Abstract
Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA-3C) is essential for EBV-mediated immortalization of human B lymphocytes and regulates both the cell cycle and transcription. Transient reporter gene assays have implicated a pivotal role for EBNA-3C in the regulation of transcription of the majority of latency-associated genes expressed during the EBV growth program, including the viral oncoprotein LMP-1. To examine the regulation of latency gene expression by EBNA-3C, we generated an EBV-positive cell line that inducibly expresses EBNA-3C. This cell line allowed us to examine expression from the endogenous latency gene promoters in the context of an actual latent infection and the presence of other EBNA proteins, in particular EBNA-2, which is presumed to coregulate transcription with EBNA-3C. EBNA-3C induced the expression of both LMP-1 and LMP-2B mRNAs from the bidirectional LMP-1/LMP-2B promoter. In contrast, no effect was seen on expression from the common EBNA promoter Cp, which is responsive to EBNA-3C in reporter assays. Activation of LMP expression was not the consequence of increases in EBNA-2, PU.1 or Spi-B transcription factors, all of which are believed to be critical for activation of LMP-1. Chromatin immunoprecipitation assays furthermore indicated that EBNA-3C is present at the bidirectional LMP-1/LMP-2B promoter. These results indicate that EBNA-3C directly activates the expression of LMP-1 and LMP-2B but is unlikely to significantly regulate EBNA expression via Cp under normal growth conditions.
Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA-3C) is one of only six viral proteins known to be essential for EBV to immortalize primary human B lymphocytes (9, 22, 33, 42, 52, 72). Although the EBNA-3 genes (3A, 3B, and 3C) probably arose through gene duplication, the sequences of the EBNA-3 proteins are highly divergent, and the fact that both EBNA-3A and EBNA-3C (but not EBNA-3B) are essential for EBV-mediated immortalization suggests that their functions have also diverged. These proteins do have related functions, however, regulating both cell cycle progression and transcription. All of the EBNA-3 proteins prevent the G2/M arrest that normally occurs in response to genotoxic or other agents, reportedly due to an association with Chk2 (38). In primary rodent cells, EBNA-3C can override the retinoblastoma protein checkpoint in the G1 phase of the cell cycle when coexpressed with Ha-Ras (60). EBNA-3C is also reported to regulate p27 stability through interactions with the E3 ubiquitin ligase SCFSkp2 (36) and to associate with cyclin A in vitro (37). The region of EBNA-3C that associates with cyclin A has been localized to a domain that is moderately conserved between the EBNA-3 proteins, and preliminary data suggest that EBNA-3B, but not EBNA-3A, may also bind cyclin A in vitro (36). Both EBNA-3A and EBNA-3C can function as immortalizing oncogenes, cooperating with activated Ras in the transformation of primary rodent fibroblasts (27, 60). This function requires a motif that mediates interaction with the transcriptional repressor CtBP (73), suggesting that the immortalizing function of the EBNA-3 proteins may not be totally distinct from their transcriptional properties.
None of the EBNA-3 proteins appears to bind DNA directly (32, 68), but all bind to a variety of cellular proteins, some of which function as general transcriptional regulatory proteins, including TATA-binding protein (TBP), histone deacetylase 1 (HDAC1), CtBP, DP103, prothymosin α, and p300 as well as SUMO-1 and SUMO-3 (3, 11, 21, 27, 35, 44, 65, 70, 73). EBNA-3C associates with the sequence-specific DNA-binding protein Jκ (also known as RBP-Jκ, CBF-1, and CSL), a downstream signaling protein in the Notch pathway, and the Ets transcription factors PU.1 and Spi-B (53, 67, 83, 84). These cellular transcription factors also interact with EBNA-2 and mediate its transcriptional activation of three EBV latency gene promoters, i.e., the LMP-1 promoter, which also serves to regulate the LMP-2B gene on the opposite DNA strand; the common promoter for the multicistronic EBNA gene, Cp; and the LMP-2A promoter (20, 30, 39, 40, 45, 47). The LMP-1 and LMP-2B transcription start sites are located 266 bases apart, and their common promoter region directs transcription in opposing directions (hence the designation bidirectional promoter) (41). These promoters presumably share upstream regulatory elements that include binding sites for both Jκ and PU.1. In reporter gene assays, the PU.1 binding site, but not the Jκ binding site, is essential for EBNA-2-mediated transactivation, although the Jκ site contributes to full activation (30). The LMP-2A promoter is located approximately 3.2 kb downstream (leftward) of the LMP-1 transcription start site and is thus distinct from the LMP-2B promoter, and it is used to transcribe an exon unique to LMP-2A (LMP-2A and LMP-2B share 11 of their 12 exons) that encodes a 118-amino acid N-terminal domain containing an immunoreceptor tyrosine-based activation motif (17, 69). Likewise, the LMP-2B promoter gives rise to the unique LMP-2B 5′ exon (4, 41). The LMP-2A and Cp promoters also contain binding sites for Jκ that can contribute to transcriptional activation by EBNA-2 (45, 86).
The domain of EBNA-3C that binds Jκ has been delineated and is conserved among the EBNA-3 proteins, all of which can bind Jκ (83). In reporter gene assays, this interaction represses EBNA-2/Jκ-mediated transcription by preventing Jκ from associating with DNA (43, 53, 64, 67, 76, 83). This finding led to the hypothesis that the EBNA-3 proteins might mediate feedback inhibition of Cp (also regulated by EBNA-2 via Jκ) to stringently regulate EBNA levels and, in turn, expression of the LMPs. The first demonstration that EBNA-3C might regulate LMP-1 came from a study in which EBNA-3C expression was restored in the EBV-positive Burkitt lymphoma cell line Raji, whose EBV genomes are deleted for most of the EBNA-3C open reading frame (1, 2). Specifically, in parental Raji cells, levels of LMP-1 fall as cells progress to G1 (5, 6), whereas upon stable expression of EBNA-3C, levels of LMP-1 remain high (2). In reporter gene assays, however, EBNA-3C can mediate either repression or activation of the LMP-1 promoter through Jκ and PU.1 sites, respectively (43, 53, 67, 76, 83, 84). These effects of EBNA-3C on the LMP-1 promoter and its associations with Jκ and PU.1 are conserved within the EBNA-3C homologues encoded by the baboon and rhesus lymphocryptoviruses (LCVs), which are genetically and biologically equivalent to EBV in their respective hosts, suggesting that they play important roles in the LCV life cycle (12, 29, 82).
