Abstract
PKC-interacting protein (PKCI), also designated histidine triad nucleotide-binding protein 1, belongs to the histidine triad (HIT) family of proteins. Its structure is highly conserved from bacteria to humans and shares homology with the tumor-suppressor gene fragile histidine triad (FHIT). Although it was originally thought to inhibit PKC, its actual physiologic function is not known. Therefore, we used the technique of homologous recombination to generate homozygous deleted PKCI-/- mice. These mice display normal fetal and adult development. However, when mouse embryo fibroblasts were established from 13.5-day embryos and serially passaged the PKCI-/- cells displayed an increase in growth rate and underwent spontaneous immortalization, whereas the PKCI+/+ cells senesced and ceased growing. Furthermore, the PKCI-/- mouse embryo fibroblasts displayed increased resistance to cytotoxicity by ionizing radiation. In view of these findings we examined possible effects of PKCI on susceptibility to carcinogenicity. Both PKCI+/+ and PKCI-/- mice were treated with the chemical carcinogen N-nitrosomethylbenzylamine (NMBA) by intragastric administration and killed 12 weeks later. As expected with this protocol, NMBA induced squamous tumors (both papillomas and carcinomas) of the forestomach. The incidence, multiplicity per mouse, volume, and degree of malignancy of these tumors were significantly greater in the PKCI-/- than in the PKCI+/+ mice. Furthermore, four adenomas and one adenocarcinoma of the glandular stomach were found in the NMBA-treated PKCI-/- mice but no tumors of the glandular stomach were found in the NMBA-treated PKCI+/+ mice or in any of the untreated mice. Taken together, these findings suggest that, like FHIT, PKCI may normally play a tumor-suppressor role. The possible role of PKCI as a tumor suppressor in humans remains to be determined.
Keywords: tumor suppressor
In previous studies from our laboratory in which we used the yeast two-hybrid system to identify proteins that interact with the regulatory domain of PKCβ1 we identified a cDNA encoding a protein that we designated human PKC-interacting protein (hPKCI). This cDNA encodes a 13.7-kDa protein that localizes in both the nucleus and cytoplasm of mammalian cells (1). A highly homologous protein bPKCI (bovine PKC inhibitor 1) was originally identified in bovine brain extracts as an in vitro inhibitor of conventional PKC isoforms, and its sequence was determined at the amino acid level (2). However, subsequent studies have questioned the physiologic significance of the ability of this protein to inhibit PKC (1, 3). The human PKCI gene was subsequently mapped by other investigators to human chromosome 5q31.2 (4). A homologous mouse gene, mPKCI, is located on mouse chromosome 11, encodes three exons, and spans 4.06 kb (see www.ensembl.org/Mus_musculus/geneview?gene=ENSMUSG00000020267). The official designation of this gene is histidine triad nucleotide-binding protein 1 (HINT1), but for consistency with our previous studies, in this paper we refer to it as PKCI. A remarkable feature of PKCI is that homologous sequences are found in a wide variety of species ranging from mycoplasma to plants and to humans (5, 6). The amino acid sequence for the entire coding region of hPKCI shares 94% identity with mouse PKCI, 53% with maize PKCI, and 31% with mycoplasma PKCI 1. Structural studies show that a possible common feature of the PKCI family of proteins is the electrostatic distribution on the molecule. Several conserved and exposed acidic residues on one face of the protein contribute to form a large negative electrostatic field (5). Crystal structures of PKCI–nucleotide complexes demonstrate that the most conserved residues in the superfamily mediate nucleotide binding and that the histidine triad (HIT) motif, described below, forms part of the phosphate-binding loop (7). Both bovine and maize PKCI specifically bind zinc (2, 8–10).
The PKCI family of proteins belongs to a larger ubiquitous superfamily, the HIT protein family, which contains a His-Xaa-His-Xaa-His motif, in which Xaa is a hydrophobic amino acid (11). An important member of this family is the “fragile locus HIT gene” (FHIT). This tumor-suppressor gene is frequently mutated in several types of human cancer (12–18), and mice deficient in FHIT are more susceptible to tumor induction by the carcinogen N-nitrosomethylbenzylamine (NMBA) (19–21). The FHIT protein sequence is 20% identical to that of hPKCI and 52% identical to that of a Schizosaccharomyces pombe HIT protein. The latter protein is a diadenosine tetraphosphate (Ap4A) asymmetric hydrolase (22). X-ray crystallography studies and in vitro enzyme assays indicate that FHIT and hPKCI share a common catalytic mechanism but have different substrates. FHIT cleaves ATP, Ap3A (diadenosine triphosphate), and Ap4A but not ADP, whereas PKCI cleaves ADP but not Ap3A or ATP (11). This substrate specificity is probably dictated by the extent of divergence in the COOH-terminal residues of these two proteins. Although the in vivo substrates of these HIT proteins are not yet known, at the biochemical level it is likely that both PKCI and FHIT function as nucleotidyl hydrolases or nucleotidyl phosphotransferases, with different substrate specificities (1, 11, 23).
