Abstract
The Epstein-Barr virus oncoprotein LMP1 has six transmembrane domains (TMs) that enable intermolecular aggregation and constitutive signaling through two C-terminal cytosolic domains. Expression of both TMs 1 and 2 without the C terminus (TM1-2ΔC) and TMs 3 to 6 fused to the C terminus (TM3-6) results in partial association, which is substantially decreased by TM1 F38WLY41 mutation to A38ALA41. We now investigate whether TM1-2ΔC can functionally interact with TM3-6. TM1-2ΔC induced TM3-6 to mediate NF-κB activation at 59% of LMP1 levels, and the effect was dependent on TM1-2 F38WLY41. TM1-2ΔC even induced TM3-4 C terminus-mediated NF-κB activation to 44% of LMP1 levels. Surprisingly, this effect was TM1 F38WLY41 independent, indicative of a role for TMs 5 and 6 in TM1 F38WLY41 effects. TM3 W98 was also important for TM1-2ΔC induction of TM3-6-mediated NF-κB activation, for association, and for TM1 F38WLY41 dependence on C-terminal NF-κB activation. These data support models in which the TM1 F38WLY41 effects are at least partially dependent on TM3 W98 and a residue(s) in TMs 5 and 6.
The Epstein-Barr virus (EBV)-encoded latent infection membrane protein 1 (LMP1) is essential for EBV-infected lymphocyte outgrowth into lymphoblastoid cell lines (15, 31). LMP1 is expressed in EBV-associated lymphoproliferative disease in immune-deficient people, Hodgkin's disease, and nasopharyngeal carcinoma (for reviews see references 33 and 49). LMP1 can cause established rodent fibroblast cells to grow with less contact inhibition, serum, or anchorage dependence and with greater tumor potential in nude mice (54). In human lymphocytes, LMP1 induces activation markers, adhesion protein expression, cell adhesion, Bcl-2 expression, and antiapoptotic effects. In transgenic mice, immunoglobulin (Ig) enhancer-and promoter-regulated LMP1 expression results in clonal B-cell proliferations (34) and polyoma promoter-regulated LMP1 expression results in epidermal hypertrophy (11). Thus, LMP1 is important for EBV effects on B-lymphocyte and epithelial cell growth and survival.
Reverse genetic analyses identify three LMP1 components that are critical for EBV-mediated transformation of human lymphocytes to lymphoblastoid cell lines: (i) six transmembrane domains (TMs) that enable intermolecular aggregation, association with lipid rafts, and patching in the lymphoblast plasma membrane (3, 9, 22, 24, 37-40, 42, 55, 58); (ii) the first 44 amino acids (aa; aa 187 to 231) of the C-terminal cytosolic domain, which interact with TNF receptor-associated factors (TRAFs) (4, 13, 14, 19, 20, 25, 30, 31, 43-46, 50, 60); and (iii) the last 36 aa (aa 351 to 386) of the C terminus, which interact with death domain proteins, including TRADD and RIP (26-28). The critical LMP1 components mimic a constitutively activated TNFR, which signals through TRAFs and TRADD. The LMP1 C-terminal signaling domains activate NF-κB, p38, and c-Jun N-terminal kinase up-regulation; PI3K and Cdc42 are also activated (12, 16, 17, 21, 35, 48, 53, 55, 56). NF-κB activation is critical for EBV-transformed-lymphoblast survival (7).
The experiments described here investigate intermolecular interactions among TMs to identify interactions that can constitutively enable C terminus-mediated activation of NF-κB. TMs 1 and 2 are critical and even partially sufficient for signaling, since expression of TMs 1 and 2 fused in frame to the C terminus (TM1-2) induces 40% of full-length LMP1 (TM1-6)-mediated NF-κB activation, whereas fused to the C terminus, TMs 3 and 4, TMs 5 and 6, or TMs 3 to 6 (TM3-4, TM5-6, and TM3-6, respectively) induce almost no signaling (10, 58). However, TMs 3, 4, 5, and 6 are also important for TM1-6 signaling, since TM1-2 induce only 40% of TM1-6-mediated NF-κB activation and TMs 1 to 4 fused to the C terminus (TM1-4) induce only 75% of TM1-6-mediated NF-κB activation (58). Within TMs 1 and 2, mutation of 7 of 11 TM1 leucines to alanines does not affect LMP1 intermolecular association but abrogates NF-κB activation (32). However, mutation of four of five TM1 leucines that are conserved in EBV and rhesus lymphocryptovirus LMP1s has no affect on NF-κB activation, indicating that the leucines have a less specific role than anticipated from the more extensive leucine mutagenesis (58). Further alanine mutational analyses of TM1 in the context of full-length LMP1 identifies TM1 residues F38WLY41, which precede the external first reverse turn (Fig. 1A), as critical for raft association, for TM1-2ΔC intermolecular association with TM3-6, and for TM1-6 NF-κB activation (58). Surprisingly, TM1-2ΔC with a mutation of F38WLY41 (TM1-2ΔC A38ALA41) can still associate with LMP1 TM1-2 (58). The potential functional role of TM1-2ΔC and TM1-2ΔC A38ALA41 in mediating NF-κB activation through intermolecular interaction with TM3-6 has not been directly investigated and is the objective of these experiments.
FIG. 1.