Though dispensable for B-cell immortalization in vitro by EBV, the LMP-2 proteins (or at least LMP-2A) are thought to promote B-cell survival in vivo and thus are critical for EBV persistence and oncogenic potential (8, 48, 49). LMP-1 is essential for EBV-mediated immortalization and functions as a constitutively active CD40-like molecule (33, 57, 74). While LMP-1 is necessary for EBV-mediated immortalization and, presumably, the establishment of a persistent latent infection, overexpression of LMP-1 results in cytostasis (15, 23). Thus, the regulation of LMP-1 expression is likely to be tightly controlled. Indeed, while EBNA-2 is clearly the major transcriptional transactivator of the LMP-1 promoter, both EBNA-LP and EBNA-3C function as coactivators, and the EBNA-3 proteins can also counter EBNA-2's effects on transcription mediated through Jκ (24, 53, 59, 67, 83, 84). Clearly, therefore, exploration of a given EBNA's contribution to transcriptional regulation must take place in the context of an infection in which the full complement of the EBV latency-associated transcription factors is present. For this reason, we have engineered Raji cells to inducibly express EBNA-3C to explore the roles that EBNA-3C plays in the regulation of viral and, potentially, cellular promoters. Using this system, we demonstrate that induction of EBNA-3C is rapidly followed by an increase in LMP-1 and LMP-2B expression and that this is concomitant with an association of EBNA-3C with the bidirectional LMP-1/2B promoter. These findings demonstrate that EBNA-3C is a bona fide transactivator of this promoter and suggest that this model system would be suitable for the analysis of potential regulation of cellular gene expression by EBNA-3C within latently infected cells.
MATERIALS AND METHODS
Cell culture.
Raji cells and their derivatives were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone) and 2 mM l-glutamine. Cells were incubated in a humidified chamber at 37°C with a 5% CO2 atmosphere.
Plasmids.
The pTRE-3C plasmid was created by ligating a SacI/EcoRI fragment of an EBNA-3C cDNA encompassing the entire open reading frame (ORF) into pTRE (Clontech). To generate a FLAG epitope-tagged protein, annealed oligonucleotides encoding a FLAG epitope (5′-CTAGGGATTACAAGGATGACGACGATAAGT-3′ and 5′-CTAGACTTATCGTCGTCATCCTTGTAATCC-3′) were inserted at the XbaI site near the 5′ end of the EBNA-3C ORF to yield pTRE-F3C. The resultant F3C protein contains a single FLAG epitope inserted between amino acids 10 and 11 of EBNA-3C.
Generation of tetracycline-regulated cell lines.
The pTet-Off plasmid (Clontech), encoding the tetracycline-responsive transcriptional activator (19) and carrying a neomycin resistance gene, was transfected into Raji cells by electroporation of 8 × 106 cells in 250 μl of complete growth medium with 10 μg plasmid DNA in a 0.4-cm-gap electroporation cuvette, using a Bio-Rad Genepulser II set at 960 μF and 250 V. After 48 h, transfected cells were transferred to 96-well plates (500 to 1,000 cells per well) and selected in 0.8 mg G418 per ml. G418-resistant cells were expanded and tested for tetracycline-regulated gene expression following transfection with a tetracycline-responsive luciferase reporter gene in the presence or absence of 5 ng doxycycline per ml. A clone with low luciferase expression in the presence of doxycycline but high expression in its absence was selected to generate a tetracycline-regulated EBNA-3C-expressing cell line by transfection with 10 μg of pTRE-3C or pTRE-F3C and 1 μg of the puromycin resistance plasmid pJ6Ωpuro (56) as described above. After 2 days, cells were selected in 0.8 mg G418, 0.5 μg puromycin, and 5 ng doxycycline per ml. Each puromycin-resistant clone was then incubated either with or without 5 ng doxycycline per ml for 2 days and analyzed for doxycycline-regulated expression of EBNA-3C by immunoblotting with an antibody against EBNA-3C. Clones with tightly regulated expression of EBNA-3C were selected for further analysis.
Immunoblotting.
Equal amounts of protein per sample were separated in a 4 to 15% gradient Criterion Tris-HCl sodium dodecyl sulfate (SDS)-polyacrylamide gel (Bio-Rad) and transferred to an Immobilon-P membrane (Millipore). For EBNA-3 detection, membranes were blocked in Tris-buffered saline containing 0.5% Tween 20 and 5% nonfat dry milk and then incubated with sheep anti-EBNA-3C or anti-EBNA-3A antibody (Ex-Alpha), followed by incubation with horseradish peroxidase (HRP)-conjugated rabbit anti-sheep secondary antibody (Chemicon). Proteins were visualized by enhanced chemiluminescence (Amersham Biotech). LMP-1 and EBNA-2 were detected with mouse anti-LMP-1 S12 antibody (51) and mouse PE2 antibody (81), respectively, followed by anti-mouse HRP-conjugated antibody. PU.1 and Spi-B were detected with antibodies T21 and N16, respectively (Santa Cruz Biotechnology). Membranes were subsequently stripped by incubation in 100 mM β-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl (pH 6.8) at 52°C for 30 min and then reprobed with mouse anti-actin (clone JLA20; Calbiochem) followed by sheep anti-mouse HRP-conjugated antibody (Amersham Biosciences).
RNA (Northern) blot analysis.
Cells were incubated in the presence or absence of doxycycline for 5 days, after which total RNA was extracted using RNA-Bee according to the manufacturer's instructions (Tel-Test). RNAs were fractionated in a 1.2% agarose gel containing formaldehyde in morpholinepropanesulfonic acid (MOPS) buffer. Following electrophoresis, RNAs were transferred to a GeneScreen Plus membrane (Perkin-Elmer) by capillary transfer in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.0]). The membrane was prehybridized in 50% formamide, 5× SSPE (1× SSPE is 150 mM NaCl, 10 mM sodium phosphate [pH 7.4], and 1 mM EDTA), 5× Denhardt's solution, 1% SDS, and 0.1 mg denatured salmon testis DNA (Sigma) per ml at 45°C for 6 h. The membrane was then hybridized in the same solution with a 32P-labeled (by nick translation) LMP-1 cDNA overnight at 45°C. The membrane was then washed at 62°C with decreasing concentrations of SSC (2× to 0.5×) containing 1% SDS and processed by phosphorimage analysis. The membrane was then reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control.
ChIP assay.