With respect to the functions of PKCI it is of interest that PKCI exists as a homodimer (5). There is evidence that the PKCI protein also interacts with other proteins. PKCI was shown to interact with an ataxia–telangiectasia (AT) candidate gene ATDC, suggesting that it might play a role in the response of cells to ionizing radiation (4, 24). However, in the interim another protein, ATM, has been identified as the true AT protein (25, 26). Therefore the physiologic function of ATDC in radiation repair is not known. PKCI also associates with the protein microphthalmia (mi), an important transcription factor that controls growth and function in mast cells and melanocytes (27). PKCI also associates with cyclin-dependent kinase 7 (CDK7), which forms a trimeric complex with cyclin H and Mat1 to form the mammalian CDK-activating kinase CAK. CDK7 is also a component of the basal transcription factor TFIIH, in which CDK7 phosphorylates the COOH-terminal domain (CTD) of the large subunit of RNA polymerase II (28). In a study of budding yeast, loss of the active site of Hnt1, the yeast homologue of PKCI protein, blocked growth on galactose as a substrate at elevated temperatures. Loss of Hnt1 enzyme activity also led to hypersensitivity to mutations in Ccl1, Tfb3, and Kin28, which constitute the TFIIK kinase subcomplex of the general transcription factor TFIIH, and to mutations in Cak1, which phosphorylates Kin28. Rabbit PKCI was able to provide functional complementation of all of the Hnt1 phenotypes, apparently because of its similar adenosine monophosphoramidase activity, despite that it shares only 22% sequence identity with yeast Hnt1 (29). Taken together, these studies suggest that PKCI may be involved in the regulation of gene transcription.
In this study, we developed mice that have a homozygous deletion in the PKCI gene and found that, although spontaneous tumorigenesis is not significantly increased in these mice, they are much more susceptible to induction of squamous carcinomas of the forestomach by the carcinogen NMBA. Furthermore, mouse embryonic fibroblast (MEF) cells isolated from the PKCI-/- mice display spontaneous immortalization during serial passage and increased resistance to ionizing radiation.
Materials and Methods
Construction of PKCI-Targeting Vector and Production of Gene-Disrupted Mice. A 13-kb fragment of DNA containing the first exon of the PKCI gene was isolated from a 129SvJ mouse genomic library (Stratagene) and mapped for suitable restriction enzyme sites. A targeting construct was generated by using plasmid pBluescript KS(-) as shown in Fig. 1A. An EcoRI site was introduced into the short arm at its 3′-end by PCR, and the EcoRI enzyme was used in Southern blot screening. The primer pairs for the short arm were 5′-ATCGAT(ClaI)ATCGAT(ClaI)ATCTCCTCTGCCTCCATC-3′ as the forward primer and 5′-GTTTAAAC(PmeI)GTTTAAAC(PmeI)GATATCGAATTC(EcoRI)ATTCGAGATCCTTATGCTC-3′ as the reverse primer. A PGKneo cassette, the selection marker, was used to replace a 1.6-kb DNA sequence including the first exon of mouse PKCI, part of the upstream sequence and part of the downstream intron.
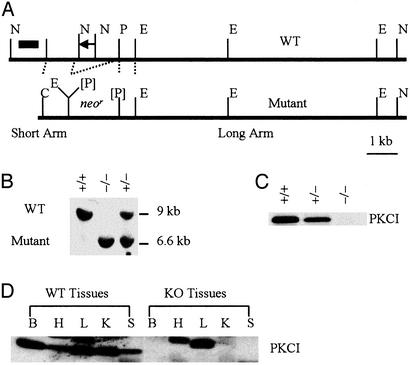
Fig. 1.
Targeted disruption of the murine PKCI gene. (A) The murine PKCI locus and the targeting construct. A 1.6-kb fragment of genomic DNA was deleted and replaced with a 1.8-kb neomycin-resistance (neor) gene. The probes used to screen ES cell clones and mice are also indicated as a short black bar. E, EcoRI; N, NotI; P, PmeI; [P], inactivated PmeI. (B) Southern blot analysis of mouse genomic DNA isolated from PKCI +/+, +/-, or -/- mice and digested with EcoRI. (C) Western blot analysis of PKCI gene expression in MEF cells by using anti-hPKCI antibody. (D) Western blot analysis of PKCI gene expression in various organs obtained from both PKCI+/+ and PKCI-/- mice. KO, knockout (PKCI-/-); B, brain; H, heart; L, liver; K, kidney; S, spleen.
The maintenance, transfections, and selection procedures used with the 129 strain embryonic stem (ES) cells were carried out as described (30). The probe for Southern blot screening was labeled by PCR using the following two primers: 5′-GGGAATTC(EcoRI)TGAATGTATGGATGAGCAAC-3′ as the forward primer and 5′-GGGAATTC(EcoRI)GTTCAGTGGCTAAGAGTAC-3′ as the reverse primer. The expected sizes for the WT and mutant PKCI DNA fragments are ≈9 and 6.6 kb, respectively.
ES cells that were heterozygous for the targeted mutation were microinjected into C57BL/6 blastocysts and implanted into pseudopregnant females. The resulting male chimeras were mated with female C57BL/6 mice. Germ-line transmission of the injected ES cells was confirmed by inheritance of the agouti coat color in the F1 mice, and the agouti offspring were tested for the presence of the mutated PKCI allele by Southern blot analysis of mouse tail DNA. These mice were backcrossed to 129 strain mice for several generations, so that in the PKCI-/- mice used in the present study over 96% of their background is from the 129 strain mice.
Immunoblot Analysis. Organs from adult mice were homogenized in cell lysis buffer (50 mM Hepes, pH 7.5/150 mM NaCl/2.5 mM EGTA/1 mM EDTA/1 mM DTT/0.1% Tween 20/10% glycerol; final pH 7.5) together with protease inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM PMSF). The lysates were incubated on ice for 15 min and then sonicated for three 10-sec bursts, followed by centrifugation at 10,000 × g at 4°C for 5 min, and the supernatant fraction was collected. A Western blot analysis was done with an anti-hPKCI antibody (1) as described (31).