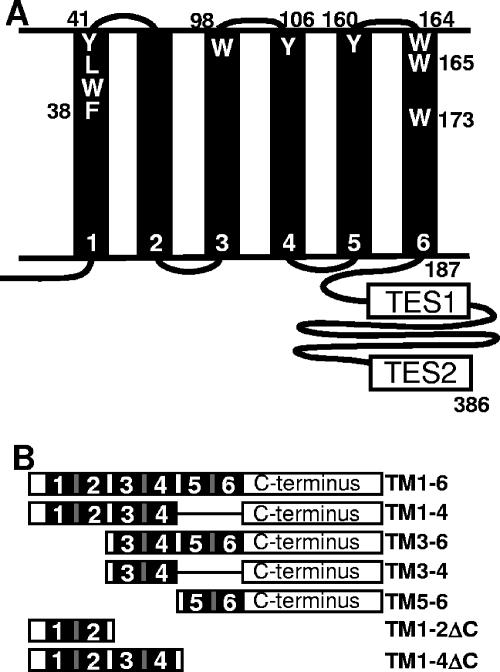
(A) Schematic depiction of LMP1 in a membrane with the critical transformation effector sites 1 and 2 (TES1 and TES2). The six TMs are shown. The locations of aromatic amino acids TM1 F38WLY41, TM3 W98, TM4 Y106, TM5 Y159, TM6 WW164-5, and TM6 W173, which were mutated, are shown. (B) Schematic diagram of LMP1 deletion mutants used in this study. TMs are shown as black, cytoplasmic domains as white, and extracellular domains as gray.
MATERIALS AND METHODS
Cell lines and antibodies.
Human embryonic kidney 293 (HEK293) cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin, and streptomycin at 37°C with 5% CO2. S12 mouse monoclonal antibody against LMP1 was purified from hybridoma supernatant (41). Antibodies to hemagglutinin (HA) (F-7) and glutathione S-transferase (GST) (B14) were purchased from Santa Cruz Biotechnology. Antibody to FLAG (M2) was purchased from Sigma.
Plasmids.
EBV B95-8 strain FLAG-tagged full-length LMP1, FLMP1 (FTM1-6), FTM5-6, and FTM3-6, GST-tagged TM1-2ΔC (GTM1-2ΔC F38WLY41) and GTM1-2ΔC F38WLY41 mutant (GTM1-2ΔC A38ALA41), and HA-tagged full-length wild-type LMP1 (HWT) and its F38WLY41 mutant (HM5) have been described (58). HA-tagged TM1-2ΔC (HTM1-2ΔC) and its mutant HTM1-2ΔC A38ALA41 were created by inserting a stop codon after the amino acid 75 codon with a QuikChange (Stratagene) kit using HWT and HM5 as templates, respectively. HA-tagged TM1-4ΔC (HTM1-4ΔC) and its mutant HTM1-4ΔC A38ALA41 were created by inserting a stop codon after the amino acid 133 codon with the QuikChange (Stratagene) kit using HWT and HM5 as templates, respectively. FTM3-6 W98A, Y106A, and W98Y106AA double mutants were created by using a QuikChange (Stratagene) kit with FTM3-6 as a template. FTM5-6 Y159A, FTM5-6 WW164-5AA, and FTM5-6 W173A mutants were created by using a QuikChange (Stratagene) kit with FTM5-6 as a template. Mutations were confirmed by nucleotide sequencing. A plasmid with five Igκ NF-κB sites preceding a minimal promoter and luciferase reporter and a pGK-β-gal reporter control plasmid were used to evaluate NF-κB activation in transient-transfection assays with HEK293 cells (58).
Transfections and reporter gene assays.
HEK293 cells, in six-well plates, were transfected with 300 ng, or as indicated in Fig. 2, of each expression construct using Effectene (QIAGEN, Chatsworth, CA). The amount of total DNA transfected was kept same by adding pSG5. Twenty-four hours after transfection, cells were lysed in reporter lysis buffer (Promega). Luciferase (Promega) and β-galactosidase (Galacton-Plus; Tropix, Bedford, MA) activities were assayed. Luciferase values were normalized for β-galactosidase activity (58).
FIG. 2.
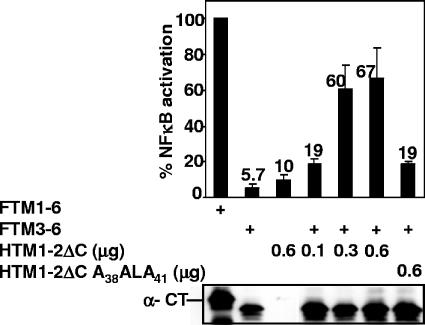
LMP1 TM1-2ΔC induction of TM3-6-mediated NF-κB activation is TM1F38WLY41 dependent. Full-length FLAG-tagged LMP1 (FTM1-6) or FTM3-6 and increasing amounts of HTM1-2ΔC or its mutant HTM1-2ΔC A38ALA41 were expressed in HEK293 cells. The HEK293 cells were also transfected with a plasmid with five Igκ NF-κB sites preceding a minimal promoter and luciferase reporter and a pGK-β-gal reporter control plasmid. Cells were lysed after 24 h and assayed for luciferase, β-galactosidase, and LMP1 C terminus expression. Luciferase values were normalized for β-galactosidase activity. FTM1-6-induced luciferase activity was set to 100% as an internal marker for each experiment. The presented data are representative of three independent transfections done in duplicate. The LMP1 C terminus (CT) was identified with S12 monoclonal antibody (41).
Immunoblotting.
Twenty-four hours after transfection, HEK293 cells were lysed in reporter lysis buffer (Promega). Lysates were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were transferred to a nitrocellulose membrane and immunoblotted with S12 antibody to the LMP1 C terminus (41) and F-7 antibody to HA (Santa Cruz Biotechnology).
GST pull-downs (GST PD) and immunoblotting.
