Abstract
Peptide methionine sulfoxide reductase (MsrA) repairs oxidative damage to methionine residues arising from reactive oxygen species and reactive nitrogen intermediates. MsrA activity is found in a wide variety of organisms, and it is implicated as one of the primary defenses against oxidative stress. Disruption of the gene encoding MsrA in several pathogenic bacteria responsible for infections in humans results in the loss of their ability to colonize host cells. Here, we present the X-ray crystal structure of MsrA from the pathogenic bacterium Mycobacterium tuberculosis refined to 1.5 Å resolution. In contrast to the three catalytic cysteine residues found in previously characterized MsrA structures, M. tuberculosis MsrA represents a class containing only two functional cysteine residues. The structure reveals a methionine residue of one MsrA molecule bound at the active site of a neighboring molecule in the crystal lattice and thus serves as an excellent model for protein-bound methionine sulfoxide recognition and repair.
Both free and protein-bound methionine (Met) residues can be oxidized to methionine sulfoxide (Met-O) by reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI) produced by aerobic metabolism. The formation of two distinct forms of Met-O, methionine-(S)-sulfoxide (Met-S-O) and methionine-(R)-sulfoxide (Met-R-O), has been observed in vitro (27, 49). Peptide methionine sulfoxide reductase A (MsrA; EC 1.8.4.6) reduces Met-S-O to Met (32, 41), while MsrB (16, 37, 42), which demonstrates no sequence identity to MsrA (21), uses Met-R-O as a substrate (27, 49). Both MsrA and MsrB proteins reduce Met-O in a manner independent of protein-bound metals or cofactors and appear to be highly conserved across living organisms ranging from bacteria to plants to mammals.
It has been proposed that Met residues in proteins can protect cells against ROS and RNI damage by acting as an ROS sink (23). Surface-exposed Met residues in proteins can scavenge ROS and RNI to generate Met-O followed by reduction of these Met residues by MsrA. The net result is a decrease in the cellular concentration of these reactive intermediates at the expense of thiol-containing reducing agents (see below). In addition, the oxidation of Met can affect the biological activity of many proteins (reviewed in reference 5), and it has been suggested that an alternative or additional role of MsrA is to act as a regulator of such proteins in a manner analogous to the removal of a phosphoryl group by phosphatases (43). For example, the rate of closing of a voltage-dependent K+ channel is modulated by the oxidation of a single Met residue. This effect is reversed when MsrA is introduced, suggesting that Met oxidation followed by reduction by MsrA could serve as an on/off mechanism to regulate the biological activity of the channel (11). Similarly, Met oxidation has been shown to affect the ability of calmodulin to bind calcium, an effect that is reversed when MsrA is added, which in turn suggests that the oxidation and reduction of Met residues may play a role in the regulation of calcium homeostasis (51).
Because Met oxidation can impair the function of proteins critical to viability, the absence of MsrA can lead to various abnormalities in different organisms. For example, Streptococcus pneumoniae, Neisseria gonorrhoeae, Escherichia coli, and Mycoplasma genitalium, which are deficient in MsrA, demonstrate altered cytoadherence patterns (50). Mice lacking MsrA develop an atypical walking pattern (tip-toe) and have a reduced life span (30). While msrA mutant mice show increased sensitivity to oxygen (30), other organisms deficient in MsrA also exhibit increased sensitivity to exogenous oxidants. For example, E. coli (33), Erwinia chrysanthemi (17), M. genitalium (12), and Staphylococcus aureus (42), which lack the msrA gene, demonstrate increased sensitivity to H2O2. Similarly, yeast strains lacking MsrA are more sensitive to oxidative stress (31). It has also been demonstrated that an msrA mutant strain of E. coli is hypersensitive to nitric oxide and that introduction of a plasmid containing msrA from Mycobacterium tuberculosis can restore this defect (45). Furthermore, MsrA has been shown to be important for the survival of E. chrysanthemi (17) and M. genitalium (12) once they have gained entry into host cells. MsrA plays a significant role in the intracellular survival of Mycobacterium smegmatis (T. Douglas, D. S. Daniel, C. Jagannath, and S. Dhandayuthapani, unpublished data). M. smegmatis has the ability to survive inside mononuclear phagocytes for several days (1), and this species has been used as a surrogate to identify genes of M. tuberculosis that are important for intracellular survival (28, 29, 34, 48). We hypothesize that by analogy, MsrA in M. tuberculosis plays a similar role. Finally, there is growing evidence that Met oxidation is likely to be important in the aging process (44) as well as in the etiology of neurodegenerative diseases (15).