MEF Cell Lines. Primary MEF cells were isolated from 13.5-days-postcoitum 129 strain control or PKCI-/- mouse embryos. Embryos were then decapitated and eviscerated. Cells were separated by passing the carcass through an 18-gauge needle 8–10 times and seeded into 10-cm cell culture dishes (one embryo per dish) and incubated at 37°C. The cells were split once at 1:4 when confluent, and then designated passage 1 and frozen or serial passaged by splitting the cultures 1:5 when they were confluent. To determine cell growth rates 4 × 104 cells were plated in triplicate, into six-well plates, and cell numbers were counted daily with a cell counter (Beckman Coulter). The medium was changed every 3 days.
Radiation Resistance Assays. Exponential-phase cultures of MEF cells were plated into six-well tissue culture dishes at a density of 0.5–12 × 103 cells per well, at 1 day before irradiation. The density was adjusted for the expected plating efficiency to yield 10–70 colonies per well. Dishes were irradiated with γ-rays from a cesium-137 irradiator at doses of 0.5, 1, 2, 4, and 6 Gy. After incubation for 7–12 days, the cells were fixed with 37% formaldehyde and stained with 5% Giemsa stain. Colonies containing more than 50 cells were counted and the plating efficiency and surviving fraction were calculated. Plating efficiency (PE) was defined as the number of colonies/number of cells seeded. Surviving fraction was defined as PE of irradiated cells/PE of control unirradiated cells.
Carcinogenicity Study. WT (PKCI+/+) 129 strain and knockout (PKCI-/-) mice were produced and maintained in the Herbert Irving Comprehensive Cancer Center animal facility. The strategy for the carcinogenicity study followed a well established mouse model of NMBA-induced squamous tumors of the forestomach (19, 32). Specifically, 39 PKCI+/+ and 71 PKCI-/- mice at ages 28–40 weeks were given eight intragastric doses (2 mg/kg body weight per dose) of NMBA (Ash Stevens, Detroit) over the course of 4 weeks. Mice in control groups (8 PKCI+/+ and 8 PKCI-/- mice) did not receive NMBA treatment. All of the mice were killed and complete autopsies were performed 12 weeks after the final NMBA dose. Tumors were examined with respect to their location, number, and volume. Tumor volume was calculated by using the formula described in previous studies (33). All of the tumors identified in the forestomach and other tissues were carefully removed and processed for histopathological examination with hematoxylin and eosin (H&E) staining. Tumors in the forestomach were histologically classified into papilloma and squamous cell carcinoma based on described criteria (34, 35). For statistical analysis, tumor incidence, multiplicity, and volume were compared between NMBA-treated mice and control mice.
Results
PKCI Gene Targeting. When we began these studies the genomic structure of the mouse PKCI gene was not known, although it has been subsequently described on the Ensembl website database supported by the European Bioinformatics Institute (see www.ensembl.org/Mus_musculus/geneview?gene=ENSMUSG00000020267; for references, see www.informatics.jax.org/searches/reference_report.cgi?_Marker_key=38789). Therefore, at that time we synthesized an oligonucleotide probe corresponding to the first 13 aa in the coding sequence of the mouse PKCI cDNA. Using this probe, we screened a 129-strain mouse genomic DNA library and found a DNA sequence corresponding to the first exon of the mouse PKCI gene, which encodes the first 37 aa of the mouse PKCI protein. We then designed a targeting construct that would delete the first exon, its 1.2-kb upstream sequence, and part of the downstream intron, and would replace this sequence with the neomycin/G418 resistance gene (neor) (Fig. 1 A). Electroporation of this linearized targeting construct into 129 strain ES cells and G418 selection for homologous recombinants yielded neomycin/ G418-resistant clones, 600 of which were picked and screened by Southern blot analysis for evidence of homologous recombination. Fourteen of the 600 clones (2.3%) were found to be homologous recombinants displaying the predicted 9-kb WT and 6.6-kb mutant fragments. Two of these 14 clones were injected into C57BL/6 blastocysts to generate chimeric mice. The high coat color male chimeric mice (ranging from 35% to 70% coating) were then bred to C57BL/6 females to obtain agouti mice in which the mutant allele had been transmitted through the germ line. Heterozygous offspring of chimeras appeared entirely normal and were fertile. Heterozygous mice were then interbred to produce homozygous deficient PKCI-/- mice, which were identified by Southern blot analysis of tail DNA (Fig. 1B). Western blot analysis of protein extracts of MEF cell cultures established from these mice by using an anti-hPKCI antibody indicated that the PKCI+/- cells had ≈50% reduction in the 13.7-kDa PKCI protein and that none of this protein was detected in the PKCI-/- cells (Fig. 1C).
Phenotype of PKCI-/- Mice. When intercrosses were set up between mice that were heterozygous for the disrupted PKCI alleles, we obtained WT (+/+), heterozygous (+/-), and nullizygous (-/-) offspring at approximately the expected Mendelian 1:2:1 ratios, indicating that there was no significant embryonic lethality. At birth, the PKCI+/- and PKCI-/- mice were indistinguishable from their WT littermates and there were no gross differences in appearance, body size, or weight at 2 months of age. After 3 months of age the body weight of the male PKCI-/- mice decreased and remained lower than that of the PKCI+/+ mice until ≈11 months of age (data not shown). This difference in body weight between months 5 and 7 was statistically significant (P < 0.01). However, no difference in body weight gain was seen between the female PKCI-/- and PKCI+/+ mice. We also prepared tissue extracts from the brain, heart, liver, kidney, and spleen obtained from 4-month-old PKCI+/+ and PKCI-/- male mice. Western blot analysis of these extracts by using an anti-hPKCI antibody indicated that all of the organs from the PKCI+/+ mice expressed a characteristic 13.7-kDa protein band, but this protein band was totally absent in all of the organs of the PKCI-/- mice (Fig. 1D). No significant histologic and morphologic differences were detected between PKCI+/+ and PKCI-/- mice in these tissues.
