Abstract
The adeno-associated viruses (AAVs) can package and deliver foreign DNA into cells for corrective gene delivery applications. The AAV serotypes have distinct cell binding, transduction, and antigenic characteristics that have been shown to be dictated by the capsid viral protein (VP) sequence. To understand the contribution of capsid structure to these properties, we have determined the crystal structure of AAV serotype 4 (AAV4), one of the most diverse serotypes with respect to capsid protein sequence and antigenic reactivity. Structural comparison of AAV4 to AAV2 shows conservation of the core β strands (βB to βI) and helical (αA) secondary structure elements, which also exist in all other known parvovirus structures. However, surface loop variations (I to IX), some containing compensating structural insertions and deletions in adjacent regions, result in local topological differences on the capsid surface. These include AAV4 having a deeper twofold depression, wider and rounder protrusions surrounding the threefold axes, and a different topology at the top of the fivefold channel from that of AAV2. Also, the previously observed “valleys” between the threefold protrusions, containing AAV2's heparin binding residues, are narrower in AAV4. The observed differences in loop topologies at subunit interfaces are consistent with the inability of AAV2 and AAV4 VPs to combine for mosaic capsid formation in efforts to engineer novel tropisms. Significantly, all of the surface loop variations are associated with amino acids reported to affect receptor recognition, transduction, and anticapsid antibody reactivity for AAV2. This observation suggests that these capsid regions may also play similar roles in the other AAV serotypes.
Adeno-associated viruses (AAVs), which are members of the Dependovirus genus of the Parvoviridae, are helper-dependent parvoviruses isolated from a number of different species, including humans (50). Despite a requirement for coinfection with a helper virus, such as adenovirus or herpesvirus, for productive replication, AAV capsids are similar to those of the autonomous parvoviruses, with T=1 icosahedral symmetry and an overall diameter of ∼260 Å that encapsulates a single-stranded-DNA (ssDNA) genome of ∼5,000 bases (9, 40, 53, 62, 70). The capsid consists of three overlapping viral proteins (VPs), namely, VP1 (90 kDa), VP2 (72 kDa), and VP3 (60 kDa), at a ratio of 1:1:10, that are generated by alternative splicing and translational initiation during productive infection. VP3 constitutes 90% of the capsid, but all three proteins contain a common C-terminal domain of about 530 amino acids.
Recombinant gene transfer vectors based on AAV serotype 2 (AAV2) have proven effective in animal models for the correction of genetic diseases of the eye, brain, muscle, liver, and lung (e.g., see references 18-20, 26, and 58), with human clinical trials now under way for the correction of several diseases. However, numerous studies, including those that use “pseudotyped” capsids, in which AAV vectors are generated using the AAV2 inverted terminal repeats to package a reporter gene (such as green fluorescent protein or luciferase) or a therapeutic gene within the capsids of other serotypes, have shown that other serotypes possess unique tissue tropisms and improved gene transfer activities compared to those of AAV2 for some cell types (7, 13, 21, 22, 31, 49, 56, 64). AAV1, for example, can transduce rodent skeletal muscle as much as a thousandfold more efficiently than AAV2 (8). AAV4 has strong tropism for ependymal cells in the central nervous systems of mice (13, 43) and for retinal pigmented epithelium in rats, dogs, and nonhuman primates (65). Among the more recently discovered serotypes, AAV7 has also been shown to have superior muscle transduction compared to AAV2, while AAV8 and AAV9 are the most efficient serotypes discovered so far for transducing the liver (e.g., see references 21 and 22).
Tropism differences dictated by the capsid sequence and the need to improve the effectiveness of AAV gene therapy applications through capsid manipulation have generated a need to understand the basic biology of the different serotypes, particularly the mechanism(s) of cellular attachment and entry, antigenic reactivity, and capsid structure. The homology between the capsid sequences of the AAV serotypes is high, although primary cell surface receptor recognition properties are dramatically different. AAV2 and AAV3, which are ∼87% identical, utilize heparin sulfate as their primary receptor (59), but with different binding affinities (24, 56). This is likely due to the absence, in AAV3, of critical arginine residues (R585 and R588) identified as being essential for heparin binding by AAV2 (35, 52). AAV1, which is ∼83% identical to AAV2, also lacks these arginine residues and does not bind heparin sulfate (56). Recent studies suggested that AAV1 utilizes sialic acid for transduction (10). Interestingly, AAV6, which is >99% identical to AAV1 and does not have residues equivalent to R585 and R588, binds heparin, but this interaction does not appear to be necessary for virus entry (23). It has also been shown that AAV4 and AAV5, which are ∼55% identical to AAV2 and to each other, bind sialic acid, not heparin sulfate (11, 56). However, their glycan specificities differ. AAV4 binds to α2,3 O-linked sialic acids, while AAV5 requires α2-3 N-linked sialic acids for cell attachment and entry (31, 63, 64).
In addition to having unique cell tropisms, several AAV serotypes are antigenically distinct. AAV2 is the antigenically best-characterized serotype, for which linear and conformational epitopes have been identified on the capsid by using peptide mapping and single amino acid mutations (44, 67). Some of the other AAV serotypes show cross-reactivity to a number of AAV2 antibodies, although the A20 conformational epitope, which maps to four different regions of the primary sequence, is specific to AAV2 and AAV3 (44, 67). AAV4, which was originally isolated from African green monkeys, is one of the most antigenically distant AAV serotypes and is unable to cross-react with antibodies generated to linear AAV2 epitopes, such as the AAV2 B1 site. The B1 antibody is able to react with most of the other AAV serotypes (e.g., see references 56 and 67).
The three-dimensional structure of AAV2 has been determined by X-ray crystallography (70) and cryo-electron microscopy (cryo-EM) with image reconstruction (39, 40), and the structures of AAV4 and AAV5 have been determined by cryo-EM (53, 62). Surprisingly, the AAV capsid surface topology is similar in architecture to those of the autonomous parvovirus Aleutian mink disease virus (47) and human parvovirus B19 virus (B19) (1, 12, 34), despite very low VP amino acid sequence identity (53). The surfaces of the AAV capsids have depressions at the icosahedral twofold axes and surrounding the fivefold axes but contain three distinct protrusions surrounding the threefold axes, unlike the single-pinwheel arrangement of surface loops at these axes for members of the Parvovirus genus (reviewed in reference 53). The protrusions surrounding the threefold axis create valley-like depressions between the two- and threefold axes, as observed in Aleutian mink disease virus and B19. The cylindrical channel at the fivefold axis of the parvovirus capsid, which is implicated in packaging of the parvovirus genome and the phospholipase activity of the VP1 unique region (4, 16, 17), is conserved.
Structural variations that cluster from symmetry-related VP monomers of autonomous parvovirus capsids determine tissue tropism, pathogenicity, and antigenic disparities between highly homologous strains (reviewed in reference 2). A comparison of the pseudoatomic models of VP3 built into the cryo-EM densities of AAV4 and AAV5 capsids to the crystal structure of AAV2 (70) provided preliminary structural information indicating that variations of surface regions between these viruses likely play a role in receptor attachment and antigenicity differences (53, 62). However, detailed information on the positions of the amino acids in these variable structural regions was beyond interpretation at the resolution of the cryo-EM structures. To facilitate this analysis, we have determined the crystal structure of AAV4 to 3.2-Å resolution, using X-ray crystallography. Comparison of the VP3 atomic models of AAV4 and AAV2 enabled accurate assignments of their variable surface amino acids, most of which are involved in receptor recognition, affect transduction, or confer antigenic specificity in AAV2. This study has also identified AAV capsid regions that can tolerate compensating structural loop insertions and deletions without detriment to capsid assembly.
MATERIALS AND METHODS
Virus production, purification, and crystallization and data collection and processing.
The production, purification, and preliminary X-ray crystallographic studies of wild-type AAV4 capsids have been reported (33). Briefly, a total of 276 0.3°-oscillation X-ray diffraction images were collected from six frozen crystals at the F1 beamline at the Cornell High Energy Synchrotron Source. The crystals belong to space group I222, with the following cell dimensions: a = 339.6, b = 319.2, and c = 285.0 Å. The final data set consisted of a total of 192,100 independent reflections for data between 50.0- and 3.2-Å resolution, with an Rsym of 16.4% and an overall completeness of 75.9% (Table 1).
TABLE 1.