HEK293 cells, in 60-mm plates, were cotransfected with 500 ng each of the expression plasmids using Effectene (QIAGEN, Chatsworth, CA). Twenty-four hours after transfection, cells were lysed in buffer containing 0.5% Nonidet P-40, 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 20% glycerol, 20 mM NaF, 1 mM Na3VO4, 10 mM Na3P2O7, 25 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. Cell lysates were incubated with glutathione-Sepharose beads (Amersham Pharmacia) at 4°C for 3 h. Precipitates were washed four times with lysis buffer and immunoblotted with S12 antibody to the LMP1 C terminus (41) and B14 antibody to GST (Santa Cruz Biotechnology) (58).
Calculation of intermolecular interaction efficiency.
Immunoblots after the GST PD were developed and analyzed on Image Station 4000R (Kodak) using molecular imaging software (Kodak). GST PD efficiency was calculated by comparing band intensities between whole-cell extract (WCE) blotted for GST and GST PD blotted for GST. Similarly efficiency of intermolecular interaction was calculated by comparing band intensities between WCE blotted for the LMP1 C terminus and GST PD blotted for the LMP1 C terminus and correcting it for GST PD efficiency. Intermolecular interaction efficiency between GTM1-2ΔC and FTM3-6 was normalized as 100%, and relative intermolecular interaction percentage was calculated accordingly.
RESULTS
F38WLY41 is critical for TM1-2ΔC induction of NF-κB activation through TM3-6.
To investigate whether N-terminally HA epitope-tagged HTM1-2ΔC, which is TM1-2 without the LMP1 C-terminal cytoplasmic signaling domain, can induce NF-κB activation through intermolecular interaction with FTM3-6, which is FLAG-tagged TM3-6 with the C-terminal cytoplasmic signaling domain, TM1-2ΔC was expressed with FTM3-6 in HEK293 cells (Fig. 1 and 2). A luciferase reporter plasmid with five Igκ NF-κB sites upstream of a minimal promoter and an NF-κB-independent pGK-β-gal reporter plasmid were cotransfected to monitor NF-κB activation and transfection efficiency, respectively. In the experiments reported here β-galactosidase level varied ±10% with no evidence for systematic bias. N-terminally FLAG epitope-tagged LMP1 (FTM1-6) activated NF-κB 30- to 150-fold and was an internal control in each experiment. HTM1-2ΔC or FTM3-6 activated NF-κB at ∼10% of FTM1-6 levels (Fig. 2). Cotransfection of FTM3-6 with increasing amounts of HTM1-2ΔC activated NF-κB at 19%, 60%, and 67% of FTM1-6 levels, respectively (Fig. 2). Thus, TM1-2ΔC can induce TM3-6 to mediate more than 60% of LMP1 NF-κB activation.
Since GST N-terminally tagged TM1-2ΔC (GTM1-2ΔC) with TM1 F38WLY41 mutated to A38ALA41 (GTM1-2ΔC A38ALA41) is deficient in association with FTM3-6 (58), HTM1-2ΔC A38ALA41 was tested for effect on FTM3-6-mediated NF-κB activation. HTM1-2ΔC and HTM1-2ΔC A38ALA41 localize similarly to the plasma membranes of B lymphoblasts in which they are transiently expressed (data not shown). In contrast with HTM1-2ΔC, which induced NF-κB activation through FTM3-6 at 67% of FTM1-6 levels, HTM1-2ΔC A38ALA41 induced NF-κB activation at only 19% of FTM1-6 levels (Fig. 2). HTM1-2ΔC A38ALA41 was expressed similarly to HTM1-2ΔC (data not shown), and FTM3-6 expression was not significantly affected by HTM1-2ΔC or HTM1-2ΔC A38ALA41 expression (Fig. 2). These data indicate that TM1-2ΔC intermolecular interaction with TM3-6 reconstitutes 67% of TM1-6-mediated NF-κB; this reconstitution is mostly dependent on TM1-2 F38WLY41.
LMP1 TM1 F38WLY41 is not critical for NF-κB activation through TM3-4.
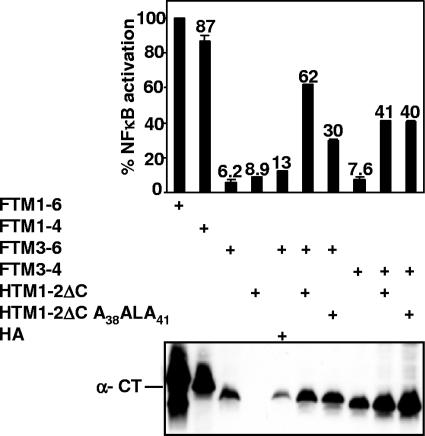
Since LMP1 TM1-4 activates NF-κB at nearly 80% of TM1-6 levels (58), we proceeded to determine whether HTM1-2ΔC can also induce TM3-4 to mediate NF-κB activation. HTM1-2ΔC or HTM1-2ΔC A38ALA41 was expressed in cells with FLAG-tagged TM3-4 (FTM3-4) or FTM3-6 (Fig. 3). Individually, HTM1-2ΔC, FTM3-6, and FTM3-4 activated NF-κB at up to ∼12% of FTM1-6 levels (Fig. 3 and Table 1). As expected from the preceding experiments, HTM1-2ΔC activated NF-κB through FTM3-6 at 62% of FTM1-6 levels and HTM1-2ΔC A38ALA41 was deficient and activated FTM3-6 to only 30% of FTM1-6 levels (Fig. 3 and Table 1). HTM1-2ΔC activated NF-κB through FTM3-4 to 41% of the FTM1-6 levels (Fig. 3 and Table 1). Surprisingly, HTM1-2ΔC A38ALA41 also activated NF-κB through FTM3-4 to 40% of FTM1-6 levels (Fig. 3 and Table 1). These data indicate that TM1-2ΔC can activate NF-κB through TM3-4 at a lower level than through TM3-6 and that TM1 F38WLY41 is unimportant for TM1-2ΔC activation of NF-κB through TM3-4. Thus, TMs 5 and 6 are important in TM1 F38WLY41-specific interaction with TM3-6.