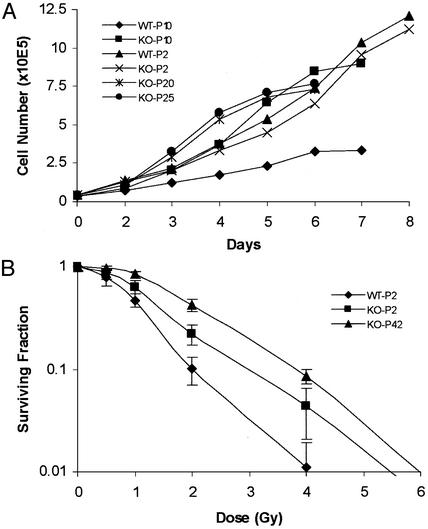
Growth of MEF Cells and Sensitivity to Radiation. MEF cultures were established from both PKCI+/+ and PKCI-/- mice by using 13.5-days-postcoitum embryos, and the cultures were then serially passaged. Both types of MEF cells grew at similar rates for the first few passages. The growth of the PKCI+/+ MEF cells began to decrease by passage 6, and the cells showed signs of crisis and their growth ceased by passage 16. The slow growth of the PKCI+/+ cells at passage 10 is shown in Fig. 2A. Growth of the PKCI-/- MEF cells slowed somewhat after passage 6 but with further passage their growth rate increased. Thus, as shown in Fig. 2 A, at passages 10, 20, and 25 they displayed rapid growth. Indeed, these cells became spontaneously immortalized and have undergone >50 serial passages. However, they do not display morphologic evidence of transformation or loss of contact inhibition of growth. To confirm these results we isolated three additional PKCI+/+ and three additional PKCI-/- MEF cultures and found that by passage 10 the growth rate of all three of the PKCI+/+ cultures had begun to decrease, but the three PKCI-/- cultures continued to grow at a rapid rate. At about passages 15–17 all of the PKCI+/+ cultures stopped growing and all of the MEF cultures continued to grow rapidly (data not shown).
Fig. 2.
Growth of MEF cells. MEF cultures were established from both PKCI+/+ (WT) and PKCI-/- (KO) mice by using 13.5-day embryos. The cells were serially passaged in DMEM with 10% FBS. (A) Growth curves were carried out with WT and KO MEF cells at passage (P) 2, 10, 20, and 25. (B) Relative sensitivity of WT and KO MEF cells to ionizing radiation. Exponentially growing early-passage 2 (P2) WT and KO MEF cells and late-passage (P42) immortalized KO MEF cells were treated with ionizing radiation ranging from 0.5 to 6 Gy. The numbers of colonies were then determined and plotted as the surviving fraction (SF). SF = PE of irradiated cells/PE of control unirradiated cells, where plating efficiency (PE) = number of colonies/number of cells seeded.
In view of the spontaneous immortalization of the PKCI-/- MEF cells, and previous evidence that PKCI might be involved in the response of cells to irradiation (24, 36), it was of interest to determine whether the PKCI-/- MEF cells displayed an altered response to radiation-induced cytotoxicity. Early-passage (passage 2) PKCI+/+ and PKCI-/- MEF cells and passage 42 immortalized and rapidly growing PKCI-/- MEF cells were seeded and irradiated with γ-rays by using a cesium-137 irradiator source at a dose range of 0.5–6 Gy (Fig. 2B). The plating efficiencies for the early nonirradiated passage PKCI+/+ MEF cells was 0.5%, for the early-passage PKCI-/- MEF cells it was 0.7%, and for the immortalized PKCI-/- MEF cells it was 7%. The surviving fraction data indicated that both the early-passage and immortalized MEF cells from PKCI-/- mice are more resistant to ionizing radiation than are the MEF cells from early-passage PKCI+/+ mice.
Carcinogenesis Study in Mice. Thus far the PKCI-/- mice have been maintained until about age 24 months without significant evidence of spontaneous tumor formation. However, the fact that PKCI-/- MEF cells underwent spontaneous immortalization in cell culture and displayed more rapid growth, as described above, suggested that mice deficient in the expression of this gene might be more susceptible to carcinogen-induced tumor formation. Therefore, WT and PKCI-/- mice were treated with the carcinogen NMBA, following a protocol in which this nitrosamine carcinogen induces squamous carcinomas of the forestomach in mice (19, 32). Thirty-nine PKCI+/+ and 71 PKCI-/- mice (both males and females average age 34 weeks) were given eight intragastric doses (2 mg/kg per dose) of NMBA over the course of 4 weeks. As controls, eight PKCI+/+ and eight PKCI-/- mice were fed the same diet but not given NMBA. All of the mice were carefully observed weekly and killed 12 weeks after the last dose of NMBA and complete autopsies were performed. Two untreated control PKCI-/- mice developed spontaneous benign tumors: one had a leiomyoma of the uterus and the other a s.c. neurofibroma. No other spontaneous tumors were observed. Treatment with NMBA led to a high incidence of tumors of the forestomach in both the PKCI+/+ and PKCI-/- mice, but the incidence, multiplicity, tumor volume, and degree of malignancy were significantly greater in the PKCI-deficient mice (Table 1). The tumors in both groups of mice were macroscopically visible and had a papillary, pedunculated, or sessile appearance. Histologic studies indicated that they could be classified as either papilloma (PAP) or well differentiated squamous cell carcinoma (SCC). Seventy-seven tumors (51 PAP and 26 SCC) were induced in NMBA-treated PKCI-/- mice and 18 tumors in treated PKCI+/+ mice. Representative histological features of each type of tumor are shown in Fig. 3. In addition, areas of squamous hyperplasia of the epithelium of the forestomach and squamous dysplasia were observed in most of the mice in both groups of the NMBA-treated mice. These lesions were accompanied by hyperkeratosis in the surface of the epithelium. The regions of hyperplasia displayed proliferating mature squamous epithelium, extending upward or downward, which was often associated with basal cell proliferation. In the PAP lesions the squamous epithelium showed papillary upward growth or nodular and irregular downward growth, and in some lesions the epithelium showed pedunculated or sessile growth. Tumors that displayed cytologic and architectural abnormalities, cellular and nuclear pleomorphism, and loss of cell polarity were classified as squamous cell carcinoma.