Summary of diffraction data and structure refinement statistics
| Parameter | Value or description |
|---|---|
| No. of crystals | 6 |
| No. of images | 276 |
| Crystal system/space group | Orthorhombic/I222 |
| Cell parameters a, b, and c (Å) | 339.6, 319.2, 285.0 |
| Resolution range (Å) | 50-3.2 |
| No. of unique reflections | 192,100 |
| Rsym (%)a | 16.4 (42.6)d |
| Completeness (%) | 75.9 (42.6)d |
| Rfactorb | 0.263 |
| Rfreec | 0.273 |
| RMSD for bond length (Å)/RMSD for angles (°) | 0.008/1.601 |
| Total no. of protein/nucleotide/ solvent atoms | 4129/22/15 |
| Average B factor for protein/nucleotide/ solvent atoms (Å2) | 30.8/54.7/8.0 |
| Ramachandran plot statistics (most favored/additionally allowed/ generously allowed) (%) | 75.8/22.6/1.6 |
Rsym = ΣI − <I>/Σ<I>, where I is the intensity of a reflection with indices h, k, and l and <I> is the average intensity of all symmetry-equivalent measurements of that reflection.
Rfactor = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated with the 5% of reflections excluded from the overall data set.
The numbers in parentheses are results for the highest-resolution shell.
Structure determination and refinement.
Molecular replacement procedures utilizing rotation and translation function searches for structure determination of the AAV4 capsid have been reported previously (33). There are two complete virus capsids in the I222 cell positioned at (0,0,0) and (1/2, 1/2, 1/2), with three orthogonal icosahedral twofold symmetry axes coincident with the three crystallographic twofold axes. Thus, there are 15 VP monomers per crystallographic asymmetric unit. Initial phases (for all reflections) were calculated using a polyalanine model generated from the VP2 coordinates of the crystal structure of feline panleukopenia virus (PDB accession no. 1C8E). The model was oriented and positioned based on the molecular replacement solution, followed by application of crystallographic symmetry operators to generate the two complete icosahedral particles in the crystal unit cell. Phases were calculated using the CNS program (6).
The refinement of the AAV4 capsid VP structure was performed by alternating cycles of refinement with the CNS program (6) and model building into averaged Fourier 2Fo-Fc and Fo-Fc electron density maps using the O program (29). A test data set of 5% was partitioned for monitoring of the refinement process (5). The initial phases were improved using simulated annealing, energy minimization, and conventional positional refinement followed by real-space electron density map averaging, using a molecular mask, while applying strict 15-fold noncrystallographic symmetry in the CNS and RAVE programs (6, 28, 36). Residues 211 to 734 of the AAV4 VP sequence were assigned to averaged sigma-weighted 2Fo-Fc electron density maps by manual substitution, insertion, and deletion of amino acids. Following this rebuilding procedure, new phases were calculated and improved by several alternating cycles of refinement, real-space electron density averaging, and rebuilding (6, 29). The refinement was deemed to have converged when there was no further improvement in the overall agreement between Fo and Fc (Rfactor) and the averaging correlation coefficient was 0.90. Water molecules were added to unassigned electron density (at 1.5σ in the Fo-Fc map) within hydrogen bond donor or acceptor distances. In addition, density inside the capsid consistent with a single DNA nucleotide was modeled as dAMP. The final AAV4 capsid VP model was then used to calculate average temperature factors and the root mean square deviation (RMSD) from ideal bond lengths and angles using the CNS program (6) and from main-chain torsion angles using the PROCHECK program (41) (Table 1). Figures were produced using the BOBSCRIPT (15), Chimera (54), GRASP (51), PyMol (14), and RASTER3d (48) programs.
Comparison of AAV4 and AAV2 VP3 structures.
The coordinates of VP3 for AAV4 and AAV2 (PDB accession no. 1LP3) were aligned with the secondary structure matching (SSM) program (38). The program superimposes C-α positions and provides information on residues that are structurally equivalent, inserts gaps when the atoms are too far apart to superimpose, and provides the distances (in Å) between the C-α positions.
Calculation of atomic contacts and buried surface area between VP monomers.
The residues involved in icosahedral two-, three-, and fivefold symmetry-related VP interactions (≤3.6 Å) were analyzed using the Contact subroutine in the CNS program (6). The buried surface areas between monomers were calculated for the AAV4 and AAV2 (PDB accession no. 1LP3) capsids at the two-, three-, and fivefold interfaces, also using the CNS program (6). This algorithm utilizes the method of buried surface calculation reported by Lee and Richards (42), assuming a probe radius of 1.4 Å.
Protein structure accession number.
The refined coordinates for AAV4 VP3 have been deposited in the Protein Data Bank (PDB accession no. 2G8G).
RESULTS
Structure of AAV4.
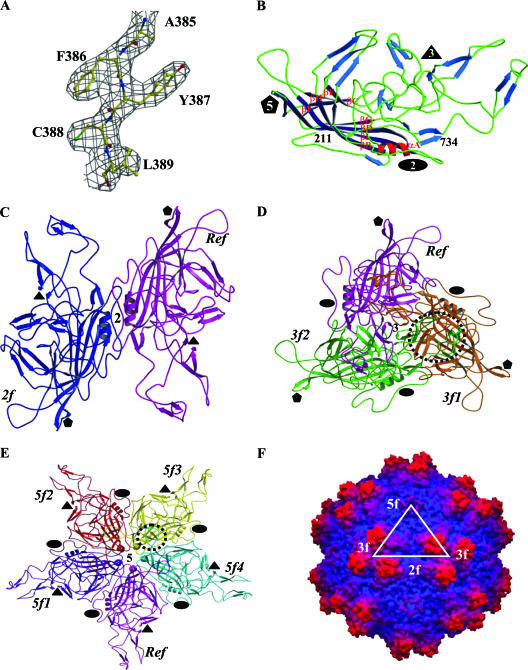
The crystal structure of AAV4 has been determined and refined to a 3.2-Å resolution. The final model consists of amino acid residues 211 to 734 (VP1 numbering), which are within the VP1/VP2/VP3 common polypeptide region (Fig. 1A and B), one DNA nucleotide (dAMP) (Fig. 2), and 15 solvent molecules (Table 1). The first 210 amino acids of AAV4, containing the VP1 unique region with the PLA2 domain, the VP1-VP2 common region (residues 137 to 195), and the first 15 residues of VP3, are not observed in the averaged density map. The lack of ordering of these N-terminal residues is consistent with reports for all parvovirus structures determined to date. This is due to the low copy numbers of VP1 and VP2 in the capsid and to the possibility that the N termini of VP1, VP2, and VP3 adopt different conformations, which is incompatible with the icosahedral symmetry assumed during structure determination. Since the AAV capsid is made up of mainly VP3 (∼90%), this protein designation will be used when describing the AAV4 VP structure for the remainder of the article. The final Rfactor, after restrained B value refinement, is 0.263 for all reflections (except for the 5% test data set) between 50- and 3.2-Å resolution. Rfree, calculated with the 5% Fo test data set, is 0.273. The similarity of Rfactor and Rfree for the structure is due to the high noncrystallographic icosahedral symmetry of the capsid. These R values are comparable to or better than those quoted for all available parvovirus capsid structures and those of viruses from other families, as detailed on the VIPER website at http://viperdb.scripps.edu (57). The stereochemical parameters and geometries of the AAV4 VP3 model (as listed in Table 1) are consistent with those reported for other virus structures at comparable resolutions.
FIG. 1.
Structure of AAV4. (A) Part of the 2Fo-Fc electron density map of AAV4 (3.2-Å resolution) (gray mesh) for residues 385 to 389, contoured at 1.5σ. (B) Ribbon diagram of AAV4 VP3 showing the core eight-stranded β-barrel strands (dark blue), stretches of antiparallel β-strands (light blue), loops (green), and the helical region (red). The first N-terminal residue observed (211), the C-terminal residue (734), the eight strands (βB to βI) that make up the core β-barrel, and a conserved alpha-helix, αA, are labeled. The secondary structure elements were assigned using the DSSP program (30). (C to E) Ribbon diagrams showing interactions of the AAV4 VP3 monomers at the two-, three-, and fivefold axes, respectively. The reference (Ref), twofold (2f), threefold (3f1 and 3f2), and fivefold (5f1 to 5f4) related monomers are labeled. The interdigitation of two VP3 monomers forming the protrusions around the threefold axis is highlighted by a dashed circle in panel D. The loop between βH and βI that contains interactions between adjacent fivefold related monomers is highlighted by a dashed circle in panel E. The approximate positions of the icosahedral two-, three-, and fivefold axes are shown in panels B to E by filled ovals, triangles, and pentagons, respectively. (F) Depth-cued surface representation of the AAV4 capsid crystal structure viewed down the icosahedral twofold axis. A viral asymmetric unit is depicted by a triangle (in white) bounded by two threefold (3f) axes, divided by a line drawn through the twofold (2f) and a fivefold (5f) axis. Panels A to E were generated using the BOBSCRIPT program (15) and rendered with the RASTER3d program (48); panel F was generated using the GRASP program (51).
