Abstract
PTEN is a tumor suppressor that reverses the action of phosphoinositide 3-kinase by catalyzing the removal of the 3′ phosphate of phosphoinositides. Despite the critical role of PTEN in cell signaling and regulation, the mechanisms of its membrane recruitment and activation is still poorly understood. PTEN is composed of an N-terminal phosphatase domain, a C2 domain, and a C-terminal tail region that contains the PSD-95/Dlg/ZO-1 homology (PDZ) domain-binding sequence and multiple phosphorylation sites. Our in vitro surface plasmon resonance measurements using immobilized vesicles showed that both the phosphatase domain and the C2 domain, but not the C-terminal tail, are involved in electrostatic membrane binding of PTEN. Furthermore, the phosphorylation-mimicking mutation on the C-terminal tail of PTEN caused an ≈80-fold reduction in its membrane affinity, mainly by slowing the membrane-association step. Subcellular localization studies of PTEN transfected into HEK293T and HeLa cells indicated that targeting of PTEN to the plasma membrane is coupled with rapid degradation and that the phosphatase domain and the C2 domain are both necessary and sufficient for its membrane recruitment. Results also indicated that the phosphorylation regulates the targeting of PTEN to the plasma membrane not by blocking the PDZ domain-binding site but by interfering with electrostatic membrane binding of PTEN. On the basis of these results, we propose a membrane-binding and activation mechanism for PTEN, in which the phosphorylation/dephosphorylation of the C-terminal region serves as an electrostatic switch that controls the membrane translocation of the protein.
The tumor suppressor gene PTEN encodes a 403-aa phosphatase with enzymatic activities toward both peptides and 3-phosphoinositides, phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3] in particular (1–4). Accumulating evidence has shown that the tumor suppressor activity of PTEN relies on its 3-phosphoinositide phosphatase activity (5–7). This activity reduces the levels of PtIns(3,4,5)P3 and other 3-phosphoinositides and thereby antagonizes the activity of growth factor-stimulated phosphoinositide 3-kinase that activates many downstream cellular processes including cell growth, apoptosis, and cell motility (2–4, 8, 9). Although much is known about the mechanism by which PTEN regulates cellular processes, less is known about the regulatory mechanism of PTEN activities.
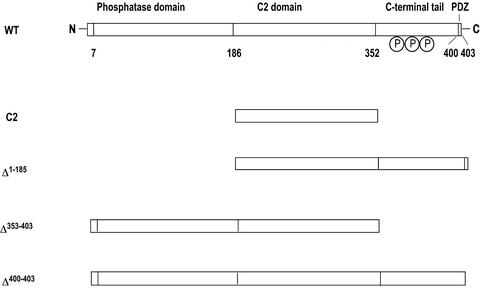
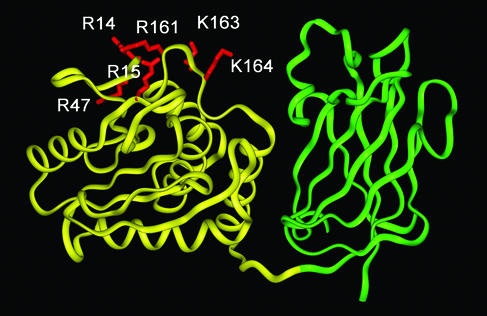
PTEN is composed of the N-terminal phosphatase domain (≈180 aa), the C2 domain (≈165 aa), and the C-terminal tail (≈50 aa) (see Fig. 1). The crystal structure of PTEN lacking the C-terminal tail showed that the phosphatase domain and the C2 domain make extensive domain contact and that the two domains contain clustered cationic residues on their putative membrane-binding surface (10) (see Fig. 2). The structure also revealed that the C2 domain lacks all but one calcium ligand of canonical calcium-binding C2 domains. On the other hand, the C-terminal tail contains several phosphorylation sites, a PSD-95/Dlg/ZO-1 homology (PDZ) domain-binding sequence (at the C-terminal end) and two possible PEST sequences.
Fig. 1.
Schematic representation of structures of PTEN and its mutants. PTEN has a N-terminal phosphatase domain, a C2 domain, and a C-terminal tail that contains multiple phosphorylation sites and a PDZ domain-binding sequence. Numbering is based on the x-ray structure of PTEN (10).
Fig. 2.
A ribbon diagram of PTEN. The phosphatase domain and the C2 domains are shown in yellow and green ribbons, respectively. The mutated residues in the phosphatase domain are highlighted in red and labeled. The molecule is oriented with its membrane-binding surface pointing upward. The coordinates are taken from the x-ray structure by Lee et al. (10).