Table 1. Tumors of the forestomach in PKCI+/+ and PKCI-/- mice treated with NMBA.
| Incidence (% mice with tumors)
|
Multiplicity, no. of tumors per mouse
|
Tumor volume, mm3
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Treatment | No. of mice | Total (PAP + SCC) | PAP | SCC | Total (PAP + SCC) | PAP | SCC | Total (PAP + SCC) | PAP | SCC |
| PKCI-/- | NMBA | 71 | 41/71 (58%)* | 33/71 (46%) | 18/71 (25%)† | 1.08 ± 1.13‡ | 0.72 ± 0.91§ | 0.37 ± 0.70¶ | 3.38 ± 4.64∥ | 2.41 ± 3.55 | 5.29 ± 5.87 |
| PKCI+/+ | NMBA | 39 | 14/39 (36%) | 13/39 (33%) | 2/39 (5%) | 0.46 ± 0.68 | 0.41 ± 0.64 | 0.05 ± 0.22 | 1.50 ± 2.27 | 1.34 ± 2.32 | 2.75 ± 1.78 |
| PKCI-/- | None | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PKCI+/+ | None | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Results are presented as mean ± SD. PAP, papilloma; SCC, squamous cell carcinoma. NMBA-treated PKCI-/- vs. PKCI+/+
, P < 0.03
, P < 0.007
, P < 0.0006
, P < 0.04
, P < 0.0007
, P < 0.02
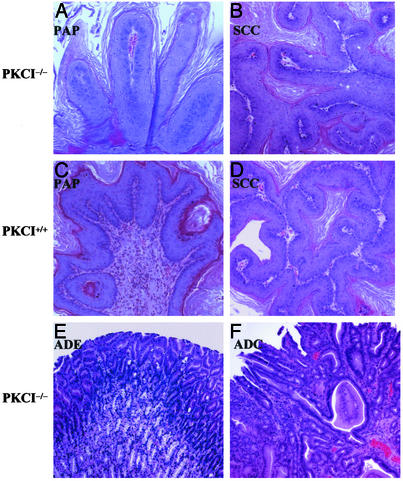
Fig. 3.
Representative histopathological features of papilloma (PAP), squamous cell carcinoma (SCC), adenoma (ADE), and adenocarcinoma (ADC). Tumors were induced in PKCI+/+ and PKCI-/- mice by treatment with NMBA. Note papillary growth in the epithelium of the forestomach (A, PKCI-/-, ×200; C, PKCI+/+, ×200). Note architectural abnormalities, cellular pleomorphism, and loss of cell polarity in the SCC lesions of the forestomach (B, PKCI-/-, ×100; D, PKCI+/+, ×100). Note back-to-back cylindrical or cuboidal crypts in the ADE (E, PKCI-/-, ×100), and the irregular shape and tubular growth of the glandular structures in the ADC (F, PKCI-/-, ×100).
The data on the forestomach tumors, with respect to incidence (number of mice with tumors), multiplicity (mean number of tumors per mouse), and mean tumor volumes are summarized in Table 1. In the NMBA-treated PKCI-/- mice the incidence of PAP plus SCC was 58%, whereas in the NMBA-treated PKCI+/+ mice it was 36% (P < 0.03). This difference was especially striking with respect to SCC, which represent more advanced lesions, because for SCC the respective incidences were 25% and 5% (P < 0.01). There were also significant increases in the numbers of SCC per mouse in the NMBA-treated PKCI-/- mice, with a mean of 0.37 per mouse compared with only 0.05 per mouse in the NMBA-treated PKCI+/+ mice. There was also an ≈2-fold increase in the average tumor volume in the NMBA-treated PKCI-/- mice when compared with the NMBA-treated PKCI+/+ mice (Table 1).
In addition to the large numbers of squamous tumors of the forestomach tabulated in Table 1, three NMBA-treated PKCI-/- mice developed a total of four adenomas (ADE) in the glandular stomach and one mouse developed an adenocarcinoma (ADC) of the glandular stomach. Representative histologic features of these lesions are shown in Fig. 3 E and F. The three adenomas displayed back-to-back cylindrical or cuboidal crypts and mild–severe atypia of the epithelial cells. The single adenocarcinoma displayed irregular tubular glandiform structures and cells with pleomorphic cytoplasm and nuclei. Therefore, this tumor was diagnosed as a tubular well differentiated adenocarcinoma, based on described criteria (37). There was an additional tumor-like lesion in the glandular stomach in one of the NMBA-treated PKCI-/- mice. Macroscopically this lesion appeared as a slight swelling of the mucosa. Microscopically, it showed elongated crypts and a villous surface lined with a regular epithelium that contained a few goblet cells. This lesion was diagnosed as a hyperplastic polyp. None of the NMBA-treated PKCI+/+ mice displayed lesions of the glandular stomach, and none of the untreated PKCI-/- or PKCI+/+ mice displayed tumors of the forestomach or glandular stomach (Table 1 and data not shown).