FIG. 3.
LMP1 TM1-2ΔC can induce TM3-4-mediated NF-κB activation, and TM1 F38WLY41 is not critical for this activation. NF-κB activation was measured after HTM1-2ΔC or its mutant HTM1-2ΔC A38ALA41 was expressed alone or with FTM3-6 or FTM3-4 in HEK293 cells. NF-κB activation was measured by cotransfection with a plasmid with five Igκ NF-κB sites preceding a minimal promoter and luciferase reporter and a pGK-β-gal reporter control plasmid and analyzed as described in the legend for Fig. 2.
TABLE 1.
Summary of NF-κB activation induced by LMP1 constructsa
| LMP1 (n) | Signaling (% of control ± SD)
|
||||
|---|---|---|---|---|---|
| Self | HTM1-2ΔC | HTM1-2ΔC A38ALA41 | HTM1-4ΔC | HTM1-4ΔC A38ALA41 | |
| FTM1-6 (4) | 100 | ||||
| FTM1-4 (4) | 78 ± 9 | ||||
| FTM3-6 (4) | 12 ± 3 | 59 ± 3 | 22 ± 5 | ||
| FTM3-6 W98A (3) | 16 ± 1 | 45 ± 8 | 47 ± 2 | ||
| FTM3-6 Y106A (3) | 16 ± 4 | 75 ± 10 | 30 ± 5 | ||
| FTM3-6 W98Y106AA (3) | 18 ± 4 | 52 ± 8 | 48 ± 3 | ||
| FTM3-4 (4) | 9 ± 2 | 44 ± 3 | 46 ± 9 | ||
| FTM5-6 (4) | 6 ± 1 | 18 ± 3 | 13 ± 4 | 20 ± 1* | 13 ± 0.2* |
| FTM5-6 Y159A (3) | 9 ± 1 | 20 ± 3 | |||
| FTM5-6 WW164-5AA (3) | 7 ± 2 | 19 ± 3 | |||
| FTM5-6 W173A (3) | 9 ± 2 | 22 ± 3 | |||
NF-κB activation by full-length FLAG-tagged LMP1 (FTM1-6) was normalized as 100%. NF-κB activation by individual transfection of a LMP1 construct is termed “self.” The values shown are averages from four or three independent experiments (n) done in duplicate, except for values shown with an asterisk, which are from three independent experiments done in duplicate.
TM1-2ΔC and TM1-4ΔC can weakly activate NF-κB through TM5-6.
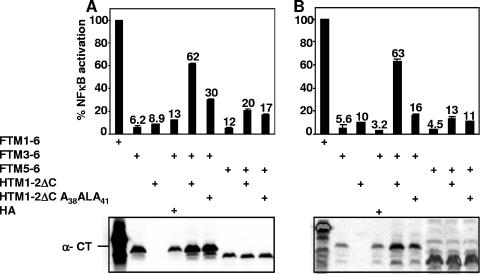
To determine whether TM1-2ΔC can activate NF-κB through TM5-6, FLAG-tagged TM5-6 (FTM5-6) was expressed in cells with HTM1-2ΔC or HTM1-2ΔC A38ALA41 (Fig. 4 and Table 1). Whereas HTM1-2ΔC induced FTM3-6-mediated NF-κB activation to 62% of FTM1-6 levels, HTM1-2ΔC-induced FTM5-6 mediated NF-κB activation to only 20% of FTM1-6 levels (Fig. 4A and Table 1). In the same experiment, HTM1-2ΔC A38ALA41 increased FTM5-6-mediated NF-κB activation to 17% of FTM1-6 levels (Fig. 4A and Table 1). Although FTM5-6 expression is lower than in comparison to FTM3-6 levels (Fig. 4A), FTM5-6 expression at a higher level than FTM3-6 with HTM1-2ΔC or HTM1-2ΔC A38ALA41 also induced NF-κB activation to only 13% and 11% of FTM1-6 levels, respectively (Fig. 4B and Table 1). These data indicate that TM1-2ΔC can induce low levels of TM5-6-mediated NF-κB activation and that TM1 F38WLY41 may have a small role in TM5-6-mediated NF-κB activation.
FIG. 4.
LMP1 TM1-2ΔC can weakly induce TM5-6-mediated NF-κB activation, and TM1 A38ALA41 has little effect. NF-κB activation was measured after HTM1-2ΔC or its mutant HTM1-2ΔC A38ALA41 was expressed alone or with FTM3-6 or FTM5-6 in HEK293 cells. NF-κB activation was measured by cotransfection with a plasmid with five Igκ NF-κB sites preceding a minimal promoter and luciferase reporter and a pGK-β-gal reporter control plasmid and analyzed as described in the legend for Fig. 2. Two independent experiments, one where expression of FTM5-6 is low in comparison to FTM3-6 (A) and one where expression of FTM5-6 is high (B), are shown.