FIG. 2.
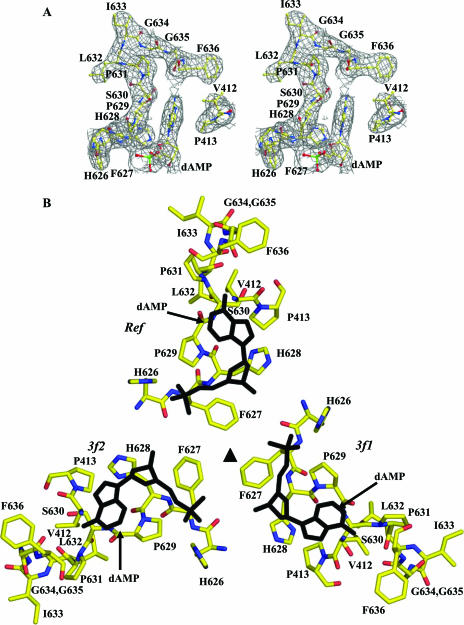
Ordered DNA nucleotide within the AAV4 capsid. (A) Stereoview of 2Fo-Fc electron density map (gray mesh) at the interior pocket containing an ordered DNA nucleotide. The amino acids and the modeled nucleotide (dAMP) are colored according to atom type and labeled. (B) Amino acid residues forming the nucleotide binding pocket, plus the nucleotide viewed through (inside to outside) the threefold axis. The amino acids are colored according to atom type, and the dAMP nucleotide is shown in black. The reference (Ref) and the two threefold related monomers (3f1 and 3f2) are labeled. This figure was generated using the BOBSCRIPT program (15) and rendered with the RASTER3d program (48).
The atomic model of AAV4 VP3 built into the X-ray density map enables the accurate assignment of amino acid side chain positions (Fig. 1A), surface loop regions, and secondary structure elements (Fig. 1B), in contrast to the lower-resolution cryo-EM structure and pseudomodel previously published (53). AAV4 VP3 contains a core eight-stranded β-barrel motif (βB to βI) decorated by loop insertions between the β-strands and a small α-helix (αA; residues 283 to 292) that is conserved in all parvovirus structures determined to date (Fig. 1B). Small stretches of antiparallel β-strands are also observed in the loops between the core strands (Fig. 1B), as previously described for AAV2 (70) and members of the autonomous parvoviruses (reviewed in reference 9).
The clustering of the loop/strand regions between the eight core β-strands from icosahedral symmetry-related VP3 monomers (Fig. 1C to E) forms the characteristic parvovirus capsid surface features of AAV4 (Fig. 1F). There is a depression at the icosahedral twofold axis, three protrusions surrounding the threefold axis, and a depression surrounding a channel at the icosahedral fivefold axis. The diameter of the capsid is 223 Å at the twofold axis, 252 Å at the icosahedral threefold axis, 279 Å at the peak of the three mounds surrounding this axis, and 246 Å at the icosahedral fivefold axis. The floor of the twofold axis is the thinnest region of the capsid shell, formed by only two symmetry-related loops (residues 695 to 704) which intertwine about each other (Fig. 1C). The highly conserved αA forms the wall of the depression at the twofold axis (Fig. 1C). The three separate mounds surrounding the icosahedral threefold axis of the AAV4 capsid (Fig. 1F) are formed from the intertwining of four loops from two threefold symmetry-related VP3 monomers (Fig. 1D). These mounds contain residues 437 to 459 and 576 to 594 from one VP3 monomer and residues 478 to 508 and 539 to 558 from another monomer. These residues are located in the large loop between strands βG and βH. Two of the small stretches of strand structure, between βD and βE, form a β-ribbon, which together with ribbons clustered from fivefold icosahedral symmetry-related monomers line the conserved cylindrical channel at this capsid region (Fig. 1E and F). The floor of the depression around this channel is lined by the loop between βH and βI (the HI loop; residues 650 to 667 [VP1 numbering]) that invades from the adjacent fivefold symmetry-related monomer (Fig. 1E).
Following the refinement of the AAV4 VP3 monomer and the addition of water molecules, a difference Fourier map showed additional density below the capsid interior at the icosahedral threefold axis, consistent with a dAMP nucleotide (Fig. 2A). The binding pocket is surrounded by two glycines, two histidines, two phenylalanines, three prolines, one valine, and a serine residue. The adenine base is stacked between two of the prolines, P413 and P629 (Fig. 2), and its N-1 and N-6 atoms are involved in a polar interaction with the main-chain atoms of residues N637 and P629 (Table 2). The phosphate group is close enough to form a hydrogen bond with one of the histidines, H626 (Table 2; Fig. 2B).
TABLE 2.
Summary of the types of interactions between symmetry-related AAV4 VP3 monomers
| Interaction residues (or nucleotide) and axisa | Interactionb | Interaction residues (or nucleotide) and axisa | Interactionb | |
|---|---|---|---|---|
| Twofold axis | *W472-P621* | W | ||
| *S283-W693* | W# | *W472-P631* | W | |
| * P284-P695* | W | *L473-P521* | W | |
| *R285-E688* | E& | *L473-L632* | W | |
| *R285-E696* | W | *E562-R383* | E | |
| *Q288-P695* | W | *E563-R383* | W | |
| *Q288-E698* | W | A565-D511 | W | |
| *R289-E688* | E | N577-H507 | W | |
| *N292-Q698* | E | L578-I478 | W | |
| *N293-N293* | E | L578-T506* | W | |
| *N293-N293* | E | L578-T595 | W | |
| *P357-W693* | W | P579-Q481 | W | |
| *E688-R285* | E | *Q583-Q481* | W | |
| *E688-R289* | E | *Q583-F483 | W | |
| *W693-S283* | W | *Q583-N490* | E | |
| *W693-S284* | W | *Q583-Y491 | W | |
| *W693-P357* | W | L588-Y491 | W | |
| *W693-Y719* | E | L588-Y504 | W | |
| *P695-P284* | W | L597-I478 | W | |
| *P695-Q288* | W | V600-P521* | W | |
| *P695-W711* | W | N607-D624* | W | |
| *E696-R285* | W | *Q606-D624* | E | |
| *Q698-Q288* | W | *S690-E340 | W | |
| *Q698-N292* | E | *K691-E340 | W | |
| *Q698-T700* | W | *K691-F392* | W | |
| F699-P695* | W | *R692-A385 | W | |
| *T700-Q698* | W | H732-H622* | W | |
| *T700-T700* | E | H733-E340 | E | |
| W711-P695* | W | H733-Y387* | W | |
| *Y719-W693* | E | dAMP-O2P-H626 | E | |
| dAMP-O2P-H626 | W | |||
| Threefold axis | dAMP-N1-N637 | E | ||
| *S416-D624* | E | dAMP-N6-N637 | E | |
| *Y418-H622* | W | dAMP-N6-P629 | E | |
| *Q422-P342* | W | |||
| *S423-L371 | W | Fivefold axis | ||
| *D425-R513* | E | *Y251-P713 | W | |
| *M428-Q347 | W | R253-S708 | W | |
| *M428-Q349 | W | *N326-K312* | E | |
| *N429-Y272* | E | *N326-N325* | E | |
| *N429-V344* | W | *T328-Q310* | E | |
| *N429-Q349 | W | *T328-N325* | W | |
| *P430-L254 | W | *Q332-W222* | W | |
| *P430-C369 | W | *Q379-Q705 | E | |
| *L431-L254 | W | Q380-L709 | W | |
| *L431-Q365* | W | D382-S701* | E | |
| * I432-P364* | W | *F386-F356* | W | |
| *Y435-R277* | W | *F386-A712 | W | |
| *Y435-P615* | W | *F386-P713 | W | |
| *L439-F483 | W | *E390-P357* | W | |
| *L439-L501 | W | *E390-N358 | W | |
| T443-K492 | W | *Y391-S283* | E | |
| L448-D499 | W | *Y391-D286* | E | |
| L448-Y504 | W | *P393-H223* | W | |
| N449-Y491 | W | *P393-D225* | W | |
| A453-K492 | W | *Q395-D221* | W | |
| T454-T551 | W | *M396-N308 | W | |
| *F457-F533 | W | *M396-Q676* | W | |
| K459-E350 | E | *M398-N308* | W | |
| R461-N548 | W | *R398-T399* | W | |
| T463-L258 | W | *K648-N358 | W | |
| F465-Y263* | W | *P653-V242 | W | |
| S466-N261* | W | *P653-T674* | W | |
| N467-I502 | W | *N655-V314* | W | |
| K469-T518 | W | *P656-Y672* | W | |
| K469-P519 | W | *S662-E350* | E | |
| K470-S515* | E | *V665-V360* | W | |
| *N471-P631* | W | *F668-V360* | W | |
| *N471-I633 | W | *I669-V323 | W |
*, residues/interactions (≤3.6Å) that are conserved between AAV4 and AAV2 (70).