Discussion
As described above, the precise function of PKCI is not known, although it shares homology with the tumor-suppressor gene FHIT, and both proteins contain a HIT sequence (5, 11). Therefore, it was of interest to use the technique of homologous recombination to develop mice that have a homozygous deletion (-/-) in the PKCI gene. We were surprised to find that these mice displayed apparently normal fetal and adult development, aside from a transient decrease in weight gain in the male PKCI-/- mice. Indeed, thus far these mice have reached an age of about 2 years and they display no evidence of increased tumor incidence or other gross pathology. Therefore, PKCI does not appear to play an essential role in development, at least in this strain of mice and under the given environmental conditions. Perhaps this is because FHIT or other members of the HIT family of proteins compensate for the missing function of PKCI. Therefore, in future studies it will be of interest to develop mice that are genetically deficient in the expression of both PKCI and FHIT.
The PKCI protein was originally identified as an in vitro inhibitor of PKC (2), and we identified PKCI through its interaction with the regulatory domain of PKCβ1 in the yeast two-hybrid system (1, 5). Western blot analysis of extracts of several tissues obtained from PKCI+/+ and PKCI-/- mice revealed no significant differences in the abundance of PKCs-α, -β1, -β2, or -γ or their state of activation as determined with a PKC phospho-specific antibody (data not shown). These negative findings are consistent with the fact that although early studies suggested that PKCI inhibits the activity of PKC, subsequent studies have given equivocal results (1, 38). However, further studies are required to determine whether PKCI plays an important physiologic role in the regulation of PKC.
Despite the absence of obvious abnormalities in growth and development in the intact PKCI-/- mice, we found that with serial in vitro passage of MEF cultures established from PKCI-/- mice the cells increased their growth rate and underwent spontaneous immortalization, whereas with serial passage MEF cells from PKCI+/+ mice underwent the expected senescence and ceased growing (Fig. 2 A). The immortalized PKCI-/- MEF cultures retained contact inhibition of growth and did not display evidence of spontaneous malignant transformation. Because of the these findings and evidence that PKCI may play a role in radiation sensitivity (24), we examined the sensitivity of these MEF cells to cytotoxicity by ionizing radiation. We found that the PKCI-/- cells were much more resistant to ionizing radiation (0.5–6 Gy) than the PKCI+/+ cells (Fig. 2B). This result is consistent with a previous study demonstrating that overexpression of PKCI in the LM217 cell line increases the sensitivity of these cells to ionizing radiation (36). The spontaneous immortalization of the PKCI-/- cells and their increased resistance to ionizing radiation suggests that these cells may have defects in the function of telomerase, p53, or other proteins involved in cellular responses to radiation damage (39–42), but this remains to be determined.
In view of the above results obtained with the PKCI-/- MEF cells it was of interest to investigate the possible role of PKCI in carcinogenesis. Therefore PKCI+/+ and PKCI-/- mice were treated with the chemical carcinogen NMBA by intragastric administration, killed 12 weeks later, and examined for gastric tumors. After bioactivation NMBA yields benzaldehyde and an electrophilic agent that methylates DNA, thus resulting in formation of the promutagenic adduct O6-methylguanine (43). As expected from previous studies using a similar carcinogenesis protocol (19), we obtained squamous tumors of the forestomach in both groups of the NMBA-treated mice, but the incidence, multiplicity per mouse, tumor volume, and degree of malignancy were significantly greater in the PKCI-/- than in the PKCI+/+ mice (Table 1). Two of the untreated PKCI-/- mice developed spontaneous nongastric benign tumors, one a leiomyoma of the uterus and the other a s.c. neurofibroma, but no spontaneous tumors were found in the PKCI+/+ mice. It is highly unlikely that the increased susceptibility of our PKCI-/- mice to tumor induction by NMBA, when compared with the control 129 strain mice, reflects residual background from the C57BL/6 mice rather than homozygous loss of the PKCI gene, because over 96% of the genetic background of our PKCI-/- mice is derived from 129 strain mice. Furthermore, 129 strain mice are much more susceptible to carcinogenesis than C57BL/6 mice or mice with a mixed 129 and C57BL/6 background (44–47).
It is of interest to compare our findings on tumorigenesis in PKCI-/- mice with those reported for FHIT-deficient mice, because of the similarities between the PKCI and FHIT proteins described above. It appears that FHIT deficiency causes a greater susceptibility to tumorigenesis than PKCI deficiency because 100% of the NMBA-treated FHIT deficient mice developed multiple tumors of the forestomach, including invasive carcinomas, as well as tumors of the sebaceous gland which were similar to those that occur in the Muir–Torre familial cancer syndrome (48). In addition multiple spontaneous sebaceous tumors, and other type of tumors, were found in the FHIT-deficient mice (19, 20). As mentioned above, we did observe a few spontaneous nongastric tumors in untreated PKCI-/- mice, but further studies with a larger group of these mice are required to determine the significance of this finding. In the previous studies on FHIT, similar results on tumorigenicity were obtained with FHIT+/- and FHIT-/- mice, which led the authors to suggest that FHIT is haploinsufficient with respect to tumorigenicity (20). The present studies were done only with PKCI-/- mice, therefore it remains to be determined whether PKCI also displays this characteristic. As discussed above, PKCI and FHIT share similar structural features and both proteins appear to have nucleotidyl hydrolase activity, although with somewhat different substrate specificities (11). However, the physiologic relevance of the latter activity and its possible role on growth control and tumor suppression is not known. Because there is also evidence that PKCI plays a role in regulating gene transcription (27, 36), it is possible that PKCI deficiency enhances tumorigenesis through alterations in profiles of gene expression.