Extensive mutational studies have indicated that the phosphatase domain and the C2 domain form a minimal catalytic unit (10–12), in which the C2 domain may play a dual role of promoting membrane recruitment and assisting a productive orientation of the phosphatase domain at the membrane surface (12). However, the role of the C-terminal tail in the function and regulation of PTEN remains controversial. Because the deletion of this region had little effect on the phosphatase activity but significantly reduces the cellular stability of PTEN (11–13), it has been thought that the C-terminal tail might regulate the cellular PTEN activities by modulating its stability. It has been proposed also that the C-terminal tail is involved in membrane localization of PTEN through the interaction with PDZ domain-containing proteins such as membrane-associated guanylate kinase with inverted orientation (MAGI) proteins (14–17). Several studies have indicated that PTEN is constitutively phosphorylated, presumably on S370, S380, T383, T383, and S385 in the C-terminal tail by casein kinase 2 (11, 18). More recently it was suggested that this phosphorylation causes a conformation change that results in the masking of the PDZ domain-binding site, thereby suppressing the recruitment of PTEN into the PTEN-associated complex in the plasma membrane (19). However, physiological relevance of the PTEN–PDZ domain binding has not been fully established yet (4). For instance, the deletion of the PDZ domain-binding site of PTEN failed to exert any effect on the cellular level of PtdIns(3,4,5)P3 (20, 21).
This study was undertaken to better understand the mechanism of membrane binding and activation of PTEN, with an emphasis on elucidating the effect of the C-terminal phosphorylation on PTEN activation. Results from biophysical and cell studies with a panel of PTEN mutants provide insight into how the membrane recruitment and activation of this important protein is regulated in the cell.
Experimental Procedures
Plasmid Construction and Protein Expression. The cDNA of PTEN was subcloned by using NdeI and EcoR1 into the pET28a vector (Novagen) that encodes an N-terminal His6 tag and a thrombin cleavage site. All mutations and truncation (see Fig. 1) were carried out by overlap-extension PCRs (22). Escherichia coli BL21 (DE3) cells were used as the host for protein expression for all PTEN constructs. All PTEN constructs except the C2 domain and Δ1–185 were expressed as soluble proteins and purified from the bacterial extracts by using a Ni2+-nitrilotriacetic acid agarose column (Qiagen, Valencia, CA) according to manufacturer protocol. The C2 domain and Δ1–185 were expressed as inclusion bodies, which then were solubilized and purified as described (23). All proteins were >90% pure electrophoretically. Protein concentration was determined by the bicinchoninic-acid method (Pierce).
Surface Plasmon Resonance (SPR) Analysis of Membrane Binding. The preparation of vesicle-coated Pioneer L1 sensor chip (Biacore, Piscataway, NJ) was described in detail elsewhere (24, 25). Plasma membrane-mimicking vesicles [1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (POPI)/cholesterol (12:35:22:9:22)] and nuclear envelope-mimicking vesicles [POPC/POPE/POPS/POPI/cholesterol (61:21:4:7:7)] were prepared as described (26). The sensor surface in the control cell was coated with 100% POPC vesicles for which all PTEN constructs have extremely low affinity. The sample cell contains the sensor surface coated with the vesicles indicated in Table 1. All kinetic experiments were performed at 23°C in 20 mM Hepes, pH 7.4, containing 0.16 M KCl and 5% glycerol unless specified otherwise. The flow rate was maintained at 60 μl/min to circumvent mass transport effects. The association was monitored for 90 s, and dissociation was monitored for 4 min. For data acquisition, five or more different concentrations (typically within a 10-fold range above or below the Kd) of each protein were used. Association and dissociation phases of all refractive index-corrected sensorgrams were globally fit to a 1:1 Langmuir-binding model [protein + (protein-binding site on the vesicle) ↔ (complex)] by using biaevaluation 3.0 software (Biacore). The association phase was analyzed by using the equation R= [kaC/(kaC + kd)]Rmax(1 – e–(kaC+kd)(t–t0)), where Rmax is the theoretical binding capacity, C is the protein concentration, ka is the association rate constant, and t0 is the initial time. The dissociation phase was analyzed by using the equation R = R0e–kd(t–t0), where kd is the dissociation rate constant, and R0 is the initial response. The dissociation constant (Kd) then was calculated from the equation, Kd = kd/ka. We have established for several peripheral proteins that kinetically determined Kd values are essentially the same as those determined from equilibrium measurements (25–27). It should be noted that in our SPR analysis Kd is defined not in terms of the molarity of phospholipids but the molarity of protein-binding sites on the vesicle. Thus, if each protein-binding site on the vesicle is composed of n lipids, nKd is the dissociation constant in terms of molarity of lipid monomer (28). Because of the difficulty involved in accurate determination of the concentration of lipids coated on the sensor chip, only Kd was determined in our SPR analysis, and the relative affinity was calculated as a ratio of Kd values assuming that n values are essentially the same for WT and mutants. This assumption was supported by the finding that Rmax values, which are proportional to [total lipid]/n (28), were similar for WT and most mutants.
