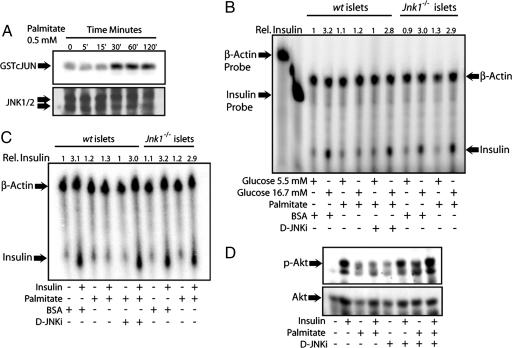
Injection (i.p.) of obese mice with the peptide inhibitor of JNK, D-JNKi, increased the pancreatic content of insulin RNA and protein (4). In addition, infection of cultured β-cells with an adenovirus expressing a catalytically inactive mutant of JNK prevented inhibition of insulin transcription by H2O2 (8). Because PA is a more natural inhibitor of insulin transcription (16–18) than H2O2, and the results shown above demonstrated it to be a potent JNK activator, we examined whether JNK mediates the inhibitory effect of PA on insulin gene transcription. As seen in hepatocytes, PA incubation of cultured mouse islets resulted in JNK activation within 30′ (Fig. 3A). Longer incubation with PA (12 h) led to an even more pronounced JNK activation, which was not affected by glucose concentrations (Fig. 10, which is published as supporting information on the PNAS web site). In the same experiment, we also measured the cellular content of insulin mRNA by S1 nuclease protection assay. Although PA did not affect insulin mRNA abundance at substimulatory glucose concentrations (5.5 mM), it severely impaired the induction of insulin gene transcription in cells exposed to high glucose (16.7 mM). Moreover, wild type islets treated with the JNK peptide inhibitor or islets from Jnk1−/− mice were protected from the inhibitory effects of PA on insulin gene (Fig. 3B).
Fig. 3.
Inhibition of insulin gene expression by PA is JNK-dependent. (A) Cultured mouse islets were incubated with PA for the indicated durations, and JNK activity and abundance were determined by a kinase assay and immunoblotting, respectively. (B) S1 nuclease-protection assay. Cytoplasmic RNA was isolated from wt or Jnk1−/− islets that were cultured in low (5.5 mM) or high (16.7 mM) glucose and treated with PA or D-JNKi as indicated for 12 h. Relative insulin mRNA levels are indicated above the panel. (C) Palmitate inhibition of insulin-induced insulin transcription is JNK-dependent. wt or Jnk1−/− islets were kept in low glucose (5.5 mM). PA with or without D-JNKi was added 12 h before the addition of insulin. RNA was prepared 2 h later. Relative insulin mRNA levels are indicated above the panel. (D) Immunoblot analysis of AKT Ser-473 phosphorylation. Primary mouse islets were treated with PA with or without D-JNKi, as above, and treated with insulin for 5 min. AKT phosphorylation and abundance were measured by immunoblotting.
It had been reported that glucose-induced insulin gene transcription requires autocrine insulin signaling (19–21). Insulin mRNA induction by either glucose or insulin is blunted upon deletion of InsR from β-cells (21). PA also inhibited insulin-induced insulin gene expression, and that effect was also prevented upon incubation with the JNK peptide inhibitor (Fig. 3C). Furthermore, no inhibition of insulin-induced insulin mRNA was seen upon treatment of Jnk1−/− islets with PA. As expected, the JNK peptide inhibitor also prevented PA-induced IRS1 Ser-307 phosphorylation (Fig. 11, which is published as supporting information on the PNAS web site) and interfered with inhibition of AKT activation (Fig. 3D).
Both IRS1 and IRS2 Are Involved in Induction of Insulin Gene Expression.