In summary, the present studies indicate that a deficiency in the expression of PKCI can enhance the immortalization of MEF cells and also their resistance to ionizing radiation. Furthermore, this deficiency enhances the susceptibility of mice to carcinogen-induced tumor formation. Taken together, these findings suggest that, like FHIT, PKCI may normally play a tumor-suppressor role, but the precise biochemical functions of both PKCI and FHIT and the possible role of PKCI in tumorigenesis in humans remain to be determined.
Acknowledgments
We thank Michael Klein, Yao Yao, Atsuko Deguchi, Christopher Lima, Tom Hei, and Hanina Hibshoosh for their valuable advice in these studies. This research was supported by National Institutes of Health/ National Cancer Institute Grant CA-02656, and awards from the National Foundation for Cancer Research and the T.J. Martell Foundation (to I.B.W.).
Abbreviations: PKCI, PKC-interacting protein; hPKCI, human PKCI; HIT, histidine triad; FHIT, fragile HIT; NMBA, N-nitrosomethylbenzylamine; MEF, mouse embryonic fibroblast; ES, embryonic stem; PAP, papilloma; SCC, squamous cell carcinoma; KO, knockout.
References
- 1.Klein, M. G., Yao, Y., Slosberg, E. D., Lima, C. D., Doki, Y. & Weinstein, I. B. (1998) Exp. Cell Res. 244, 26-32. [DOI] [PubMed] [Google Scholar]
- 2.McDonald, J. R. & Walsh, M. P. (1985) Biochem. J. 232, 559-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser, E. D. & Walsh, M. P. (1991) FEBS Lett. 294, 285-289. [DOI] [PubMed] [Google Scholar]
- 4.Brzoska, P. M., Chen, H., Levin, N. A., Kuo, W. L., Collins, C., Fu, K. K., Gray, J. W. & Christman, M. F. (1996) Genomics 36, 151-156. [DOI] [PubMed] [Google Scholar]
- 5.Lima, C. D., Klein, M. G., Weinstein, I. B. & Hendrickson, W. A. (1996) Proc. Natl. Acad. Sci. USA 93, 5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson, K. & Aitken, A. (1994) Biochem. J. 304, 662-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner, C., Garrison, P., Gilmour, J., Peisach, D., Ringe, D., Petsko, G. A. & Lowenstein, J. M. (1997) Nat. Struct. Biol. 4, 231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson, J. D., DeWald, D. B., Mathews, W. R., Mozier, N. M., Zurcher-Neely, H. A., Heinrikson, R. L., Morris, M. A., McCubbin, W. D., McDonald, J. R., Fraser, E. D., et al. (1990) J. Biol. Chem. 265, 4583-4591. [PubMed] [Google Scholar]
- 9.Mozier, N. M., Walsh, M. P. & Pearson, J. D. (1991) FEBS Lett. 279, 14-18. [DOI] [PubMed] [Google Scholar]
- 10.Robinson, K., Jones, D., Howell, S., Soneji, Y., Martin, S. & Aitken, A. (1995) Biochem. J. 307, 267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima, C. D., Klein, M. G. & Hendrickson, W. A. (1997) Science 278, 286-290. [DOI] [PubMed] [Google Scholar]
- 12.Ohta, M., Inoue, H., Cotticelli, M. G., Kastury, K., Baffa, R., Palazzo, J., Siprashvili, Z., Mori, M., McCue, P., Druck, T., et al. (1996) Cell 84, 587-597. [DOI] [PubMed] [Google Scholar]
- 13.Croce, C. M., Sozzi, G. & Huebner, K. (1999) J. Clin. Oncol. 17, 1618-1624. [DOI] [PubMed] [Google Scholar]
- 14.Virgilio, L., Shuster, M., Gollin, S. M., Veronese, M. L., Ohta, M., Huebner, K. & Croce, C. M. (1996) Proc. Natl. Acad. Sci. USA 93, 9770-9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gemma, A., Hagiwara, K., Ke, Y., Burke, L. M., Khan, M. A., Nagashima, M., Bennett, W. P. & Harris, C. C. (1997) Cancer Res. 57, 1435-1437. [PubMed] [Google Scholar]
- 16.Hibi, K., Taguchi, M., Nakamura, H., Hirai, A., Fujikake, Y., Matsui, T., Kasai, Y., Akiyama, S., Ito, K. & Takagi, H. (1997) Jpn. J. Cancer Res. 88, 385-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, P. M., Liu, T. C., Chang, J. G., Chen, T. P. & Lin, S. F. (1997) Br. J. Haematol. 99, 612-617. [DOI] [PubMed] [Google Scholar]
- 18.Ahmadian, M., Wistuba, II, Fong, K. M., Behrens, C., Kodagoda, D. R., Saboorian, M. H., Shay, J., Tomlinson, G. E., Blum, J., Minna, J. D. & Gazdar, A. F. (1997) Cancer Res. 57, 3664-3668. [PubMed] [Google Scholar]