Raji Tet-Off-EBNA-3C cells (5 × 107 to 8 × 107) were cultured in the presence or absence of doxycycline for 48 h. Chromatin immunoprecipitation (ChIP) was then performed using a ChIP-IT chromatin immunoprecipitation kit (Active Motif). Briefly, protein-DNA complexes were cross-linked for 10 min at room temperature in RPMI 1640 medium containing 1.1% formaldehyde. Cross-linking was terminated by the addition of glycine, cells were pelleted by centrifugation and then lysed, and the extract was sonicated to shear the DNA to sizes ranging between 200 and 1,000 base pairs in length. Total cell extracts were precleared with protein G agarose beads and then subjected to immunoprecipitation with an antibody to EBNA-3C (ExAlpha) or the immunoglobulin G (IgG) and anti-TFIIB controls provided with the kit. A fraction of the precleared chromatin samples was saved to represent input DNA. Immunoprecipitated DNA was then purified following reversal of cross-linkage to protein according to the manufacturer's instructions and was amplified by PCR using primers for the Raji LMP-1 promoter (GenBank accession no. M20868). The forward and reverse primer sequences were 5′-CTACGGTGAACCCACATCCT-3′ and 5′-CGGTGTGTGTGTGCATGTAA-3′, respectively, and yielded a product of 365 bp. GAPDH primers were provided with the ChIP-IT kit. PCR was performed using ReddyMix PCR master mix (ABgene) containing 1.25 units Taq DNA polymerase, 75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% Tween 20, a 200 μM concentration of each deoxynucleoside triphosphate, dye for electrophoresis, a 500 nM concentration of each primer, and 3.75 μCi [α-32P]dCTP (10 mCi/ml; NEN/Perkin-Elmer) per reaction mixture. Following an initial denaturation step at 94°C for 2 min, DNA was amplified by a variable number of cycles of 94°C for 20 seconds, 61.9°C for 1 min, and 72°C for 60 seconds, followed by a final extension step at 72°C for 7 min. PCR products were fractionated by electrophoresis in a nondenaturing 5% acrylamide gel that was then vacuum dried and processed by phosphorimage analysis for quantification (STORM 860; Molecular Dynamics).
Reverse transcription-PCR (RT-PCR) analysis of LMP-2B and EBNA-3C RNAs.
Raji Tet-Off-EBNA-3C cells were cultured in the presence or absence of 5 ng doxycycline per ml for 24 or 48 h. For each experimental time point, approximately 107 cells were washed twice with ice-cold phosphate-buffered saline, and total RNA was isolated with RNA-Bee according to the manufacturer's protocol (Tel-Test). RNAs were treated with 10 U of RNase-free DNase (Promega) and 100 U of RNasin (Promega) for 15 min at 37°C, followed by extraction in phenol-chloroform and precipitation in ethanol. RNAs were reverse transcribed to cDNAs by using an Advantage RT-for-PCR kit (BD Biosciences) according to the manufacturer's instructions. Primers for GAPDH were included in the kit. The forward and reverse primers used to amplify LMP-2B cDNA (GenBank accession no. AJ507799) had the sequences 5′-CAACGTTGGGAGGTCGTTGG-3′ and 5′-AAGCCAGTAGCAGCAGCGTC-3′, corresponding to sequences in exons 1 and 4, respectively, and yielded a 528-bp product. The forward and reverse primers for EBNA-3C (GenBank accession no. AJ507799) had the sequences 5′-CTGGCAAAACTTGCTCCA-3′ and 5′-GTGCTTCTGCCTTATCAGA-3′, respectively, and yielded a 499-bp product. PCR was performed as indicated above for ChIP assays, except that the annealing step in each cycle was 50.5°C for 90 seconds. The mRNA level of each sample was normalized to the signal obtained from GAPDH mRNA products.
RESULTS
EBNA-3C activates LMP-1 protein expression.
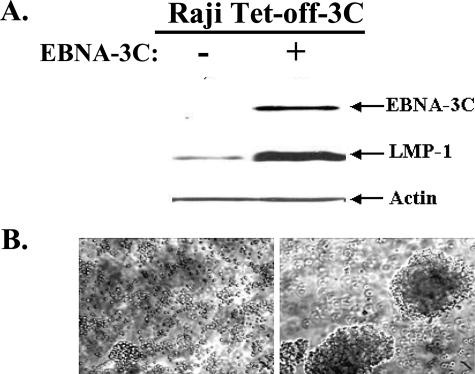
To examine the transcriptional properties of EBNA-3C in a biologically relevant setting, a Raji BL cell system was selected because stable expression of EBNA-3C (whose gene is deleted from EBV genomes in Raji cells) increases the expression of LMP-1 from its endogenous viral promoter in the presence of the complete complement of latency proteins (1, 2). We engineered an inducible EBNA-3C expression cassette in Raji cells that is controlled by a tetracycline-regulated promoter. Several clones were selected in which EBNA-3C expression was undetectable in the presence of the tetracycline derivative doxycycline (i.e., Tet off) but was readily induced upon removal of doxycycline. To determine whether induction of EBNA-3C expression affected the level of LMP-1, an immunoblot was stripped and reprobed with an LMP-1-specific monoclonal antibody. Clearly, LMP-1 levels were substantially increased in the EBNA-3C-expressing cells (Fig. 1A). Furthermore, cells expressing EBNA-3C and LMP-1 underwent a dramatic phenotypic change: whereas parental Raji or uninduced cells grew as single cells or in small clusters, EBNA-3C-expressing Raji cells grew in large clumps (Fig. 1B), a characteristic feature of lymphoblastoid cell lines (LCLs) and LMP-1-expressing cells. These results were consistent with those of Allday et al. for stable expression of EBNA-3C (1) and suggested that the increase in LMP-1 was likely to be a specific effect of EBNA-3C and not a compensatory change that occurred to promote cell survival during selection for a cell line that stably expressed EBNA-3C. The doxycycline-regulated expression of EBNA-3C in Raji cells therefore represents an ideal system with which to further explore EBNA-3C-regulated gene expression.
FIG. 1.
EBNA-3C activates expression of the LMP-1 protein. Expression of EBNA-3C was induced by growing cells in the absence of doxycycline for 5 days. (A) Expression of EBNA-3C, LMP-1, and actin was determined by immunoblot analysis. (B) Cells that were not induced (left) or induced to express EBNA-3C (right) were examined by light microscopy.
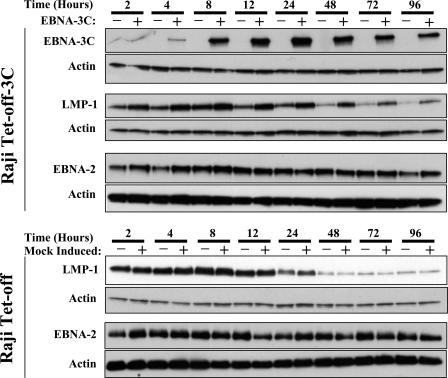
To determine the time course of EBNA-3C expression, EBNA-3C was monitored by immunoblot analysis at various times after the removal of doxycycline. EBNA-3C was detectable 4 h following the removal of doxycycline and attained maximal expression at approximately 12 h (Fig. 2). To evaluate the effect of EBNA-3C on LMP-1 expression, LMP-1 levels were monitored at the same time points. In the parental cell line, Raji-Tet-off, LMP-1 levels were relatively low and exhibited a reproducible decrease after 24 to 48 h. Similar decreases have been reported previously for Raji cells; LMP-1 levels are increased in the presence of serum and have been demonstrated to fall with time in culture (5, 6). As expected, doxycycline had no effect on the level of LMP-1. For the EBNA-3C-inducible cell line analyzed in parallel in the presence of doxycycline (i.e., in the absence of EBNA-3C), the levels of LMP-1 were low and comparable to those in the parental Raji cell line at each time point, exhibiting a similar decrease after 24 h. Following the induction of EBNA-3C expression, some increases in LMP-1 levels could be seen at early times (i.e., 2 to 4 h) that were coincident with changes in EBNA-2 that may not be explained entirely by differences in protein loading. The most striking differences, however, were seen beginning at 12 h, when EBNA-3C is maximally expressed, and continuing through the later time points. By 12 h, LMP-1 levels were significantly increased in cells expressing EBNA-3C, independent of any changes in EBNA-2. In cells that did not express EBNA-3C, LMP-1 levels began to decrease after this time, whereas LMP-1 levels were at least partially sustained in cells that expressed EBNA-3C. As the levels of EBNA-3C began to decline with time, there was a corresponding decrease in the levels of LMP-1, providing further evidence that increases in LMP-1 are dependent on EBNA-3C.
