Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen 1 (LANA-1) is required for the replication of episomal viral genomes. Regions in its N-terminal and C-terminal domains mediate the interaction with host cell chromatin. Several cellular nuclear proteins, e.g., BRD2/RING3, histones H2A and H2B, MeCP2, DEK, and HP1α, have been suggested to mediate this interaction. In this work, we identify the double-bromodomain proteins BRD4 and BRD3/ORFX as additional LANA-1 interaction partners. The carboxy-terminal region of the short variant of BRD4 (BRD4S) containing the highly conserved extraterminal domain directly interacts with an element in the LANA-1 carboxy-terminal domain. We show that ectopically expressed BRD4S and BRD2/RING3 delay progression into the S phase of the cell cycle in epithelial and B-cell lines and increase cyclin E promoter activity. LANA-1 partly releases epithelial and B cells from a BRD4S- and BRD2/RING3-induced G1 cell cycle arrest and also promotes S-phase entry in the presence of BRD4S and BRD2/RING3. This is accompanied by a reduction in BRD4S-mediated cyclin E promoter activity. Our data are in keeping with the notion that the direct interaction of KSHV LANA-1 with BRD4 and other BRD proteins could play a role in the G1/S phase-promoting functions of KSHV LANA-1. Further, our data support a model in which the LANA-1 C terminus contributes to a functional attachment to acetylated histones H3 and H4 via BRD4 and BRD2, in addition to the recently demonstrated direct interaction (A. J. Barbera, J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye, Science 311:856-861, 2006) of the LANA-1 N terminus with histones H2A and H2B.
Worldwide, Kaposi's sarcoma (KS) is today the most common malignancy in the AIDS setting. Kaposi's sarcoma-associated herpesvirus (KSHV) (6) is the major etiologic agent for KS (52). Further, KSHV is etiologically linked to two lymphoproliferative diseases: primary effusion lymphoma (PEL) and the plasmablastic variant of multicentric Castleman's disease (5, 47). Similar to other herpesviruses, one characteristic biological feature of KSHV is a latent state of infection, with the expression of only a small subset of viral genes (35, 48, 57). KSHV can infect and establish latency in a variety of cells in vitro, including cells of epithelial, endothelial, and mesenchymal origin (4). One of the KSHV latent gene products is the latency-associated nuclear antigen 1 (LANA-1) encoded by KSHV open reading frame (ORF) 73 (27, 29, 39). LANA-1 is a nuclear protein expressed in the tumor cells of KS, PEL, and multicentric Castleman's disease and is ubiquitously expressed in the vast majority, if not all, of the infected cells (12, 17, 18, 28, 39). In the nuclei of KSHV-infected PEL and KS tumor cells, LANA-1 is located to subnuclear bodies (27, 39).
The protein sequence of LANA-1 can be divided into three domains: a proline- and serine-rich N-terminal region, a central region that is variable in length between different viral isolates and composed of several acidic repeat elements, and a C-terminal domain containing a proline-rich region and a region rich in charged and hydrophobic amino acids (aa) (41), which shares sequence similarity with homologous ORF 73 proteins of other rhadinoviruses (21), with the highest degree of similarity in the region of the LANA-1 amino acids 1128 to 1143 (our own analyses).
LANA-1 attaches to interphase chromatin as well as to mitotic chromosomes (36). Both the N-terminal and the C-terminal domains of LANA-1 localize to the nucleus when expressed separately. However, only the C-terminal domain accumulates as subnuclear speckles characteristic of the intact protein (42). The N-terminal 15 amino acids of LANA-1 as well as the C-terminal region encompassing aa 1128 to 1143 mediate the interaction with chromatin and are required for LANA-1 functions, such as the replication of a latent episome and activation of heterologous promoters (31, 36, 44, 50, 51, 54). The N-terminal domain of LANA-1 directly binds to histones H2A and H2B (3), while several chromatin-associated proteins, including BRD2/RING3 (37, 51), DEK (30), and HP1 (31), have been suggested to mediate the attachment of the LANA-1 C-terminal domain to chromatin.
Recent work shows that a number of LANA-1-associated proteins, such as the chromatin binding protein, the double-bromodomain-containing protein BRD2/RING3, and the origin recognition complex 2 protein, bind to the latent origin of KSHV, which is enriched in hyperacetylated histones H3 and H4, relative to other regions of the latent KSHV genome (49). BRD2/RING3, an interaction partner of the LANA-1 C-terminal domain (37, 51), is a member of the BET/fsh (bromodomain and extraterminal domain/female sterile homeotic) protein family. Bromodomains are conserved sequence elements identified in several protein families and constitute chromatin targeting modules that mediate attachment to acetylated histones (22, 25, 53). Mammalian BET proteins may be involved in certain aspects of oncogenesis: for example, BRD2/RING3 showed elevated kinase activity in leukemic lymphoblasts of the B lineage and BRD2 transgenic mice with B-cell-specific expression of the transgene develop B-cell lymphomas (9, 20). BRD4, another mammalian BET protein, appears to play a role in cell cycle regulation and was found to be chromosomally translocated in certain carcinomas (11, 14, 15, 32).
Due to the high degree of sequence similarity of BET/fsh family members, especially in their ET domains, which we have previously identified in BRD2/RING3 to constitute one site of LANA-1 interaction, we wondered whether KSHV LANA-1 would interact with other members of the BET/fsh protein family (37, 51). Besides BRD2/RING3, we focused on BRD4, as mouse BRD4/mitotic chromosome-associated protein (MCAP), the longer one of two alternatively spliced BRD4 variants, is associated with interphase and mitotic chromosomes via acetylated histones H3 and H4 (10, 11). Therefore, BRD4 could potentially provide an interaction partner to mediate the chromosome association and other functions of LANA-1. While the longer BRD4 variant MCAP of mice has been studied in some detail, no functional studies have so far been reported on the short BRD4 variant, also termed HUNK, which we refer to as BRD4S throughout this study.
We show the expression of full-length BRD4S transcripts in a variety of KSHV-negative and KSHV-positive cells. Further, KSHV LANA-1 interacted in vivo with endogenous BRD4 proteins, as shown by coimmunoprecipitation studies using the KSHV-infected PEL cell line BCBL-1. Purified BRD4S and LANA-1 interacted directly in vitro. A sequence element in the LANA-1 C terminus (aa 1133 to 1143) was indispensable for the interaction with BRD4S. The highly conserved ET domain in BRD4 was sufficient for LANA-1 binding. The ectopic expression of BRD2/RING3 and BRD4S in epithelial and B-cell lines prevented entry into the S phase of the cell cycle, and the coexpression of LANA-1 was able to release cells from this cell cycle arrest. In line with this, LANA-1 inhibited BRD4S-mediated activation of the cyclin E promoter. Since BRD4 has previously been shown to directly interact with acetylated histones H3 and H4 via the BRD4 bromodomains, our data support a model in which the LANA-1 C terminus contributes to a functional nucleosome attachment via the direct interaction of LANA-1 with BRD4 in addition to the attachment of the LANA-1 N terminus to histones H2A and H2B (3). LANA-1 therefore appears to rely on diverse interaction partners to assure its association with chromatin, a prerequisite for the vast majority of known LANA-1 functions.
MATERIALS AND METHODS
Cell culture methods, transient transfections, and electroporation.
The two epithelial cell lines HEK 293T (human embryonic kidney) and HeLa were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% heat-inactivated fetal calf serum, 50 IU/ml penicillin, 50 μg/ml streptomycin, and 200 μg/ml l-glutamine at 37°C with 5% CO2. For transfections, 293T and HeLa cells were grown to subconfluence in six-well plates and transfected with FuGENE 6 transfection reagent (FuGENE-to-DNA ratio of 3 μl:1 μg; Roche) according to the manufacturer's instructions. Total amounts of transfected DNA were adjusted by using the respective empty vector DNA or salmon sperm DNA (Sigma). The KSHV- and EBV-negative Burkitt's lymphoma cell line BJAB and the KSHV-positive primary effusion lymphoma cell line BCBL-1 (40) were maintained in RPMI 1640 with 10 and 20% bovine growth serum, respectively, plus 50 IU/ml penicillin and 50 μg/ml streptomycin. A total of 8 × 106 BJAB or BCBL-1 cells were resuspended in 400 μl medium without antibiotics. After 10 min of incubation at room temperature, 15 μg of endotoxin-free DNA, as indicated in the figure legends, was added to the cells. Cells were electroporated in 0.4-cm cuvettes using a Gene Pulser Xcell (Bio-Rad, Hercules, CA) with 220 V and 950 μF. After electroporation, cells were cultured in 25 ml of medium plus bovine growth serum for 40 h before the bromodeoxyuridine (BrdU) incorporation was performed, followed by cell cycle analysis (see “Flow cytometry-based cell cycle analyses”).
Cloning of the short transcript variant of BRD4, BRD4S.
Nested reverse transcriptase PCR (RT-PCR) was performed on RNA of different human cell lines, including KSHV-positive PEL B-cell lines Jsc-1, BCP-1, and CroAP-5. The KSHV-negative, EBV-positive B-cell line Raji, HeLa cells, and peripheral blood lymphocytes were included. Total RNA was prepared with the RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. The RT reaction was performed with the Expand reverse transcriptase kit (Roche), using the BRD4S-specific primer BRD4S R NEST (5′-GGAATCTGGAACTGAAGACC-3′) annealing downstream of the stop codon in exon 11. A total of 1 to 1.5 μg of total RNA and 10 picomoles of specific reverse primer were denatured at 65°C and cooled on ice. The RT reaction was carried out at 42°C for 1 h following the manufacturer's instructions. Subsequently, we performed the first-round PCR on the generated cDNA molecules using primer pairs BRD4S F NEST (5′-CCTGGTGAAGAATGTGATGG-3′) and BRD4S R NEST and the second-round PCR with the primers BRD4S FOR B (5′-AGAGGATCCTCTGCGGAGAGCGGC-3′) and BRD4S REV X (5′-AGACTCGAGTTAGGCAGGACCTGTTTC-3′). The obtained 2,169-bp coding fragment was cloned into the expression vector pcDNA3 9E10 using the endonucleases BamHI/XhoI. The generated construct was named BRD4S pcDNA3 9E10, coding for an amino-terminally myc epitope-tagged full-length BRD4S protein. The construct used for further experiments originated from Raji cell cDNA and was completely sequenced and found to be identical to the already deposited sequence entry NM_014299.
DNA constructs.