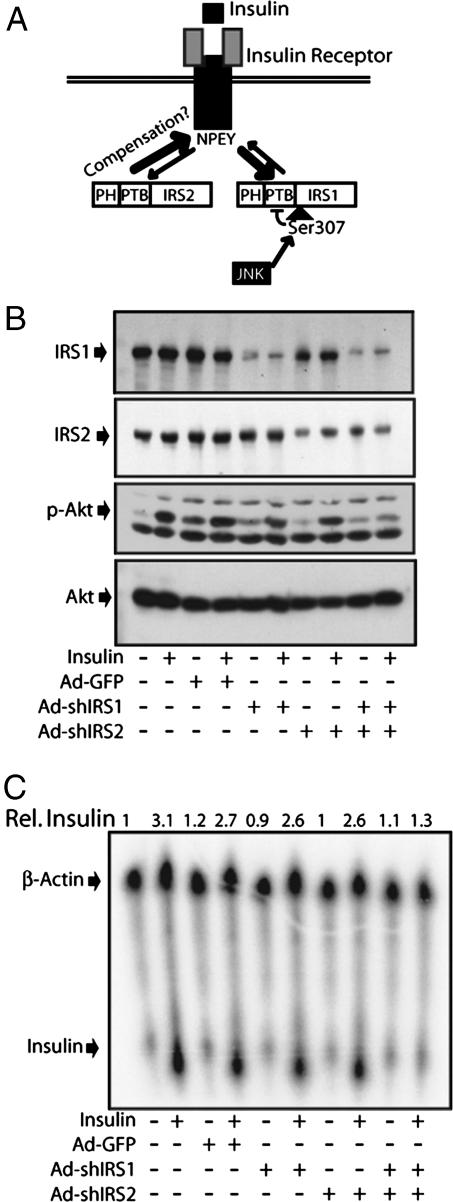
Currently, the main molecular mechanism for JNK-induced insulin resistance entails the phosphorylation of IRS1 at Ser-307, which is located next to its PTB motif (5). The PTB motif increases the affinity of IRS1 for the activated InsR (22–24), and Ser-307 phosphorylation interferes with this function (7, 22), thereby decreasing binding of IRS1 for the activated receptor (Fig. 4A). However, a recent study using adenoviral vectors expressing small hairpin interfering RNAs for IRS1 (shIRS1) or IRS2 (shIRS2) showed that an 80% decrease in IRS1 expression did not inhibit insulin-induced PI3K and AKT activation, whereas a concomitant knockdown of both IRS1 and IRS2 resulted in a substantial inhibition of PI3K and AKT activation (13).
Fig. 4.
Both IRS1 and IRS2 are involved in insulin-induced insulin gene transcription in β-cells. (A) The currently proposed mechanism of JNK action, IRS1 Ser-307 phosphorylation, decreases the affinity of the IRS1 PTB domain for activated InsR, thereby decreasing its tyrosine phosphorylation and ability to activate various effector functions. IRS2 may compensate for loss of IRS1 function. (B) IRS1 and IRS2 are required for insulin-induced AKT phosphorylation in β-cells. Cultured mouse islets were infected or not with Ad-shIRS1, Ad-shIRS2, or an Ad-GFP control virus as indicated. After 24 h, the cells were treated with insulin (200 microunits/ml) for 5 min. AKT phosphorylation and IRS1 and IRS2 expression were analyzed by immunoblotting. (C) IRS1 and IRS2 are required for insulin-induced insulin gene expression. Islets were treated as above, and insulin mRNA abundance was examined 2 h after insulin addition. Relative insulin mRNA levels are indicated above the panel.
To examine the contribution of IRS1 and IRS2 to insulin signaling and insulin gene induction in β-cells, we infected these cells with adenoviruses expressing shIRS1, shIRS2, or GFP. Infection of β-cells with shIRS1 reduced IRS1 protein levels by 80%, whereas shIRS2 decreased the IRS2 amount by 60% (Fig. 4B). Infection of primary β-cells with either the GFP virus or the shIRS1 or shIRS2 viruses alone did not modulate insulin-induced AKT phosphorylation and insulin gene induction (Fig. 4 B and C). However, coinfection with both shIRS1 and shIRS2 led to severe defects in insulin-induced AKT phosphorylation and insulin gene induction (Fig. 4 B and C). These results strongly suggest that IRS1 phosphorylation alone cannot account for the inhibitory effect of JNK on InsR signaling and that an additional inhibitory effect is likely to be extended to IRS2.