To evaluate whether TM1-4ΔC can better induce TM5-6-mediated NF-κB activation, HTM1-4ΔC or HTM1-4ΔC A38ALA41 were assayed for induction of FTM5-6-mediated NF-κB activation (Table 1). HTM1-4ΔC was similar to HTM1-2ΔC in inducing FTM5-6-mediated NF-κB activation at 20% of FTM1-6 levels (Table 1). However, HTM1-4ΔC A38ALA41 was more deficient in this context and induced FTM5-6-mediated NF-κB activation to only 13% of the FTM1-6 levels (Table 1). These data indicate that TMs 3 to 6 are an important functional unit in these assays and that TM1 F38WLY41 may have a role in NF-κB activation through TM5-6.
TM5 Y159, TM6 WW165, and TM6 W173 are not critical for TM1-4ΔC induction of TM5-6-mediated NF-κB activation.
The possibility that F38WLY41 in TM1-4ΔC interacts with similarly positioned tyrosines or tryptophans at the external side of TM5 or TM6 was evaluated by comparing NF-κB activation induced by HTM1-4ΔC through FTM5-6 or FTM5-6 mutants (Fig. 1 and Table 1). HTM1-4ΔC activated FTM5-6, FTM5-6 Y159A, FTM5-6 WW164-5AA, and FTM5-6 W173A to similar levels of ∼20% of FTM1-6 levels (Table 1). Thus, TM5-6 Y159, WW164-5, and W173 individually are not critical for TM1-4ΔC-induced TM5-6-mediated NF-κB activation.
TM3 W98 is important for TM1-2ΔC induction of TM3-6-mediated NF-κB activation.
The possibility that TM1 F38WLY41 may interact with similarly positioned aromatic amino acids at the external side of TM3 or TM4 was also evaluated by comparing levels of HTM1-2ΔC and HTM1-2ΔC A38ALA41 activation of NF-κB through FTM3-6 or FTM3-6 mutated for W98A, Y106A, or W98Y106AA (Fig. 1 and 5 and Table 1). In the experiment shown in Fig. 5, HTM1-2ΔC-induced FTM3-6 mediated NF-κB activation to 55% of FTM1-6 levels and FTM3-6 Y106A mediated NF-κB activation to 82% of FTM1-6 levels (Fig. 5), although the average in repeated experiments was 59% ± 3% and 75% ± 10%, respectively (Table 1). As expected, HTM1-2ΔC A38ALA41 was markedly deficient in FTM3-6- and FTM3-6 Y106A-mediated NF-κB activation, with overall inductions of 22% and 30%, respectively, much less than the 59% and 75% with HTM1-2ΔC and marginally above the 12% induction by FTM3-6 alone (Table 1).
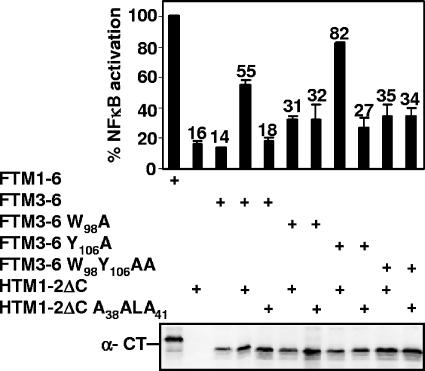
FIG. 5.
LMP1 TM1-2ΔC induction of NF-κB activation through TM3-6 is partially dependent on TM3 W98. HTM1-2ΔC or its mutant HTM1-2ΔC A38ALA41 were expressed in HEK293 cells alone or with FTM3-6 or its mutant FTM3-6 W98A, FTM3-6 Y106A, or FTM3-6 W98Y106AA. NF-κB activation was measured by cotransfection with a plasmid with five Igκ NF-κB sites preceding a minimal promoter and luciferase reporter and a pGK-β-gal reporter control plasmid and analyzed as described in the legend for Fig. 2.
Most interestingly, HTM1-2ΔC induced lower NF-κB levels through FTM3-6 mutants W98A and W98Y106AA than through FTM3-6 or FTM3-6 mutant Y106A (Fig. 5 and Table 1). HTM1-2ΔC-induced FTM3-6 W98A and FTM3-6 W98Y106AA mediated NF-κB activation to only 45% and 52%, respectively, versus 59% for FTM3-6 levels (Table 1 and Fig. 5). These data indicate a role for TM3 W98 in TM1-2ΔC induction of TM3-6-mediated NF-κB activation.
Equally as surprising, HTM1-2ΔC A38ALA41 did not differ from HTM1-2ΔC and induced FTM3-6 W98A- and FTM3-6 W98Y106AA-mediated NF-κB activations to 47% and 48%, respectively, of the FTM1-6 levels (Table 1 and Fig. 5). Thus, TM3 W98 is important for TM1-2ΔC induction of TM3-6-mediated NF-κB activation and specifically for TM1 F38WLY41 effective induction of TM3-6-mediated NF-κB activation.
TM3 W98A is important for intermolecular association with TM1-2ΔC.