The KSHV LANA-1 expression constructs in pcDNA3.1 encoding full-length LANA-1 or carboxy-terminally truncated versions of LANA-1 as well as the glutathione S-transferase (GST)-LANAC construct for the expression of amino acids 951 to 1162 of KSHV LANA-1 in bacteria were described previously (50, 51). The construct GST-RING3 B in pGEX-1, which expresses the C-terminal domain of RING3 (aa 601 to 801), including the ET domain (aa 640 to 703), was also previously described (37). GST-BRD4S in pGEX-1 expresses the C-terminal domain of BRD4S (aa 607 to 722), including the ET domain (aa 608 to 671) amino-terminally fused to GST, and was generated by RT-PCR on HeLa cell RNA using primers BRD4S F (5′-GAGGGATCCGTGCAAGCCTATGTCCTAT-3′) and BRD4S R (5′-GAGGAATTCTTAGGCAGGACCTGTTCC-3′). GST-ORFX in pGEX-1 expresses the C-terminal domain of BRD3/ORFX (aa 569 to 726), including the ET domain (aa 569 to 633) fused to the C terminus of GST, and was generated by RT-PCR using a leukocyte cDNA library as a template using the primers ORFX F (5′-GAGGGATCCGGGCCTGCCCATGAGCTAC-3′) and ORFX R (5′-GAGGAATTCTCATTCTGAGTCACTGCT-3′). The full-length (aa 2 to 801) construct BRD2/RING3 in EGFPC1 codes for full-length RING3 fused with its N terminus to enhanced green fluorescent protein (EGFP) and was generated by PCR from the full-length (aa 2 to 801) template BRD2/RING3 in pcDNA3 9E10 (37) with primers RING3 GFPC1 FOR (5′-ACTAGATCTCTGCAAAACGTGACTCCCCAC-3′) and RING3 GFPC1 REV (5′-AGACCCGGGATTAGCCTGAGTCTGAATCACTG-3′), followed by BglII/SmaI insertion into EGFPC1 (Clontech). The vector BRD2/RING3, aa 640 to 801 in EGFPC1, codes for RING3, initiating at the ET domain (aa 640 to 703) fused to the C terminus of EGFP, and was generated by PCR using the full-length (aa 2 to 801) template BRD2/RING3 in pcDNA3 9E10 (37) with primers RING3 NETF (5′-AGAAGATCTAGGCCCATGAGTTACGATG-3′) and GS23 (5′-CTCGAATCCTTAGCCTGAGTCTGAATCACT-3′). The PCR product was inserted into EGFPC1 using BglII/EcoRI. The construct BRD3/ORFX, aa 569 to 726 in EGFPC1, codes for the BRD3/ORFX carboxy-terminal fragment beginning at the ET domain (aa 569 to 633) fused to the C terminus of EGFP and was generated by PCR on the template GST-ORFX (see above) with primers ORFX NETF (5′-AGAAGATCTGGCCTGCCCATGAGC-3′) and ORFX R and subsequent insertion of the PCR product into EGFPC1 using BglII/EcoRI. A BRD4S full-length (aa 2 to 722) construct in EGFPC1, coding for full-length BRD4S fused with its N terminus to EGFP, was generated by PCR using the full-length (aa 2 to 722) template BRD4S in pcDNA3 9E10 (see above) with primers BRD4S GFPC1 FOR (5′-ACTAGATCTTCTGCGGAGAGCGGCCCTG-3′) and BRD4S GFPC1 REV (5′-AGACCCGGGTTAGGCAGGACCTGTTTCGG-3′), followed by BglII/SmaI insertion of the PCR product into EGFPC1. Further, the vector BRD4S aa608-722 in EGFPC1, coding for EGFP C-terminally fused to the BRD4S carboxy-terminal domain starting at the ET domain (aa 608 to 671), was cloned by PCR using primers BRD4S NET F (5′-TAGAGATCTAAGCCTATGTCCTATGAGG-3′) and BRD4S R (5′-GAGGAATTCTTAGGCAGGACCTGTTCC-3′) with the full-length (aa 2 to 722) template BRD4S in pcDNA3 9E10 (see above), followed by BglII/SalI insertion of the PCR product into EGFPC1. The cyclin E reporter construct used in this study contains the 2.2-kb upstream region of the human cyclin E coding sequence and was generated in Robert Weinberg's laboratory (19). All constructs generated by PCR were completely sequenced.
SF9 insect cell culture, baculoviruses, and the generation of a BRD4S aa444-722 baculovirus.
Spodoptera frugiperda (SF9) insect cells were maintained at room temperature in Grace's insect medium supplemented with 10% fetal calf serum, 50 IU/ml penicillin, and 50 μg/ml streptomycin in spinner flasks. For routine production of recombinant proteins, SF9 cells in logarithmic growth phase were infected at a cell density of 1.5 × 106 cells ml−1 with 1 to 10 infectious units per cell, cultured at 27°C, and harvested 72 h postinfection. The recombinant baculovirus for the expression of the carboxy-terminal domain of KSHV LANA-1 (aa 951 to 1162) has been previously described (37). We generated a recombinant baculovirus for the expression of an amino-terminally Flag-tagged and carboxy-terminally hexahistidine-tagged fragment of BRD4S, initiating at aa 444 (downstream of the second bromodomain) and ending at the natural C terminus (aa 722) of BRD4S (aa444-722) using the BaculoDirect kit (Invitrogen). PCR was performed on the full-length BRD4S cDNA in pcDNA3.1 and primers BRD4Saa444BACF (5′-AGAGGATCCACCATGGACTACAAGGACGACGACGACAAGCGCAAGCTCCAGGATG-3′[nucleotides introducing the Flag epitope are underlined]) and BRD4S BAC R (5′-AGACTCGAGGCAGGACCTGTTTCGG-3′). The fragment was introduced into vector pENTR1A (Invitrogen), followed by in vitro recombination with BaculoDirect C-Term linear DNA (Invitrogen) and the transfection of SF9 cells, following the protocol recommended by the manufacturer. The hexahistidine tag is coupled to amino acids 444 to 722 of BRD4S by a 37-amino-acid-long linker element.
Expression and purification of GST and GST-fusion proteins.
GST alone or the GST-fusion protein GST-RING3 B, GST-BRD4S, or GST-ORFX was expressed in Escherichia coli strain M15. E. coli was grown at 30°C in LB medium supplemented with 100 μg/ml of ampicillin, induced at an optical density at 600 nm (OD600) of 0.2 to 0.3 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and harvested 6 h after induction by centrifugation. The pellet was resuspended in 1,000 μl phosphate-buffered saline (PBS) plus protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 50 μM leupeptin, 100 U/ml aprotinin, 200 μM benzamidine, and 1 μM pepstatin A) per 10 ml of culture and an OD600 of 2.5. After sonication on ice (3 times for 20 s with 20-s breaks), Triton X-100 was added to a final concentration of 1% and cells were kept on ice for 10 min and centrifuged for 15 min at 13,000 × g at 4°C. For GST pull-down experiments, the supernatant was adsorbed onto 20 to 50 μl of glutathione-Sepharose beads for 1 h at 4°C and then beads were washed twice in PBS and twice in 1% NP-40 lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, protease inhibitors). A total of 10 μl of beads with bound GST proteins were then used for each sample in the GST pull-down experiments.
For the purification of GST-LANAC, the system was scaled up starting with 400 ml of bacterial culture with slight changes to the protocol. Cells were induced at an OD600 of 0.7 with 0.4 mM IPTG and grown for 4 h at 30°C. Pellets were lysed on ice in 8 ml GST lysis buffer (PBS, 1% Triton X-100, 10 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride), sonicated, and centrifuged at 4°C for 20 min at 13,000 × g. The supernatants were incubated with 700 μl glutathione beads for 1.5 h at room temperature. Following centrifugation at 500 × g for 5 min, the beads were washed four times with 5 ml GST lysis buffer and twice with 5 ml of TBS (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA). Subsequently, bound GST or GST-LANAC was eluted off the beads in five subsequent elution steps, each of them with a volume of 300 μl GST elution buffer (50 mM Tris-HCl, pH 8.0, 0.1% Triton X-100, 10 mM glutathione). Elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining for purity of the protein. Protein concentrations were determined by Coomassie staining with a bovine serum albumin standard and by Bradford assay. The specificity of the protein was controlled by anti-GST immunoblotting.
Purification of recombinant BRD4S aa444-722 from SF9 insect cells.
The amino-terminally Flag-epitope-tagged and carboxy-terminally hexahistidine-tagged BRD4S aa444-722 protein was purified using Ni2+ affinity chromatography. A total of 4 × 108 SF9 insect cells in the logarithmic phase of a suspension culture was infected with the BRD4S aa444-722 baculovirus at a high multiplicity of infection of 10. Ninety-six hours after infection, cells were pelleted, lysed on ice in 40-ml hypotonic lysis buffer (50 mM Tris-HCl, pH 8.0, 1% NP-40, protease inhibitors), and sonicated (six times for 20 s with 20-s breaks). After centrifugation at 13,000 × g for 20 min at 4°C, supernatants were incubated with 2 ml prewashed Ni-nitrilotriacetic acid for 3 h at 4°C. Beads with bound proteins were then washed as a batch at 4°C twice for 30 min with 30 ml wash buffer I (50 mM Tris-HCl, pH 8.0, 500 mM NaCl), followed by three washes for 20 min with 20 ml wash buffer II (wash buffer I plus 20 mM imidazole). Subsequently, the beads were resuspended in 10 ml wash buffer II, transferred to a column, and eluted with an imidazole gradient. A total of 1 ml of each imidazole concentration of elution buffers (50 mM Tris, pH 8.0, 100 mM NaCl, and 50 to 300 mM imidazole in 50 mM increments) was applied to the column. Five-hundred-microliter fractions were collected. BRD4S aa444-722 was eluted efficiently at an imidazole concentration of 150 mM. Protein concentrations were determined by Coomassie staining with a bovine serum albumin standard and by Bradford assay. The specificity of the eluted protein was controlled by anti-Flag and anti-His immunoblotting.
GST pull-down assay with SF9 cell lysates or purified proteins.
GST, GST-RING3 B, GST-BRD4S, or GST-ORFX was bound to Sepharose beads as described above. A total of 3.5 × 107 SF9 insect cells infected with a LANAC baculovirus (see above) or noninfected control SF9 cells were washed once in PBS, subsequently lysed in 1 ml of 1% NP-40 lysis buffer (see above) on ice for 10 min, and subsequently cleared by centrifugation (13,000 × g for 1 min). Supernatants of infected as well as uninfected cells were kept as input controls. Eight microliters of supernatant was diluted in 300 μl GST binding buffer (“lysis buffer” with only 0.1% NP-40) and incubated with GST proteins immobilized to Sepharose beads for 1 h at 4°C. Subsequently, the beads were washed seven times in lysis buffer, boiled in 20 μl of SDS electrophoresis sample buffer, and analyzed by SDS-PAGE and immunoblotting for bound LANA protein. The GST pull-down assay with purified GST or GST-LANAC and purified BRD4S aa444-722 as a binding partner was performed as follows. Equal amounts of GST or GST-LANAC were bound to 10 μl glutathione-Sepharose beads for 1 h at room temperature to saturate the GST binding capacity. Beads were washed three times with GST binding buffer to remove unbound protein. Subsequently, purified BRD4S aa444-722 was incubated in a 1:2.5 dilution series (3 μg, 1.2 μg, 480 ng, 190 ng, and 75 ng) in a volume of 300 μl GST binding buffer with the beads for 1 h at room temperature. Beads were washed seven times in GST binding buffer. Samples were analyzed for bound protein by SDS-PAGE and anti-Flag or anti-His immunoblotting. Equal loading was controlled by anti-GST immunoblotting of the stripped membranes.
Direct interaction of purified BRD4S aa444-722 with GST-LANAC as determined by ELISA.