Table 1. Binding parameters for PTEN constructs determined from SPR analysis.
| Proteins | Lipids | ka, M-1·s-1 | kd, s-1 | Kd, M | Fold increase in Kd* |
|---|---|---|---|---|---|
| Wild type | POPC/POPS (8:2) | (5.2 ± 0.4) × 105 | (1.5 ± 0.1) × 10-3 | (2.9 ± 0.3) × 10-9 | 1 |
| Wild type | POPC/POPG (8:2) | (5.0 ± 0.5) × 105 | (1.6 ± 0.1) × 10-3 | (3.2 ± 0.4) × 10-9 | 1.1 |
| C2 | POPC/POPS (8:2) | (2.5 ± 0.2) × 104 | (2.1 ± 0.2) × 10-3 | (8.4 ± 0.9) × 10-8 | 29 |
| Δ1-185 | POPC/POPS (8:2) | (2.3 ± 0.2) × 104 | (2.1 ± 0.2) × 10-3 | (9.1 ± 1.1) × 10-8 | 31 |
| Δ350-403 | POPC/POPS (8:2) | (5.7 ± 0.1) × 105 | (2.4 ± 0.2) × 10-3 | (4.2 ± 0.4) × 10-9 | 1.5 |
| R11A/K13A/R14A/R15A | POPC/POPS (8:2) | (5.0 ± 0.2) × 103 | (1.1 ± 0.1) × 10-3 | (2.2 ± 0.2) × 10-7 | 76 |
| R161A/K163A/K164A | POPC/POPS (8:2) | (2.8 ± 0.1) × 104 | (1.8 ± 0.1) × 10-3 | (6.4 ± 0.4) × 10-8 | 22 |
| S380A/T382A/T383A | POPC/POPS (8:2) | (4.8 ± 0.1) × 105 | (1.7 ± 0.2) × 10-3 | (3.5 ± 0.5) × 10-9 | 1.2 |
| S380E/T382E/T383E | POPC/POPS (8:2) | (5.3 ± 0.2) × 103 | (1.0 ± 0.1) × 10-3 | (2.4 ± 0.2) × 10-7 | 83 |
| R161A/K163A/K164A/S380E/T382E/T383E | POPC/POPS (8:2) | (5.5 ± 0.1) × 103 | (1.0 ± 0.1) × 10-3 | (2.3 ± 0.3) × 10-8 | 80 |
| Wild type | Plasma membrane mimic† | (1.2 ± 0.2) × 106 | (1.5 ± 0.2) × 10-3 | (1.3 ± 0.2) × 10-9 | 0.45 |
| Wild type | Nuclear membrane mimic‡ | NM | NM | NM | — |
All values represent means and standard deviations from triplicate determinations. All measurements for PTEN constructs were performed in 20 mM Hepes, pH 7.4, containing 0.16 M KCI and 5% glycerol. All mutants except R11A/K13A/R14A/R15A have wild-type-like specific activity values toward monomeric PtdIns(3,4,5)P3. NM, not measurable.
Fold increase in Kd relative to the binding of PTEN to POPC/POPS (8:2) vesicles.
POPC/POPE/POPS/POPI/cholesterol (12:35:22:9:22).
POPC/POPE/POPS/POPI/cholesterol (61:21:4:7:7).
Phosphatase Assay. Phosphatase assays were performed in 50 μl of assay buffer (100 mM Tris·HCl, pH 8.0/10 mM DTT) containing 100 μM monomeric 1,2-dioctanoyl derivative of PtdIns(3,4,5)P3 (Echelon, Salt Lake City). The phosphatase reaction was initiated by adding ≈2.5 μg of purified PTEN or mutant and quenched by adding the malachite green reagent containing 1 M HCl after a 40-min incubation at 37°C. The release of the inorganic phosphate then was quantified by the malachite green calorimetric assay as described (13).
Cell Culture, Transfection, and Confocal Microscopy. WT PTEN and its mutants were subcloned in frame with a C-terminal enhanced GFP (EGFP) into the pIND (for HEK293T cells) or pcDNA3.1 (for HeLa cells) vectors (29). Stable HEK293T cells expressing ecdysone receptor (Invitrogen) or HeLa cells were grown in DMEM containing 10% FBS at 37°C with 5% CO2 and 98% humidity. The cell transfection and protein expression were performed as described (30). Confocal imaging was performed by using a four-channel Zeiss LSM 510 laser scanning microscope. EGFP was excited by using the 488-nm line of an argon/krypton laser. A 505-nm line-pass filter and a ×63, 1.2-numerical aperture water-immersion objective were used for all experiments. Immediately before imaging, induction medium was removed, and the cells were washed twice with 1 mM Hepes, pH 7.4, containing 2.5 mM MgCl2, 1 mM NaCl, 0.6 mM KCl, 0.67 mM d-glucose, and 6.4 mM sucrose. The cells then were overlaid with the same buffer and imaged.
Antibodies and Immunoblotting. The PTEN antibody and S380/T382/T383-specific phospho-PTEN antibodies were purchased from Cell Signaling Technologies (Beverly, MA). Lysis of HEK293T cells and immunoblotting of PTEN proteins were performed as described (31).