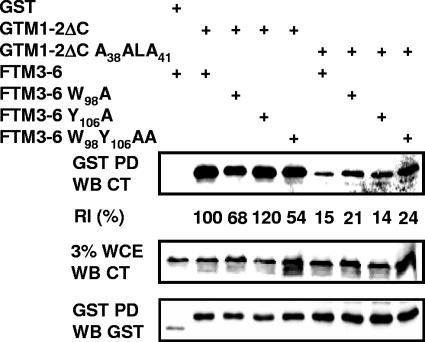
The potential effect of TM3 W98A on TM1-2ΔC association with TM3-6 was investigated by transfecting GST-tagged TM1-2ΔC (GTM1-2ΔC) or GTM1-2ΔC A38ALA41 and FTM3-6, FTM3-6 W98A, FTM3-6 Y106A, or FTM3-6 W98Y106AA into HEK293 cells. The transfected cells were lysed, and GST-tagged proteins were pulled down with glutathione-conjugated Sepharose beads. The lysates and the immunoprecipitates were immunoblotted for GST and TM3-6 and analyzed with Kodak Imager (Fig. 6). In general, GTM1-2ΔC associated with ∼24% (normalized to 100%) of FTM3-6, whereas GST did not associate with FTM3-6 (Fig. 6). GTM1-2ΔC associated with 120% of FTM3-6 Y106A (Fig. 6), consistent with the slightly higher NF-κB activation than with FTM3-6 (Fig. 5 and Table 1). Interestingly, GTM1-2ΔC associated with 68% and 54% of FTM3-6 W98A and W98Y106AA, respectively, indicative of slight deficiency relative that of FTM3-6 levels (Fig. 6). Furthermore, GTM1-2ΔC A38ALA41 was substantially deficient in intermolecular association with FTM3-6 at 15% in comparison to 100% with GTM1-2ΔC (Fig. 6). GTM1-2ΔC A38ALA41 was similarly deficient in association with FTM3-6 Y106A and was slightly less deficient in association with FTM3-6W98A and W98Y106AA, with relative intermolecular associations of 21% and 24%, respectively (Fig. 6). These data are consistent with a model in which TM1 F38WLY41 and TM3 W98 are important for TM1-2 association with and signaling from TM3-6 and TM3 W98 is important for TM1 F38WLY41.
FIG. 6.
LMP1 TM1-2ΔC interaction with TM3-6 is partially dependent on TM3W98. GST alone, GST-tagged TM1-2ΔC (GTM1-2ΔC), or its mutant GTM1-2ΔC A38ALA41 was expressed in HEK293 cells with FTM3-6, FTM3-6 W98A, FTM3-6 Y106A, or FTM3-6 W98Y106AA. After 24 h, GST, GTM1-2ΔC, or GTM1-2ΔC A38ALA41 was adsorbed to glutathione-conjugated Sepharose beads. The proteins pulled down by the beads were eluted with sodium dodecyl sulfate gel loading buffer (GST PD) and were identified by polyacrylamide gel electrophoresis and immune blotted for the LMP1 C terminus (CT) (top) or GST (bottom). Three percent of the WCE was also immune blotted for the LMP1 C terminus (middle). The data were analyzed by using Molecular Imaging software (Kodak). The band intensity for wild-type intermolecular association with GTM1-2ΔC was normalized as 100%, and relative intermolecular association percentages were computed accordingly. The data shown are representative of three independent experiments.
DISCUSSION
These experiments further investigate the role of LMP1 TMs in intermolecular interactions that induce C-terminal signaling as measured by NF-κB activation. The data confirm a special role for TMs 1 and 2, for TM1-2ΔC interaction with TM3-6, and for TM1 F38WLY41 in LMP1 intermolecular interactions and provide a surprisingly robust functional dimension to these interactions. TM1-2ΔC induced TM3-6 C terminus-mediated NF-κB activation to 59% of the LMP1 levels, and the effect was critically dependent on TM1 F38WLY41. TM1-2ΔC even induced TM3-4-mediated NF-κB activation at a reduced level of 44%, and this effect was TM1 F38WLY41 independent, indicative of an important role for TMs 5 and 6 in TM1 F38WLY41 effects. Furthermore, TM3 W98 was deficient in TM1-2ΔC induction of NF-κB activation and association and was not afffected by TM1 F38WLY41 mutation.
The LMP1 TMs are remarkably hydrophobic. TM1, -2, -3, -4, -5, and -6 have 15, 17, 14, 17, 17, and 13 leucines, isoleucines, phenylalanines, and valines, respectively (18), whereas 6 hydrophobic residues can be adequate for membrane insertion (23, 29). These very hydrophobic domains are likely to engage in intramolecular interactions between antiparallel adjacent α-helices, perhaps similar to the interactions described for rhodopsin alpha-helical transmembrane domains (36, 47). Interactions between antiparallel alpha helices may have contributed to the evolution of TM3 W98 interaction with TM1 F38WLY41 in intermolecular aggregation and signaling, since TM1 and TM3 are both linked to TM2 and are antiparallel to TM2.
Accumulated data are consistent with models in which constitutive LMP1 signaling is due to intermolecular interactions among LMP1 TM segments. TM1-2 can intermolecularly aggregate and constitutively signal (10, 58). Furthermore, TM1-2ΔC can intermolecularly associate with and induce TM3-6 to activate NF-κB; these effects are TM1 F38WLY41 dependent (58). Moreover, the studies presented here define a role for TM3 W98 in effecting the intermolecular interaction of TM1 F38WLY41 with TM3-6 and constitutive signaling through TM3-6. Tryptophan and tyrosines are frequently at the outer margin of TMs and together with phenyalanines are implicated in hydrophobic stacking interactions, including interactions among aromatic residues and cholesterol-rich membrane lipid raft microdomains (1, 2, 8, 51, 52, 57-59); TM1 F38WLY41 is critical for stable LMP1 interaction with lipid rafts (58). The epistatic effect between mutations in TM1 F38WLY41 and TM3 W98 is consistent with direct or indirect physical interaction of TM1 F38WLY41 and TM3 W98. This stacking interaction could stabilize LMP1 intermolecular associations and contribute to signaling complex stability, consistent with the importance of TM3 W98 in TM1 F38WLY41 association with TM3-6 and in signaling through TM3-6. However, in the context of transient LMP1 expression in 293 cells, as described here, LMP1 with the W98A mutation is not significantly different from full-length wild-type LMP1 (TM1-6) in signaling and LMP1 with the W98Y106AA mutation is only marginally decreased (data not shown). The data are therefore most consistent with a model in which other similarly positioned aromatic residues in LMP1 can substitute for TM3W98 in supporting intermolecular interactions of TMs 3 to 6 with TMs 1 and 2.