FIG. 2.
Time course of EBNA-3C and LMP-1 induction. Raji Tet-off-3C cells (upper panel) were cultured in the presence or absence of doxycycline and harvested at different time points. SDS-polyacrylamide gel electrophoresis was performed in triplicate. Each immunoblot was stripped and reprobed for actin. Parallel experiments were performed with the Raji Tet-off parental cell line (lower panel). Mock induction refers to the absence of doxycycline (+), which would induce EBNA-3C in Raji Tet-off-3C cells; cells incubated in the presence of doxycycline, which would not induce EBNA-3C, are represented by “−.”
EBNA-3C does not overtly regulate expression from Cp.
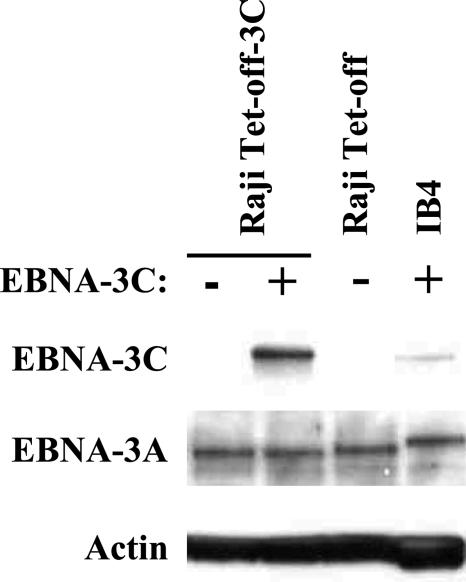
Previous studies based on transient reporter assays have demonstrated an ability of EBNA-3C to down-regulate expression from the EBNA promoter Cp (64). The mRNAs encoding the six EBNA proteins are generated from a large primary transcript originating at Cp that is processed into the individual mRNAs. It has therefore been suggested that EBNA-3C might control overall EBNA gene expression through feedback repression. A previous study that analyzed promoter usage in various cell lines suggested that Raji cells might not use Cp (78); an alternative explanation, however, is that the oligonucleotides used did not hybridize to RNAs in Raji cells due to differences in sequence from the prototypical EBV, B95-8. Using RT-PCR analysis, we have clearly demonstrated that Cp is used in Raji cells (D. Hughes, J. Sample, and C. Sample, unpublished observations). To determine whether EBNA-3C regulated Cp-driven EBNA expression in the context of a latent infection, the levels of EBNA-2 were examined by immunoblotting 2 to 96 h after the induction of EBNA-3C expression. As shown in Fig. 2 (bottom panels), although we observed some variations in EBNA-2 levels in the presence and absence of doxycycline at some time points (i.e., 4 h) that did not appear to be due to unequal protein loading, overall we did not see a consistent effect of EBNA-3C on EBNA-2 levels, and specifically saw no decrease in EBNA-2 levels associated with EBNA-3C expression, throughout the time course experiment. We also examined whether EBNA-3C affected the expression of a second protein expressed from Cp, EBNA-3A. Consistent with our analysis of EBNA-2, EBNA-3C had no effect on the level of EBNA-3A, even when overexpressed relative to EBNA-3C levels in the LCL IB4 (Fig. 3). Taken together, these findings suggest that in the presence of the other EBNA proteins in an actual latent infection, EBNA-3C expression has little or no negative effect on Cp, as predicted from earlier studies measuring EBNA-3C's influence on Cp within a reporter plasmid and in the presence of EBNA-2 in EBV-negative cells.
FIG. 3.
EBNA-3C does not affect the level of EBNA-3A. Raji Tet-off-3C cells were cultured for 48 h in the presence or absence of doxycycline. A sample of IB4 cells (an LCL) was included as a control. Immunoblots were performed for the indicated proteins and reprobed for actin.
EBNA-3C expression results in increased levels of LMP-1 and LMP-2B mRNAs.
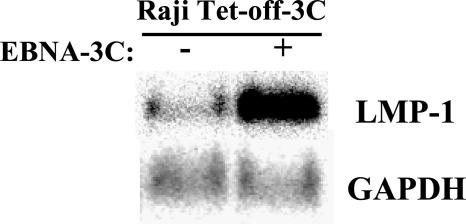
To determine whether the increase in LMP-1 protein was the result of an increased level of LMP-1 mRNA, we performed Northern blot analysis of RNAs from EBNA-3C-expressing versus non-EBNA-3C-expressing Raji cells. In the absence of EBNA-3C, LMP-1 mRNA levels were low. Following induction of EBNA-3C expression, levels of LMP-1 mRNA were increased approximately fivefold, whereas there was no change in the GAPDH mRNA level (Fig. 4). Thus, an increase in LMP-1 protein correlates with an increase in mRNA, as one would expect if EBNA-3C transactivates the endogenous LMP-1 promoter.
FIG. 4.
EBNA-3C increases expression of LMP-1 mRNA. LMP-1 mRNA was detected by Northern blot analysis of total RNA isolated from Raji Tet-off cells expressing (+) or not expressing (−) EBNA-3C. The blot was stripped and reprobed for GAPDH as a loading control.