Results
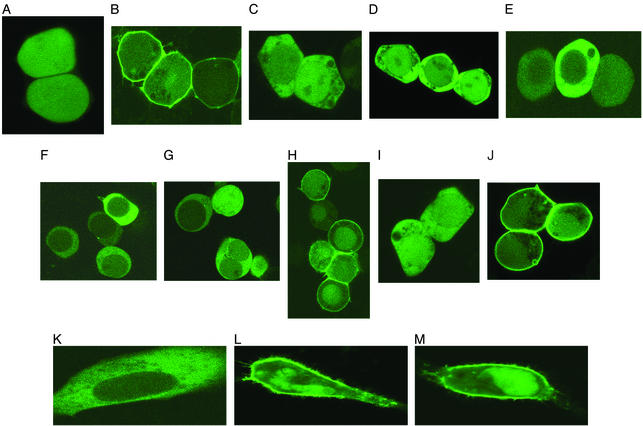
Role of C2 Domain in Membrane Binding. C2 domains are Ca2+-dependent membrane-targeting modules found in many cellular proteins involved in cell signaling and membrane trafficking. Although the C2 domain of PTEN lacks most Ca2+-binding ligands, it has been implicated in Ca2+-independent membrane recruitment of the protein (10, 12). To quantitatively assess the contribution of the C2 domain to overall membrane binding of PTEN, we measured the in vitro membrane-binding parameters of PTEN and its isolated C2 domain. Our initial SPR measurements showed that both the full-length PTEN and the C2 domain had extremely low affinity for zwitterionic POPC vesicles. Further measurements using mixed vesicles of POPC/POPS (8:2), which roughly approximate the lipid composition of the inner plasma membrane of mammalian cells to which PTEN is targeted, showed that both the full-length PTEN and the C2 domain had higher affinity (in terms of Kd) for these anionic vesicles (see Table 1). The affinity for anionic vesicles was mainly attributed to nonspecific electrostatic interactions, because they did not distinguish POPC/POPS (8:2) from POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) (8:2) vesicles (Table 1) and the affinity was reduced greatly by 0.5 M KCl [Kd = (2.2 ± 0.4) × 10–7 M; ka = (9.5 ± 1.2) × 103 M–1·s–1; kd = (2.1 ± 0.3) × 10–3·s–1]. Most importantly, the affinity of the C2 domain for POPC/POPS (8:2) vesicles was ≈30 times lower than that of the full-length PTEN, suggesting that the C2 domain itself is not critically involved in membrane binding of PTEN. Consistent with this finding, the EGFP-tagged PTEN C2 domain did not show localization at the plasma membrane when expressed in HEK293T cells (Fig. 3A). Taken together, it would seem that the membrane affinity of the PTEN C2 domain is not high enough to drive the membrane recruitment of the PTEN molecule.
Fig. 3.
Subcellular localization of PTEN and its mutants in HEK293T and HeLa cells. PTEN and its mutants tagged with EGFP at their C termini were transiently transfected into HEK293T (A–G) and HeLa (H–J) cells, and their subcellular localization was monitored by confocal microscopy. (A) C2 domain. (B) Δ353–403/C124A. (C) Δ1–185. (D) Δ353–403/C124A/R161A/K163A/R164A. (E) Phosphatase domain (C124A). (F) C124A–PTEN. (G) C124A/S380E/T382E/T383E. (H) C124A/S380A/T382A/T382A. (I) C124A/Δ400–403. (J) C124A/S380A/T382A/T383A/Δ400–403. (K) C124A (HeLa). (L) C124A/S380A/T382A/T382A (HeLa). (M) Δ400–403/C124A/S380A/T382A/T383A (HeLa).
Roles of Phosphatase Domain and the C-terminal Tail in Membrane Binding. The finding that the C2 domain cannot drive the membrane recruitment of PTEN suggests that either the N-terminal phosphatase domain or the C-terminal tail (or both) is involved more directly in membrane targeting of PTEN. Evaluation of the contribution of either the isolated phosphatase domain (residues 1–185) or the isolated C-terminal tail (residues 353–403) to membrane targeting of PTEN was hampered by extremely low yields of their bacterial expression, presumably due to low protein stability. Alternatively, we prepared truncated mutants, Δ1–185 and Δ353–403, which lack the phosphatase domain and the C-terminal tail, respectively, and measured their membrane binding in comparison with the full-length PTEN.
As shown in Table 1, Δ1–185 and Δ353–403 had 31- and 1.5-fold lower affinity for POPC/POPS (8:2) vesicles than WT, respectively. This suggests that the phosphatase domain is important for the membrane binding of PTEN, whereas the C-terminal tail plays practically no role in membrane binding. In particular, Δ353–403, which includes both the phosphatase and C2 domains, showed ≈20-fold higher membrane affinity than the C2 domain alone, indicating that either the phosphatase domain is much more important than the C2 domain for membrane binding or the synergy between the two domains is crucial for effective membrane binding of PTEN. To distinguish between these possibilities, we mutated residues in two prominent cationic patches (R11/K13/R14/R15 and R161/K163/K164) on the putative membrane-binding surface of the phosphatase domain (see Fig. 2) and measured their membrane binding. As shown in Table 1, the mutation of R161/K163/K164 to alanine caused a large 22-fold decrease in affinity primarily by reducing the association rate constant (ka). This large drop in affinity was not due to deleterious conformational changes, because the mutant demonstrated the phosphatase activity (35 ± 5 nmol/mg per min) toward a water-soluble PtdIns(3,4,5)P3 substrate that was comparable to that of the WT PTEN (37 ± 5 nmol/mg per min). We previously showed that long-range (i.e., nonspecific) electrostatic interactions mainly affect ka for protein-membrane binding, whereas the short-range (i.e., specific) interactions influence the dissociation rate constant (kd) (24, 32). Thus, our results indicate that R161, K163, and K164 in the phosphatase domain play a key role in nonspecific electrostatic membrane binding of PTEN. R11A/K13A/R14A/R15A also caused a drastic 76-fold decrease in affinity, but in this case the mutant exhibited <1% of the WT activity toward the monomeric substrate, which made it difficult to sort out the effects of the mutation on membrane binding and on protein stability. Taken together, these results indicate that the phosphatase domain is essential for the electrostatic membrane binding of PTEN, whereas the C2 domain plays an indirect role.