An enzyme-linked immunosorbent assay (ELISA) was established in a 96-well format using Immulon 4 HBX microtiter plates to quantify the direct interaction of purified BRD4S aa444-722 with purified GST-LANAC. Purified GST or GST-LANAC proteins were diluted in coating buffer (150 mM NaHCO3) to a final concentration of 10 μg ml−1 and incubated overnight at 4°C. All incubation steps were carried out in a final volume of 100 μl per well of a 96-well plate. The next day, plates were washed five times with PBS-T (PBS-0.1% [vol/vol] Tween 20) and blocked with 250 μl blocking buffer (PBS-T-5% [wt/vol] fat-free milk powder) for 90 min at 37°C. Following three washes with PBS-T, purified BRD4S aa444-722 was incubated with GST or GST-LANAC in a 1:3 dilution series in binding buffer (50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 0.5 mM EDTA, 0.1% [vol/vol] Nonidet P-40), starting with a concentration of 160 μg ml−1 for 90 min at 37°C. Subsequently, plates were washed five times with PBS-T. To detect bound amino-terminally Flag epitope-tagged BRD4S aa444-722 protein, an alkaline phosphatase-conjugated mouse monoclonal anti-Flag M2 antibody (Sigma) was diluted in blocking buffer to a final concentration of 2 μg ml−1. Wells were incubated with the antibody dilution for 30 min at 37°C. After five washes with PBS-T, 100 μl per well of substrate solution (p-nitrophenylphosphate [1 mg ml−1], diethanolamine [1 M], 0.02% [wt/vol] NaN3, 0.01% [wt/vol] MgCl2, pH 9.8) was incubated for 30 min at 37°C in the dark. The reaction was stopped by adding 50 μl 3 N NaOH. The absorption was measured at a wavelength of 405 nm against the reference wavelength of 450 nm.
Coimmunoprecipitation, immunoblotting, and antibodies.
Coimmunoprecipitation with an anti-green fluorescent protein (GFP) monoclonal antibody (BD Biosciences) was performed as previously described (51). Briefly, lysates of EGFP and LANA-1 or EGFP-BRD4S and LANA-1 coexpressing cells were incubated with anti-GFP-coupled Sepharose A beads. After washing steps, SDS-PAGE, and immunoblotting with the GFP-specific antibody (1:4,000; BD) to detect immunoprecipitated EGFP and EGFP-BRD4S or KS patient serum (1:300) to detect KSHV LANA-1 was performed. The Flag- and His-tagged BRD4S aa444-722 protein produced in insect cells was detected with either a monoclonal mouse anti-Flag antibody (M2, 1:2,000; Sigma) or an anti-tetra-His monoclonal antibody (1:2,000; QIAGEN). GST and GST-fusion proteins were detected with a “homemade” polyclonal anti-GST rabbit antibody (1:5,000) or by Coomassie staining. Endogenous BRD4 proteins were detected with an affinity-purified rabbit anti-BRD4 antibody (directed against the BRD4 N terminus and kindly provided by Peter Howley's laboratory) in a 1:2,000 dilution. Cellular β-actin was detected with a monoclonal anti-actin antibody (used 1:200,000; Chemicon).
The coimmunoprecipitation of endogenous LANA-1 with endogenous BRD4 proteins was performed with KSHV-positive BCBL-1 cells and KSHV-negative BJAB cells as a negative control. A total of 15 μg of the affinity-purified rabbit anti-BRD4 antibody or 15 μg of normal rabbit immunoglobulin G (IgG) (Upstate) was immobilized on 50 μl of protein A agarose beads. Per immunoprecipitation (IP) reaction, 2 × 107 BJAB or BCBL-1 cells were lysed in 1 ml TBS-T (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitors [Roche Complete]) and centrifuged at 16,000 × g for 1 min and supernatants were incubated with the beads overnight, rolling at 4°C. The next day, beads were washed nine times with 1 ml TBS-T, samples were boiled in loading buffer, and 4 to 12% Bis-Tris PAGE was performed, followed by the transfer of proteins to a polyvinylidene difluoride membrane. The immunodetection of proteins was performed as previously described (51) with antibodies as indicated in the figure legends.
Luciferase-based reporter assays.
HEK 293T or HeLa cells were plated at a density of 4 × 105 or 8 × 104 cells per well of a six-well plate, respectively. Twenty hours postseeding, cells were transiently cotransfected with 50 ng of the cyclin E reporter plasmid or the empty reporter plasmid pGL2 basic (Promega) and different amounts of LANA-1 expression constructs or BRD protein expression constructs as described in the figure legends. Forty hours posttransfection, cells were washed with PBS and lysed in reporter lysis buffer (Promega). Luciferase activities were measured in cleared lysates according to the manufacturer's instructions (Promega). Luciferase activity is either given as raw luciferase activity or calculated as a severalfold induction compared to that for mock (empty expression vector)-transfected controls as indicated in the figure legends. Standard deviations were depicted as error bars. Experiments were performed in duplicate or triplicate and repeated independently at least three times.
Flow cytometry-based cell cycle analyses.
A total of 4 × 105 293T cells were seeded per well of a six-well plate. Briefly, 24 h later, 293T cells were cotransfected with 1 μg of the plasmid EGFPC1, EGFPC1-BRD4S, or EGFPC1-BRD2/RING3, together with 1 μg KSHV LANA-1 expression plasmid (GSLANA) or its empty vector pcDNA3. The total amount of transfected DNA was kept constant at 2 μg per well. Forty hours posttransfection, BrdU was added to the medium of the growing population of cells for 1 h to a final concentration of 10 μM. Subsequently, cells were trypsinized, washed in PBS, and washed twice with PBS plus 1% bovine serum albumin (BSA) (wt/vol). Cells were fixed with 4% paraformaldehyde, pH 7.4, for 15 min and permeabilized with 0.1% Triton X-100 in PBS plus 1% BSA at room temperature for 10 min. After a wash in PBS, cells were resuspended in 1 ml of PBS plus RNase A (20 μg/ml, DNAse free) and incubated at 37°C for 30 min. Cells were then subjected to acidic DNA denaturation with 1 ml 2 N HCl plus 0.5% Triton X-100 for 30 min at room temperature, followed by a neutralization step with 1 ml of 0.1 M Na2B4O7, pH 8.5. Subsequently, cells were labeled with a primary mouse monoclonal anti-BrdU antibody (final concentration, 500 ng/ml) (BD Biosciences) in 450 μl of PBS plus 0.5% Tween 20 (vol/vol) plus 1% BSA (wt/vol) for 30 min at room temperature, followed by incubation with a secondary Cy5-conjugated affinity-purified goat anti-mouse IgG (H+L) antibody (final concentration, 15 μg/ml) (Jackson Immuno Research/Dianova) in 100 μl of PBS plus 0.5% Tween 20 (vol/vol) plus 1% BSA (wt/vol) for 30 min at room temperature. After two washing steps with PBS plus 1% BSA, cells were resuspended in PBS containing 100 μg/ml propidium iodide to stain DNA. Flow cytometric cell cycle analysis with a total number of 105 events per sample was performed on a BD FACSCalibur. Raw data were recorded and analyzed using the software Windows Multiple Document Interface for Flow Cytometry 2.8 (WinMDI2.8) available at http://facs.scripps.edu/software.html. Briefly, a first region was defined to gate for EGFP-positive cells, followed by a second gate to exclude cell clusters. For cell cycle analysis, the BrdU content of a cell was quantified in relation to its DNA content.
For cell cycle analyses in BJAB or BCBL-1 cells, 8 × 106 cells were electroporated in duplicate with 15 μg plasmid DNA as described above. Forty hours postelectroporation, cells were BrdU pulsed for 1.5 h and fixed and cell cycle analysis was performed as with 293T cells. A total of 2 × 105 events per sample were analyzed.
RESULTS
BRD4S transcripts were identified in KSHV-positive and KSHV-negative cells.
Having shown previously that KSHV LANA-1 interacts with BRD2/RING (37), we wondered whether other BRD family members (13) (Fig. 1A) might also interact with LANA-1. The human BRD4 gene is located on chromosome 19 and codes for two predicted alternatively spliced transcripts (Fig. 1B). The shorter transcript, BRD4S, codes for a 2,169-bp (722 aa) ORF encompassing 11 exons. The longer transcript, BRD4L, codes for a 4,089-bp (1,362 aa) ORF encompassing 19 exons. In mice, this long BRD4 variant is also referred to as MCAP. Most functional studies relating to BRD4 have so far been concentrated on the long variant and mainly on the mouse protein MCAP (10, 11). To investigate the expression of BRD4S in human cell lines, we performed nested RT-PCR on RNA from KSHV-positive B-cell lines Jsc-1, BCP-1, and CroAP-5 as well as the KSHV-negative, EBV-positive B-cell line Raji, HeLa cells, and peripheral blood lymphocytes. We obtained specific PCR products of the expected molecular size (2,169 bp) from cDNA of all investigated cell lines (not shown), indicating wide expression of BRD4S. The PCR product derived from Raji cells was cloned downstream of a c-myc tag (BRD4S pcDNA3 9E10) and used for later functional studies.
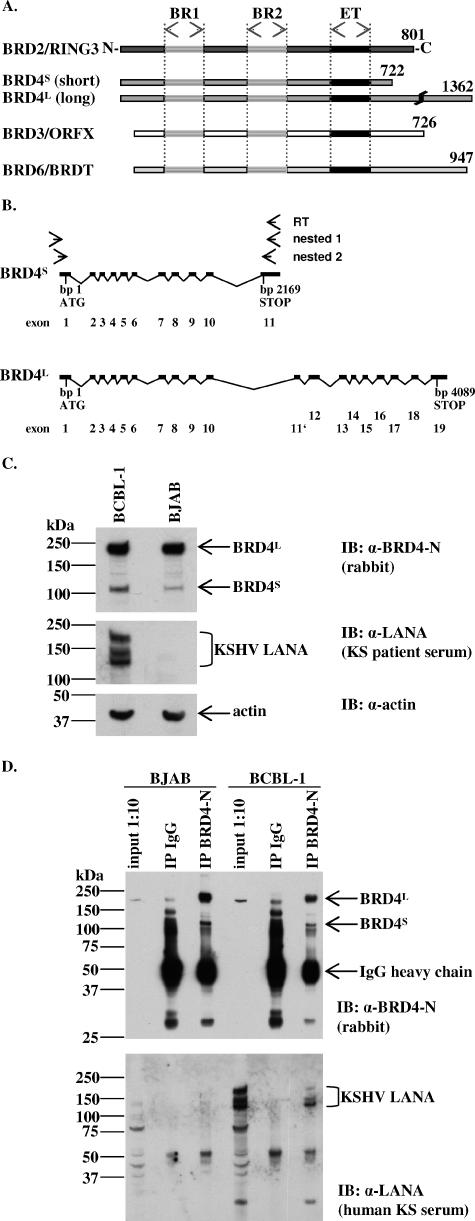
FIG. 1.
KSHV LANA-1 forms complexes with endogenous BRD4 proteins in the PEL cell line BCBL-1 as shown by coimmunoprecipitation. (A) Schematic diagram of human BET/fsh proteins. Amino acid positions of the highly conserved, ∼64 aa long, ET domains of RING3 (aa 640 to 703), BRD4 (aa 608 to 671), ORFX (aa 569 to 633), and BRDT (aa 509 to 571). BR1, bromodomain 1; BR2, bromodomain 2. (B) Schematic depiction of the BRD4S and BRD4L exon-intron structure. BRD4 is localized on human chromosome 19 (19p13.1). BRD4S is composed of 11 exons and gives rise to a 2,169-bp ORF, while BRD4L is composed of 19 exons with an alternative exon (11′) and gives rise to a 4,089-bp ORF. (C) Immunoblots (IB) showing the expression of endogenous BRD4 short and long variant proteins in the KSHV-negative Burkitt's lymphoma cell line BJAB and the PEL cell line BCBL-1. α, anti. (D) Detection of BRD4/LANA complexes in BCBL-1 cells by coimmunoprecipitation (co-IP) using a polyclonal anti-BRD4 antibody to precipitate endogenous BRD4S and BRD4L proteins from BJAB cells (negative control) or BCBL-1 lysates. Normal IgG served as a negative control. Four to 12% Bis-Tris PAGE and immunoblotting (IB) were performed. The upper panel shows a rabbit anti-BRD4 immunoblot. The lower panel shows a blot of KS patient serum to detect LANA in BCBL-1 cells. One-tenth of the lysates used for the co-IP was loaded as input control. With longer exposures, the long and the short forms of BRD4 become visible in the input lanes (compare with panel C, top panel). α, anti.