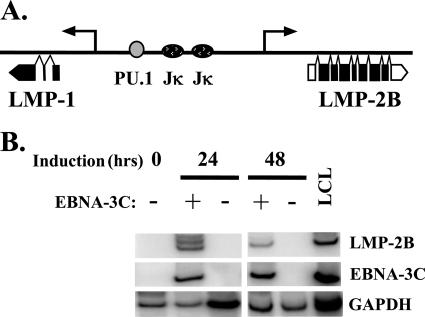
EBNA-2 has been shown to activate the expression of both LMP-2A and LMP-2B in reporter gene assays as well as by exogenous expression in the EBV-positive cell line P3HR1, which lacks EBNA-2 as a consequence of deletion (40, 55, 87). For EBV-negative cells, reporter gene assays suggest that the EBNA-3 proteins repress expression from the LMP-2A promoter (43), but whether these proteins also affect LMP-2B expression has not been examined. In Raji cells, expression from the endogenous LMP-2A promoter cannot be examined due to the deletion of the promoter and the first exon (25, 62). The LMP-1/2B bidirectional promoter (diagrammed in Fig. 5A), however, is intact, and we reasoned that EBNA-3C could coordinately regulate the expression of both genes or might preferentially regulate LMP-1 expression. Because there are no antibodies available with which to effectively monitor LMP-2B, we employed an RT-PCR-based approach to determine whether EBNA-3C also influences LMP-2B expression. In the absence of EBNA-3C (in the presence of doxycycline), LMP-2B mRNA was undetectable but was easily amplified upon induction of EBNA-3C both 24 and 48 h after the removal of doxycycline (Fig. 5B). Note that the multiple bands appearing at the 24-h time point are not reproducible and thus are not reflective of distinct RNA structures present at this time point. The increases in LMP-2B mRNA ranged from 3.4- to 6.1-fold in four separate experiments. This clearly demonstrated that EBNA-3C activates the expression of both genes from the LMP-1/2B bidirectional promoter, suggesting that its effect is likely exerted on the promoter itself.
FIG. 5.
EBNA-3C activates expression of LMP-2B mRNA. (A) Schematic of LMP-1/LMP-2B bidirectional promoter. Binding sites for the transcription factors Jκ and PU.1 are shown. Boxes below the map depict the mRNAs for LMP-1 and LMP-2B (not to scale), showing coding (black boxes) and noncoding (white boxes) exons. (B) Raji Tet-off EBNA-3C cells were induced to express EBNA-3C, and cells were harvested 24 or 48 h later. Total RNA was isolated, and primers specific for LMP-2B, EBNA-3C, and GAPDH were used to determine the level of each mRNA by RT-PCR, using [α-32P]dCTP in the reaction mix for quantification.
EBNA-3C does not induce expression of known cellular transactivators of the LMP-1 promoter.
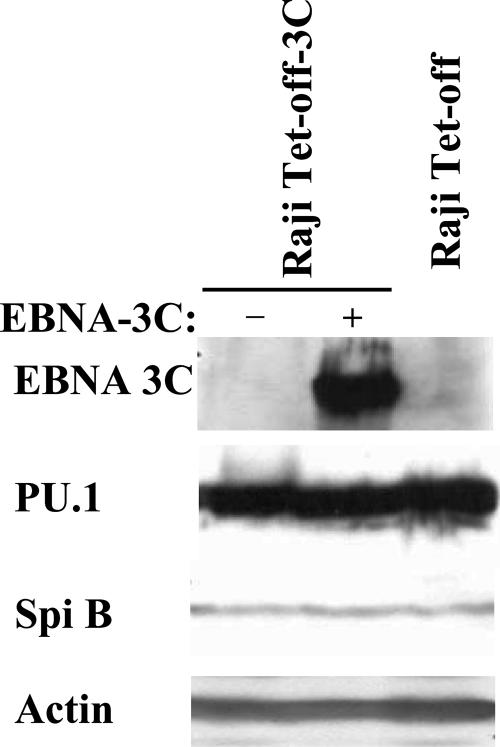
Having shown that EBNA-3C can clearly up-regulate the expression of LMP-1 and LMP-2B mRNAs in the context of a latent EBV infection, we next began to address the most likely mechanism of EBNA-3C action in this regard. One possibility is that EBNA-3C increases the levels of viral or cellular transcription factors that activate expression from the bidirectional promoter. Clearly, the level of the major viral transactivator EBNA-2, which is expressed from Cp, is not notably affected by EBNA-3C (Fig. 2). In reporter gene assays, the PU.1 binding site is essential for EBNA-2-mediated activation of LMP-1 and also for the activation mediated through EBNA-3C in conjunction with EBNA-2 (30, 84). Thus, PU.1 and the closely related Ets family member Spi-B are the cellular factors that bind to the LMP-1 promoter, and they are believed to be critical for transactivation of LMP-1 expression (30, 39). The levels of PU.1 and Spi-B were therefore analyzed by immunoblotting following the induction of EBNA-3C expression. As shown in Fig. 6, EBNA-3C had no effect on the level of either PU.1 or Spi-B.
FIG. 6.
EBNA-3C does not affect the level of PU.1 or Spi-B. Extracts from Raji Tet-off cells expressing (+) or not expressing (−) EBNA-3C and from the parental cell line Raji Tet-off were analyzed for levels of the transcription factors PU.1 and Spi-B, which are key activators of the LMP-1 promoter.
EBNA-3C interacts with the bidirectional LMP-1/2B promoter.
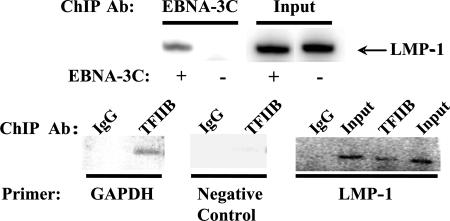
Although EBNA-3C alone has no known specific DNA-binding capability, EBNA-3C expressed in cellular extracts can bind to DNA (32, 68). This result suggests that EBNA-3C might be targeted indirectly to DNA, and in support of this possibility, EBNA-3C associates with a variety of transcription factors, including PU.1 (3, 65, 67, 83, 84). Therefore, we next examined whether the induction of LMP-1 and LMP-2B expression by EBNA-3C was a result of its interaction with the bidirectional promoter via such a protein-protein interaction. To investigate this possibility, we performed a ChIP assay to determine whether EBNA-3C is indeed present on the promoter following its induction of expression. As shown in Fig. 7, an antibody to EBNA-3C or TFIIB (positive control), but not an irrelevant IgG (negative control), immunoprecipitated DNA from the LMP-1 promoter. Furthermore, LMP-1 promoter DNA was enriched 10.9-fold in immunoprecipitates from cells in which EBNA-3C expression had been induced relative to those from cells in which EBNA-3C had not been induced, whereas DNA from the EBNA-1-specific promoter Qp, used as a negative control, was only enriched 1.7-fold. Thus, the activation of LMP-1 and LMP-2B expression by EBNA-3C is most likely mediated through direct activation of transcription from the LMP-1/2B bidirectional promoter.
FIG. 7.