We then measured the subcellular localization of EGFP-tagged phosphatase domain (residues 1–185), Δ1–185, and Δ350–403 transiently transfected in HEK293T cells. It was reported that the deletion of the C-terminal tail of PTEN decreased the cellular half-life of the protein due to rapid protein degradation (11, 13). Indeed, EGFP-tagged Δ353–403 was not expressed in detectable amounts in HEK293T cells. Serendipitously, we found that a catalytically inactive mutant of PTEN (C124A) had higher cellular stability than the WT (data not shown) while having the same affinity for POPC/POPS (8:2) vesicles as the WT (data not shown). This implied that the cellular degradation of PTEN might represent a negative feedback control of its lipid phosphatase activities. We thus introduced the C124A mutation to all the phosphatase domain-containing constructs used in cell studies. As shown in Fig. 3, Δ350–403/C124A–EGFP and the phosphatase domain (C124A)–EGFP were successfully expressed in HEK293T cells. The fact that the EGFP-tagged phosphatase domain (C124A) was expressed without the C2 domain in HEK293T cells suggests that its C-terminal EFGP might stabilize the protein. Most importantly, Δ353–403/C124A–EGFP was prelocalized to the plasma membrane (Fig. 3B), whereas Δ1–185–EGFP (Fig. 3C) and the R161A/K163A/K164A mutant of Δ353–403/C124A–EGFP (Fig. 3D) were distributed in the cytoplasm. These localization data indicate that the phosphatase domain plays a key role in cellular membrane targeting of PTEN, whereas the C-terminal tail is not required for the process. Furthermore, the finding that the isolated phosphatase domain (C124A) was found in the cytoplasm (Fig. 3E) supports the notion that the presence of the C2 domain is required for the effective membrane binding of the phosphatase domain.
Effect of the C-terminal Tail Phosphorylation on Membrane Binding and Enzyme Activation. The effect of the phosphorylation of the C-terminal tail on the membrane binding of PTEN has not been investigated thus far. To address the question as to whether the phosphorylation has a direct impact on membrane binding of PTEN, we prepared a dephosphorylation-mimicking mutant (S380A/T382A/T383A) and a phosphorylation-mimicking mutant (S380E/T382E/T383E) of PTEN and measured their in vitro membrane binding. It was shown that the phosphorylation of these three sites had the largest effect on cellular PTEN activities (11). Table 1 shows that S380A/T382A/T383A has WT-like affinity for POPC/POPS (8:2) vesicles, which is expected from the fact that the bacterially expressed WT would not have a posttranslational modification. In contrast, S380E/T382E/T383E exhibited ≈83-fold reduced affinity mainly due to a smaller ka, which suggests that the phosphorylation of the C-terminal tail might directly interfere with the electrostatic membrane binding of PTEN. Furthermore, introducing the S380E/T382E/T383E mutation to the R161A/K163A/K164A mutant reduced its affinity by only 4-fold, suggesting that the phosphoryl groups on S380/T382/T383 might interact with these cationic residues and thereby block their interaction with the anionic membrane surface.