- 19.Fong, L. Y., Fidanza, V., Zanesi, N., Lock, L. F., Siracusa, L. D., Mancini, R., Siprashvili, Z., Ottey, M., Martin, S. E., Druck, T., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanesi, N., Fidanza, V., Fong, L. Y., Mancini, R., Druck, T., Valtieri, M., Rudiger, T., McCue, P. A., Croce, C. M. & Huebner, K. (2001) Proc. Natl. Acad. Sci. USA 98, 10250-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumon, K. R., Ishii, H., Fong, L. Y., Zanesi, N., Fidanza, V., Mancini, R., Vecchione, A., Baffa, R., Trapasso, F., During, M. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Y., Garrison, P. N. & Barnes, L. D. (1995) Biochem. J. 312, 925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima, C. D., D'Amico, K. L., Naday, I., Rosenbaum, G., Westbrook, E. M. & Hendrickson, W. A. (1997) Structure 5, 763-774. [DOI] [PubMed] [Google Scholar]
- 24.Brzoska, P. M., Chen, H., Zhu, Y., Levin, N. A., Disatnik, M. H., Mochly-Rosen, D., Murnane, J. P. & Christman, M. F. (1995) Proc. Natl. Acad. Sci. USA 92, 7824-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savitsky, K., Sfez, S., Tagle, D. A., Ziv, Y., Sartiel, A., Collins, F. S., Shiloh, Y. & Rotman, G. (1995) Hum. Mol. Genet. 4, 2025-2032. [DOI] [PubMed] [Google Scholar]
- 26.Savitsky, K., Bar-Shira, A., Gilad, S., Rotman, G., Ziv, Y., Vanagaite, L., Tagle, D. A., Smith, S., Uziel, T., Sfez, S., et al. (1995) Science 268, 1749-1753. [DOI] [PubMed] [Google Scholar]
- 27.Razin, E., Zhang, Z. C., Nechushtan, H., Frenkel, S., Lee, Y. N., Arudchandran, R. & Rivera, J. (1999) J. Biol. Chem. 274, 34272-34276. [DOI] [PubMed] [Google Scholar]
- 28.Korsisaari, N. & Makela, T. P. (2000) J. Biol. Chem. 275, 34837-34840. [DOI] [PubMed] [Google Scholar]
- 29.Bieganowski, P., Garrison, P. N., Hodawadekar, S. C., Faye, G., Barnes, L. D. & Brenner, C. (2002) J. Biol. Chem. 277, 10852-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan, B., Beddington, R., Costantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 31.Jiang, W., Kahn, S. M., Zhou, P., Zhang, Y. J., Cacace, A. M., Infante, A. S., Doi, S., Santella, R. M. & Weinstein, I. B. (1993) Oncogene 8, 3447-3457. [PubMed] [Google Scholar]
- 32.Fong, L. Y. & Magee, P. N. (1999) Cancer Lett. 143, 63-69. [DOI] [PubMed] [Google Scholar]
- 33.Yanase, T., Tamura, M., Fujita, K., Kodama, S. & Tanaka, K. (1993) Cancer Res. 53, 2566-2570. [PubMed] [Google Scholar]
- 34.Hirose, M., Inoue, T., Asamoto, M., Tagawa, Y. & Ito, N. (1986) Carcinogenesis 7, 1285-1289. [DOI] [PubMed] [Google Scholar]
- 35.Hirose, M., Inoue, T., Masuda, A., Tsuda, H. & Ito, N. (1987) Carcinogenesis 8, 1555-1558. [DOI] [PubMed] [Google Scholar]
- 36.Choi, E. K., Rhee, Y. H., Park, H. J., Ahn, S. D., Shin, K. H. & Park, K. K. (2001) Int. J. Radiat. Oncol. Biol. Phys. 49, 397-405. [DOI] [PubMed] [Google Scholar]
- 37.Pozharisski, K. M. (1990) Pathology of Tumors in Laboratory Animals (Intl. Agency Res. Cancer, Lyon, France).
- 38.Robinson, K., Jones, D., Howell, S., Soneji, Y., Martin, S. & Aitken, A. (1995) Biochem. J. 307, 267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gollahon, L. S., Kraus, E., Wu, T. A., Yim, S. O., Strong, L. C., Shay, J. W. & Tainsky, M. A. (1998) Oncogene 17, 709-717. [DOI] [PubMed] [Google Scholar]
- 40.McIlrath, J., Bouffler, S. D., Samper, E., Cuthbert, A., Wojcik, A., Szumiel, I., Bryant, P. E., Riches, A. C., Thompson, A., Blasco, M. A., et al. (2001) Cancer Res. 61, 912-915. [PubMed] [Google Scholar]
- 41.Muschel, R. J., Soto, D. E., McKenna, W. G. & Bernhard, E. J. (1998) Oncogene 17, 3359-3363. [DOI] [PubMed] [Google Scholar]
- 42.Gewirtz, D. A. (2000) Breast Cancer Res. Treat. 62, 223-235. [DOI] [PubMed] [Google Scholar]
- 43.Fong, L. Y., Lin, H. J. & Lee, C. L. (1979) Int. J. Cancer 23, 679-682. [DOI] [PubMed] [Google Scholar]
- 44.Malkinson, A. M. & Thaete, L. G. (1986) Cancer Res. 46, 1694-1697. [PubMed] [Google Scholar]
- 45.Donehower, L. A., Harvey, M., Vogel, H., McArthur, M. J., Montgomery, C. A., Jr., Park, S. H., Thompson, T., Ford, R. J. & Bradley, A. (1995) Mol. Carcinog. 14, 16-22. [DOI] [PubMed] [Google Scholar]
- 46.Reiners, J. J., Jr., & Singh, K. P. (1997) Carcinogenesis 18, 593-597. [DOI] [PubMed] [Google Scholar]
- 47.Justice, M. J., Carpenter, D. A., Favor, J., Neuhauser-Klaus, A., Hrabe de Angelis, M., Soewarto, D., Moser, A., Cordes, S., Miller, D., Chapman, V., et al. (2000) Mamm. Genome 11, 484-488. [DOI] [PubMed] [Google Scholar]
- 48.Lynch, H. T., Lynch, P. M., Pester, J. & Fusaro, R. M. (1981) Arch. Intern. Med. 141, 607-611. [PubMed] [Google Scholar]