The BRD4 short and long protein variants (BRD4S and BRD4L) are expressed in KSHV-positive PEL cells.
Since BRD proteins are known to be differentially expressed (43), we wondered whether both BRD4 protein forms would be expressed in KSHV-positive and -negative B cells, a prerequisite for relevant interaction with KSHV or one of its gene products in those cells. Both forms of BRD4, BRD4S and BRD4L, are readily detectable in cell lysates of the KSHV-negative Burkitt's lymphoma cell line BJAB and the PEL cell line BCBL-1. BRD4S migrates at an apparent molecular mass of ∼110 kDa, and BRD4L migrates at an apparent molecular mass of ∼220 kDa. The relative expression level of endogenous BRD4, particularly of the short variant, was moderately higher in BCBL-1 cells compared to that in BJAB cells (Fig. 1C).
KSHV LANA-1 of BCBL-1 cells forms complexes with endogenous BRD4 proteins.
We have recently shown that the C-terminal domain of LANA-1 interacts with a region in BRD2/RING3 that contains the ET domain (37, 51). The ET domain is a conserved sequence element that is characteristic for all members of the BRD family (13) (Fig. 1A). Due to the high degree of amino acid conservation of the ET domain, we wondered whether other BRD family members (Fig. 1A) might also interact with LANA-1. Coimmunoprecipitation studies, in which we immunoprecipitated endogenous BRD4 from BCBL-1 and BJAB cells with a BRD4-specific antibody but not with control IgG (Fig. 1D, upper panel), revealed the existence of a complex in BCBL-1 cells containing LANA-1 and BRD4 (Fig. 1D, lower panel, right lane). Although the normal rabbit IgG control immunoprecipitated many unspecific bands, no LANA-1 was detectable in this control (Fig. 1D), demonstrating the LANA-1 interaction to be specific for BRD4. Since the overall structure of the smaller BRD4S protein is more similar to that of BRD2/RING3 and due to the high degree of conservation of the ET domain, we hypothesized that the protein interaction between LANA-1 and BRD4, and possibly other BRD family members, might take place via the BRD proteins' ET domains (Fig. 1A). Therefore, and because the short variant of BRD4 was uncharacterized until now, we decided to focus our studies on BRD4S. The transfection of different cell lines with our newly generated BRD4S pcDNA3 9E10 construct resulted in the expression of a myc epitope-tagged protein with an apparent molecular mass of 110 kDa by SDS-PAGE (not shown), consistent with the molecular mass of the endogenous BRD4S protein (Fig. 1C, upper panel). In HeLa cells and 293 cells, transfected BRD4S showed an intranuclear localization (data not shown).
Notably, BRD4S protein was detectable with a BRD4 monoclonal antibody raised against the C terminus of BRD4S as well as with a BRD2/RING3 polyclonal antibody raised against the C terminus of RING3 (33, 37), indicating antigenic cross-reactivity of the polyclonal RING3 antiserum used in previous studies (33, 37) with BRD4.
KSHV LANA-1 interacted with BRD4S and BRD3/ORFX proteins via the carboxy-terminal part of BRD proteins, including the ET domain.
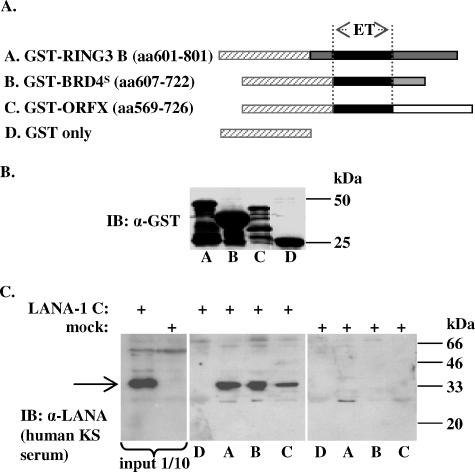
To investigate the interaction of LANA-1 with BRD4 and possibly other members of the BRD family in detail, we employed ectopic expression strategies. We expressed the C-terminal regions of BRD2/RING3, BRD4S, and BRD3/ORFX, including their respective ET domains, as GST-fusion proteins in bacteria (Fig. 2B) to perform in vitro GST-fusion protein binding assays (Fig. 2C). GST-RING3 B (aa 601 to 801), GST-BRD4S (aa 607 to 722), and GST-ORFX (aa 569 to 726) fusion proteins or GST alone (schematically depicted in Fig. 2A) was bound to glutathione-Sepharose beads to pull down the C-terminal domain of LANA-1 expressed in SF9 insect cells. We observed binding of the C-terminal domain of LANA-1 to GST-RING3 B, GST-BRD4S, and GST-ORFX, but not to GST (Fig. 2C).
FIG. 2.
KSHV LANA-1 interacts with the C termini of BRD4S and BRD3/ORFX in vitro in GST pull-down assays. (A) Schematic representation of the GST-BET fusion proteins used in panels B and C. GST-RING3 B (aa 601 to 801), GST-BRD4S (aa 607 to 722), and GST-ORFX (aa 569 to 726) all contain the carboxy-terminal region of the respective BET protein. (B) Expression levels of GST fusion proteins used in panel C and as schematically depicted in panel A determined by anti-GST immunoblotting (IB). α, anti. (C) GST pull-down experiment showing the interaction of the KSHV LANA-1 carboxyterminal domain expressed in SF9 insect cells (arrow) with GST fusion proteins GST-RING3 B (middle panel, lane A), GST-BRD4S (middle panel, lane B), and GST-ORFX (middle panel, lane C) but not with GST alone (middle panel, lane D). The interaction of LANA-1 C with GST-RING3 B (37) served as positive control. One-tenth of the lysates used for the interaction assay were used as input control (left panel). +, presenceof.
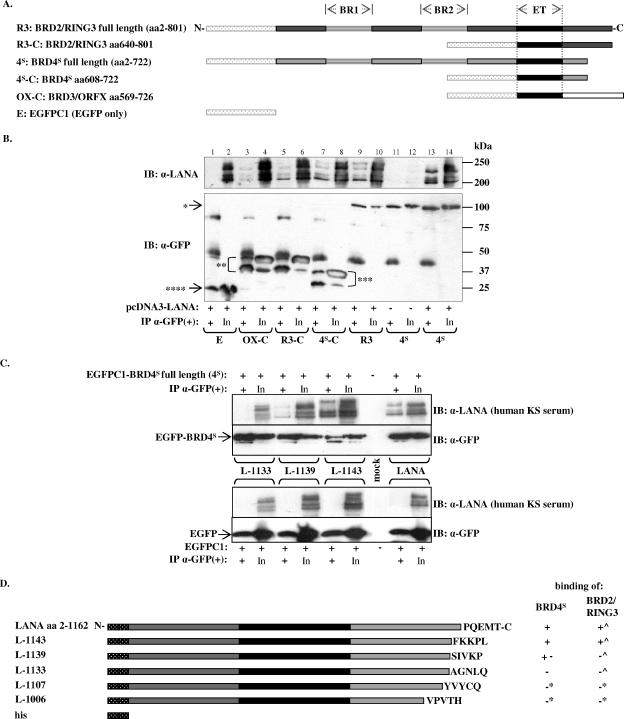
To confirm and extend this observation with proteins expressed in mammalian cells, we cotransfected epithelial HEK 293T cells with a full-length LANA-1 expression plasmid (GSLANA) and one of five different EGFP-tagged BET protein fusion constructs, full-length BRD2/RING3 and BRD4S, deletion construct BRD2/RING3 (aa 640 to 801), BRD4S (aa 608 to 722), or BRD3/ORFX (aa 569 to 726), all encompassing the C-terminal regions of the respective protein, including the ET domains (Fig. 3A). As a control, HEK 293T cells were cotransfected with GSLANA and EGFPC1. EGFP-fusion proteins or EGFP alone was immunoprecipitated with an anti-GFP antibody. Subsequent SDS-PAGE and immunoblotting with an anti-GFP antibody revealed efficient immunoprecipitation of all EGFP-fusion proteins and EGFP alone (Fig. 3B, lower panel). BRD2/RING3 and BRD4S full-length constructs had apparent molecular masses of ∼120 kDa as expected. BRD2/RING3 aa 640 to 801, BRD4S aa 608 to 722, and BRD3/ORFX aa 569 to 726 had apparent masses of 50 kDa, 38 kDa, and 51 kDa, respectively. BRD2/RING3 aa 640 to 801, BRD4S aa 608 to 722, and BRD3/ORFX aa 569 to 726 showed additional smaller bands that were also efficiently immunoprecipitated with the anti-GFP antibody (Fig. 3B, lower panel). The molecular nature of these lower-molecular-mass bands is not known.
FIG. 3.
KSHV LANA-1 coimmunoprecipitates with BRD4S and BRD3/ORFX in cotransfected cells. (A) Schematic diagram of the EGFP fusion proteins of full-length BRD2/RING3 and BRD4S and the C-terminal regions of BRD2/RING3, BRD4S, and BRD3/ORFX used in this experiment. (B) Coimmunoprecipitation experiments demonstrating the interaction of full-length LANA-1 with different EGFP-BRD fusion proteins. 293T cells were cotransfected with a full-length LANA-1 expression construct and one of the EGFP-tagged BRD expression constructs as depicted in panel A. Immunoprecipitation was performed using an anti (α)-GFP monoclonal antibody. After extensive washing, SDS-PAGE and immunoblotting (IB) were performed by probing the lower part of the membrane with an anti-GFP antibody (lower panel) to monitor the precipitation of EGFP-fusion proteins BRD2/RING3 (*, “R3”) and BRD4S (*, “4S”) or the carboxy-terminal domains of BRD2/RING3 (**, “R3-C”), BRD4S (***, “4S-C”), or BRD3/ORFX (**, “OX-C”) or EGFPC1 (****, “E”). The carboxy-terminal domains of the BRD proteins were consistently detected as two distinct bands (double and triple asterisks). The upper part of the membrane was probed with human KS patient serum to detect coimmunoprecipitated LANA-1 (upper panel). One-tenth of the lysate used for the IP is shown next to each IP lane as input (In). Bands corresponding to the immunoglobulin heavy chains are visible in all IP lanes around 50 kDa. +, presence of; −, absence of. (C) Coimmunoprecipitation experiments investigating the binding of LANA-1 full-length and carboxy-terminally truncated LANA-1 mutants L-1143, L-1139, and L-1133 to EGFP-tagged BRD4S (“4S”) full-length protein (upper panel) or EGFP alone (lower panel). +, presence of; −, absence of. IB, immunoblot; In, input; α, anti. (D) Schematic representation of C-terminally truncated LANA-1 mutants as used in panel C and a summary of their binding properties to BRD4S (this study) and BRD2/RING3 (^) (51). *, Binding data for L-1107 and L-1006 are not shown. The carboxy-terminal five amino acids of the respective LANA-1 construct are depicted.