Biochemical data suggest a catalytic mechanism for MsrA that involves the formation of a sulfenic acid intermediate followed by thiol-disulfide exchange between three conserved cysteine residues and thioredoxin to regenerate the active site (3, 4, 25, 32). Here we present the crystal structure of M. tuberculosis MsrA (MsrAMtb), an MsrA that contains only two cysteine residues, in complex with protein-bound Met. The structure reveals differences relative to the Bos taurus (MsrABt) and E. coli (MsrAEcoli) structures that may assist in the understanding of observed differences in the ability of thioredoxin to efficiently regenerate the active site in these two classes of proteins.
MATERIALS AND METHODS
Preparation and crystallization of MsrA.
The gene rv0137c (msrA) was amplified by PCR using primers TBMSRAEX1 (5′-GGGAGGTCCGCATATGACGAGCAATCAG-3′; NdeI site underlined) and TBMSRAEX2 (5′-CGGTCGGGTGGGGATCCGCCGACGCTG-3′; BamHI site underlined) and M. tuberculosis H37Rv chromosomal DNA, prepared according to published procedures (13). The gene was inserted into the NdeI and BamHI sites of the pET16b vector (Novagen) to create recombinant MsrAMtb protein with a factor Xa-cleavable N-terminal His10 tag. Recombinant protein was overexpressed in E. coli BL21(DE3) and purified from cell lysates using nickel affinity chromatography. The purified protein was dialyzed overnight into 25 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM tris(2-carboxyethyl)phosphine hydrochloride, a non-thiol-containing reducing agent, followed by concentration to 30 mg/ml for crystallization trials. The N-terminal His10 tag was not removed prior to crystallization screening experiments. Polycrystalline clusters of MsrAMtb were obtained initially from Hampton Research Crystal Screen I condition 34 using the sitting drop vapor diffusion method. Optimization of these crystallization conditions using hanging drop methods produced suitable single crystals in 2.0 M sodium formate-0.1 M sodium citrate, pH 6.0, after 1 week of incubation at 4°C. Prior to data collection, crystals were soaked in 6.3 M sodium formate-0.1 M sodium citrate, pH 6.0, for 2 min and flash-cooled in liquid nitrogen.
Data collection.
Data were acquired using a Rigaku FR-D rotating copper anode X-ray generator (Rigaku Corporation) equipped with an R-AXIS IV image plate system (Molecular Structure Corporation). Crystals were mounted on a single phi-axis goniostat and cooled during data collection using an X-STREAM low-temperature system (Molecular Structure Corporation). One-degree oscillation images were collected over a total sweep of 150° with a 4-min exposure time per image. The crystal-to-detector distance was 70 mm. The data were processed with HKL2000 (38) using all data in the 50.0 to 1.5 Å resolution range. The MsrA crystals belong to space group P212121 with unit cell dimensions a = 40.55 Å, b = 64.11 Å, and c = 73.56 Å and contain one MsrA molecule per asymmetric unit. Diffraction data statistics are summarized in Table 1.
TABLE 1.
Statistics for MsrAMtb X-ray diffraction data and model refinement
| Parameter (units) | Value(s) |
|---|---|
| Unit cell dimensions (Å) | a = 40.55, b = 64.11, c = 73.56 |
| Space group | P212121 |
| Resolution range (Å) | 50.0-1.5 |
| No. of reflections | 30,657 |
| Completeness (%) | 97.6 (94.8)a |
| Redundancy | 5.8 (5.5) |
| Rsymb | 0.060 (0.576) |
| I/σ(I) | 27.8 (3.1) |
| Rcrystc | 0.160 |
| Rfreed | 0.177 |
| No. of amino acids | 168 |
| No. of protein atoms | 1380 |
| No. of water atoms | 231 |
| RMS deviatione of: | |
| Bond lengths (Å) | 0.014 |
| Angles (°) | 1.529 |
| Mean B factor of: | |
| Protein atoms (Å2) | 14.8 |
| Water atoms (Å2) | 28.9 |
| B factor from Wilson plot (Å2) | 17.3 |
Statistics in the highest-resolution shell (1.55 to 1.50 Å) are in parentheses.
Rsym = Σ|I − <I>|/ΣI, where I is the observed intensity and <I> is the average intensity of multiple symmetry-related observations of that reflection.
Rcryst = Σ||Fobsw| − |Fcalcw||/Σ|Fobsw|, where |Fobsw| is from a working set used in the structural refinement.
Rfree = Σ||Fobst| − |Fcalct||/Σ|Fobst|, where |Fobst| is from a test set not used in the structural refinement.
RMS deviation values are deviations from ideal values.
Structure determination.