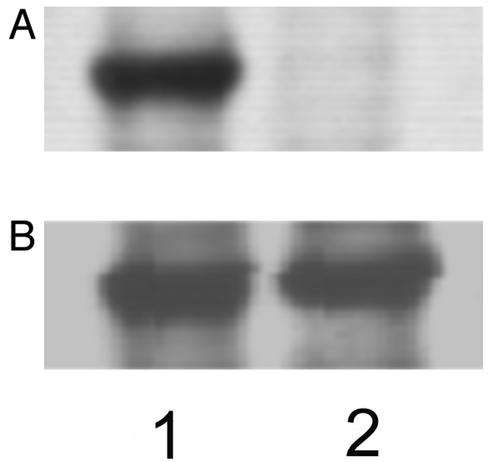
To test this notion further, PTEN and phosphorylation site mutants (C124A, C124A/S380A/T382A/T383A, and C124A/S380E/T382E/T383E) were transiently transfected into HEK293T cells as EGFP fusion proteins. C124A (Fig. 3F) and C124A/S380E/T382E/T383E (Fig. 3G) were distributed in the cytoplasm, whereas C124A/S380A/T382A/T383A (Fig. 3H) was prelocalized to the plasma membrane. Consistent with previous reports (11, 18), immunoblotting with S380/T382/T383-phosphopeptide-specific antibody (see Fig. 4) showed that C124A expressed in HEK293T cells was phosphorylated on S380/T382/T383. Importantly, the removal of the PDZ domain-binding site from C124A and C124A/S380A/T382A/T383A, respectively, had no effect on their subcellular localization; i.e., C124A/Δ400–403 (Fig. 3I) was distributed in the cytoplasm, whereas C124A/S380A/T382A/T383A/Δ400–403 (Fig. 3J) was localized to the plasma membrane. These results thus provide direct evidence that the phosphorylation of C-terminal tail interferes with the cellular membrane targeting of PTEN not by blocking its PDZ domain binding but by interfering with its electrostatic membrane binding. To preclude the possibility that our cell data are attributed to the lack of PDZ domain-containing proteins such as membrane-associated guanylate kinase with inverted orientation 2 (MAGI-2) in HEK293T cells, we measured the subcellular localization of C124A, C124A/S380A/T382A/T383A, and C124A/S380A/T382A/T383A/Δ400–403 in another cell line, HeLa cells. As shown in Fig. 3K–M, the same pattern was seen in these cells, thereby supporting the notion that the PDZ domain-binding site is not directly involved in the regulation of membrane targeting of PTEN by phosphorylation. Last, it was found that for both PTEN and S380E/T382E/T383E, their EGFP-fusion proteins were expressed successfully with and without the C124A mutation. In the case of the dephosphorylation-mimicking mutant (S380A/T382A/T383A), however, the EGFP-fusion protein was expressed only with the C124A mutation. In conjunction with the rapid degradation of constitutively active Δ353–403, these data underscore that the degradation of PTEN is a negative feedback regulatory mechanism of PTEN activities.
Fig. 4.
Phosphorylation of PTEN and mutants in HEK293T cells. (A) Immunostaining of PTEN-transfected cell extracts with the S380/T382/T383-phospho-specific antibody. Lane 1, cells transfected with PTEN–EGFP; lane 2, cells transfected with S380A/T382A/T383A–EGFP. (B) Immunostaining of the same cell extracts with the anti-PTEN antibody shows that total PTEN amounts are comparable for the two lanes.
Plasma Membrane Specificity of PTEN. If the membrane recruitment of PTEN is mainly driven by nonspecific electrostatic membrane binding, a question rises as to how it achieves the specific targeting to the plasma membrane. In the case of C2 domains, we were able to demonstrate that their subcellular localization depends on their lipid specificity (26). In particular, C2 domains showed high in vitro selectivity for vesicles with lipid compositions that approximate those of the cell membranes to which they are targeted (26). We thus measured the in vitro binding of PTEN to vesicles with compositions that mimic the cytoplasmic plasma membrane [i.e., POPC/POPE/POPS/POPI/cholesterol (12:35:22:9:22)] and the cytoplasmic nuclear membrane [i.e., POPC/POPE/POPS/POPI/cholesterol (61:21:4:7:7)], respectively. As shown in Table 1, PTEN had 2-fold higher affinity for the plasma membrane mimic than for POPC/POPS (8:2) vesicles, presumably because the former has a higher anionic lipid content. The 2-fold enhanced binding to the plasma membrane mimic is not directly related to cholesterol content, because PTEN showed essentially the same affinity for POPC/POPS (8:2) and POPC/POPS/cholesterol (6:2:2) vesicles (data not shown). In contrast, PTEN showed no detectable affinity for the nuclear membrane mimic with protein concentration up to 1 μM (i.e., Kd > μM). As a result, PTEN exhibited strong preference for the plasma membrane mimic over the nuclear membrane mimic. As was the case with POPC/POPS (8:2) vesicles, PTEN showed much reduced (i.e., ≈80-fold) affinity for the plasma membrane mimic at 0.5 M KCl (data not shown). Thus, PTEN has strong preference for the plasma membrane mimic (and the plasma membrane) because of its high anionic lipid content.
Discussion
This work describes a systematic study on the mechanism of membrane binding and activation of PTEN. Because the main physiologic substrate of PTEN is the membrane lipid PtdIns(3,4,5)P3 and because PTEN is normally a cytosolic protein, the activation of PTEN should involve its membrane recruitment. The present study indicates that the membrane recruitment of PTEN is controlled by phosphorylation-induced conformational change of the protein, which in turn directly modulates membrane–protein interactions. The phosphatase and C2 domains of PTEN have clustered cationic residues that can interact favorably with anionic membranes through long-range (nonspecific) electrostatic interactions. We have shown for many peripheral proteins that long-range electrostatic interactions mainly accelerate the membrane-association kinetics (24, 32). Because cationic residues on the putative membrane-binding surface of the phosphatase domain greatly enhance the membrane-association rate with a much smaller effect on the membrane dissociation rate, they should play a critical role in long-range electrostatic membrane interactions. Although the phosphatase domain is involved most directly in electrostatic membrane binding, our in vitro and cell studies indicate that the presence of the C2 domain is essential for high-affinity membrane binding. This is consistent with the previous finding that the C2 domain assists the productive membrane binding of the phosphatase domain (12). Most importantly, the mutation of three residues (S380, T382, and T383) in the C-terminal tail to glutamic acid, which simulates the effect of phosphorylation, dramatically lowers the membrane-association rate of PTEN, presumably by interacting directly with the cationic residues in the phosphatase domain. If the main effect of phosphorylation is electrostatic shielding, incorporation of dianionic phosphoryl groups into these sites would have an even larger effect than the introduction of single anions by the glutamic acid mutations. As such, the three-site (or more extensive) phosphorylation would serve as an electrostatic switch that spontaneously turns the membrane binding of PTEN on and off.