As shown in Fig. 3B (upper panel), KSHV LANA-1 was efficiently coimmunoprecipitated with full-length BRD4S and full-length BRD2/RING3. Further, LANA-1 coimmunoprecipitated with BRD2/RING3 aa 640 to 801, BRD4S aa 608 to 722, and BRD3/ORFX aa 569 to 726 but not with the EGFP protein alone (Fig. 3B, upper panel). Reproducibly, the LANA-1-specific signal was strongest in the coimmunoprecipitations with full-length BRD4S and moderately weaker with full-length BRD2/RING3 and BRD4S aa608-722 proteins. Relatively weak LANA-1 signals were detectable with BRD2/RING3 aa 640 to 801 and BRD3/ORFX aa 569 to 726 (Fig. 3B). Further, LANA-1 signals were reproducibly stronger when coimmunoprecipitated with full-length BRD4S than when coimmunoprecipitated with BRD4S aa 608 to 722. Similarly, LANA-1 coimmunoprecipitated more efficiently with full-length BRD2/RING3 than with BRD2/RING3 aa 640 to 801. These differences in the efficiency of coimmunoprecipitation when comparing full-length proteins with constructs starting at the ET domain suggest the presence of a second LANA-1 binding site upstream of the ET domain of BRD2 and BRD4, as we have previously reported for BRD2/RING3 (37).
The highly conserved C-terminal domain of LANA-1 was indispensable for the binding to BRD4, resembling the interaction pattern of LANA-1 with BRD2/RING3.
To identify regions in LANA-1 that are required for the in vivo interaction in transfected cells with BRD4S, EGFP-tagged BRD4S or EGFP was transiently coexpressed with full-length LANA-1 or the carboxy-terminally truncated KSHV LANA-1 mutant LANA Δ1161-1162 (L-1160), LANA Δ1144-1162 (L-1143), LANA Δ1140-1162 (L-1139), LANA Δ1134-1162 (L-1133), LANA Δ1129-1162 (L-1128), LANA Δ1108-1162 (L-1107), LANA Δ1056-1162 (L-1055), or LANA Δ1007-1162 (L-1006) (Fig. 3C and D). We used an anti-GFP antibody to immunoprecipitate EGFP-tagged BRD4S or EGFP from 293T lysates. As expected, full-length LANA-1 (aa 2 to 1162) coimmunoprecipitated with BRD4S (Fig. 3C, upper panel, second lane from right) but not the EGFP control (Fig. 3C, lower panel). Further, LANA mutants L-1160 (not shown), and L-1143 coimmunoprecipitated with BRD4 but not EGFP (Fig. 3C). L-1139 showed residual binding to BRD4S (Fig. 3C). No shorter LANA-1 constructs, namely L-1133 (Fig. 3C), L-1128, L-1107, L-1055, and L-1006 (not shown), coimmunoprecipitated with BRD4S. Interestingly, this loss of LANA-1 BRD4 interaction when deleting LANA-1 from the carboxy-terminal end by 29 amino acids or more almost exactly resembles the binding pattern of KSHV LANA-1 with BRD2/RING3 (51). These data clearly demonstrate that deleting the C-terminal 29 amino acids of KSHV LANA-1 (LANA mutant L-1133) results in a complete loss of BRD2/RING3 binding (51) as well as in a complete loss of BRD4S binding (present study) in transfected cells. Notably, the same LANA-1 mutants that are no longer capable of binding to BRD2/RING3 or BRD4S cannot act as transcriptional activators, as assayed by the use of the human cyclin E promoter in luciferase-based reporter assays (51). Further, LANA Δ1129-1162 (L-1128) or shorter mutants no longer interact with chromatin (50).
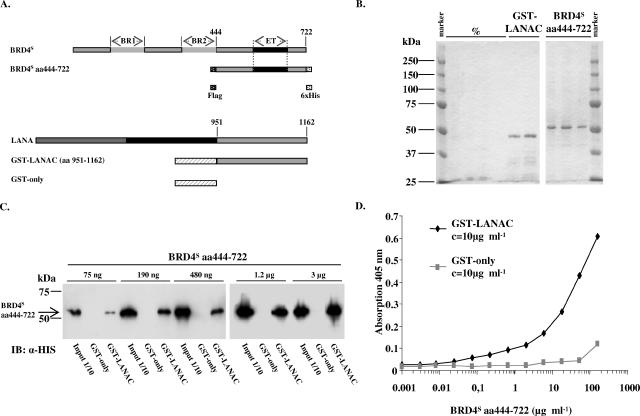
Purified BRD4S aa444-722 and purified GST-LANAC interact directly.
So far, our findings show the carboxy-terminal region of BRD4S as well as the KSHV LANA-1 carboxy-terminal region to be involved in the interaction between the two proteins but do not exclude an indirect interaction of BRD4 and LANA-1 via one or more cellular proteins. To address this question, we purified both proteins and subsequently investigated their interaction. The carboxy-terminal LANA-1 fragment (aa 951 to 1162) was expressed as a GST fusion protein (GST-LANAC) in bacteria and affinity purified via the interaction of GST with glutathione G-Sepharose beads. The carboxy-terminal fragment of BRD4S (aa 444 to 722) was expressed in insect cells with the help of a newly generated recombinant baculovirus and affinity purified via the hexahistidine tag with a Ni2+ affinity column. The proteins used for this direct interaction study are schematically depicted in Fig. 4A. As shown on the Coomassie-stained gels in Fig. 4B, the ∼46-kDa GST-LANAC protein and the ∼53-kDa BRD4S aa444-722 protein were obtained at high purity. These purified proteins and purified GST protein alone were now used for a semiquantitative GST pull-down assay (Fig. 4C). We immobilized constant amounts of either GST or GST-LANAC and incubated them with increasing amounts of BRD4S aa444-722. BRD4S aa444-722 bound to GST-LANAC but not to the GST control. We performed the assay with small bead volumes, which led to a saturation of the system using high amounts of BRD4 (3 μg and 1.2 μg). Reducing the amount of BRD4 resulted in a dose-dependent decrease of the amount of BRD4 which bound to LANA-1 (Fig. 4C). We further established an ELISA-based binding assay using the same purified proteins. Constant amounts of GST or GST-LANAC were immobilized in 96-well plates and incubated with defined amounts of BRD4S aa444-722. As depicted in Fig. 4D, BRD4S aa444-722 specifically interacted with GST-LANAC in a dose-dependent manner but not with GST alone. These results show the interaction of the KSHV LANA-1 C terminus with the carboxy-terminal part of BRD4S (aa 444 to 722) to be direct.
FIG. 4.
Purified BRD4S aa444-722 and purified KSHV LANA-1 C-terminal domain directly interact in a GST pull-down assay and in an ELISA. (A, upper panel) Schematic depiction of the full-length BRD4S with its two bromodomains (BR1 and BR2) and the ET domain in comparison with the recombinant BRD4S fragment (amino acids 444 to 722) with an N-terminal Flag tag and a C-terminal six-histidine tag expressed in SF9 cells using a recombinant baculovirus. (Lower panel) Schematic representation of full-length KSHV LANA-1 and GST-LANAC protein representing the C-terminal domain of LANA-1 with amino acids 951 to 1162 N-terminally fused to GST. (B) Coomassie-stained SDS-PAGE gels depicting selected elution fractions of glutathione-Sepharose G affinity-purified recombinant GST-LANAC (left panel, right two lanes) expressed in E. coli or the Ni2+ affinity-purified recombinant BRD4S aa444-722 protein expressed in SF9 insect cells (right panel). The shown elution fractions of GST-LANAC and BRD4S aa444-722 were pooled, protein concentrations were determined, and these proteins and purified GST protein alone (not shown) were used for the GST pull-down (C) and ELISA (D) experiments. %, empty lanes. (C) Semiquantitative GST pull-down experiment with purified proteins from panel B and as depicted in panel A. Identical amounts of purified GST or GST-LANAC proteins were immobilized on glutathione 4B beads for 2 h at 4°C, washed, and incubated for 1 h at room temperature with increasing amounts of purified BRD4S aa444-722 protein as indicated in the figure. After extensive washing, SDS-PAGE and immunoblotting (IB) with an anti (α)-His antibody were performed. One-tenth of the input amounts were loaded as input control. Lanes were assembled from two gels (left and right). The figure shows one representative of three independent experiments. (D) ELISA with a fixed concentration of purified GST or GST-LANAC coated onto ELISA plates as described in Material and Methods. After washing steps, coated proteins were incubated with increasing concentrations of purified BRD4S aa444-722 protein. Unbound protein was washed off, and bound BRD4S aa444-722 was detected by using an anti-Flag monoclonal antibody and a secondary alkaline phosphatase-conjugated antibody. The figure shows one representative experiment out of four.
BRD2/RING3 and BRD4S induce cell cycle arrest in HeLa cells.
After establishing the direct interaction between KSHV LANA-1 and BRD4, in addition to the previously described interaction of KSHV LANA-1 with BRD2/RING3 (37, 51), we sought to address the possible functional implications of these interactions.
The longer BRD4 splice variant MCAP identified in mice codes for a protein of 1,401 aa, which is chromatin associated during mitosis and, when ectopically expressed, delays the cell cycle in the transition from G1 to S phase (32). Further, cells injected with a BRD4/MCAP-specific antibody showed a higher proportion of cells in G2/M, pointing to a role for BRD4/MCAP in G2/M transition and/or mitosis (11). The long human BRD4 variant BRD4L is identical to the short variant BRD4S in the amino-terminal part, including both bromodomains and the ET domain (Fig. 1A), but in addition has a carboxy-terminal tail (about 640 amino acids long) of unknown physiological function. The second bromodomain of the murine BRD4/MCAP and its direct upstream region have been shown to be critical for the BRD4/MCAP-induced cell cycle block (32). For BRD2/RING3, there is evidence of an impact on the expression of E2F-dependent promoters (9) and some evidence suggests BRD2/RING3 to be involved in the recruitment of E2F to the cyclin A promoter, thereby possibly promoting the cell cycle in fibroblasts under certain conditions but delaying it under other conditions (46). However, BRD4S has so far not been investigated.
We tested a possible role of the human BRD4S and BRD2/RING3 proteins in cell cycle regulation using BrdU (an artificial thymidine analog) incorporation assays. In an initial experiment, we found that transiently transfected HeLa cells were effectively prevented from entering the S phase of the cell cycle by the overexpression of EGFP-tagged BRD4S and BRD2/RING3 but not by BRD4S aa 608 to 722, BRD2/RING3 aa 640 to 801, or EGFP alone (data not shown).
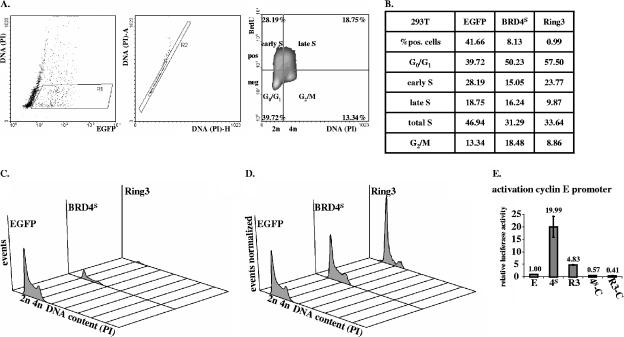
BRD4S and BRD2/RING3 increase the proportion of cells in G1 phase in epithelial 293T cells and decrease the S-phase population.