JNK Phosphorylates IRS2.
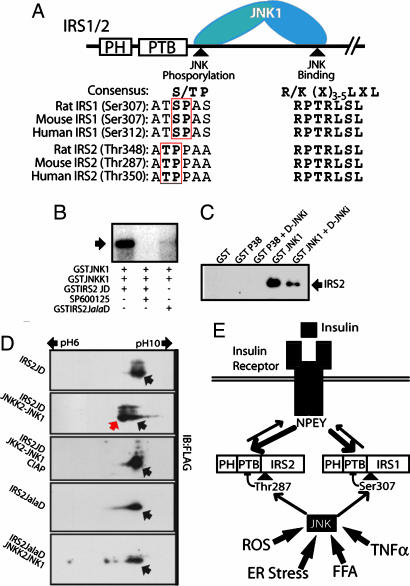
We next tested whether JNK can directly phosphorylate IRS2 at a site that is functionally equivalent to Ser-307 of IRS1. JNK-mediated phosphorylation depends on a specific interaction with a docking site on its substrates called the D domain and the presence of a Pro at the P + 1 position relative to the phosphoacceptor (P) site (25). Sequence alignment shows that the D domain of IRS1 is fully conserved in IRS2 (Fig. 5A), Although Ser-307 on IRS1 is not conserved in IRS2, sequence alignment reveals a potential JNK phosphoacceptor site in IRS2 one amino acid before the one corresponding to IRS1 Ser-307; in rat IRS2, this site corresponds to Thr-348 (Fig. 5A). Using lysates of HEK293 expressing rat IRS2, we found that JNK1, but not p38α, bound to IRS2 in a manner sensitive to the JNK peptide inhibitor, whose sequence corresponds to the D domain of the JNK-interacting protein JIP1 (Fig. 5C). To test whether JNK1 phosphorylates Thr-348, we subcloned a fragment of the IRS2 cDNA containing the D domain and the putative JNK phosphoacceptor site (IRS2JD), or a mutant thereof in which the Thr-348 codon was replaced by an Ala codon (IRS2JalaD), into a bacterial GST-fusion protein expression vector. Whereas JNK1 readily phosphorylated the wild-type IRS2JD polypeptide, and the phosphorylation was inhibited by the JNK inhibitor SP600125, no phosphorylation of the IRS2JalaD mutant was observed (Fig. 5B). To examine phosphorylation of IRS2JD in living cells, we have cloned Flag-tagged IRS2JD and IRS2JalaD in a mammalian expression vector. Coexpression of IRS2JD with a constitutively active JNK1-JNKK2 fusion protein (26) resulted in a leftward shift of IRS2JD on a 2D gel electrophoresis system, indicating a decrease in its isoelectric point, a change that is consistent with phosphorylation (Fig. 5D). Treatment of cell lysates with calf intestinal phosphatase increased the isoelectric point of the protein, consistent with its dephosphorylation. No shift in isoelectric point was observed upon coexpression of IRS2JalaD with the activated JNK1 (Fig. 5D).
Fig. 5.
JNK1 binds to and phosphorylates rat IRS2 on Thr-348. (A) Schematic representation of JNK-mediated IRS phosphorylation. Specificity is achieved by specific docking and phosphorylation sites that are conserved between IRS1 and IRS2. (B) JNK1 phosphorylates IRS2 at Thr-348 in vitro. JNK kinase assay was performed by coincubation of purified GSTJNK1 with its upstream kinase GSTJNKK1 and either purified GSTIRS2JD or GSTIRS2JalaD as substrates. The addition of the JNK inhibitor SP600125 was used as a specificity control. (C) JNK1 specifically binds to IRS2. Lysates from HEK293T cells transfected with pcDNA3-IRS2 were coincubated with purified GST-JNK1 in the presence or absence of D-JNKi or with GST-p38α as a control. Glutathione-bead “pull-downs” were analyzed by immunoblotting with IRS2-specific antibodies. (D) JNK1 phosphorylates IRS2 at Thr-348 in living cells. HEK293T cells were transfected with mammalian expression vectors expressing Flag-tagged IRS2JD or IRS2JalaD with a vector encoding the constitutive active JNKK2–JNK1 fusion. Protein extracts were analyzed by 2D gel electrophoresis and immunoblotting with Flag antibodies. (E) A model illustrating the role of JNK in insulin resistance caused by fatty acids and other stressors involved in obesity-induced diabetes. Phosphorylation of IRS1 at Ser-307 and IRS2 at Thr-348 decreases their binding and phosphorylation by the activated InsR. This decrease attenuates InsR signaling and prevents activation of effector functions.