EBNA-3C interacts with the bidirectional LMP-1/LMP-2B promoter. Raji Tet-off-3C cells were induced to express EBNA-3C by removal of doxycycline from the growth medium (+) for 48 h or were kept under doxycycline repression (−). A ChIP assay was performed to examine the association of EBNA-3C with the LMP-1/LMP-2B promoter (upper panel). The chromatin was immunoprecipitated using an antibody specific for EBNA-3C. As a positive control, the chromatin was immunoprecipitated with an antibody to TFIIB, a known element of the basal transcriptional machinery which associates with the GAPDH promoter (lower panel) and also with the LMP-1 promoter. As a negative control, the chromatin was immunoprecipitated with an irrelevant IgG. The GAPDH and LMP-1 primers correspond to PCR primers to amplify the promoter sequences of these genes. Negative control primers were designed to span sequences outside the GAPDH promoter area with which TFIIB is associated.
DISCUSSION
Although EBNA-3C is essential for EBV-mediated immortalization of B cells, its specific contribution is not fully understood. The first function reported for EBNA-3C was the ability to increase LMP-1 protein levels in clones of the EBV-infected cell line Raji that stably expressed EBNA-3C (1, 2). These and other studies (77) suggested that EBNA-3C could function as a transcription factor, which was later confirmed (43, 44, 53, 67, 76, 83, 84). However, given anecdotal evidence that EBNA-3C-expressing cell lines are difficult to obtain, suggesting that the protein might be toxic, it was possible that a compensatory increase in LMP-1, which prevents apoptosis (26), had been selected for during establishment of the cell lines. By using a cell line that expressed EBNA-3C in an inducible fashion, we were able to determine that the increase in LMP-1 protein occurred only hours after EBNA-3C expression, suggesting that this is a specific effect of EBNA-3C and demonstrating that this occurred not only in growth-arrested cells, as previously described, but also in proliferating cells. As previously reported, LMP-1 levels are increased by serum and fall over time in culture (5, 6). The increases seen with EBNA-3C are largely due to sustained levels of LMP-1 at later time points. Our inducible cell line therefore offered a unique opportunity to further explore the mechanism through which EBNA-3C functions in the presence of the other latency-associated proteins. The inclusion of all latency-associated proteins is essential because recent studies indicated that EBNA-LP, like EBNA-3C, can serve as a coactivator of EBNA-2-mediated transcription; for EBNA-LP, coactivation is most likely mediated through interactions with Sp100 (24, 46, 59). While EBNA-2 activates expression from the common EBNA (Cp), LMP-2A, and LMP-1/2B promoters, EBNA-LP selectively regulates a subset of these promoters, i.e., Cp and LMP-1/2B but not LMP-2A (61). As shown here, EBNA-3C activates expression from the LMP-1/2B promoter in the presence of EBNA-2 and -LP, but not that from Cp. Thus, although both EBNA-LP and EBNA-3C serve as coactivators of EBNA-2, they can regulate different subsets of promoters. Moreover, EBNA-3C activated expression from the LMP-1/2B promoter even in the presence of EBNA-LP, suggesting that each operates through a distinct mechanism.
Given the essential contribution of LMP-1 to EBV-mediated transformation, the ability to regulate LMP-1 is likely an important function of EBNA-3C. While reporter gene assays with EBV-negative cell lines have yielded conflicting results as to whether EBNA-3C activates or represses LMP-1 expression (53, 67, 83), EBNA-3C expression led to a substantial increase in the amount of LMP-1 protein in Raji cells. Furthermore, there was a corresponding increase, not only in the LMP-1 message but also in the amount of LMP-2B message, which is transcribed in the opposite direction and presumably controlled by the same regulatory elements in the LMP-1/2B bidirectional promoter. This coordinated regulation, together with the results of reporter gene assays, strongly suggests that EBNA-3C regulates the transcription of these two distinct latency-associated genes. Further evidence to support this conclusion was obtained from our ChIP assays, whose results suggested that EBNA-3C is associated with the LMP-1/2B bidirectional promoter when LMP-1 and LMP-2B are expressed. Because EBNA-3C has no demonstrable specific DNA-binding capability (68), it is likely that its interaction with the promoter is mediated through interactions with cellular transcription factors. Indeed, our previous studies have demonstrated that EBNA-3C can interact with the transcription factor PU.1, known to be essential for activation of the LMP-1 promoter by EBNA-2 (30), and can activate the expression of reporter genes controlled solely through PU.1 binding sites (84). Although other mechanisms could be envisioned to account for this observation (such as the removal of a repressor from PU.1), these data collectively suggest that EBNA-3C is targeted to DNA through associations with PU.1. Like that of other Ets family members, the activity of PU.1 is regulated through associations with other transcription factors (58). The most notable examples are the Ig κ and λ 3′ enhancers, to which PU.1 recruits IRF-4 to activate transcription (13, 63). This activity is repressed by the association of PU.1 with BSAP, a protein essential for proper B-cell development (50). More recently, it was reported that PU.1 associates with HDAC1, which leads to a repression of transcription (34). EBNA-3C might be targeted to DNA solely through associations with PU.1 (or other cellular proteins) or may make contacts with the DNA through, for example, the basic domain located in its amino terminus. These associations may perturb the interaction of PU.1 with other cellular transcription factors, known or novel, that regulate its function or may recruit additional transcription factors to the promoter.
EBNA-3C's function as a transcription factor has been studied previously using reporter gene assays, and the results demonstrated that EBNA-3C (in addition to EBNA-3A and EBNA-3B) can repress activation mediated through EBNA-2/Jκ. Jκ is a highly conserved protein, and the activity of the Drosophila melanogaster homologue, Suppressor of Hairless [Su(H)], is controlled by Hairless, a protein that regulates Su(H) activity in a manner similar to that of the EBNA-3 proteins observed in reporter gene assays, i.e., Hairless represses Su(H) activity (7). Given the precedent for this type of regulation, the hypothesis was developed that EBNA-3C prevents overexpression of the latency-associated proteins by repression of the EBNA (Cp), LMP-1, and LMP-2A promoters. Since all of the EBNAs are expressed from Cp, a widely accepted model arose in which the EBNA-3 proteins regulated Cp by a feedback repression mechanism, thereby preventing overexpression of LMP-1, which would lead to cytostasis. Although this is an appealing hypothesis, no evidence for repression of either Cp or the bidirectional LMP-1/2B promoter was seen in our system. Although EBNA-3C is not present in Raji cells, EBNA-3A and EBNA-3B are expressed, so it is possible that some repression is indeed exerted by these other members of the EBNA-3 family. However, studies of EBV-immortalized cell lines suggest that the level of Jκ exceeds that of the EBNA proteins (31). Thus, in an EBV-infected cell, the EBNAs may not compete for Jκ, whereas in transient transfection assays (where the highly overexpressed EBNA proteins are likely to be in considerable excess of endogenous Jκ) the substantially higher levels of EBNA proteins may result in competition for the available Jκ. In support of the supposition that EBNA-3C does not routinely regulate expression from Cp, recombinant EBV strains carrying a mutation of the Jκ sites in Cp or a deletion of Cp are able to immortalize B lymphocytes, and the resulting LCLs express normal levels of EBNA proteins, as do two naturally occurring lymphoblastoid cell lines that have deleted Cp (14, 71, 79, 80). It is possible that repression through Jκ might be a fail-safe mechanism for controlling Cp, such that if transcription reached an extremely high level, the EBNA-3 proteins could restore transcription to a suitable level.