To characterize the impact of BRD4S and BRD2/RING3 on the cell cycle in more detail, we established a fluorescence-activated cell sorter-based cell cycle assay that enabled us to determine the proportion of cells in each phase of the cell cycle. We transfected 293T cells with an EGFP expression vector or with expression vectors for EGFP-RING3 or EGFP-BRD4S and analyzed the cell cycle distribution of the transgene-expressing cells. Forty hours posttransfection, cells transfected with the EGFP plasmid were identified at a proportion of ∼40% in G1/G0 phase, ∼28% in early S phase, ∼19% in late S phase, and ∼13% in G2/M phase (Fig. 5A). Nontransfected, EGFP-negative cells showed cell cycle distributions similar to those of EGFP-positive cells (not shown), implying that the EGFP plasmid does not affect the cell cycle distribution of 293T cells. In cells transfected with EGFP-BRD2/RING3 and EGFP-BRD4S, the proportion of EGFP-expressing cells was much lower than that in EGFP-transfected cells (Fig. 5B and C). While ∼42% of the cells in the EGFP-transfected sample were EGFP positive, only ∼8% of cells transfected with EGFP-BRD4S were EGFP-BRD4S positive and ∼1% of cells transfected with EGFP-RING3 were EGFP-RING3 positive. Assuming that the transfection efficiency was similar with the three different plasmids, these data suggest that the expression of BRD4S or BRD2/RING3 can efficiently slow down or arrest the proliferation of the transfected cell. Thereby, the proportion of EGFP-BRD4S or EGFP-RING3-positive cells would decline over time after transfection in relation to the population of untransfected cells that continue to proliferate. Supporting this notion, when we analyzed cells at earlier time points (20 h after transfection), we observed ∼42% of positive cells in the EGFP sample, ∼25% of positive cells in the EGFP-BRD4S sample, and ∼15% of positive cells in the EGFP-RING3 sample (not shown).
FIG. 5.
BRD4S and BRD2/RING3 induce G1/G0 cell cycle arrest and increase the activity of the cyclin E promoter in 293T cells. (A to D) Cell cycle analysis of EGFP (“E”), EGFP-BRD4S (“4S”), or EGFP-RING3 (“R3”) expressing cells using flow cytometry. Briefly, 293T cells in six-well plates were transfected with 1 μg of the respective expression plasmid and 1 μg of pcDNA3. Forty hours posttransfection, cells were BrdU pulsed for 1 h by adding BrdU to a final concentration of 10 μM. Subsequently, cells were paraformaldehyde fixed, permeabilized, and labeled with a primary BrdU-specific antibody and a secondary Cy5-conjugated antibody. Finally, cells were resuspended in PBS containing propidium iodide (PI) and flow cytometric analysis was performed. A total of 105 cells were measured in each sample by using a BD FACSCalibur. Data were analyzed using the software Windows Multiple Document Interface for Flow Cytometry 2.8 (WinMDI2.8). Four independently performed experiments showed similar results. pos, positive; neg, negative. (A) Example of gating procedure and cell cycle analysis for 293T cells transfected with EGFPC1. (A, left panel) Dot plot depicting 2,000 out of a total of 105 analyzed EGFP-transfected cells with the EGFP signal on the x axis (logarithmic scale) and the DNA content of the cells on the y axis (linear scale). With the gate R1, EGFP-positive cells with a DNA content ≥2n (diploid) and ≤4n (tetraploid) were included in subsequent analyses. (Middle panel) Dot plot. With the gate R2, exclusive analysis of single cells was assured. (Right panel) Cells within gates R1 and R2 were analyzed for BrdU positivity (y axis, log scale) in relation to their DNA content (x axis, linear scale) depicted as density plot. Quadrants were defined as shown and differentiate cells in the depicted cell cycle states. The percentage of cells in each individual cell cycle state is listed in each quadrant. (B) Table summarizing the analysis as outlined in panel A for EGFP-, EGFP-BRD4S-, or EGFP-RING3-expressing cells. (C) Histogram with the DNA content on the x axis and the number of cells on the y axis for EGFP-, EGFP-BRD4S-, or EGFP-RING3-expressing cells. (D) Same data as depicted in panel C, normalized to the total number of EGFP-positive cells. (E) Luciferase-based reporter assay. 293T cells in six-well plates were cotransfected with 50 ng of a human cyclin E promoter pGL2 basic reporter plasmid, together with 1 μg of the respective expression construct as depicted in Fig. 3A. Forty-eight hours posttransfection, cells were lysed and lysates were assayed for luciferase activity. Activities are depicted as relative activities relative to the empty vector EGFPC1 control. One representative of four independent experiments performed in duplicate is shown. Standard deviations (error bars) are depicted. No activation was observed with pGL2 basic as control reporter vector (data not shown).
We next looked at the cell cycle distribution of the transfected cells (Fig. 5B, C, and D). Around 40% of EGFP-expressing cells were in G1/G0 phase, whereas ∼50 and ∼58% of cells were found to be in G1/G0 phase in the EGFP-BRD4S and EGFP-RING3-positive cells, respectively. Accordingly, ∼47% of EGFP-positive cells were in S phase, in contrast to a significantly smaller proportion, ∼31 and ∼34%, respectively, of EGFP-BRD4S and EGFP-RING3-positive cells. In addition, a larger proportion, ∼18%, of BRD4S-positive cells were in G2 relative to ∼13% in the EGFP control. In contrast, BRD2/RING3 expression did not increase the fraction of cells in G2 (∼9%).
These data indicate that transient expression of BRD4S and BRD2/RING3 in a rapidly proliferating cell population causes a G1/G0 arrest under the conditions of our assay, with RING3 being more efficient than BRD4S. In line with the G1/G0 arrest, a smaller proportion of cells were in S phase in BRD4S- and BRD2/RING3-expressing cells compared to that for EGFP-expressing cells.
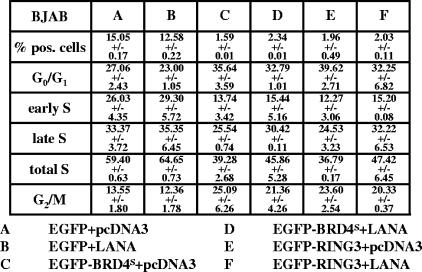
BRD4S and BRD2/RING3 increase the proportion of cells in G1 phase in the B-cell line BJAB and decrease the S-phase population.
When we performed the cell cycle assay in BJAB cells with a 1.5-h BrdU pulse, observations similar to those with 293T cells were made. In the EGFP control population, 27% of cells were in G1/G0 phase, while 59% were in S phase (see Fig. 8A). The expression of BRD4S and RING3 increased the G1/G0 population from 27 to 36 and 40%, respectively, while the fraction of cells in S phase decreased from 59 to 39% in BRD4S-positive cells and to 37% in RING3-positive cells (see Fig. 8C and E). Notably, BRD4S and RING3 expression resulted in more cells in G2/M in BJAB cells, while in 293T cells, only BRD4S increased this fraction (see Fig. 8C and E and compare with Fig. 5B). In BJAB cells, the electroporation of BRD4S and RING3 resulted in lower numbers of transgene-expressing cells compared to that in EGFP control cells, most likely reflecting a growth disadvantage of the cells overexpressing either BRD4S or RING3, as described above for 293T cells (Fig. 5 and see Fig. 8). Therefore, as in 293T cells, BRD4S or RING3 overexpression results in a delay in G1/S transition in BJAB cells.
FIG. 8.
KSHV LANA-1 releases BJAB cells from the BRD4S- or the BRD2/RING3-induced G1 cell cycle arrest. BJAB cells were electroporated with 7.5 μg plus 7.5 μg of the plasmids as indicated in the legend to Fig. 7 (samples A, B, C, D, E, and F). Flow cytometric cell cycle analysis was performed as described in the legend for Fig. 5 and Material and Methods. Briefly, 40 h postelectroporation, BJAB cells were pulsed with 10 μM BrdU for 1.5 h before fixing and labeling. A total of 2 × 105 cells was analyzed per sample, and the cell cycle data are given. The numbers represent means ± standard deviations of two independent experiments.
BRD2/RING3 and BRD4S transactivate the human cyclin E promoter.
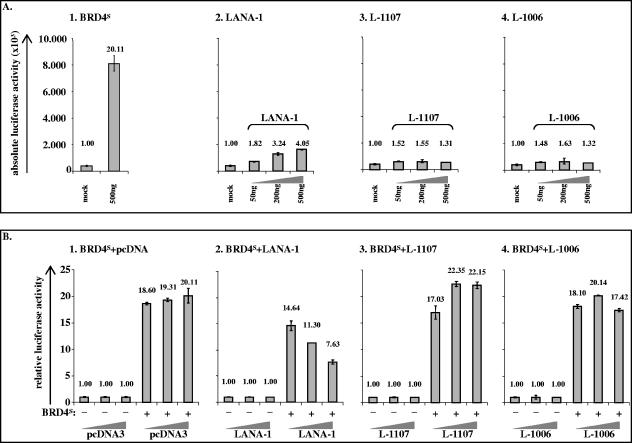
Cyclin E is a critical player at the G1/S transition of the eukaryotic cell cycle with particularly high promoter activity and an increase of cyclin E transcription in late G1 and early S phase (19). BRD2/RING3 has been suggested to transactivate cell cycle promoters, including the cyclin E promoter, in an E2F-dependent manner (9). Since our cell cycle data show that BRD4S and BRD2/RING3 delay the transition from G1 to S phase, we were interested in studying whether BRD2/RING3 and BRD4S expression would have an effect on cyclin E promoter activity. We performed luciferase-based reporter assays in 293T and HeLa cells with transient expression of the EGFP-tagged BET protein constructs (Fig. 3A) used in the BrdU incorporation assays in HeLa, 293T, and BJAB cells (Fig. 5 and see Fig. 8; data not shown). The EGFP-tagged BRD4S full-length protein strongly activated the cyclin E promoter in 293T and HeLa cells (Fig. 5E and data not shown) approximately 20-fold, similar to the myc-tagged BRD4S protein (Fig. 6A1). Highly reproducibly, the BRD2/RING3 protein also activated the cyclin E promoter around fivefold (Fig. 5E and data not shown). The activation of the cyclin E promoter by BRD2/RING3 was in line with a previously reported four- to fivefold activation of the cyclin E promoter by BRD2/RING3 (9). In contrast, BRD4S aa 608 to 722 and BRD2/RING3 aa 640 to 801 did not activate the cyclin E reporter construct significantly (Fig. 5E and data not shown). Protein expression levels were controlled by SDS-PAGE and immunoblotting and revealed comparable levels of expression for BRD4S, BRD2/RING3, BRD4S aa 608 to 722, and BRD2/RING3 aa 640 to 801 (not shown). A series of BRD4S and BRD2/RING3 expression constructs tagged with an N-terminal myc epitope instead of EGFP behaved identically with regard to the transactivation of the human cyclin E promoter in both tested cell lines HeLa and HEK 293T (Fig. 6A, panel 1, and data not shown), while neither BRD2 nor BRD4S with a carboxy-terminal EGFP tag was functional in these assays (not shown).
FIG. 6.