Discussion
The homeostatic control of blood glucose is determined by two major factors: the concentration of insulin in the circulation and insulin sensitivity of target organs (e.g., muscle, adipose tissue, and liver). It is well accepted that, in obesity-induced diabetes, insulin resistance progresses toward glucose intolerance and type-2 diabetes because of increased demand for insulin and eventual failure of pancreatic β-cells to meet this demand (27). Studies had demonstrated that JNK deficiency is protective in different mouse models of obesity-induced glucose intolerance because it increases the sensitivity of target tissues to insulin (2, 3). Yet, despite improved glucose tolerance and increased peripheral insulin sensitivity, pharmacological intervention with JNK activity was also found to increase the amount of insulin mRNA and protein in pancreatic β-cells (4, 8). The latter results suggested that, in addition to its insulin tolerizing activity in peripheral tissues, JNK might also be involved in negative regulation of insulin synthesis or signaling within the pancreatic β cell, the central site of blood glucose regulation. In this study, we examined the basis for JNK activation during obesity-induced insulin resistance and the role of JNK in perturbation of β-cell function. Two major adverse outcomes of obesity-induced insulin resistance are glucotoxicity and lipotoxicity. Our results indicate that lipotoxicity because of increased blood FFA levels or ectopic fat deposition is the most likely mediator of sustained JNK activation in obese mice. Elevated blood glucose and glucotoxicity alone do not result in JNK activation in peripheral insulin-responsive tissues, such as the liver. Sustained JNK activation in such tissues is of pathophysiological importance because it interferes with insulin signaling, thereby leading to insulin resistance and glucose intolerance (2, 4). Our results show that, among different FFA, long-chain-saturated fatty acids, such as PA, are the most potent activators of JNK in cultured hepatocytes and β-cells. These data are consistent with recently published results showing that a mixture of FFA activates JNK in adipocytes (28) and that PA and stearic acid induce JNK phosphorylation in primary hepatocytes (29).
Although the mechanisms leading to JNK activation by FFA are unknown, its consequences are clear. Sustained JNK activation inhibits insulin signaling not only in hepatocytes and other peripheral sites but also in pancreatic β-cells, where it interferes with glucose and insulin-induced insulin gene transcription. Recently, JNK activation was also shown to be responsible for the effects of H2O2 on insulin gene transcription by a mechanism that involves inhibition of AKT activation, which is required for phosphorylation of the stress- and nutrient-responsive transcription factor Foxo1 (8, 9), resulting in enhanced nuclear retention of Foxo-1, which negatively affects insulin gene transcription by inhibiting PDX-1, a major activator of the insulin promoter (9). Interestingly, PA treatment of β-cells also decreases the nuclear pool of PDX-1 (17, 18). We suggest that most of these effects are consequences of JNK-mediated phosphorylation of IRS1 and IRS2, which prevents InsR-proximal events. Using adenovirus-mediated delivery of shRNA, we show that both IRS1 and IRS2 are required for insulin-induced AKT activation and insulin gene transcription. Because the knockdown of IRS1 alone does not disrupt either AKT activation or insulin gene induction, it seems unlikely that IRS1 phosphorylation at Ser-307 accounts for the full inhibitory effect of PA on insulin gene expression. The shRNA-knockdown experiments, which are consistent with gene-disruption experiments that have revealed a critical role for IRS2 in InsR signaling (11), highlighted an important function for IRS2 in glucose-induced insulin expression and led us to examine the possibility that JNK also phosphorylates IRS2. Indeed, we found that IRS2 is an efficient substrate for JNK1, which phosphorylates it at Thr-348. Based on the location of this site, we suggest that it is the functional homolog of Ser-307 on IRS1 and that its phosphorylation is likely to disrupt the interaction between the PTB motif of IRS2 and activated InsR. This suggestion is consistent with earlier results, which demonstrate that treatment of Chinese hamster ovary cells with the potent JNK activator anisomycin, inhibited insulin-induced tyrosine phosphorylation of both IRS1 and IRS2 (5). In addition, we observed that PA treatment inhibited insulin-induced IRS1- and IRS2-associated PI3K activity in a JNK-dependent manner (Figs. 2B and 8).