#, electrostatic interactions (H bonds, salt bridges, and polar interactions); &, hydrophobic and van der Waals contacts.
Structural comparison of AAV4 and AAV2 VP3 proteins.
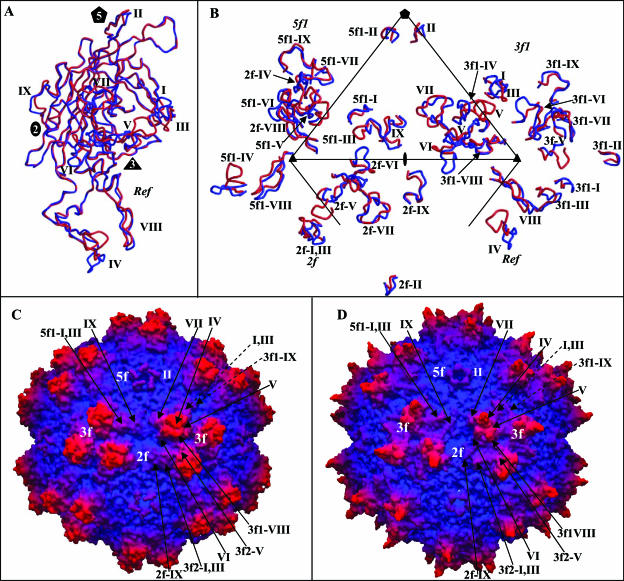
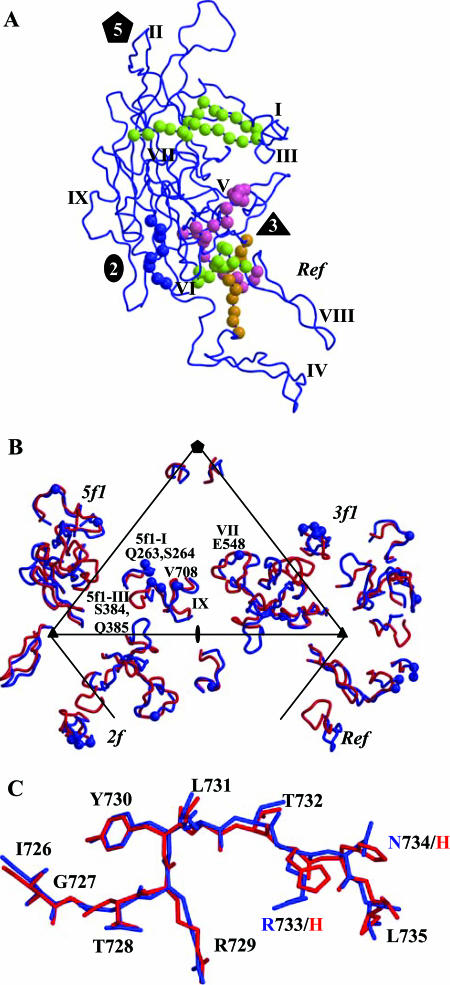
A superimposition of the AAV2 VP3 model (70) onto the AAV4 electron density map identified main- and side-chain regions that were structurally different between the two serotypes (Fig. 3A and B). Using the SSM program (38), 497 of 524 residues of AAV4 VP3 were superimposable onto the equivalent VP3 residues of AAV2, with an overall RMSD of 1.0 Å. The remaining C-α positions were too variable to be compared. This comparison highlighted nine regions (I to IX) that are conformationally distinct (defined as stretches of more than two amino acids with C-α positions that are >1.0 Å apart) between the two serotypes (Fig. 3C). The majority of the variable regions (IV to VIII) are located in the stretch of amino acids between strands βG and βH (Fig. 3C). All of the regions are close to or on the capsid surface (Fig. 4). Structural alignment of the AAV4 and AAV2 VP3 proteins shows that their ordered regions are ∼75% identical, which is ∼20% greater than their primary amino acid sequence identity. The percentage of structural identity is lower than that previously reported (98%) for comparison of the pseudoatomic model of AAV4 VP3 built into a 13-Å-resolution cryo-EM and image-reconstructed map with the AAV2 VP3 atomic model (53). This is to be expected because the pseudoatomic model was built from the AAV2 VP3 atomic coordinates and docked by rigid-body rotation and translation into a cryo-EM envelope, with limited adjustment of main-chain regions (53).
FIG. 3.
Structural superimposition and alignment of AAV4 and AAV2 VP3 sequences. (A and B) Atomic models of AAV4 (gray) and AAV2 (black) variable loop regions VI and IX superimposed onto the respective pieces of 2Fo-Fc electron density maps of AAV4 (gray mesh). (C) Structural alignment of the ordered regions of AAV4 and AAV2 VP3 proteins. The alignment was generated by superimposition of the C-α positions of the VP3 atomic models of AAV4 (residues 211 to 734 [VP1 numbering]) and AAV2 (residues 217 to 735 [VP1 numbering]) (PDB accession no. 1LP3). Identical residues and structural regions are shown as white characters in black boxes, superimposable nonidentical residues are shown in black, and regions that adopt a different conformation (nonsuperimposable, with C-α differences of >1.0 Å) are shown in black above (AAV2) and below (AAV4) the aligned residues. The variable regions are labeled I to IX based on the comparison of AAV4 and AAV2 alone and are thus refined from the original definition of Padron et al. (53).
FIG. 4.
Comparison of AAV4 and AAV2. (A) Superimposition of coil representations of atomic models of AAV4 (in red) and AAV2 (in blue) (PDB accession no. 1LP3) VP3 monomers. The positions of variable regions I to IX, defined in Fig. 3, are labeled. The approximate two-, three-, and fivefold axes are indicated as in Fig. 1. (B) Positions of variable loop regions I to IX within a viral asymmetric unit. The asymmetric unit is defined in the legend to Fig. 1. The viral asymmetric unit contains contributions from the reference and a twofold (2f), a threefold (3f1), and a fivefold (5f1) VP3 monomer. The prefixes on the labels for the variable regions indicate the contributing monomers. (C and D) Depth-cued surface representations of the AAV4 and AAV2 capsid crystal structures, respectively, viewed down the icosahedral twofold axis. The clustered locations of variable regions I to IX are shown for a viral asymmetric unit of each virus and some of the adjacent regions; the dashed arrows indicate variable regions of the reference outside the asymmetric unit. Panels A and B were generated using the BOBSCRIPT program (15) and rendered with the RASTER3d program (48); panels C and D were generated using the GRASP program (51).