The structure of MsrAMtb was determined by the molecular replacement method (40) using the program AMORE (36). MsrAEcoli (Protein Data Bank accession code 1FF3) was used as the search model because it has the highest sequence identity (44%) to MsrAMtb of the two MsrA structures available in the Protein Data Bank (2). After the rotation, translation, and rigid body refinement functions were applied to the search model against the X-ray data in the 50 to 3 Å resolution range, the resulting solution had a correlation coefficient of 0.408 and an R factor of 0.518. Manual rebuilding was performed using O (19) followed by simulated annealing refinement using CNS (7, 8). Further manual rebuilding was alternated with computational refinement, including individual isotropic B factor refinement and a bulk solvent correction using REFMAC (35). A test set containing 5% of the observed reflections was reserved for cross-validation during refinement (6). The final model has Rcryst and Rfree values of 0.160 and 0.177, respectively, with no amino acid residues found in the disallowed regions of a Ramachandran plot calculated using PROCHECK (22). Model refinement statistics are summarized in Table 1. Structural alignments were performed using DALI (18) and LSQMAN (20). Solvent-accessible surface area calculations were made using CNS with a probe radius of 1.4 Å.
Protein structure accession number.
The structure factors and refined coordinates of MsrAMtb have been deposited in the Protein Data Bank with accession code 1NWA.
RESULTS AND DISCUSSION
MsrAMtb structure.
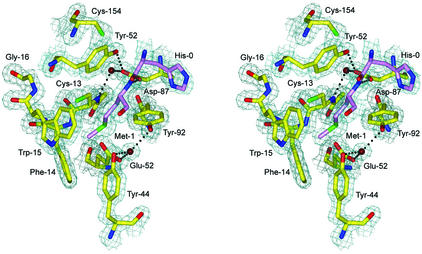
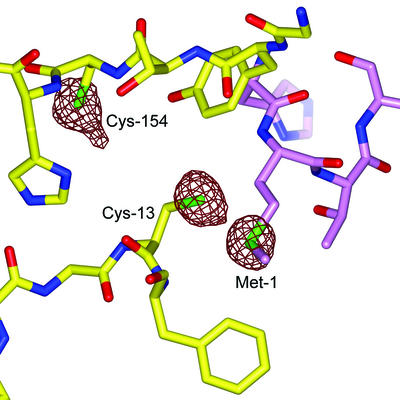
The MsrA fold has been previously characterized as a member of the α/β class in MsrABt (Protein Data Bank accession code 1FVA) (26) and MsrAEcoli (Protein Data Bank accession code 1FF3) crystal structures (46). The MsrAMtb fold resembles these structures as expected based on the similarity of their primary sequences with 38 and 44% identity, respectively (Fig. 1). The electron density of the MsrAMtb structure is generally very well defined (Fig. 2). Two residues at the N terminus that are part of the linker region between the His10 tag and the MsrAMtb sequence are clearly observed in addition to 166 residues of the MsrAMtb protein. Sixteen residues at the C terminus were not modeled due to disorder in this region. Cys-154 near the active site has extra electron density extending from the sulfur position, which creates the appearance of a modified cysteine residue. However, calculation of an anomalous difference Fourier map shows the same extension at the sulfur position of Cys-154, while the peaks for Cys-13 and Met-1 are roughly spherical (Fig. 3). Additionally, the isotropic thermal parameter for the Cys-154 sulfur atom (27.5 Å2) is about twice that of both Cys-13 (13.3 Å2) and Met-1 (13.2 Å2). These results are suggestive of small local motion for the sulfur atom of Cys-154.
FIG. 1.
Sequence alignment of MsrAs from B. taurus, E. coli, and M. tuberculosis. Residues that are identical in these proteins are boxed. CysA, CysB, and CysC (see text) are indicated above the alignment as bold letters A, B, and C, respectively. The consensus sequence found in all known MsrAs is shown in bold lettering. The alignment was generated using the CLUSTAL W method (47).
FIG. 2.
Stereo view of a 2Fo-Fc SIGMAA-weighted electron density map (39) calculated at a contour level of 1.4 σ at the active-site region containing the consensus sequence GCFWG. His-0 and Met-1 from a neighboring MsrAMtb molecule are shown in violet. Water molecules are shown in dark red, and hydrogen bonds at a distance of 3.2 Å or less are shown as dashed lines. This figure was generated using BOBSCRIPT (14), GLR (version 0.5.0; National Institutes of Health, Bethesda, Md.; http://convent.nci.nih.gov/∼web/glr/glrhome.html), and POV-Ray (version 3.5; POV-Team, Williamstown, Victoria, Australia; http://www.povray.org).
FIG. 3.