Strikingly, the JNK docking site, the D domain, is perfectly conserved between IRS1 and IRS2, and the sequence surrounding the JNK phosphoacceptor sites is also quite well conserved, suggesting that IRS1 and IRS2 have coevolved not only as InsR substrates and signal transducers but also as targets for JNK-mediated inhibitory signaling. Thus, JNK activation may be part of a homeostatic mechanism that controls the output from activated InsR. Because it had been observed that JNK is activated in the livers of rats during repeated fasting–refeeding cycles (30), and because FFA levels are particularly elevated during fasting, one possibility is that this pathway might have evolved as negative regulator of insulin signaling during starvation. Because de novo lipogenesis (fatty acid synthesis) is one of the major effects of insulin, excessive insulin signaling will not only lower blood glucose below a critical threshold but will also increase the circulating levels of FFA. Increased levels of saturated FFA lead to JNK activation, IRS1 and IRS2 Ser/Thr phosphorylation, and down-regulation of InsR signaling. The latter results in insulin-stimulated metabolic processes in peripheral tissues including de novo lipogenesis. At the same time, JNK-mediated down-regulation of InsR signaling in pancreatic β-cells would decrease the synthesis and eventual release of insulin. This hypothesis is consistent with previous studies showing that an adipose tissue-specific InsR knockout protects from obesity and obesity-induced glucose intolerance (31), whereas a knockout of the InsR gene in the β-cell results in an insulin secretion defect similar to that in type-2 diabetes (32).
In addition to further explaining the intricate involvement of JNK in the regulation of insulin signaling, our findings underscore the importance of JNK inhibition as a therapeutic strategy for prevention of glucose intolerance and increasing both insulin sensitivity and insulin synthesis. Although this study has focused on saturated FFA as JNK activators, other studies have shown that inflammatory cytokines, such as TNFα (33) and endoplasmic reticulum stress (34), which also inhibit insulin signaling, do so through JNK activation. Thus, JNK inhibition should provide a general strategy for increasing insulin sensitivity and availability.
Materials and Methods
Mice and Models of Diabetes.
We used male mice of C57BL/6J background including ob/ob and Jnk1−/− mice. For STZ-induced diabetes, mice were fasted overnight, followed by i.p. injection of STZ in pH 4.5 citrate buffer at a concentration of 200 mg/kg. For alloxan-induced hyperglycemia, 8-week-old mice fasted for 6 h were i.p. injected with alloxan in PBS at a concentration of 200 mg/kg. Blood glucose was measured before and at 3 and 10 days after injection. Mice were killed, and their livers were immediately collected and frozen in liquid nitrogen. For diet-induced obesity, 6-week-old mice were given a high-fat diet (Diet F3282; Bioserve, Frenchtown, NJ) for 8 weeks. Liver perfusion was conducted on 8- to 16-week-old anesthetized mice. The portal vein was exposed by abdominal incision and cannulated. After a brief (30 sec) perfusion with sterile PBS, livers were perfused at 2 ml/min for 30 sec with either delipidated BSA (0.5%) or PA-loaded BSA (0.5 mM PA) in PBS. The perfusion rate was decreased to 0.5 ml/min, and perfusion continued for 1 h, after which the liver was isolated and snap-frozen in liquid nitrogen.