Superimposition of the AAV4 and AAV2 VP3 proteins shows the distribution of AAV4/AAV2 variable regions in the contexts of the monomer and the viral asymmetric unit (Fig. 4A and B, respectively). While they are distributed throughout the VP3 primary sequence and monomer structure (Fig. 3C and 4A), these variable regions are clustered from icosahedral symmetry-related monomers to create local variations on the AAV capsid (Fig. 4C and D). Variable regions I, III, VII, and IX contribute to differences between the two viruses in the raised capsid surface region between the depressions at the icosahedral twofold axis and surrounding the fivefold axis (Fig. 4C and D). This region is more pronounced in AAV4, resulting in a deeper twofold axis than that of AAV2, which has an almost continuous depression between the two- and fivefold axes (Fig. 4D). Variable region IV is “folded” down toward the interface between threefold monomers containing variable region V in AAV4 (Fig. 4B), resulting in the rounder appearance of the protrusions surrounding its icosahedral threefold axis than the finger-like projections in AAV2 (Fig. 4C and D). Variable regions V and VII result in a wider base for the AAV4 protrusion. The conformational difference at variable region VI is located on the wall surrounding the twofold depression/base of the threefold protrusions, also adding to the width of the AAV4 protrusions. The larger protrusions surrounding the threefold axis of AAV4 result in a narrowing of the “valley” that runs from the threefold axis to the twofold axis of its capsid compared to that in AAV2 (Fig. 4C and D). The clustering of variable region II results in a noticeable structural difference between the AAV4 and AAV2 capsids at the top of the channel formed by fivefold symmetry-related VP3 monomers (Fig. 4C and D). Structural variations similar to the AAV4/AAV2 loop differences are observed in analogous capsid regions in structures for members of the Parvovirus genus, including the closely related strains of Minute virus of mice (MVM) (reviewed in references 2 and 37).
This comparison of the crystal structures of AAV4 and AAV2 VP3 topologies, along with earlier comparisons of parvovirus structures, indicates that their VPs can tolerate variation in equivalent surface regions that cluster to create three main groups of capsid topologies (reviewed in references 9, 37, and 53). Notably, for AAV4 and AAV2, despite the surface loop variations described above, adjacent regions of compensating structural deletions and insertions (e.g., regions III and VI or regions I and V in Fig. 4B) cluster on the capsid surface to form similar overall parvovirus capsid topologies.
AAV subunit interface interactions.
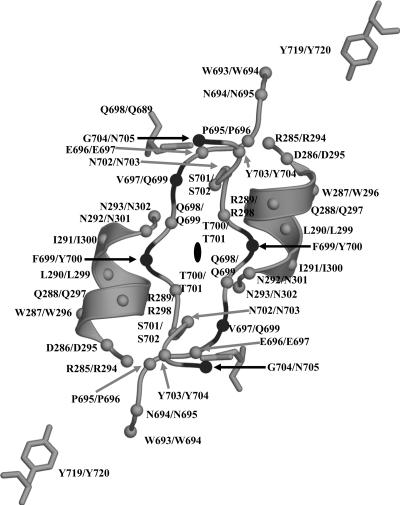
The results of analysis of the contact residues and their interactions at the two-, three-, and fivefold capsid interfaces are shown in Table 2 for AAV4. The data for AAV2 are similar (not shown). The majority of the interface interactions involve hydrophobic and van der Waals contacts, although there is also a significant number of electrostatic and hydrogen bonding interactions (11 of 29) at the twofold axis (Table 2). The threefold interactions involve 9 electrostatic and 57 hydrophobic contacts, while the fivefold interface is formed by 8 electrostatic and 25 hydrophobic contacts.
The residues involved in the twofold (dimer) and fivefold (pentamer) interactions are almost completely conserved between AAV4 and AAV2, while those creating the threefold mounds are more extensive and varied (Table 2). The two- and fivefold interactions utilize residues in the core eight-stranded β-barrel domain, and the conserved αH helix forms the majority of the twofold interface contacts (19/29) (Table 2; Fig. 3C and 5). This is consistent with a direct comparison of the AAV capsid interface residues and those forming the variable surface loop regions described above, which shows that the most structurally conserved interface is between twofold-related VP monomers, with only variable region IX involved in dimer formation (Table 2; Fig. 1, 3C, 4, and 5). The pentamer interface is also formed mainly from structurally conserved regions but does involve interactions between residues in variable regions III and IX and in region II at the top of the fivefold channel (Table 2; Fig. 4B, C, and D). The threefold interactions mostly involve residues within the elaborate loop between βG and βH (the G-H loop) (Table 2; Fig. 3C). Trimer formation involves interactions between amino acids in six of the variable regions, namely, regions III to VIII (Table 2; Fig. 4B and D). As mentioned above, complementary loop conformations (similar to deletions/insertions) are juxtaposed to assemble the AAV-like parvovirus capsid topology.
FIG. 5.
AAV4 twofold interface. The figure shows a coil representation of the residues lining the depression at the icosahedral twofold axis of the AAV4 capsid and the conserved helix αA on the wall of the depression. The C-α positions are shown as small gray and black balls for amino acids that are conserved and different, respectively, between AAV4 and AAV2. The residue numbers and types are labeled for AAV4/AAV2. These residues form the twofold interface interactions listed in Table 2. The approximate twofold axis is shown as a filled oval. This figure was generated with the BOBSCRIPT program (15) and rendered with the RASTER3d program (48).
Calculation of the buried surface areas between the AAV4 and AAV2 VP3 monomers at the dimer, trimer, and pentamer interfaces showed that the most extensive interactions are between threefold-related monomers, while the least extensive interactions are at the twofold axis. In AAV4, the dimer interface buries 4.4% (2,479 Å2) of the VP3 surface, the trimer interface buries 21% (10,030 Å2), and the pentamer interface buries 8.6% (4,690 Å2). For AAV2, the buried surface areas are 5.3% (2,895 Å2), 20% (9,653 Å2), and 8.6% (4,690 Å2) for the dimer, trimer, and pentamer interfaces, respectively. A similar ranking of buried surface areas is observed for interface formation in autonomous parvovirus capsids (68, 71). Calculation of the total buried surface area for capsid formation showed that the AAV4 capsid buries 17,664 Å2 more VP surface than does AAV2. The larger buried surface area for AAV4 capsid formation is due largely to the conformational difference at variable region IV, which folds this loop back onto the interface between the threefold monomers (Fig. 4A and B), suggesting that this capsid might be more stable than AAV2.
DISCUSSION
“Pseudotyping” and “cross packaging” strategies (25, 27, 55, 56) that have enabled direct comparison of the capsids of representative serotypes of the distinct AAV clades, as well as mutagenesis studies on AAV2, the best-studied serotype, have shown that the capsid sequence determines tissue tropism, transduction efficiency, and antigenic reactivity (21). In addition, the available cryo-EM and crystal structures of AAV2 have enabled the mapping of these functional properties onto the capsid (4, 35, 40, 44, 52, 70). In efforts to correlate capsid structure to the basic biology of the other AAV serotypes, including interactions for infection, uncoating, assembly, and antibody reactivity, we have determined the crystal structure of AAV4. This virus has a distinct tissue tropism from those of the other AAV serotypes and is the most antigenically different. A detailed comparison of the AAV4 and AAV2 VP3 structures showed that the core β-barrel motif is highly conserved, while variable regions map to surface loops between the β-strands (Fig. 1, 3, and 4). A similar arrangement of conserved and variable regions has also been observed for crystal structures of the other parvovirus capsids (reviewed in references 9 and 37).
AAV subunit interactions and buried surface area: implications for capsid assembly and stability.
The amino acids and structural regions forming the AAV4 and AAV2 VP3 twofold and fivefold interactions are more conserved than those utilized for threefold interactions. Attempts to mix entire AAV1-AAV5 VPs to form mosaic capsids in an effort to produce novel tropisms showed the VPs of AAV4 to be the most incompatible with those of the other AAVs (56). In addition, alanine scanning mutagenesis of charged residues in the AAV2 VPs has shown that mutations of two-, three-, and fivefold-interface amino acid residues can result in no-capsid phenotypes (69). There is limited information on the assembly pathway of the AAV capsid with respect to the required intermediates, and the above observations (56, 69) suggest that fidelity in the formation of all three icosahedral interfaces is important. Thus, it is probable that structural incompatibility of the interface regions, particularly at the threefold axes since these are the most varied, is responsible for the inability of the AAV4 VPs to mix with those of AAV2 and other serotypes for the formation of mosaic capsids, as observed by Rabinowitz et al. (56). Future attempts to generate novel tropisms by mixing AAV VPs could be aided by consideration of potential structural variations, as described above for AAV4 and AAV2, which could be circumvented by engineering amino acid stretches that satisfy structurally compensating regions of surface loop deletions/insertions.
The larger buried surface area calculated for the formation of the AAV4 capsid suggests that is likely to be more stable than that of AAV2. Recently, the AAV2 capsid was reported to be uncoated at a lower rate than those for AAV6 and AAV8, which results in slower cellular transduction than that of the latter viruses due to a lag in conversion of the genome to double-stranded DNA forms (60). While a similar comparative study of AAV2 and AAV4 is not yet available, the prediction is that AAV4 capsids would be uncoated even slower than AAV2 capsids.