Intermolecular associations among LMP1 TMs 1 and 2 have been previously described (58), and LMP1 TMs 1 to 6 without the C-terminal cytoplasmic signaling domains have also been reported to activate a signaling-deficient LMP1 with seven TM1 leucines mutated to alanines (32). The data presented here are the first demonstration that TM1-2ΔC can induce robust NF-κB activation through intermolecular interactions with TMs 3 to 6 and the induction is TM1 F38WLY41 and TM3 W98 dependent.
These LMP1 TM intermolecular interactions and constitutive signaling effects begin to explain the role of the TMs in enabling constitutive LMP1 signaling. Lymphoblastoid cell lines are dependent on LMP1-mediated NF-κB activation (5-7), and inhibition of TM1-2 intermolecular interactions with FTM3-6 is a potential therapeutic target for EBV-associated lymphoproliferative disease, Hodgkin's disease, and nasopharyngeal carcinoma.
Acknowledgments
This work was supported by grant R01CA85086 from the National Cancer Institute of the USPHS.
We are grateful for the advice of Yoon-Jae Song and Daniela Böhm.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Albert, A. D., J. E. Young, and P. L. Yeagle. 1996. Rhodopsin-cholesterol interactions in bovine rod outer segment disk membranes. Biochim. Biophys. Acta 1285:47-55. [DOI] [PubMed] [Google Scholar]
- 2.Anzenbacher, P., J. Hudecek, S. Vajda, V. Fidler, C. Larroque, and R. Lange. 1991. Nanosecond fluorescence of tryptophans in cytochrome P-450scc (CYP11A1): effect of substrate binding. Biochem. Biophys. Res. Commun. 181:1493-1499. [DOI] [PubMed] [Google Scholar]
- 3.Ardila-Osorio, H., B. Clausse, Z. Mishal, J. Wiels, T. Tursz, and P. Busson. 1999. Evidence of LMP1-TRAF3 interactions in glycosphingolipid-rich complexes of lymphoblastoid and nasopharyngeal carcinoma cells. Int. J. Cancer 81:645-649. [DOI] [PubMed] [Google Scholar]
- 4.Brodeur, S. R., G. Cheng, D. Baltimore, and D. A. Thorley-Lawson. 1997. Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J. Biol. Chem. 272:19777-19784. [DOI] [PubMed] [Google Scholar]
- 5.Cahir-McFarland, E., and E. Kieff. 2002. NF-κB inhibition in EBV-transformed lymphoblastoid cell lines. Recent Results Cancer Res. 159:44-48. [DOI] [PubMed] [Google Scholar]
- 6.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahir-McFarland, E. D., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff. 2000. NF-κB inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 97:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarti, A. C., I. Clark-Lewis, and P. R. Cullis. 1994. Influence of charge, charge distribution, and hydrophobicity on the transport of short model peptides into liposomes in response to transmembrane pH gradients. Biochemistry 33:8479-8485. [DOI] [PubMed] [Google Scholar]
- 9.Clausse, B., K. Fizazi, V. Walczak, C. Tetaud, J. Wiels, T. Tursz, and P. Busson. 1997. High concentration of the EBV latent membrane protein 1 in glycosphingolipid-rich complexes from both epithelial and lymphoid cells. Virology 228:285-293. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, W. F., III, T. R. Geiger, and J. M. Martin. 2003. Transmembrane domains 1 and 2 of the latent membrane protein 1 of Epstein-Barr virus contain a lipid raft targeting signal and play a critical role in cytostasis. J. Virol. 77:3749-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran, J. A., F. S. Laverty, D. Campbell, J. Macdiarmid, and J. B. Wilson. 2001. Epstein-Barr virus encoded latent membrane protein-1 induces epithelial cell proliferation and sensitizes transgenic mice to chemical carcinogenesis. Cancer Res. 61:6730-6738. [PubMed] [Google Scholar]
- 12.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 13.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devergne, O., E. C. McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dirmeier, U., B. Neuhierl, E. Kilger, G. Reisbach, M. L. Sandberg, and W. Hammerschmidt. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982-2989. [PubMed] [Google Scholar]
- 16.Eliopoulos, A. G., N. J. Gallagher, S. M. Blake, C. W. Dawson, and L. S. Young. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085-16096. [DOI] [PubMed] [Google Scholar]
- 17.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731-1742. [DOI] [PubMed] [Google Scholar]
- 18.Fennewald, S., V. van Santen, and E. Kieff. 1984. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J. Virol. 51:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerisation for NF-κB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 20.Franken, M., O. Devergne, M. Rosenzweig, B. Annis, E. Kieff, and F. Wang. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 70:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammarskjold, M. L., and M. C. Simurda. 1992. Epstein-Barr virus latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF-κB activity. J. Virol. 66:6496-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennessy, K., S. Fennewald, M. Hummel, T. Cole, and E. Kieff. 1984. A membrane protein encoded by Epstein-Barr virus in latent growth-transforming infection. Proc. Natl. Acad. Sci. USA 81:7207-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessa, T., H. Kim, K. Bihlmaier, C. Lundin, J. Boekel, H. Andersson, I. Nilsson, S. H. White, and G. von Heijne. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377-381. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi, M., K. M. Izumi, and E. Kieff. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. USA 98:4675-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 26.Izumi, K. M., E. D. Cahir-McFarland, E. A. Riley, D. Rizzo, Y. Chen, and E. Kieff. 1999. The residues between the two transformation effector sites of Epstein-Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation. J. Virol. 73:9908-9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi, K. M., E. D. Cahir-McFarland, A. T. Ting, E. A. Riley, B. Seed, and E. D. Kieff. 1999. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol. Cell. Biol. 19:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayasinghe, S., K. Hristova, and S. H. White. 2001. Energetics, stability, and prediction of transmembrane helices. J. Mol. Biol. 312:927-934. [DOI] [PubMed] [Google Scholar]