Primary Cell Cultures.
Mouse islets were isolated as described (35). Briefly, 8- to 16-week-old mice were killed, and 2.5 ml of 40 mg/ml collagenase P (Roche, Indianapolis, IN) in Hanks' balanced salt solution (HBSS) were slowly infused into the common bile duct. The pancreas was collected and incubated with collagenase solution at 37°C for 13 min with agitation. Digestion was stopped by addition of ice-cold HBSS–5% FBS. Pancreas was filtered trough a 400-μm mesh, and islets were purified on a four-layer HBSS–Ficoll gradient. Islets were collected at the interphases between 11.5–21.5% and 21.5–23%. Islets were washed twice with HBSS–5% FBS and cultured in RPMI medium 1640–10% FBS with antibiotics.
Hepatocytes were isolated and cultured as described (36) with minor modifications. Briefly, mice were anesthetized, and the livers were perfused with calcium-free HBSS with EGTA (5 mM) via the portal vein, followed by perfusion with HBSS (with calcium) containing 500 mg/liter collagenase (Sigma). Digested livers were transferred to cold DMEM plus 10% FCS and agitated. The slurry was strained and spun at 50 × g for 30 sec. Hepatocytes were then plated in DMEM–10% FCS containing 10 mg/liter insulin (Sigma) on collagen-coated dishes. The next morning, the medium was replaced with fresh medium without insulin.
Biochemical Assays.
JNK solid-state kinase assays using GST-cJun (1–79) as a substrate were performed as described (37). PI3K assays and phospho-AKT Ser-473 immunoblots were also performed as described (38). For PI3K assays, 300 μg of protein was immunoprecipitated with the relevant antibodies, and reaction products were separated on gel 60 TLC plates (Whatman, Clifton, NJ). IRS1 Ser-307 phosphorylation was assessed by immunoblotting with antiphospho-IRS1 Ser-307 antibodies (Upstate Biotechnology, Lake Placid, NY). After stripping, the membrane was reprobed with total IRS1 antibodies (Upstate Biotechnology). Binding of IRS2 to JNK1 was assessed as described for IRS1 binding to JNK1 (3). The source of IRS2 were lysates of HEK293T cells transfected with pcDNA3IRS2 (38), provided by Luciano Pirola (University of Lyon, Lyon, France). To examine IRS2 phosphorylation in vitro, a fragment of IRS2 including the phosphoacceptor site and the D domain was PCR cloned by using the following primers: forward (5′-actgaattcctggcggccacccccccagcagcc-3′) and reverse (5′-tgctgggaagggtctcgagtccctc-3′). To convert Thr-348 to Ala, we used a forward primer (5′-actgaattcctggcggccgcccccccagcagcc-3′) containing an Ala instead of a Thr codon at position 348. PCR products were cloned in pET-41(+) (Novagen, San Diego, CA) to express either GSTIRS2JD or GSTIRS2JalaD. Kinase reactions were performed by using 10 μg of partially purified IRS2. To examine IRS2 phosphorylation in cells, we have cloned IRS2JD and IRS2JalaD fused to an N-terminal Flag epitope into pcDNA3. All of the resultant constructs were transfected into HEK293T cells. Phosphatase inhibitors were added to all samples except those treated with alkaline phosphatase. Proteins were precipitated with 20% trichloracetic acid (TCA) and dissolved in sample buffer: 9 M urea, 2% Triton X-100, 0.5% ampholytes, 50 mM DTT, and 0.002% bromophenol blue. Isoelectrofocusing was performed on Zoom Strips (Invitrogen, Carlsbad, CA). Second-dimension SDS/PAGE was performed by using NuPAGE 4–12% Bis-Tris Zoom gels (Invitrogen).
Viral Transductions.
Lentiviruses expressing eGFP or JNK1 were described (39). Primary hepatocytes were infected at MOI = 20 transduction units per cell, sufficient to transduce >95% of hepatocytes. Adenoviruses vectors expressing shIRS1 or shIRS2 were provided by M. Montminy at the Salk Institute for Biological Studies (La Jolla, CA) (40). Primary mouse islets were infected at MOI = 2,000 plaque-forming units per islet, resulting in the infection of nearly all of the cells.