Putative nucleotide binding site.
The ordering of a dAMP nucleotide inside the AAV4 capsid, directly under the icosahedral threefold axis, was unexpected. Significantly, the adenine base was unambiguously assigned into the Fo-Fc density (at a 4σ density cutoff), as defined by the density for the amino group at ring position C-6. The density interpreted as the base could not accommodate an NH2 group at the C-2 position of a guanine nucleotide. The AAVs package single copies of either the plus- or minus-sense strand of their ssDNA genomes inside different capsids. Therefore, the AAV4 crystals were obtained from two different populations of capsids. In addition, the application of noncrystallographic symmetry operators during the structure refinement and electron density map averaging procedures assumed icosahedral symmetry, and as such the resultant observed structure is compliant with this constraint. Thus, the assumption is that dAMP, which was refined with an occupancy of 1.0 and a B factor of 54.7 Å2 (Table 1), is bound in the majority of the 60 binding pockets inside the capsid. This specificity of nucleotide binding suggests either that the genome is structurally ordered to place the same nucleotide into the 60 equivalent pockets or that the configuration of the pocket is highly specific for interaction with this nucleotide. The mechanism driving this interaction is not known.
Interestingly, the amino acids that form the nucleotide binding pocket are highly conserved between all AAVs (53), and there are other reports of amino acid-base interactions involving proline residues and adenine (46). However, there was no report of a similar nucleotide ordered in the crystal structure of AAV2 (70). This binding pocket is not conserved in the capsid structures of members of the Parvovirus genus that package only the minus-sense strand of their ssDNA genomes, and the ordered nucleotide is not in an analogous region to the ssDNA nucleotides ordered inside the capsid in the crystal structures of canine parvovirus (CPV) (71) and MVM (3). Mutagenesis studies are under way to investigate the significance of the conserved amino acid residues in AAV genomic DNA packaging, capsid stability, assembly, and disassembly.
Functional comparison of AAV4 and AAV2 with respect to regions involved in heparin binding and transduction efficiency.
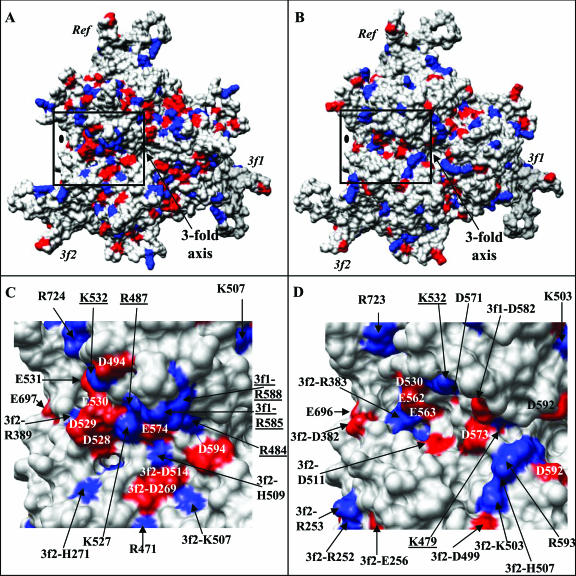
Although it is known that AAV4 binds O-linked sialic acids for cellular entry and infection, the capsid residues/regions required for this interaction are not known. There is no consensus amino acid sequence known to recognize sialic acid. On the other hand, heparin sulfate, utilized by AAV2 as a primary receptor, is known to interact with patches of basic amino acids. Residues contributing to the heparin binding site of AAV2, namely, R484, R487, K532, R585, and R588, have been identified by mutagenesis (35, 52) and are clustered from threefold symmetry-related VP3 monomers on the walls and bases of the mounds surrounding the icosahedral threefold axes (Fig. 6A and C). The two critical residues, R585 and R588, are not conserved in AAV4, but AAV4 K479, which aligns with AAV2 R484 in the primary sequence, is located in a structurally equivalent position in both capsids (Fig. 3 and 6C and D). AAV4 K485 and R532, which align with AAV2 R487 and K532, respectively, in the primary sequence, are located in variable regions V and VI. The clustering of variable regions V, VI, and VIII from symmetry-related VP3 monomers buries AAV4 K485 and alters the surface position of AAV4 K532 on the wall of the threefold protrusion/valley between the two- and threefold axes (Fig. 6C and D). These differences also bury most of the side chain of AAV4 K479 (Fig. 6D). Interestingly, AAV4 contains a number of basic surface residues, including R383, K503, H507, and R593, close to the AAV2 basic patch (Fig. 6D). The C-α position of AAV4 R383 is structurally equivalent to that of AAV2 R389, although the arginine side chain is mostly buried in AAV2 because of adjacent variable regions (Fig. 6C). The C-α position of AAV4 H507 is equivalent to that of AAV2 K507, but the surface exposures of their side chains are different due to their proximity to variable region V (Fig. 6C and D). There are no basic residues in AAV2 that are structurally equivalent to AAV4 K503 or R593; instead, three acidic residues, D269, D514, and D594, occupy the equivalent surface region (Fig. 6C). Thus, while both viruses have basic residues in this capsid region, a continuous appropriately disposed minimum basic surface required to bind heparin sulfate is missing in AAV4, resulting in its negative heparin binding phenotype.
FIG. 6.
Charge distributions on the AAV2 and AAV4 capsid surfaces. (A and B) Distributions of basic (blue) and acidic (red) residues on theVP3 trimers (Ref, 3f1, and 3f2) of AAV2 and AAV4, respectively, viewed down the icosahedral threefold axis. The approximate twofold axis is indicated with a filled oval. (C and D) Close-up views of the distributions of basic and acidic residues in the AAV2 capsid region (boxed in panel A) mapped as the heparin binding site by mutagenesis (35, 52) and the structurally equivalent region in AAV4 (boxed in panel B), respectively. The residue positions are either labeled on top of the respective surface region or indicated by black arrows. The underlined residues were shown to affect heparin binding in AAV2. This figure was generated using the Chimera program (54).
A further observation in comparing the heparin binding region of AAV2 and the equivalent capsid surface region of AAV4 is that the basic patch required for AAV2 heparin binding is in contact with an acidic region (Fig. 6C). This acidic patch, next to AAV2 R487, K527, and K532, is formed by residues AAV2 D528, D529, E530, E531, and E574 (Fig. 6C), which are conserved in most of the AAV serotypes, except for AAV4 and AAV5 (53). AAV6 contains lysine at the primary sequence position equivalent to position 530 in AAV2. AAV4 D573, which is structurally equivalent to AAV2 E574, is separated from the acidic patch by a hydrophobic region (Fig. 6D). However, the AAV4/AAV2 surface variations due to regions V and VI expose residues D530, E562, and E563 to create an acidic patch next to K532 in AAV4 (Fig. 6D). Mutagenesis and structural studies of CPV and MVM, respectively, showed that residues on the walls of the twofold axes at the bases of their threefold protrusions which are structurally equivalent or close to the acidic/basic residues in the AAVs function as sialic acid binding regions (45, 61). Interestingly, CPV and MVM also have similar distributions of acidic/basic residues on the walls of their twofold axes extending toward the threefold axis (data not shown). Thus, the local loop variations and the resulting charge distributions in this region of the AAV4/AAV2 capsid surface may play a part in their ability to recognize and bind different cell surface carbohydrates. The significance of the structural juxtaposition of the acidic and basic residues in heparin binding, and possibly sialic acid binding, for the AAV capsids awaits structural determination of these capsids with their respective receptors.
A cell transduction “dead zone” which is unrelated to heparin binding was recently delineated on the AAV2 capsid by mutagenesis of capsid surface residues followed by transduction experiments with human HepG2 cells (44). Mutation of residues in this capsid region affects the transduction properties of AAV2. Six of the nine AAV4/AAV2 variable regions (I, II, III, V, VI, and IX) contain residues in this dead zone, which is located in the raised capsid surface region between the twofold and fivefold depressions and the wall of the twofold depression/base of the threefold protrusions (Table 3; Fig. 4D). It was suggested that the dead zone could be a coreceptor attachment site for AAV2 and that such an interaction could be mediated by heparin since the region is close to the AAV2 basic patch (44). Notably, AAV2 residue K532 (in variable region VI), originally mapped as part of the heparin binding region (discussed above), was shown to affect AAV2 transduction rather than heparin binding in this study and was classed as a dead zone residue (44). A basic residue is conserved at this amino acid position in most of the human and nonhuman primate AAV serotypes isolated to date, except for AAV5, which has an asparagine (53). This correlation of AAV4/2 conserved and variable regions with the transduction dead zone defined for AAV2 suggests that analogous capsid regions of the other AAV serotypes may also affect their transduction properties.