KSHV LANA-1 decreases BRD4S-induced cyclin E promoter activity. Luciferase based reporter assay in HEK 293T cells. (A) HEK 293T cells in six-well plates were transfected with 50 ng of reporter plasmid (human cyclin E promoter) and 500 ng of expression plasmid for the myc-tagged BRD4S full-length protein (1) or different amounts of KSHV LANA-1 full-length (2), LANA Δ1108-1162 (L-1107) (3) or LANA Δ1007-1162 (L-1006) (4) expression constructs and lysed at confluence 48 h later. The total amount of DNA per transfection was adjusted to 1.05 μg using salmon sperm DNA. For details see Material and Methods. Absolute luciferase activities are shown with standard deviations (error bars). Additionally, mean relative activities are given as numbers on top of each column. (B) HEK 293T cells were cotransfected with 50 ng of reporter plasmid (human cyclin E promoter) and constant amounts of 500 ng of either empty vector pcDNA3 (mock) or BRD4S full-length expression plasmid in pcDNA3, together with increasing amounts (50 ng, 200 ng, and 500 ng) of pcDNA3 (1), full-length KSHV LANA-1 (2), LANA Δ1108-1162 (L-1107) (3), or LANA Δ1007-1162 (L-1006) (4) expression constructs, and lysed at confluence 48 h later. Mean luciferase activities in the absence of BRD4S expression were set at 1 for each amount (50 ng, 200 ng, or 500 ng) of the ORF 73 constructs or pcDNA3 (left three bars in panels 1, 2, 3, and 4). The relative activities in the presence of BRD4S are shown in the three right bars in panels 1, 2, 3, and 4. Mean relative luciferase activities are depicted and given as numbers on top of each column. Standard deviations (error bars) are shown. +, presence of; −, absence of.
Coexpression of KSHV LANA-1 reduced the ability of BRD4S to induce cyclin E promoter activity.
We reported earlier that KSHV LANA-1 induces cyclin E promoter activity, whereas carboxy-terminal LANA-1 mutants shorter than L-1133 do not (51). We wondered whether LANA-1 and BRD4S would influence each other with regard to their abilities to activate the cyclin E promoter. We cotransfected HEK 293T cells with the cyclin E reporter construct, together with one of the expression constructs for BRD4S, KSHV LANA-1, L-1107, or L-1006. BRD4S activated the human cyclin E promoter 20-fold, as expected (Fig. 6A, panel 1, compare with 5E). The full-length LANA-1 protein activated the cyclin E promoter dose dependently up to fourfold, whereas the LANA-1 mutants L-1107 and L-1006 did not activate the cyclin E promoter, as expected (Fig. 6A). However, when coexpressing BRD4S, together with different amounts of KSHV LANA-1, L-1107, or L-1006, we found that LANA-1 reduced the BRD4S-induced cyclin E activation in a dose-dependent manner (Fig. 6B, panel 2), whereas the LANA-1 mutants L-1107 and L-1006, which are defective with respect to several LANA-1 functions, including the interaction with BRD2/RING3 (50, 51) and BRD4S (Fig. 3C and D), did not show this inhibition (Fig. 6B, panels 3 and 4). The observed phenotype was not due to lower BRD4S expression levels in the presence of LANA as controlled by Western blotting (Fig. 3B, lower panel, compare lanes 12 and 14, and data not shown).
KSHV LANA-1 partially compensated a BRD2/RING3- or BRD4S-induced G1 cell cycle arrest in 293T and BJAB cells.
Physiologically, cyclin E promoter activity increases at the G1/S transition (19). The activation of the cyclin E promoter by BRD4S and BRD2/RING3 could therefore be linked to their abilities to prevent entry into the S phase (Fig. 5 and see Fig. 8; data not shown), rather than to a direct effect on the cyclin E promoter. We hypothesized that KSHV LANA-1, which has previously been shown to promote S-phase entry in cell cycle-arrested cells (1, 16) and to provide a growth advantage to HeLa or Rat1 cells stably expressing LANA-1 (45), might have released cells from the BRD4S-induced cell cycle arrest at G1/S and that, subsequently, cyclin E promoter activity would be decreased.
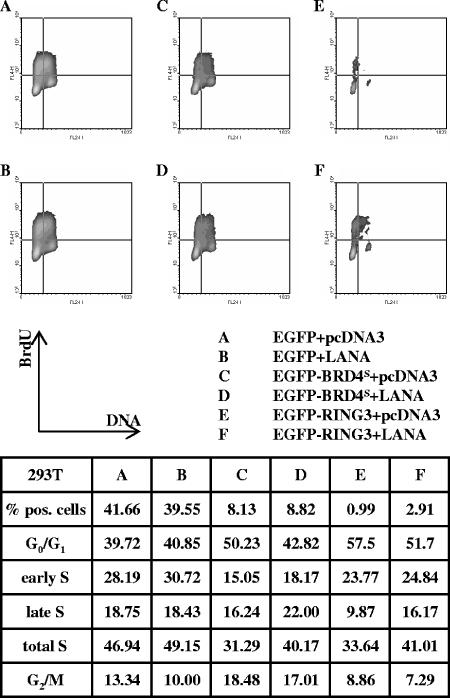
To test this hypothesis, we studied the cell cycle distributions of 293T cells as well as those of BJAB cells transiently expressing KSHV LANA-1 with or without BRD4S and BRD2/RING3 (Fig. 7 and 8). To define a possible effect of LANA-1 expression alone, 293T cells were cotransfected and BJAB cells electroporated with an EGFP expression plasmid, together with a LANA-1 construct or the empty vector (pcDNA3) (Fig. 7A and B and 8A and B). The cell cycle analysis was performed as for Fig. 5. The expression of KSHV LANA-1 increased the proportion of EGFP-transfected cells in S phase moderately in 293T cells (49.15 versus 46.94% in the control population) or BJAB cells (64.65 versus 59.40%). The proportion of EGFP-positive cells in the total population was not increased by LANA-1 (41.66% [15.05%] in the control cells versus 39.55% [12.58%] in the LANA-1-transfected 293T [BJAB] cells). Therefore, the proliferation of LANA-1 transiently expressing cells was not increased relative to the nontransfected cells of the same sample during the short time interval (40 h) of our assay. Notably, in our experimental setting, the cells are nonsynchronized and proliferate rapidly, making it easier to detect a growth-delaying effect as observed with BRD4S and BRD2/RING3 (Fig. 5 and 8A, C, and E) than a growth-promoting effect. LANA-1 did not significantly affect the relative expression levels of EGFP, EGFP-BRD4S, or EGFP-BRD2/RING3 at the single-cell level as assessed by flow cytometry (not shown) or the bulk population as regularly controlled by Western blotting (Fig. 3B, lower panel, compare lanes 12 and 14; data not shown).
FIG. 7.
KSHV LANA-1 releases 293T cells from the BRD4S- or the BRD2/RING3-induced G1 cell cycle arrest. Flow cytometric cell cycle analysis was performed as described in the legend for Fig. 5 and Material and Methods. Cells were cotransfected with equal amounts (1 μg each) of EGFPC1 and pcDNA3 (A), EGFPC1 and a LANA-1 expression construct in pcDNA3 (LANA-pcDNA3) (B), EGFPC1-BRD4S and pcDNA3 (C), EGFPC1-BRD4S and LANA-pcDNA3 (D), EGFPC1-RING3 and pcDNA3 (E), or EGFPC1-RING3 and LANA-pcDNA3 (F). Briefly, 40 h posttransfection, 293T cells were pulsed with 10 μM BrdU for 1 h before fixing and labeling the cells. The upper part depicts density blots comparing EGFP-signal-positive cells from samples A, B, C, D, E, and F. DNA content measured as propidium iodide signal is depicted on the x axis. The y axis represents the BrdU signal. A total of 105 cells was analyzed per sample, and the cell cycle data are given in the table. The density blots are generated based on 2,000 analyzed cells of the initial population. The assay was repeated independently three times with similar results. pos, positive.
Comparing EGFP-BRD4S and LANA-1 cotransfected cells (Fig. 7D) with cells transfected with EGFP-BRD4S and empty vector (Fig. 7C), we observed fewer 293T cells in G1/G0 in the presence of LANA-1 (∼43 versus ∼50%) and more cells in S phase in the presence of LANA-1 (∼40 versus ∼31%). This finding indicates that LANA-1 shifted a proportion of EGFP-BRD4S-expressing cells from the G1/G0 to the S phase of the cell cycle. Likewise, LANA-1 coexpression with BRD2/RING3 increased the proportion of cells in S phase (∼41% with LANA-1 compared to ∼34% with RING3 alone) while reducing the proportion of cells in G1/G0 phase (∼52% with LANA-1 compared to ∼57% with RING3 alone). In addition, the proportion of cells expressing EGFP-RING3 was almost tripled in EGFP-RING3 and LANA-1 cotransfected cells (Fig. 7F) relative to that in cells transfected with EGFP-RING3 and a control plasmid (2.91 versus 0.99%). Similarly, LANA-1 increased the S-phase proportion of BJAB cells expressing EGFP-BRD4S or EGFP-RING3 from 39.28 to 45.86% and 36.79 to 47.42%, respectively (Fig. 8).
These observations show that LANA-1 can promote S-phase entry, even in the presence of overexpressed BRD2/RING3 or BRD4S, which in the absence of LANA-1 significantly delays S-phase entry. Our data are therefore in keeping with the notion that LANA-1 releases cells from a BRD2/RING3- or BRD4S-induced cell cycle arrest in epithelial 293T cells as well as in BJAB cells.
Overexpression of BRD4S in BCBL-1 cells still induced a G1 cell cycle arrest.
We wondered whether the overexpression of BRD4S would also induce a G1 cell cycle arrest in BCBL-1 PEL cells or whether endogenous LANA-1 in these cells could compensate the G1 effect of BRD4S. EGFP-BRD4S overexpression increased the G1/G0 population in BCBL-1 cells from 11.92 to 20.75% in the control, while reducing the S-phase population from 23.6 to 12.03% (data not shown), resembling the relative changes in BJAB cells in the absence of LANA-1 (Fig. 8). Therefore, BRD4S is also able to impose a significant G1/G0 arrest in the presence of endogenous LANA-1 in BCBL-1, at least under the conditions of our experiment.
DISCUSSION
BET proteins are a class of highly conserved bromodomain-containing proteins involved in fundamental cellular processes, such as meiosis, embryonic development, cell cycle regulation, and transcription (7, 13, 23, 24, 55). Via its bromodomains, BRD2/RING3 interacts with acetylated histone H4 and BRD4 interacts with acetylated histones H3 and H4, thereby providing a docking station for other proteins to attach to chromatin (10, 26, 34, 56). We have previously shown KSHV LANA-1 to interact with the human BET family member BRD2/RING3 and suggested a role for BRD2/RING3 in LANA-1 functions (37, 51). Here we investigate the potential of LANA-1 to bind to other human BET family members, since the ET domain of BRD2/RING3, which we have identified to constitute one binding site for LANA-1 (37), is characteristic of and highly conserved within BET proteins (Fig. 1) (13).
We found endogenous BRD4 protein to form complexes with KSHV LANA-1 in the KSHV-positive PEL cell line BCBL-1 (Fig. 1D). Using GST pull-down and coimmunoprecipitation approaches, we firstly confirmed the LANA-1 interaction with BRD4 and secondly established LANA-1 to also bind to BRD3/ORFX. Together with the previously shown interaction of LANA-1 with BRD2/RING3 (37, 51), we established the interaction of LANA-1 with three different BET family members, namely BRD2/RING3, BRD3/ORFX, and BRD4.