Anomalous difference Fourier map indicating peaks for sulfur atoms, the only significant anomalous scatterers in the MsrAMtb structure. The peaks for Cys-13 and Met-1 are roughly spherical, while Cys-154 shows an extended peak suggestive of local motion at this position. The sulfur anomalous difference peaks were contoured at 3.0 σ. This figure was generated using BOBSCRIPT (14), GLR (version 0.5.0; National Institutes of Health; http://convent.nci.nih.gov/∼web/glr/glrhome.html), and POV-Ray (version 3.5; POV-Team; http://www.povray.org).
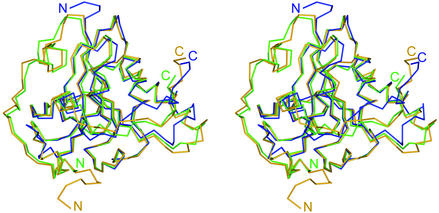
MsrAMtb Cα atoms align with those of MsrABt with a root mean square distance (RMSD) of 1.5 Å for 161 target pairs and with those of MsrAEcoli with an RMSD of 1.6 Å for 161 target pairs (Fig. 4). MsrAMtb differs from these molecules in that it possesses a significantly truncated N-terminal region and in that it lacks a cysteine residue conserved in the C-terminal region (Cys-227 in MsrABt and Cys-206 in MsrAEcoli). MsrAMtb has only 11 residues preceding the conserved GCFWG active-site motif, while the bovine and E. coli forms have 70 and 49 residues, respectively. The N-terminal extension in MsrAEcoli has been proposed to have a role in folding efficiency in vivo (3), but its absence in MsrAMtb suggests that such an extension is unnecessary for proper folding and catalysis in MsrAMtb.
FIG. 4.
Stereoview of alignment of the Cα backbones of the MsrAMtb, MsrABt, and MsrAEcoli crystal structures shown in blue, green, and orange, respectively. N and C termini are designated according to the same color code. MsrAMtb Cα atoms align to those of MsrABt with an RMSD of 1.5 Å for 161 target pairs and to MsrAEcoli with an RMSD of 1.6 Å for 161 target pairs. The C terminus of MsrAMtb exhibits the largest structural variation when compared to MsrABt and MsrAEcoli (see text). This figure was generated using BOBSCRIPT (14), GLR (version 0.5.0; National Institutes of Health; http://convent.nci.nih.gov/∼web/glr/glrhome.html), and POV-Ray (version 3.5; POV-Team; http://www.povray.org).
The MsrAMtb active site and implications for substrate recognition and catalysis.
The proposed catalytic mechanism for MsrABt and MsrAEcoli, which involves three cysteine residues designated CysA, CysB, and CysC, is summarized as follows (3, 4): (i) CysA carries out a nucleophilic attack on the sulfur of Met-S-O, thereby forming a trigonal-bipyramidal intermediate; (ii) a rearrangement occurs, producing a proposed sulfenic acid moiety at CysA and a Met leaving group; (iii) acid catalysis facilitates the release of the hydroxyl group of the sulfenic acid as a water molecule, followed by attack of CysB on CysA leading to the formation of a disulfide bond; and (iv) CysC similarly attacks CysB, forming a disulfide bond which in turn is ultimately reduced by the thioredoxin/thioredoxin reductase system or dithiothreitol (DTT) to recycle the active site.
MsrAMtb Cys-13 and Cys-154 are the only cysteine residues in the molecule and are equivalent to CysA and CysB, respectively. The lack of CysC in MsrAMtb means that the mechanism of regeneration of the active site must be different from what has been proposed for the previously characterized MsrABt and MsrAEcoli proteins. Mutation and truncation experiments eliminating CysC performed on MsrABt and MsrAEcoli produced Met-O reductase activity with little or no ability to regenerate the active site using the thioredoxin/thioredoxin reductase system (3, 4, 25). Thus, the CysA-CysB disulfide bond described above that is proposed to be formed during catalysis in MsrABt and MsrAEcoli is not directly reduced by thioredoxin. In contrast, it has been shown that recombinant MsrAMtb is active in vitro and has a comparable specific activity to unmutated MsrAEcoli using the thioredoxin/thioredoxin reductase system (45). These results suggest that in MsrAMtb, thioredoxin can directly reduce the CysA-CysB disulfide bond formed after product release (see below).