Together, these results raise the question of the function of the Jκ binding sites in Cp and of the Jκ binding domain in each EBNA-3 protein. Clearly, the Jκ protein plays a prominent role in EBV biology. EBNA-2 is targeted to DNA through its interactions with Jκ, converting an inhibitory protein-DNA complex into an activating one (28, 75, 85). Binding sites for Jκ are conserved in the Cp, LMP-1/2B, and LMP-2A promoters of EBV-like LCVs (16, 18, 66). Likewise, the Jκ binding domain is conserved in the EBNA-3 homologues of these LCVs, suggesting that this domain serves an important function (12, 29, 82). Two other studies support this conclusion. In an EBV-transformed B-cell line, three- to fivefold overexpression of EBNA-3A results in growth arrest at the G0/G1 stage of the cell cycle, whereas EBNA-3A containing an inactivating mutation in the Jκ binding domain failed to arrest cells (10). More significantly, studies in an LCL containing a conditional EBNA-3A mutant demonstrated that wild-type EBNA-3A could support continued proliferation after inactivation of the endogenous EBNA-3A protein, whereas EBNA-3A with a mutation in the Jκ binding domain could not (54). Collectively, these data suggest an important function of the Jκ binding domain in each EBNA-3 protein. The interaction with Jκ may normally be involved in the activation of LMP-1 in a way that is not manifested in reporter gene assays, but such a function would have to involve a mechanism that is unique to EBNA-3C because we have previously demonstrated that neither EBNA-3A nor EBNA-3B can activate the LMP-1 promoter (84). The association with Jκ may also be involved in the regulation of cellular genes by EBNA-3C. Alternatively, the interaction with Jκ may serve an as yet unknown purpose. Our model system provides an ideal model for investigating these possibilities.
Acknowledgments
This research was supported by Public Health Service grant CA56645, Cancer Center Support (CORE) grant CA21765, and the American Lebanese Syrian Associated Charities (ALSAC).
We thank Evelyn Stigger-Rosser for technical assistance.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Allday, M. J., D. H. Crawford, and J. A. Thomas. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361-369. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkenbach, M., D. Liebowitz, F. Wang, J. Sample, and E. Kieff. 1989. Epstein-Barr virus latent infection membrane protein increases vimentin expression in human B-cell lines. J. Virol. 63:4079-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boos, H., R. Berger, C. Kuklik-Roos, T. Iftner, and N. Mueller-Lantzsch. 1987. Enhancement of Epstein-Barr virus membrane protein (LMP) expression by serum, TPA, or n-butyrate in latently infected Raji cells. Virology 159:161-165. [DOI] [PubMed] [Google Scholar]
- 6.Boos, H., M. Stoehr, M. Sauter, and N. Mueller-Lantzsch. 1990. Flow cytometric analysis of Epstein-Barr virus (EBV) latent membrane protein expression in EBV-infected Raji cells. J. Gen. Virol. 71:1811-1815. [DOI] [PubMed] [Google Scholar]
- 7.Brou, C., F. Logeat, M. Lecourtois, J. Vandekerckhove, P. Kourilsky, F. Schweisguth, and A. Israel. 1994. Inhibition of the DNA-binding activity of Drosophila suppressor of hairless and of its human homolog, KBF2/RBP-J kappa, by direct protein-protein interaction with Drosophila hairless. Genes Dev. 8:2491-2503. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell, R. G., J. B. Wilson, S. J. Anderson, and R. Longnecker. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405-411. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, A., E. Johannsen, S. Maruo, E. Cahir-McFarland, D. Illanes, D. Davidson, and E. Kieff. 2003. EBNA3A association with RBP-Jkappa down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J. Virol. 77:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, M. A., and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalbies-Tran, R., E. Stigger-Rosser, T. Dotson, and C. E. Sample. 2001. Amino acids of Epstein-Barr virus nuclear antigen 3A essential for repression of Jkappa-mediated transcription and their evolutionary conservation. J. Virol. 75:90-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenbeis, C. F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9:1377-1387. [DOI] [PubMed] [Google Scholar]
- 14.Evans, T. J., P. J. Farrell, and S. Swaminathan. 1996. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J. Virol. 70:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 16.Franken, M., B. Annis, A. N. Ali, and F. Wang. 1995. 5′ Coding and regulatory region sequence divergence with conserved function of the Epstein-Barr virus LMP2A homolog in herpesvirus papio. J. Virol. 69:8011-8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fruehling, S., and R. Longnecker. 1997. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology 235:241-251. [DOI] [PubMed] [Google Scholar]
- 18.Fuentes-Panana, E. M., S. Swaminathan, and P. D. Ling. 1999. Transcriptional activation signals found in the Epstein-Barr virus (EBV) latency C promoter are conserved in the latency C promoter sequences from baboon and rhesus monkey EBV-like lymphocryptoviruses (cercopithicine herpesviruses 12 and 15). J. Virol. 73:826-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jk recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 22.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 23.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatfull, G., A. Bankier, B. G. Barrell, and P. J. Farrell. 1988. Sequence analysis of Raji Epstein-Barr virus DNA. Virology 164:334-340. [DOI] [PubMed] [Google Scholar]
- 26.Henderson, S., M. Rowe, C. Gregory, D. Croom-Carter, F. Wang, R. Longnecker, E. Kieff, and A. Rickinson. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107-1115. [DOI] [PubMed] [Google Scholar]
- 27.Hickabottom, M., G. A. Parker, P. Freemont, T. Crook, and M. J. Allday. 2002. Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP). J. Biol. Chem. 277:47197-47204. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh, J. J., and S. D. Hayward. 1995. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science 268:560-563. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, H., Y. G. Cho, and F. Wang. 2000. Structural, functional, and genetic comparisons of Epstein-Barr virus nuclear antigen 3A, 3B, and 3C homologues encoded by the rhesus lymphocryptovirus. J. Virol. 74:5921-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johannsen, E., C. L. Miller, S. R. Grossman, and E. Kieff. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJkappa in Epstein-Barr virus-transformed B lymphocytes. J. Virol. 70:4179-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallin, B., J. Dillner, I. Ernberg, B. Ehlin-Henriksson, A. Rosen, W. Henle, G. Henle, and G. Klein. 1986. Four virally determined nuclear antigens are expressed in Epstein-Barr virus-transformed cells. Proc. Natl. Acad. Sci. USA 83:1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihara-Negishi, F., H. Yamamoto, M. Suzuki, T. Yamada, T. Sakurai, T. Tamura, and T. Oikawa. 2001. In vivo complex formation of PU.1 with HDAC1 associated with PU.1-mediated transcriptional repression. Oncogene 20:6039-6047. [DOI] [PubMed] [Google Scholar]