S1 Nuclease Protection Assay.
S1 nuclease protection was performed as described (41) by using the following single-strand probes: preproinsulin, 5′-TGACAAAAGCCTGGGTGGGTTTGGGCTCCCAAAGGGCAAGCAGGGaagtttcca-3′; and β-actin, 5′-TACGTACATGGCTGGGGTGTTGAAGGTCTCAAACATGATCTGGGTCATCTTTTCACGGTTGGCCTTCcaaagttag-3′. The uppercase sequence is complementary to the target mRNA, and the 3′ lowercase sequence is a random sequence serving as a specificity control.
Supplementary Material
Acknowledgments
We thank Dr. Marc Montmini for shIRS1 and shIRS2 adenoviruses and Dr. Luciano Pirola for pcDNA3IRS2. This work was supported by National Institutes of Health Grant ES06376. G.S. was supported by a fellowship of the Swiss National Science Foundation and a mentor-based postdoctoral fellowship from the American Diabetes Association. M.K. is an American Cancer Society Research Professor.
Abbreviations
- D-JNKi
JNK-inhibitory peptide
- FFA
free fatty acid(s)
- InsR
insulin receptor
- IRS
InsR substrate
- PA
palmitic acid
- PTB
phosphotyrosine-binding motif.
Footnotes
The authors declare no conflict of interest.
References
- 1.Zimmet P, Alberti KG, Shaw J. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 2.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 3.Tuncman G, Hirosumi J, Solinas G, Chang L, Karin M, Hotamisligil GS. Proc Natl Acad Sci USA. 2006;103:10741–10746. doi: 10.1073/pnas.0603509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneto H, Nakatani Y, Miyatsuka T, Kawamori D, Matsuoka TA, Matsuhisa M, Kajimoto Y, Ichijo H, Yamasaki Y, Hori M. Nat Med. 2004;10:1128–1132. doi: 10.1038/nm1111. [DOI] [PubMed] [Google Scholar]
- 5.Aguirre V, Uchida T, Yenush L, Davis R, White MF. J Biol Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi CM, Emanuelli B, Kahn CR. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 7.Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF. J Biol Chem. 2002;277:1531–1537. doi: 10.1074/jbc.M101521200. [DOI] [PubMed] [Google Scholar]
- 8.Kaneto H, Xu G, Fujii N, Kim S, Bonner-Weir S, Weir GC. J Biol Chem. 2002;277:30010–30018. doi: 10.1074/jbc.M202066200. [DOI] [PubMed] [Google Scholar]
- 9.Kawamori D, Kaneto H, Nakatani Y, Matsuoka TA, Matsuhisa M, Hori M, Yamasaki Y. J Biol Chem. 2006;281:1091–1098. doi: 10.1074/jbc.M508510200. [DOI] [PubMed] [Google Scholar]
- 10.Araki E, Lipes MA, Patti ME, Bruning JC, Haag B, III, Johnson RS, Kahn CR. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 11.Withers DJ, Gutierrez JS, Towery H, Burks DJ, Ren JM, Previs S, Zhang Y, Bernal D, Pons S, Shulman GI, Bonner-Weir S, White MF. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 12.Patti ME, Sun XJ, Bruening JC, Araki E, Lipes MA, White MF, Kahn CR. J Biol Chem. 1995;270:24670–24673. doi: 10.1074/jbc.270.42.24670. [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi CM, Ueki K, Kahn R. J Clin Invest. 2005;115:718–727. doi: 10.1172/JCI23187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Stumvoll M, Goldstein BJ, van Haeften TW. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 15.Szkudelski T. Physiol Res. 2001;50:537–546. [PubMed] [Google Scholar]
- 16.Kelpe CL, Moore PC, Parazzoli SD, Wicksteed B, Rhodes CJ, Poitout V. J Biol Chem. 2003;278:30015–30021. doi: 10.1074/jbc.M302548200. [DOI] [PubMed] [Google Scholar]