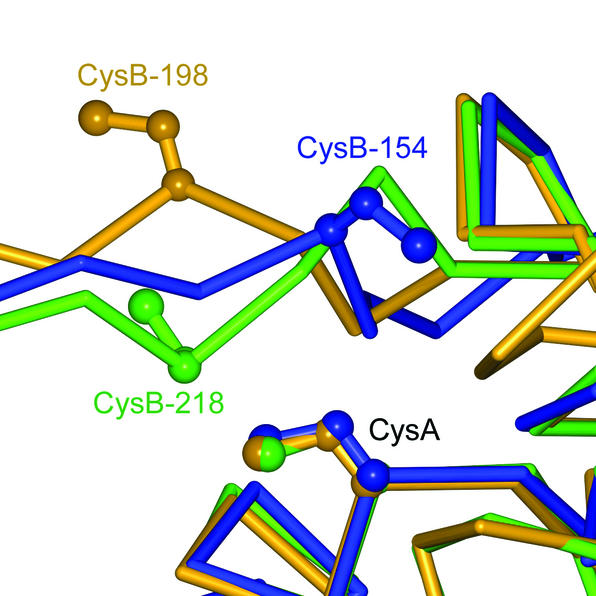
Examination of the three available MsrA structures indicates a structurally conserved core, including a well-conserved motif at the active site containing the consensus sequence GCFWG. However, two observations in the MsrAMtb crystal structure suggest possible reasons for the difference in mechanisms for recycling the active-site nucleophilic cysteine residue among these MsrAs. First, structural differences are evident by visual inspection of the C-terminal region containing CysB in MsrAMtb and that containing CysB and CysC in MsrABt and MsrAEcoli (Fig. 4). In a stretch of 18 residues at the C terminus which have observable electron density in all three known MsrA structures, the RMSD for Cα atoms in MsrAMtb residues 146 to 163 is 2.9 Å when compared to the equivalent MsrABt residues 211 to 228 and 3.3 Å when compared to the equivalent MsrAEcoli residues 191 to 208. These differences are greater than the RMSD for this region comparing MsrABt to MsrAEcoli (2.2 Å) and the average RMSD comparing the entire MsrAMtb structure to the others (1.5 to 1.6 Å). Comparing the primary sequence for this 18-residue region shows only 3 identities between MsrAMtb and MsrABt or MsrAEcoli, while the latter two share 11 identities. Second, Fig. 5 shows that the position of CysB in MsrAMtb is shifted one residue relative to the positions of CysB in the bovine or E. coli forms, which must lead to a different position for the CysA-CysB disulfide bond formed during the reaction cycle and in turn could lead to variation in accessibility and/or reactivity of this disulfide bond toward thioredoxin (3). Evidence for flexibility of the C terminus has been observed as a conformational change in this region in crystal structures of a free form of MsrABt compared to a DTT-bound form, where a DTT molecule bridges the sulfur atoms of CysA and CysB (26). By analogy, the C-terminal region of MsrAMtb is presumed to be flexible and, furthermore, with the shifted CysB relative to the bovine and E. coli forms, it may adopt a conformation that is amenable to direct reduction of the CysA-CysB disulfide bond by thioredoxin. Such a conformation might be disfavored or disallowed in MsrABt and MsrAEcoli to account for the low reactivity toward thioredoxin observed when CysC is inactivated by mutation or truncation. Examination of distances between CysA and CysB reveals that the positions of their side chains in MsrAMtb can be altered such that formation of a disulfide bond is possible. The Cα-Cα atom distance for CysA and CysB in MsrAMtb is 7.8 Å. The rotamers of the cysteine side chains can be modeled such that the closest distance between sulfur atoms is 3.4 Å. In contrast, the Cα-Cα atom distance for CysA and CysB in MsrAEcoli is 11.0 Å, too long for disulfide bond formation (46). In MsrABt this distance is 7.1 Å, but the side chain rotamers of CysA and CysB in MsrABt cannot be reoriented in as favorable a manner for disulfide bond formation as is observed in MsrAMtb. This is due to the fact that CysB in MsrABt is shifted one residue toward the N terminus relative to the position of CysB in MsrAMtb. Thus, a more substantial conformational change to the protein backbone would be necessary to form the bond (Fig. 5).
FIG. 5.
Alignment of the Cα backbones of the MsrAMtb, MsrABt, and MsrAEcoli crystal structures shown in blue, green, and orange, respectively, illustrating the relative positions of CysB. CysB of MsrAMtb (CysB-154) structurally aligns to residues shifted one position towards the C terminus relative to MsrABt CysB-218 and MsrAEcoli CysB-198, which may play a role in the observed difference in reactivity of CysA-CysB disulfide bonds toward thioredoxin in these enzymes. This figure was generated using BOBSCRIPT (14), GLR (version 0.5.0; National Institutes of Health; http://convent.nci.nih.gov/∼web/glr/glrhome.html), and POV-Ray (version 3.5; POV-Team; http://www.povray.org).