- 30.Kaye, K. M., O. Devergne, J. N. Harada, K. M. Izumi, R. Yalamanchili, E. Kieff, and G. Mosialos. 1996. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc. Natl. Acad. Sci. USA 93:11085-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaykas, A., K. Worringer, and B. Sugden. 2002. LMP-1's transmembrane domains encode multiple functions required for LMP-1's efficient signaling. J. Virol. 76:11551-11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr Virus and its replication, p. 2511-2574. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 34.Kulwichit, W., R. H. Edwards, E. M. Davenport, J. F. Baskar, V. Godfrey, and N. Raab-Traub. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. USA 95:11963-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laherty, C. D., H. M. Hu, A. W. Opipari, F. Wang, and V. M. Dixit. 1992. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor κB. J. Biol. Chem. 267:24157-24160. [PubMed] [Google Scholar]
- 36.Li, J., P. C. Edwards, M. Burghammer, C. Villa, and G. F. Schertler. 2004. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 343:1409-1438. [DOI] [PubMed] [Google Scholar]
- 37.Liebowitz, D., and E. Kieff. 1989. Epstein-Barr virus latent membrane protein: induction of B-cell activation antigens and membrane patch formation does not require vimentin. J. Virol. 63:4051-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebowitz, D., R. Kopan, E. Fuchs, J. Sample, and E. Kieff. 1987. An Epstein-Barr virus transforming protein associates with vimentin in lymphocytes. Mol. Cell. Biol. 7:2299-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebowitz, D., J. Mannick, K. Takada, and E. Kieff. 1992. Phenotypes of Epstein-Barr virus LMP1 deletion mutants indicate transmembrane and amino-terminal cytoplasmic domains necessary for effects in B-lymphoma cells. J. Virol. 66:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liebowitz, D., D. Wang, and E. Kieff. 1986. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J. Virol. 58:233-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann, K. P., D. Staunton, and D. A. Thorley-Lawson. 1985. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J. Virol. 55:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, J., and B. Sugden. 1991. The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ. 2:653-660. [PubMed] [Google Scholar]
- 43.Miller, W. E., J. L. Cheshire, A. S. Baldwin, Jr., and N. Raab-Traub. 1998. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene 16:1869-1877. [DOI] [PubMed] [Google Scholar]
- 44.Miller, W. E., G. Mosialos, E. Kieff, and N. Raab-Traub. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J. Virol. 71:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389-399. [DOI] [PubMed] [Google Scholar]
- 47.Palczewski, K., T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima, B. A. Fox, I. Le Trong, D. C. Teller, T. Okada, R. E. Stenkamp, M. Yamamoto, and M. Miyano. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739-745. [DOI] [PubMed] [Google Scholar]
- 48.Puls, A., A. G. Eliopoulos, C. D. Nobes, T. Bridges, L. S. Young, and A. Hall. 1999. Activation of the small GTPase Cdc42 by the inflammatory cytokines TNF-α and IL-1, and by the Epstein-Barr virus transforming protein LMP1. J. Cell Sci. 112(Pt. 17):2983-2992. [DOI] [PubMed] [Google Scholar]
- 49.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2628. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 50.Sandberg, M., W. Hammerschmidt, and B. Sugden. 1997. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J. Virol. 71:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimada, Y., M. Maruya, S. Iwashita, and Y. Ohno-Iwashita. 2002. The C-terminal domain of perfringolysin O is an essential cholesterol-binding unit targeting to cholesterol-rich microdomains. Eur. J. Biochem. 269:6195-6203. [DOI] [PubMed] [Google Scholar]
- 52.Ulmschneider, M. B., and M. S. Sansom. 2001. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta 1512:1-14. [DOI] [PubMed] [Google Scholar]
- 53.Wan, J., L. Sun, J. W. Mendoza, Y. L. Chui, D. P. Huang, Z. J. Chen, N. Suzuki, S. Suzuki, W. C. Yeh, S. Akira, K. Matsumoto, Z. G. Liu, and Z. Wu. 2004. Elucidation of the c-Jun N-terminal kinase pathway mediated by Estein-Barr virus-encoded latent membrane protein 1. Mol. Cell. Biol. 24:192-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 55.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 58.Yasui, T., M. Luftig, V. Soni, and E. Kieff. 2004. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc. Natl. Acad. Sci. USA 101:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yau, W. M., W. C. Wimley, K. Gawrisch, and S. H. White. 1998. The preference of tryptophan for membrane interfaces. Biochemistry 37:14713-14718. [DOI] [PubMed] [Google Scholar]
- 60.Ye, H., Y. C. Park, M. Kreishman, E. Kieff, and H. Wu. 1999. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell 4:321-330. [DOI] [PubMed] [Google Scholar]