Our cell studies not only confer physiological relevance on our in vitro membrane-binding data but also provide further insight into how the membrane recruitment of PTEN is coupled to its cellular function and regulation. It should be noted that the subcellular localization of PTEN has not been studied in detail mainly because of rapid degradation of the protein after membrane binding. In this regard, the C124A mutants with greatly improved cellular stability provided us with a unique opportunity to monitor the effects of various mutations and the C-terminal tail phosphorylation on the subcellular localization of PTEN. Our results show that the N-terminal region containing the phosphatase domain and the C2 domain is both necessary and sufficient for the targeting of PTEN to the plasma membrane in HEK293T cells. The presence of the C-terminal tail inhibits the membrane targeting of this region, because S380, T382, and T383 seem constitutively phosphorylated in these cells. As evidenced by the prelocalization of C124A/S380A/T382A/T383A to the plasma membrane, the dephosphorylation of these sites promotes the membrane recruitment of PTEN by accelerating its membrane-association step. In that the membrane-bound PTEN seems to be degraded rapidly by a putative negative feedback mechanism, the phosphorylation-mediated control of the membrane-association rate makes better sense than the control of the dissociation rate in terms of cellular regulation of PTEN activities. Intriguingly, it has been proposed that the phosphorylation primarily regulates the membrane dissociation rate of group IVA cytosolic phospholipase A2, the lipolytic activity of which would be benefited greatly from its processive action on the membrane surface (S.D., K. Kim, S. Gigy, and W.C., unpublished data). Thus, it would seem that the mechanism by which phosphorylation mediates the membrane binding of peripheral proteins varies depending on their cellular functions and regulation. Our results also dispute the notion that the C-terminal tail regulates the PTEN activity by directly modulating its stability, because the reduced cellular stability of the C-terminal truncated PTEN derives not from its reduced protein stability but from its enhanced membrane binding, which leads to rapid degradation.
The nonspecific nature of its electrostatic membrane binding raises a question as to whether the specific targeting of PTEN to the plasma membrane is driven primarily by its lipid selectivity or requires a specific adaptor protein present in the plasma membrane. The cytoplasmic distribution of the Δ350–403/C124A/R161A/K163A/K164A mutant argues against the latter possibility. Notice that the R161A/K163A/K164A mutant has 22-fold lower affinity for POPC/POPS (8:2) vesicles than the WT PTEN due primarily to a lower ka, which indicates that these cationic residues are directly involved in electrostatic membrane binding. Because the same three residues are not likely to be involved in both membrane binding and binding to an adaptor protein, the cytoplasmic localization of Δ350–403/C124A/R161A/K163A/K164A should be attributed to low membrane affinity. High selectivity of PTEN for the plasma membrane mimicking vesicles over the nuclear membrane mimicking vesicles lends further support to the notion that the specific targeting of PTEN to the plasma membrane is attributed to the high anionic lipid composition of the inner leaflet of the plasma membrane.
In summary, our studies indicate that the dephosphorylation of the C-terminal tail residues triggers the targeting of PTEN to the plasma membrane by exposing its surface cationic residues, which resulted in rapid electrostatic membrane binding. Once bound to the plasma membrane, PTEN would bind and dephosphorylate 3-phosphorylated phosphoinositides, most notably PtdIns(3,4,5)P3, which in turn induces the degradation of PTEN. Further studies are needed to elucidate how phosphorylation on other sites (e.g., S370 and S385) affects the membrane binding of PTEN, the protein phosphatase of which is involved in PTEN dephosphorylation, how the membrane-bound PTEN is degraded, and what the exact role of the putative PDZ domain-binding site in PTEN regulation is. Our proposed model of PTEN activation should provide a framework within which these questions can be investigated systematically.
Acknowledgments
This work was supported by National Institutes of Health Grants GM52598 and GM53987 (to W.C.) and grants from the Walther Cancer Institute and the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases Grant 18849 to J.E.D.).
Abbreviations: PtdIns(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; PDZ, PSD-95/Dlg/ZO-1 homology; SPR, surface plasmon resonance; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; POPI, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol; EGFP, enhanced GFP.