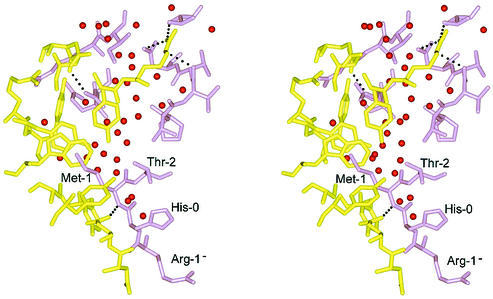
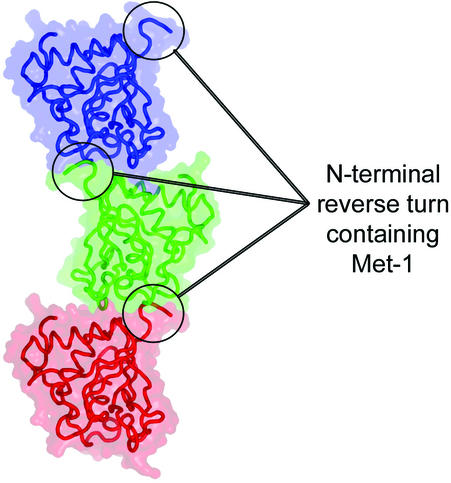
A fortuitous crystal-packing contact is observed in that Met-1 of one MsrAMtb molecule binds to the active-site pocket of a neighboring MsrAMtb molecule in the crystal lattice. The interaction between MsrA molecules at this crystal contact buries approximately 587 Å2 of solvent-accessible surface area per polypeptide. The specificity of the interaction appears to be mediated primarily through Met binding to the active site. In addition to this specific interaction, there exist a large number of water-mediated contacts (Fig. 6), consistent with the ability of MsrA to act on Met-O, found in a wide variety of protein molecules. Met-1 is positioned at the surface of the protein in a type I reverse turn formed by His-0 (part of the linker region of the His10 tag), Met-1, Thr-2, and Ser-3 and binds in the active-site pocket of a neighboring MsrA molecule related by crystal symmetry. This interaction forms a crystal lattice comprised of continuous chains of MsrA molecules stacking upon one another (Fig. 7). This crystal contact at the active site does not appear to be favored per se by residues near Met-1 in the polypeptide chain. Electron density is observed for only two other residues N-terminal to Met-1, Arg-1− and His-0, both of which come from the His10 tag. The side chains of these residues do not interact with the neighboring MsrA molecule in the crystal (Fig. 6). As an exercise, we substituted Arg-1−, His-0, and Thr-2 by using a database of the most commonly observed side chain rotamers (24). The results suggest that permissible rotamers for almost all amino acid substitutions could occur as judged by the resulting van der Waals interactions and potential hydrogen bonds that can be formed (data not shown). Bulky aromatic residues at these positions are the notable exception, however, as they appear sterically incompatible with the observed crystal packing pattern.
FIG. 6.
Stereoview of residues involved in the crystal contact interface at the MsrA active site. The active-site region is shown in yellow, and the region of the neighboring molecule containing Met-1 is shown in violet. Hydrogen bonds at a distance of 3.2 Å or less between the two protein molecules are shown as dashed lines, and water molecules are shown as red spheres. This figure was generated using BOBSCRIPT (14), GLR (version 0.5.0; National Institutes of Health; http://convent.nci.nih.gov/∼web/glr/glrhome.html), and POV-Ray (version 3.5; POV-Team; http://www.povray.org).
FIG. 7.
Crystal packing showing three MsrAMtb molecules related by the crystallographic twofold screw axis parallel to the c-axis of the unit cell. Each molecule is presented as a Cα backbone trace surrounded by a transparent rendering of its molecular surface. The N termini containing the Met-1 residue that interacts with the active site of their neighboring molecules are circled. The crystal lattice is composed of continuous chains of this packing pattern extending in the vertical direction of the graphic. This figure was generated using PyMOL (version 0.86; DeLano Scientific, San Carlos, Calif.; http://www.pymol.org).