TABLE 3.
AAV2-AAV4 variable regions and reported functional roles in AAV2
| Variable region (amino acid residues)a | Functional role(s) in AAV2b | Reference(s) |
|---|---|---|
| I (262-268)a | Transduction, A20 and IVIG neutralization | 44, 67, 69 |
| II (326-330) | Transduction | 44 |
| III (380-388) | Transduction and A20 neutralization | 44, 67, 69 |
| IV (449-468) | HS and IVIG neutralization | 44, 69 |
| V (487-504) | Transduction, HB, HS and IVIG neutralization | 35, 44, 52, 69 |
| VI (525-541) | Transduction, HS and IVIG neutralization | 35, 44, 52, 69 |
| VII (544-556) | A20, HS, and IVIG neutralization | 44 |
| VIII (579-594) | HB, HS, and IVIG neutralization | 35, 44, 52 |
| IX (704-711) | Transduction, HB, A20, HS, and IVIG neutralization | 44, 67, 69 |
AAV2 VP1 numbering.
A20, AAV2 capsid antibody; IVIG, intravenous immunoglobulin G; HB, heparin binding; HS, human serum.
Functional comparison of AAV4 and AAV2 with respect to antigenic reactivity.
In efforts to generate second-generation AAV vectors that conserve their original tissue tropism but can evade neutralization by preexisting host immune antibodies, information on antigenic regions of the capsid is valuable. However, the only antibody available for AAV4 was generated to a predicted surface peptide (residues 455 to 467 [AAV4 VP1 numbering]) (32). Polyclonal antibodies generated against this peptide are useful for AAV4 VP Western blot analysis but are unreactive against all other AAV serotypes. This peptide is located in a region of high amino acid variability between the AAV serotypes (53) and is structurally located in variable region IV at the top of the protrusions surrounding the threefold axis (Fig. 3C and 4B and C). Thus, the lack of reactivity of other AAV serotypes to polyclonal antibodies generated against it is not surprising.
As discussed above for capsid regions controlling receptor recognition and transduction efficiency, AAV2 is the antigenically best-characterized AAV serotype. Peptide mapping has been used to identify both linear and conformation-dependent epitopes on the AAV2 capsid (66, 67). Three linear epitopes (A1, A69, and B1) and four conformational epitopes (A20, C24-B, C37-B, and D3) have been reported, although the residues for the C24-B site have not been mapped. For the linear epitopes, A1 antibody recognizes VP1 only, while A69 antibody recognizes a peptide in the VP1/VP2 common region and B1 antibody recognizes all three capsid proteins via an epitope at their C termini (67). The C24-B and C37-B epitopes elicit antibodies that neutralize by inhibiting heparin sulfate receptor attachment, while antibodies to the A20 site neutralize at a postattachment step and D3 antibodies are nonneutralizing.
AAV serotype reactivity to antibodies to the A20 epitope is limited to AAV2 and AAV3. The A20 epitope, which was mapped by peptide antibodies (Fig. 7A, residues 272 to 281, 369 to 378, and 566 to 575) (67) and, more recently, fine mapped by single-amino-acid mutagenesis (residues 263, 264, 384, 385, 548, and 708) (Fig. 7B) (44), contains residues from four different regions of the primary VP3 sequence. Structurally, these regions are clustered together on the capsid surface ridge between the two-, three-, and fivefold axes in the AAV2 structure, at or close to variable regions I, III, VII, and IX (Table 3; Fig. 7B). Interestingly, the study describing the AAV2 transduction dead zone (discussed above) also identified residues in variable regions I and IV to IX as affecting binding and neutralization by pooled human sera and intravenous immunoglobulin G (44) (Table 3). Thus, if the dead zone is involved in coreceptor recognition, as Lochrie et al. proposed (44), then the A20 antibody, pooled sera, and immunoglobulin G may block this interaction and inhibit subsequent steps required for trafficking.
FIG. 7.
Antigenic variation between AAV4 and AAV2. (A) The conformational antigenic sites defined for AAV2 using peptide mapping are shown on a VP3 monomer (blue coil). The C-α positions for the A20 epitope (VP1 numbering; residues 272 to 281, 369 to 378, and 566 to 575) are shown in lime green balls, the C37-B epitope (residues 493 to 502 and 602 to 610) is shown in magenta, the D3 epitope (residues 474 to 483) is shown in orange, and the linear B1 epitope (residues 726 to 733) is shown in blue. The two-, three-, and fivefold axes are depicted as described in the legend to Fig. 1, and variable regions I to IX are labeled. (B) C-α positions (in blue balls) of residues involved in A20 antibody binding and neutralization, as defined by single amino acid mutations mapped onto the AAV4/AAV2 variable regions. The mutated residues and the variable regions in which they are located are labeled. The viral asymmetric unit, as defined in the legend to Fig. 1F, is shown. (C) Superimposition of the AAV2 (blue) and AAV4 (red) structures at the amino acid stretch that forms the B1 epitope. The two amino acids that differ in this region are labeled with the same color as that used for the parent virus structure. This figure was generated using the BOBSCRIPT program (15) and rendered with the RASTER3d program (48).
Antibodies to the C37-B epitope specifically recognize AAV2, not AAV1, -3, -4, or -5 (67). This is consistent with this epitope (Fig. 7A, residues 493 to 502 and 602 to 610 [VP1 numbering]) containing residues in variable region V and being close to region VIII, containing the critical R585 and R588 AAV2 heparin binding residues. Thus, the phenotype of its antibodies is consistent with their inhibiting receptor attachment. D3 antibodies recognize serotypes AAV1, AAV3, and AAV5 but not AAV4 (67). The recognition of AAV1, AAV3, and AAV5 is consistent with the D3 epitope (Fig. 7A, residues 474 to 483) being fairly conserved among these AAV serotypes (53). The lack of AAV4 recognition by D3 antibodies is consistent with the proximity of this epitope's residues to variable region V and with the other structural differences between AAV4 and AAV2 that cluster at the mounds surrounding the icosahedral threefold axes (Fig. 4).
The AAV4 capsid has a unique phenotype among the AAVs in that it is not recognized by the B1 antibody, which is directed to a linear epitope (residues 726 to 733) at the conserved C-terminal end of the AAV VP3 protein (67). Comparison of the AAV2 and AAV4 VP3 crystal structures at the B1 epitope amino acids showed no major conformational differences, with the C-α atoms being superimposable (Fig. 7C). This suggests that H732 in AAV4 (Fig. 7C), located at the amino acid position equivalent to AAV2 R733, is involved in the inhibition of the AAV4 capsid-antibody interaction since most other AAVs recognized by the B1 antibody have an arginine or lysine at this position.
In summary, this study has identified regions of the AAV capsid that are structurally conserved between two distinct serotypes that can be probed further by mutagenesis to determine their functional roles in capsid assembly and genome packaging, which require concerted interactions of high fidelity. Significantly, all of the AAV4/AAV2 variable regions identified in this study coincide with surface regions associated with the functionality of the AAV2 capsid protein with respect to receptor binding, transduction efficiency, and antigenic reactivity (Table 3). This observation suggests an evolutionary divergence resulting in distinct phenotypes for the two viruses. It also suggests that these variable regions likely play similar roles for the other AAV serotypes. Finally, this AAV4/AAV2 comparison identified capsid surface regions that can tolerate structural insertions and deletions, which can be utilized to rationally engineer specific cell/tissue-targeted vectors for improved applications by peptide insertion.
Acknowledgments
We thank the MACCHESS staff for their help during X-ray diffraction data collection at the Cornell High Energy Synchrotron Source (CHESS), Ithaca, N.Y. We especially thank Lana Walsh for help in obtaining beamline time (EM 407 and 477).
This project was supported by NIH/NHLBI grant P01 HL51811 (M.A.-M. and N.M.) and by NIH intramural support (J.A.C.).