References
- 1.Maehama, T. & Dixon, J. E. (1999) Trends Cell Biol. 9, 125–128. [DOI] [PubMed] [Google Scholar]
- 2.Maehama, T., Taylor, G. S. & Dixon, J. E. (2001) Annu. Rev. Biochem. 70, 247–279. [DOI] [PubMed] [Google Scholar]
- 3.Wishart, M. J. & Dixon, J. E. (2002) Trends Cell Biol. 12, 579–585. [DOI] [PubMed] [Google Scholar]
- 4.Leslie, N. R. & Downes, C. P. (2002) Cell. Signalling 14, 285–295. [DOI] [PubMed] [Google Scholar]
- 5.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 6.Myers, M. P., Pass, I., Batty, I. H., Van der Kaay, J., Stolarov, J. P., Hemmings, B. A., Wigler, M. H., Downes, C. P. & Tonks, N. K. (1998) Proc. Natl. Acad. Sci. USA 95, 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stambolic, V., Suzuki, A., de la Pompa, J. L., Brothers, G. M., Mirtsos, C., Sasaki, T., Ruland, J., Penninger, J. M., Siderovski, D. P. & Mak, T. W. (1998) Cell 95, 29–39. [DOI] [PubMed] [Google Scholar]
- 8.Cantley, L. C. & Neel, B. G. (1999) Proc. Natl. Acad. Sci. USA 96, 4240–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Cristofano, A. & Pandolfi, P. P. (2000) Cell 100, 387–390. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J. O., Yang, H., Georgescu, M. M., Di Cristofano, A., Maehama, T., Shi, Y., Dixon, J. E., Pandolfi, P. & Pavletich, N. P. (1999) Cell 99, 323–334. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez, F., Ramaswamy, S., Nakamura, N. & Sellers, W. R. (2000) Mol. Cell. Biol. 20, 5010–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgescu, M. M., Kirsch, K. H., Kaloudis, P., Yang, H., Pavletich, N. P. & Hanafusa, H. (2000) Cancer Res. 60, 7033–7038. [PubMed] [Google Scholar]
- 13.Georgescu, M. M., Kirsch, K. H., Akagi, T., Shishido, T. & Hanafusa, H. (1999) Proc. Natl. Acad. Sci. USA 96, 10182–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu, Y., Dowbenko, D., Spencer, S., Laura, R., Lee, J., Gu, Q. & Lasky, L. A. (2000) J. Biol. Chem. 275, 21477–21485. [DOI] [PubMed] [Google Scholar]
- 15.Wu, X., Hepner, K., Castelino-Prabhu, S., Do, D., Kaye, M. B., Yuan, X. J., Wood, J., Ross, C., Sawyers, C. L. & Whang, Y. E. (2000) Proc. Natl. Acad. Sci. USA 97, 4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adey, N. B., Huang, L., Ormonde, P. A., Baumgard, M. L., Pero, R., Byreddy, D. V., Tavtigian, S. V. & Bartel, P. L. (2000) Cancer Res. 60, 35–37. [PubMed] [Google Scholar]
- 17.Yu, Z., Fotouhi-Ardakani, N., Wu, L., Maoui, M., Wang, S., Banville, D. & Shen, S. H. (2002) J. Biol. Chem. 277, 40247–40252. [DOI] [PubMed] [Google Scholar]
- 18.Torres, J. & Pulido, R. (2001) J. Biol. Chem. 276, 993–998. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez, F., Grossman, S. R., Takahashi, Y., Rokas, M. V., Nakamura, N. & Sellers, W. R. (2001) J. Biol. Chem. 276, 48627–48630. [DOI] [PubMed] [Google Scholar]
- 20.Leslie, N. R., Bennett, D., Gray, A., Pass, I., Hoang-Xuan, K. & Downes, C. P. (2001) Biochem. J. 357, 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolkacheva, T. & Chan, A. M. (2000) Oncogene 19, 680–689. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 23.Bittova, L., Sumandea, M. & Cho, W. (1999) J. Biol. Chem. 274, 9665–9672. [DOI] [PubMed] [Google Scholar]
- 24.Stahelin, R. V. & Cho, W. (2001) Biochemistry 40, 4672–4678. [DOI] [PubMed] [Google Scholar]
- 25.Stahelin, R. V., Long, F., Diraviyam, K., Bruzik, K. S., Murray, D. & Cho, W. (2002) J. Biol. Chem. 277, 26379–26388. [DOI] [PubMed] [Google Scholar]
- 26.Stahelin, R. V., Rafter, J. D., Das, S. & Cho, W. (2003) J. Biol. Chem. 278, 12452–12460. [DOI] [PubMed] [Google Scholar]
- 27.Karathanassis, D., Stahelin, R. V., Bravo, J., Perisic, O., Pacold, C. M., Cho, W. & Williams, R. L. (2002) EMBO J. 21, 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho, W., Bittova, L. & Stahelin, R. V. (2001) Anal. Biochem. 296, 153–161. [DOI] [PubMed] [Google Scholar]
- 29.Kulkarni, S., Das, S., Funk, C. D., Murray, D. & Cho, W. (2002) J. Biol. Chem. 277, 13167–13174. [DOI] [PubMed] [Google Scholar]
- 30.Das, S. & Cho, W. (2002) J. Biol. Chem. 277, 23838–23846. [DOI] [PubMed] [Google Scholar]
- 31.Kim, Y. J., Kim, K. P., Rhee, H. J., Das, S., Rafter, J. D., Oh, Y. S. & Cho, W. (2002) J. Biol. Chem. 277, 9358–9365. [DOI] [PubMed] [Google Scholar]
- 32.Cho, W. (2001) J. Biol. Chem. 276, 32407–32410. [DOI] [PubMed] [Google Scholar]