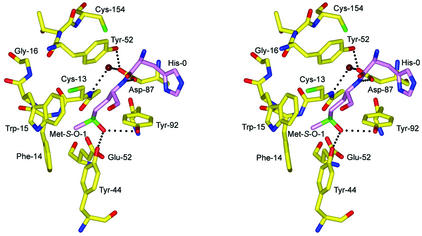
Although the binding of Met-1, the product of MsrA catalysis, may be an artifact of crystallization, the specific contacts involving Met-1 in the active site provide a plausible model for the mode of substrate binding and recognition by MsrA when modeling Met-1 as Met-S-O (Fig. 8). The sulfur atom of Met-1 is located in proximity (3.4 Å) to the sulfur atom of Cys-13 (CysA), which provides a view of Cys-13 poised for nucleophilic attack on Met-S-O. The ɛ-methyl group of Met-1 is found in the hydrophobic pocket formed by Phe-14 and Trp-15 in the GCFWG active-site consensus sequence. A side chain oxygen (Oδ1) of Asp-87 forms a hydrogen bond to the backbone amide of Met-1, possibly contributing stability during substrate binding. The other oxygen (Oδ2) of the Asp-87 side chain is involved in a hydrogen bond with conserved Tyr-152 and with a water molecule (Fig. 8) that forms a bridge to the backbone amide of Cys-13. This water molecule is conserved in the MsrABt and MsrAEcoli crystal structures and is therefore likely to play a structural role in properly orienting the side chain of Asp-87 to interact with the backbone amide of incoming substrate Met-S-O in proteins. The aromatic ring of Tyr-152 also forms a stacking interaction with the peptide bond between His-0 and Met-1. Two conserved acidic residues in MsrABt and MsrAEcoli, Glu and Asp in the vicinity of CysA, were previously identified as potential acid catalysts (26, 46). Based on the structure of MsrAMtb with Met-1 bound in the active site, Glu-52 seems a more feasible acid catalyst due to its orientation and proximity to the Met-1 and Cys-13 sulfur positions, while Asp-87 serves to stabilize substrate binding. Conserved residues Tyr-44 and Tyr-92, in addition to Glu-52, form hydrogen bonds to an additional structurally conserved water molecule common to the three known MsrA structures (Fig. 2). Assuming this water is displaced during substrate binding, one can envision how the Tyr and Glu residues might form a recognition site for the oxygen atom of the Met-S-O side chain by contributing hydrogen bonds in a similar fashion to a proposed model for an MsrABt transition-state intermediate (26). To further account for substrate specificity, it appears that binding of the R form of Met-O would be disfavored due to steric clash. In fact, a crystal structure of the MsrB domain from the N. gonorrhoeae pilB protein reveals a mirrored active-site arrangement when compared to MsrA, illustrating that the architecture of the active sites has evolved to act on only one enantiomer of Met-O (27).
FIG. 8.
Stereoview of a putative model of Met-S-O binding to the active site prior to nucleophilic attack by Cys-13. Met-S-O-1, shown in violet, is modeled in the position of Met-1 of the crystal structure. Hydrogen bonds at a distance of 3.2 Å or less are shown as black dashed lines. This figure was generated using BOBSCRIPT (14), GLR (version 0.5.0; National Institutes of Health; http://convent.nci.nih.gov/∼web/glr/glrhome.html), and POV-Ray (version 3.5; POV-Team; http://www.povray.org).
A dimethyl arsenate moiety is covalently bound to the sulfur atom of CysA in the crystal structure of MsrAEcoli (46) and may serve as a model for an enzymatic intermediate or transition state. This is similar to a model described for the MsrB domain of N. gonorrhoeae pilB based on the position of a cacodylate ion noncovalently bound in its active site (27). Structural alignment of MsrAMtb and MsrAEcoli proteins based on the minimization of differences between Cα positions of residues of the GCFWG motif superimposes elements of the Met-1 side chain from MsrAMtb onto the dimethyl arsenate atoms in the active site of the MsrAEcoli structure. The Met-1 Cγ, Sδ, and Cɛ atoms occupy positions similar to the Cɛ1, Asδ, and Cɛ2 atoms of the dimethyl arsenate moiety, respectively, with an RMSD of 0.4 Å. Thus, the similarity of the positions of MsrAMtb Met-1 and the putative transition state or intermediate analog dimethyl arsenate in MsrAEcoli lends further support to the notion that the protein-bound Met-1 found in the active site serves as a model for protein-bound substrate binding.
Genes associated with bacterial virulence and survival in hosts are gaining increased recognition as potentially more-effective antimicrobial targets than current therapeutic targets treating the growing problem of multidrug-resistant bacteria (9, 10). Since M. tuberculosis MsrA and MsrB are likely factors in intracellular survival in humans, it will be desirable to obtain crystal structures for M. tuberculosis MsrB and the human forms of these enzymes to combine with the MsrAMtb data toward the prospect of producing a new class of antituberculosis drugs.
Acknowledgments
This work was supported by funding from the R. A. Welch Foundation (AQ-1399; P.J.H.) and the Howard Hughes Medical Institute (pilot research grant; S.D.). A.B.T. and P.J.H. are affiliated with the Mycobacterium tuberculosis Structural Genomics Consortium (Structural Organization and Proteomics of TB; P50 GM62410).