- 35.Knight, J. S., K. Lan, C. Subramanian, and E. S. Robertson. 2003. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 77:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight, J. S., and E. S. Robertson. 2004. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J. Virol. 78:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight, J. S., N. Sharma, D. E. Kalman, and E. S. Robertson. 2004. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells. J. Virol. 78:12857-12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krauer, K. G., A. Burgess, M. Buck, J. Flanagan, T. B. Sculley, and B. Gabrielli. 2004. The EBNA-3 gene family proteins disrupt the G2/M checkpoint. Oncogene 23:1342-1353. [DOI] [PubMed] [Google Scholar]
- 39.Laux, G., B. Adam, L. J. Strobl, and F. Moreau-Gachelin. 1994. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laux, G., F. Dugrillon, C. Eckert, B. Adam, U. Zimber-Strobl, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laux, G., A. Economou, and P. J. Farrell. 1989. The terminal protein gene 2 of Epstein-Barr virus is transcribed from a bidirectional latent promoter region. J. Gen. Virol. 70:3079-3084. [DOI] [PubMed] [Google Scholar]
- 42.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Roux, A., B. Kerdiles, D. Walls, J. F. Dedieu, and M. Perricaudet. 1994. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205:596-602. [DOI] [PubMed] [Google Scholar]
- 44.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling, P. D., J. J. Hsieh, I. K. Ruf, D. R. Rawlins, and S. D. Hayward. 1994. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 68:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling, P. D., R. S. Peng, A. Nakajima, J. H. Yu, J. Tan, S. M. Moses, W. H. Yang, B. Zhao, E. Kieff, K. D. Bloch, and D. B. Bloch. 2005. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 24:3565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Longnecker, R., C. L. Miller, X. Q. Miao, A. Marchini, and E. Kieff. 1992. The only domain which distinguishes Epstein-Barr virus latent membrane protein 2A (LMP2A) from LMP2B is dispensable for lymphocyte infection and growth transformation in vitro; LMP2A is therefore nonessential. J. Virol. 66:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longnecker, R., C. L. Miller, X. Q. Miao, B. Tomkinson, and E. Kieff. 1993. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J. Virol. 67:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maitra, S., and M. Atchison. 2000. BSAP can repress enhancer activity by targeting PU.1 function. Mol. Cell. Biol. 20:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mann, K. P., D. Staunton, and D. A. Thorley-Lawson. 1985. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J. Virol. 55:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mannick, J. B., J. I. Cohen, M. Birkenbach, A. Marchini, and E. Kieff. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 65:6826-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maruo, S., E. Johannsen, D. Illanes, A. Cooper, and E. Kieff. 2003. Epstein-Barr virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J. Virol. 77:10437-10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meitinger, C., L. J. Strobl, G. Marschall, G. W. Bornkamm, and U. Zimber-Strobl. 1994. Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element. J. Virol. 68:7497-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morgenstern, J. P., and H. Land. 1990. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 18:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389-399. [DOI] [PubMed] [Google Scholar]
- 58.Nagulapalli, S., J. M. Pongubala, and M. L. Atchison. 1995. Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3). J. Immunol. 155:4330-4338. [PubMed] [Google Scholar]
- 59.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA) 3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541-2549. [PubMed] [Google Scholar]
- 61.Peng, R., S. C. Moses, J. Tan, E. Kremmer, and P. D. Ling. 2005. The Epstein-Barr virus EBNA-LP protein preferentially coactivates EBNA2-mediated stimulation of latent membrane proteins expressed from the viral divergent promoter. J. Virol. 79:4492-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polack, A., H. Delius, U. Zimber, and G. W. Bornkamm. 1984. Two deletions in the Epstein-Barr virus genome of the Burkitt lymphoma nonproducer line Raji. Virology 133:146-157. [DOI] [PubMed] [Google Scholar]
- 63.Pongubala, J. M., S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol. Cell. Biol. 12:368-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rivailler, P., C. Quink, and F. Wang. 1999. Strong selective pressure for evolution of an Epstein-Barr virus LMP2B homologue in the rhesus lymphocryptovirus. J. Virol. 73:8867-8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sample, C. E., and B. D. Parker. 1994. Biochemical characterization of the Epstein-Barr virus nuclear antigen 3A and 3C proteins. Virology 205:535-539. [DOI] [PubMed] [Google Scholar]
- 69.Sample, J., D. Liebowitz, and E. Kieff. 1989. Two related Epstein-Barr virus membrane proteins are encoded by separate genes. J. Virol. 63:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swaminathan, S. 1996. Characterization of Epstein-Barr virus recombinants with deletions of the BamHI C promoter. Virology 217:532-541. [DOI] [PubMed] [Google Scholar]
- 72.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Touitou, R., M. Hickabottom, G. Parker, T. Crook, and M. J. Allday. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 75:7749-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uchida, J., T. Yasui, Y. Takaoka-Shichijo, M. Muraoka, W. Kulwichit, N. Raab-Traub, and H. Kikutani. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300-303. [DOI] [PubMed] [Google Scholar]
- 75.Waltzer, L., P. Y. Bourillot, A. Sergeant, and E. Manet. 1995. RBP-J kappa repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 23:4939-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J. Virol. 70:5909-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woisetschlaeger, M., J. L. Strominger, and S. H. Speck. 1989. Mutually exclusive use of viral promoters in Epstein-Barr virus latently infected lymphocytes. Proc. Natl. Acad. Sci. USA 86:6498-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woisetschlaeger, M., C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. USA 87:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yandava, C. N., and S. H. Speck. 1992. Characterization of the deletion and rearrangement in the BamHI C region of the X50-7 Epstein-Barr virus genome, a mutant viral strain which exhibits constitutive BamHI W promoter activity. J. Virol. 66:5646-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 82.Zhao, B., R. Dalbies-Tran, H. Jiang, I. K. Ruf, J. T. Sample, F. Wang, and C. E. Sample. 2003. Transcriptional regulatory properties of Epstein-Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J. Virol. 77:5639-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao, B., D. M. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jk. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing a Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou, S., M. Fujimuro, J. J. Hsieh, L. Chen, and S. D. Hayward. 2000. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol. 74:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila suppressor of hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zimber-Strobl, U., K. O. Suentzenich, G. Laux, D. Eick, M. Cordier, A. Calender, M. Billaud, G. M. Lenoir, and G. W. Bornkamm. 1991. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J. Virol. 65:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]