Fragments of BRD3/ORFX and BRD4 encompassing their ET domains are sufficient for LANA-1 binding (Fig. 2, 3, and 4). The interaction between LANA-1 and a region of BRD4 encompassing the ET domain is direct (i.e., does not require any additional cellular proteins), as shown by GST pull-down assays and a newly established ELISA with purified recombinant proteins (Fig. 4). The stronger binding of LANA-1 to full-length BRD2/RING3 and BRD4S relative to BRD2/RING3 and BRD4S truncation constructs lacking the region upstream of the ET domain (Fig. 3) suggests the presence of an additional binding site (or sites) upstream of the ET domain. Such a second LANA-1 binding site in BRD2/RING3 has previously been demonstrated by our group (37). We also show the C-terminal domain of LANA-1 to be indispensable for the interaction with BRD4, since C-terminally truncated versions of LANA-1 no longer coimmunoprecipitate with BRD4 when truncated beyond LANA-1 amino acid residue 1139 (Fig. 3). The loss of BRD4 binding to C-terminal LANA-1 deletion mutants truncated beyond aa 1139 mirrors the interaction pattern of LANA-1 with BRD2/RING3 (51). A LANA-1 mutant, L-1133, which does not interact with BRD2/RING3 (51) or with BRD4 (Fig. 3), loses the major known LANA-1 functions, i.e., the ability to bind to the LANA-binding sites within the terminal repeat of KSHV and support the replication of KSHV terminal repeat-containing plasmids, the activity to act as a transcriptional regulator, and the ability to dimerize (51). The loss of these functions of L-1133 coincides with the loss of chromatin or nuclear matrix association (50). Taken together, these observations suggest that the interaction with members of the BRD/fsh family, such as BRD4, BRD2, and BRD3, could contribute to the attachment of the C-terminal domain of LANA-1 to chromatin and to the function of LANA-1 in latent episomal replication and transcriptional activation. In addition, another nuclear chromatin binding protein, DEK-1, also binds to the LANA-1 C-terminal domain and could contribute to functions of LANA-1 (30). To date, we cannot definitively link the chromatin association of the LANA-1 C terminus to one specific LANA-1 binding partner, since LANA-1 mutants described so far lose multiple interactions and functions (16, 50, 51). Given the wide expression of BRD2 and BRD4 in many cell lineages (not shown) it is conceivable that there is substantial redundancy in cellular proteins that contributes to the attachment of the C-terminal domain of LANA-1 to nuclear chromatin.
In addition to this C-terminal domain, the N-terminal domain of LANA-1 is also crucial for a fully functional LANA-1 protein. Amino acids 5 to 22 mediate the binding to histones H2A and H2B and thereby the attachment to mitotic chromosomes; they are also required for the LANA-1-mediated replication of a viral episome and transcriptional regulator functions (2, 3, 36, 42, 54). Our data, together with the work of several other groups, strongly support a model in which LANA-1 is only fully functional when attached to chromatin via both the N-terminal and the C-terminal domains (2, 3, 36, 50, 51, 54).
However, the presently available experimental data are also compatible with different and/or additional roles of individual BRD proteins in LANA-1-mediated functions. Using CHIP assays, BRD2/RING3 has been shown to be associated with the terminal repeat region of the KSHV genome, to which LANA-1 binds (49), and we have previously shown a relocalization of BRD2/RING3 to KSHV genome- and LANA-1-containing nuclear speckles in PEL cell lines (33). The cross-reactivity with BRD4S of our polyclonal antibody raised against BRD2/RING3 (not shown) which was used in previous studies (33, 37) could suggest that, in addition to BRD2/RING3, shown by the results of Stedman et al. (49) to be present on viral genomes, BRD4S or BRD4L could be recruited to viral episomes via their interaction with LANA-1. We therefore investigated whether LANA-1 could modify the functions of interacting BET proteins.
Several studies link members of the BET protein family to cell cycle regulation or the activation of E2F-dependent promoters (11, 32, 46). KSHV LANA-1 has been demonstrated to positively regulate cell cycle-dependent promoters and to promote the G1/S transition of cells either arrested by serum starvation or by the overexpression of p16INK4a (1, 16, 38). We therefore hypothesized that the direct interaction of KSHV LANA-1 with BET proteins might play a role in cell cycle regulation. The ectopic expression of BRD4S and BRD2/RING3 in 293T, BJAB, or HeLa cells severely impaired the progression of cells through the cell cycle (Fig. 5 and 8 and data not shown). We found BRD4S to increase the proportion of cells in G1 and in G2 (Fig. 5 and 8). The long variant of BRD4 in mice, MCAP, has previously been shown to delay G1/S transition when overexpressed (32). Further, depleting cells of BRD4/MCAP using a microinjected BRD4 antibody results in an impaired G2/M transition (11). Our data show, for the first time, that BRD4S negatively regulates G1/S and G2/M checkpoint transitions in epithelial 293T and BJAB B cells when overexpressed (Fig. 5 and 8) and that BRD2/RING3 delays G1/S transition (Fig. 5 and 8). Notably, around 80% of 293T cells ectopically expressing BRD2/RING3 were found in either G1 or early S phase (Fig. 5), in keeping with a recent report from another group on the effect of high BRD2/RING3 levels on the G1/S checkpoint (46). In addition, both BRD2/RING3 and BRD4S activate the cyclin E promoter (Fig. 5 and data not shown), an observation previously reported for BRD2/RING3 (9) but not for BRD4. This might be linked to the ability of BRD2/RING3 to form complexes with transcriptional activators and repressors (8) or to the G1/S block induced by both BRD2/RING3 and BRD4S by increasing the number of cells at the stage of the cell cycle when cyclin E activity is strongest, namely the transition from G1 to S phase.
When exploring the effect of LANA-1 on these BRD2/RING3- and BRD4S-mediated cell cycle effects, we noticed that LANA-1, although itself an activator of the cyclin E promoter (Fig. 6A, panel 2) (51), inhibits the BRD4S-induced activation of cyclin E transcription (Fig. 6B, panel 2). LANA-1 mutants that are defective in their abilities to interact with BRD2/RING3 (51) or BRD4S (Fig. 4) do not show this inhibition. We hypothesized that this effect could be explained by LANA-1 releasing cells from a BRD4S-induced G1 arrest. We found that LANA-1 could indeed in part compensate the BRD4S- and BRD2/RING3-induced G1/S block in both tested cell lines, 293T and BJAB (Fig. 7 and 8). Whether this requires the physical association of LANA-1 with BRD4S and/or BRD2/RING3, which we have demonstrated in this and earlier reports (37, 51), is not yet resolved and will require the generation of LANA-1 mutants that are defective only in their abilities to interact with BET proteins. Interestingly, the overexpression of BRD4S in BCBL-1 cells also resulted in a higher proportion of cells in G0/G1 at the cost of the S-phase population (data not shown). Therefore, endogenous LANA-1 in BCBL-1 cells could not neutralize the G1/S cell cycle effect of overexpressed BRD4S. Whether this is due to the relative expression levels of LANA-1 and BRD4S in our specific experimental setting or whether other viral factors present in BCBL-1 cells are involved needs further investigation.
At present it is also conceivable that previously reported mechanisms of LANA-1-induced G1/S progression, such as the ability of LANA-1 to interact with GSK-3β (16) and the retinoblastoma protein (38), are involved in the ability of LANA-1 to overcome the BRD4S-and BRD2/RING3-induced G1/S arrest. The fact that LANA-1 can promote S-phase entry in different experimental settings in a variety of different cell types, e.g., in serum-starved cells (16) or in cells arrested by the expression of p16INK4a (1) or, as shown here, in cells of epithelial or B-cell origin arrested by the expression of BRD2/RING3 or BRD4S, underlines the potency of LANA-1 to promote S-phase entry, a property that is presumably linked to the role of LANA-1 in the replication of episomal latent viral DNA in concert with the host cell cycle.
Acknowledgments
We thank Matthias Ballmeier from the FACS sorting facility of the Hannover Medical School for assistance with the flow cytometric data aquisition. We thank Julie Sheldon for expert technical assistance and Viswanathan Srinivasan and Kenneth Kaye for kindly providing BJAB and BCBL-1 cells. We are very grateful to Michal-Ruth Schweiger at Harvard Medical School for providing the affinity-purified rabbit anti-BRD4-N antibody.
M.O. was funded by the grant DFG GRK 745 ‘Mucosal Host-Pathogen Interactions’ and by DFG SPP1130. Some of the later experiments were carried out by M.O. in Peter Howley's laboratory at Harvard Medical School and were supported by grant R01CA116720 (to Peter Howley) from the National Cancer Institute.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862-3874. [DOI] [PubMed] [Google Scholar]
- 2.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856-861. [DOI] [PubMed] [Google Scholar]
- 4.Bechtel, J. T., Y. Liang, J. Hvidding, and D. Ganem. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Chua, P., and G. S. Roeder. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 15:3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis, G. V., M. E. McComb, D. V. Faller, A. Sinha, P. B. Romesser, and C. E. Costello. 2006. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 5:502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, G. V., C. Vaziri, N. Guo, and D. V. Faller. 2000. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 11:417-424. [PMC free article] [PubMed] [Google Scholar]
- 10.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florence, B., and D. V. Faller. 2001. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 6:D1008-D1018. [DOI] [PubMed] [Google Scholar]
- 14.French, C. A., I. Miyoshi, J. C. Aster, I. Kubonishi, T. G. Kroll, P. Dal Cin, S. O. Vargas, A. R. Perez-Atayde, and J. A. Fletcher. 2001. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 159:1987-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French, C. A., I. Miyoshi, I. Kubonishi, H. E. Grier, A. R. Perez-Atayde, and J. A. Fletcher. 2003. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63:304-307. [PubMed] [Google Scholar]
- 16.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 17.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. S. Moore. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335:233-241. [DOI] [PubMed] [Google Scholar]
- 18.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 19.Geng, Y., E. N. Eaton, M. Picon, J. M. Roberts, A. S. Lundberg, A. Gifford, C. Sardet, and R. A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12:1173-1180. [PubMed] [Google Scholar]
- 20.Greenwald, R. J., J. R. Tumang, A. Sinha, N. Currier, R. D. Cardiff, T. L. Rothstein, D. V. Faller, and G. V. Denis. 2004. Eμ-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood 103:1475-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haynes, S. R., C. Dollard, F. Winston, S. Beck, J. Trowsdale, and I. B. Dawid. 1992. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20:2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 25.Jeanmougin, F., J. M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson. 1997. The bromodomain revisited. Trends Biochem. Sci. 22:151-153. [DOI] [PubMed] [Google Scholar]
- 26.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 13:33-43. [DOI] [PubMed] [Google Scholar]
- 27.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 29.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 30.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama, T., A. Farina, A. Dey, J. Cheong, V. P. Bermudez, T. Tamura, S. Sciortino, J. Shuman, J. Hurwitz, and K. Ozato. 2002. A mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 22:6509-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattsson, K., C. Kiss, G. M. Platt, G. R. Simpson, E. Kashuba, G. Klein, T. F. Schulz, and L. Szekely. 2002. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 83:179-188. [DOI] [PubMed] [Google Scholar]
- 34.Pamblanco, M., A. Poveda, R. Sendra, S. Rodriguez-Navarro, J. E. Perez-Ortin, and V. Tordera. 2001. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 496:31-35. [DOI] [PubMed] [Google Scholar]
- 35.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 39.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 41.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang, E., G. Salazar, T. E. Crowley, X. Wang, R. A. Lopez, X. Wang, and D. J. Wolgemuth. 2004. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr. Patterns. 4:513-519. [DOI] [PubMed] [Google Scholar]
- 44.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Si, H., and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinha, A., D. V. Faller, and G. V. Denis. 2005. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem. J. 387:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, and L. Degos. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 48.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stedman, W., Z. Deng, F. Lu, and P. M. Lieberman. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566-12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viejo-Borbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. A domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 77:7093-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viejo-Borbolla, A., M. Ottinger, E. Bruning, A. Burger, R. Konig, E. Kati, J. A. Sheldon, and T. F. Schulz. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618-13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, and A. S. Denton. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 53.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 54.Wong, L. Y., G. A. Matchett, and A. C. Wilson. 2004. Transcriptional activation by the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J. Virol. 78:10074-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 56.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]
- 57.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]