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Agbandje, M., S. Kajigaya, R. McKenna, N. S. Young, and M. G. Rossmann. 1994. The structure of human parvovirus B19 at 8 Å resolution. Virology 203:106-115. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., and M. S. Chapman. 2006. Correlating structure with function in the viral capsid, p. 125-139. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, N.Y.
- 3.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 4.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunger, A. T. 1992. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Burger, C., O. Gorbakyuk, M. J. Velardo, C. Peden, P. Williams, S. Zolotukhin, P. J. Reier, R. J. Mandel, and N. Muzyczka. 2004. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 10:302-317. [DOI] [PubMed] [Google Scholar]
- 8.Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, M. S., and M. Agbandje-McKenna. 2006. Atomic structure of viral particles, p. 107-123. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, N.Y.
- 10.Chen, S., M. Kapturczak, S. A. Loiler, S. Zolotukhin, O. Y. Glushakova, K. M. Madsen, R. J. Samulski, W. W. Hauswirth, M. Campbell-Thompson, K. I. Berns, T. R. Flotte, M. A. Atkinson, C. C. Tisher, and A. Agarwal. 2005. Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Hum. Gene Ther. 16:235-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiorini, J. A., F. Kim, L. Yang, and R. M. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chipman, P. R., M. Agbandje-McKenna, S. Kajigaya, K. E. Brown, N. S. Young, T. S. Baker, and M. G. Rossmann. 1996. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc. Natl. Acad. Sci. USA 93:7502-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, B. L., C. S. Stein, J. A. Heth, I. Martins, R. M. Kotin, T. A. Derksen, J. Zabner, A. Ghodsi, and J. A. Chiorini. 2000. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 97:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLano, W. L. 2002. The PyMol molecular graphics system. DeLano Scientific, San Carlos, Calif.
- 15.Esnouf, R. M. 1997. Bobscript: an extensively modified version of MOLSCRIPT that includes greatly enhanced coloring capabilities. J. Mol. Graph Model. 15:132-143. [DOI] [PubMed] [Google Scholar]
- 16.Farr, G. A., S. F. Cotmore, and P. Tattersall. 2006. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J. Virol. 80:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flannery, J. G., S. Zolotukhin, M. I. Vaquero, M. M. LaVail, N. Muzyczka, and W. W. Hauswirth. 1997. Efficient photoreceptor-targeted gene expression in vivo by recombinant adeno-associated virus. Proc. Natl. Acad. Sci. USA 94:6916-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flotte, T., A. Agarwal, J. Wang, S. Song, E. S. Fenjves, L. Inverardi, K. Chesnut, S. Afione, S. Loiler, C. Wasserfall, M. Kapturczak, T. Ellis, H. Nick, and M. Atkinson. 2001. Efficient ex vivo transduction of pancreatic islet cells with recombinant adeno-associated virus vectors. Diabetes 50:515-520. [DOI] [PubMed] [Google Scholar]
- 20.Flotte, T. R., M. L. Brantly, L. T. Spencer, B. J. Byrne, C. T. Spencer, D. J. Baker, and M. Humphries. 2004. Phase I trial of intramuscular injection of a recombinant adeno-associated virus alpha 1-antitrypsin (rAAV2-CB-hAAT) gene vector to AAT-deficient adults. Hum. Gene Ther. 15:93-128. [DOI] [PubMed] [Google Scholar]
- 21.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halbert, C. L., J. M. Allen, and A. D. Miller. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handa, A., S. Muramatsu, J. Qiu, H. Mizukami, and K. E. Brown. 2000. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J. Gen. Virol. 81:2077-2084. [DOI] [PubMed] [Google Scholar]
- 25.Hauck, B., L. Chen, and W. Xiao. 2003. Generation and characterization of chimeric recombinant AAV vectors. Mol. Ther. 7:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzog, R. W., J. N. Hagstrom, S.-H. Kung, S. J. Tai, J. M. Wilson, K. J. Fisher, and K. A. High. 1997. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc. Natl. Acad. Sci. USA 94:5804-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hildinger, M., A. Auricchio, G. Gao, L. Wang, N. Chirmule, and J. M. Wilson. 2001. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J. Virol. 75:6199-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, T. A. 1992. A, yaap, asap, @#*? A set of averaging programs, p. 92-105. In E. J. Dodson, S. Gover, and W. Wolf (ed.), Molecular replacement. SERC Daresbury Laboratory, Chilton, United Kingdom.
- 29.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 31.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaludov, N., B. Handelman, and J. A. Chiorini. 2002. Scalable purification of adeno-associated virus type 2, 4, or 5 using ion-exchange chromatography. Hum. Gene Ther. 13:1235-1243. [DOI] [PubMed] [Google Scholar]
- 33.Kaludov, N., E. Padron, L. Govindasamy, R. McKenna, J. A. Chiorini, and M. Agbandje-McKenna. 2003. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 4. Virology 306:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann, B., A. A. Simpson, and M. G. Rossmann. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. USA 101:11628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleywegt, G. J., and T. A. Jones. 1994. Halloween…masks and bones, p. 59-66. In S. Bailey, R. Hubbard, and D. Waller (ed.), From first map to final model. SERC Daresbury Laboratory, Chilton, United Kingdom.
- 37.Kontou, M., L. Govindasamy, H. J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M. P. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 79:10931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60:2256-2268. [DOI] [PubMed] [Google Scholar]
- 39.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskowski, R. A., J. A. Rullmannn, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 42.Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55:379-400. [DOI] [PubMed] [Google Scholar]
- 43.Liu, G., I. H. Martins, J. A. Chiorini, and B. L. Davidson. 2005. Adeno-associated virus type 4 (AAV4) targets ependyma and astrocytes in the subventricular zone and RMS. Gene Ther. 12:1503-1508. [DOI] [PubMed] [Google Scholar]
- 44.Lochrie, M. A., G. P. Tatsuno, B. Christie, J. W. McDonnell, S. Zhou, R. Surosky, G. F. Pierce, and P. Colosi. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Bueno, A., M. P. Rubio, N. Bryant, R. McKenna, M. Agbandje-McKenna, and J. M. Almendral. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80:1563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luscombe, N. M., R. A. Laskowski, and J. M. Thornton. 2001. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29:2860-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J. Virol. 73:6882-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merritt, E. A., and M. E. Murphy. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50:869-873. [DOI] [PubMed] [Google Scholar]
- 49.Mori, S., L. Wang, T. Takeuchi, and T. Kanda. 2004. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330:375-383. [DOI] [PubMed] [Google Scholar]
- 50.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2360. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, New York, N.Y.
- 51.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 52.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 79:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 55.Rabinowitz, J. E., D. E. Bowles, S. M. Faust, J. G. Ledford, S. E. Cunningham, and R. J. Samulski. 2004. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J. Virol. 78:4421-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy, V. S., P. Natarajan, B. Okerberg, K. Li, K. V. Damodaran, R. T. Morton, C. L. Brooks III, and J. E. Johnson. 2001. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J. Virol. 75:11943-11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snyder, R. O., S. K. Spratt, C. Lagarde, D. Bohl, B. Kaspar, B. Sloan, L. K. Cohen, and O. Danos. 1997. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum. Gene Ther. 8:1891-1900. [DOI] [PubMed] [Google Scholar]
- 59.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas, C. E., T. A. Storm, Z. Huang, and M. A. Kay. 2004. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 78:3110-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tresnan, D. B., L. Southard, W. Weichert, J. Y. Sgro, and C. R. Parrish. 1995. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology 211:123-132. [DOI] [PubMed] [Google Scholar]
- 62.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 78:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walters, R. W., J. M. Pilewski, J. A. Chiorini, and J. Zabner. 2002. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 277:23709-23713. [DOI] [PubMed] [Google Scholar]
- 64.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 65.Weber, M., J. Rabinowitz, N. Provost, H. Conrath, S. Folliot, D. Briot, Y. Cherel, P. Chenuaud, J. Samulski, P. Moullier, and F. Rolling. 2003. Recombinant adeno-associated virus serotype 4 mediates unique and exclusive long-term transduction of retinal pigmented epithelium in rat, dog, and nonhuman primate after subretinal delivery. Mol. Ther. 7:774-781. [DOI] [PubMed] [Google Scholar]
- 66.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, H., and M. G. Rossmann. 1993. The canine parvovirus empty capsid structure. J. Mol. Biol. 233:231-244. [DOI] [PubMed] [Google Scholar]
- 69.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]