- 17.Hagman DK, Hays LB, Parazzoli SD, Poitout V. J Biol Chem. 2005;280:32413–32418. doi: 10.1074/jbc.M506000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gremlich S, Bonny C, Waeber G, Thorens B. J Biol Chem. 1997;272:30261–30269. doi: 10.1074/jbc.272.48.30261. [DOI] [PubMed] [Google Scholar]
- 19.Leibiger IB, Leibiger B, Moede T, Berggren PO. Mol Cell. 1998;1:933–938. doi: 10.1016/s1097-2765(00)80093-3. [DOI] [PubMed] [Google Scholar]
- 20.Leibiger B, Moede T, Schwarz T, Brown GR, Kohler M, Leibiger IB, Berggren PO. Proc Natl Acad Sci USA. 1998;95:9307–9312. doi: 10.1073/pnas.95.16.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leibiger B, Leibiger IB, Moede T, Kemper S, Kulkarni RN, Kahn CR, de Vargas LM, Berggren PO. Mol Cell. 2001;7:559–570. doi: 10.1016/s1097-2765(01)00203-9. [DOI] [PubMed] [Google Scholar]
- 22.White MF. Science. 2003;302:1710–1711. doi: 10.1126/science.1092952. [DOI] [PubMed] [Google Scholar]
- 23.Yenush L, Makati KJ, Smith-Hall J, Ishibashi O, Myers MG, Jr, White MF. J Biol Chem. 1996;271:24300–24306. doi: 10.1074/jbc.271.39.24300. [DOI] [PubMed] [Google Scholar]
- 24.Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 25.Kallunki T, Deng T, Hibi M, Karin M. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C, Xiang J, Hunter T, Lin A. J Biol Chem. 1999;274:28966–28971. doi: 10.1074/jbc.274.41.28966. [DOI] [PubMed] [Google Scholar]
- 27.Prentki M, Nolan CJ. J Clin Invest. 2006;116:1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen MT, Satoh H, Favelyukis S, Babendure JL, Imamura T, Sbodio JI, Zalevsky J, Dahiyat BI, Chi NW, Olefsky JM. J Biol Chem. 2005;280:35361–35371. doi: 10.1074/jbc.M504611200. [DOI] [PubMed] [Google Scholar]
- 29.Malhi H, Bronk SF, Werneburg NW, Gores GJ. J Biol Chem. 2006;281:12093–12101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- 30.Nishio H, Kuwabara H, Mori H, Suzuki K. Hepatology. 2002;36:72–80. doi: 10.1053/jhep.2002.34131. [DOI] [PubMed] [Google Scholar]
- 31.Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni RN, Bruning JC, Winnay JN, Postic C, Magnuson MA, Kahn CR. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 33.Hotamisligil GS, Murray DL, Choy LN, Spiegelman BM. Proc Natl Acad Sci USA. 1994;91:4854–4858. doi: 10.1073/pnas.91.11.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 35.Chang I, Kim S, Kim JY, Cho N, Kim YH, Kim HS, Lee MK, Kim KW, Lee MS. Diabetes. 2003;52:1169–1175. doi: 10.2337/diabetes.52.5.1169. [DOI] [PubMed] [Google Scholar]
- 36.Seglen PO. Exp Cell Res. 1972;74:450–454. doi: 10.1016/0014-4827(72)90400-4. [DOI] [PubMed] [Google Scholar]
- 37.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 38.Solinas G, Summermatter S, Mainieri D, Gubler M, Montani JP, Seydoux J, Smith SR, Dulloo AG. Endocrinology. 2006;147:31–38. doi: 10.1210/en.2005-1033. [DOI] [PubMed] [Google Scholar]
- 39.Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Canettieri G, Koo SH, Berdeaux R, Heredia J, Hedrick S, Zhang X, Montminy M. Cell Metab. 2005;2:331–338. doi: 10.1016/j.cmet.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Westin G, Gerster T, Muller MM, Schaffner G, Schaffner W. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.