REFERENCES
- 1.Barker, K., H. Fan, C. Carroll, G. Kaplan, J. Barker, W. Hellmann, and Z. A. Cohn. 1996. Nonadherent cultures of human monocytes kill Mycobacterium smegmatis, but adherent cultures do not. Infect. Immun. 64:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boschi-Muller, S., S. Azza, and G. Branlant. 2001. E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide. Protein Sci. 10:2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boschi-Muller, S., S. Azza, S. Sanglier-Cianferani, F. Talfournier, A. Van Dorsselear, and G. Branlant. 2000. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J. Biol. Chem. 275:35908-35913. [DOI] [PubMed] [Google Scholar]
- 5.Brot, N., and H. Weissbach. 2000. Peptide methionine sulfoxide reductase: biochemistry and physiological role. Biopolymers 55:288-296. [DOI] [PubMed] [Google Scholar]
- 6.Brünger, A. T. 1992. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 7.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Brünger, A. T., and L. M. Rice. 1997. Crystallographic refinement by simulated annealing: methods and applications. Methods Enzymol. 277:243-269. [DOI] [PubMed] [Google Scholar]
- 9.Chopra, I., J. Hodgson, B. Metcalf, and G. Poste. 1996. New approaches to the control of infections caused by antibiotic-resistant bacteria. An industry perspective. JAMA 275:401-403. [PubMed] [Google Scholar]
- 10.Chopra, I., J. Hodgson, B. Metcalf, and G. Poste. 1997. The search for antimicrobial agents effective against bacteria resistant to multiple antibiotics. Antimicrob. Agents Chemother. 41:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciorba, M. A., S. H. Heinemann, H. Weissbach, N. Brot, and T. Hoshi. 1997. Modulation of potassium channel function by methionine oxidation and reduction. Proc. Natl. Acad. Sci. USA 94:9932-9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhandayuthapani, S., M. Mudd, and V. Deretic. 1997. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J. Bacteriol. 179:2401-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 15.Gabbita, S. P., M. Y. Aksenov, M. A. Lovell, and W. R. Markesbery. 1999. Decrease in peptide methionine sulfoxide reductase in Alzheimer's disease brain. J. Neurochem. 73:1660-1666. [DOI] [PubMed] [Google Scholar]
- 16.Grimaud, R., B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, and F. Barras. 2001. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 17.Hassouni, M. E., J. P. Chambost, D. Expert, F. Van Gijsegem, and F. Barras. 1999. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. USA 96:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 20.Kleywegt, G. J., and T. A. Jones. 1997. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 277:525-545. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, R. A., A. Koc, R. L. Cerny, and V. N. Gladyshev. 2002. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 277:37527-37535. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 23.Levine, R. L., L. Mosoni, B. S. Berlett, and E. R. Stadtman. 1996. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 93:15036-15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovell, S. C., J. M. Word, J. S. Richardson, and D. C. Richardson. 2000. The penultimate rotamer library. Proteins 40:389-408. [PubMed] [Google Scholar]
- 25.Lowther, W. T., N. Brot, H. Weissbach, J. F. Honek, and B. W. Matthews. 2000. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 97:6463-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowther, W. T., N. Brot, H. Weissbach, and B. W. Matthews. 2000. Structure and mechanism of peptide methionine sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry 39:13307-13312. [DOI] [PubMed] [Google Scholar]
- 27.Lowther, W. T., H. Weissbach, F. Etienne, N. Brot, and B. W. Matthews. 2002. The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat. Struct. Biol. 9:348-352. [DOI] [PubMed] [Google Scholar]
- 28.Miller, B. H., and T. M. Shinnick. 2000. Evaluation of Mycobacterium tuberculosis genes involved in resistance to killing by human macrophages. Infect. Immun. 68:387-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, B. H., and T. M. Shinnick. 2001. Identification of two Mycobacterium tuberculosis H37Rv ORFs involved in resistance to killing by human macrophages BMC. Microbiology 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moskovitz, J., S. Bar-Noy, W. M. Williams, J. Requena, B. S. Berlett, and E. R. Stadtman. 2001. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 98:12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moskovitz, J., B. S. Berlett, J. M. Poston, and E. R. Stadtman. 1997. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc. Natl. Acad. Sci. USA 94:9585-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskovitz, J., J. M. Poston, B. S. Berlett, N. J. Nosworthy, R. Szczepanowski, and E. R. Stadtman. 2000. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 275:14167-14172. [DOI] [PubMed] [Google Scholar]
- 33.Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner, N. Brot, and H. Weissbach. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mundayoor, S., and T. M. Shinnick. 1994. Identification of genes involved in the resistance of mycobacteria to killing by macrophages. Ann. N. Y. Acad. Sci. 730:26-36. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 36.Navaza, J., and P. Saludjian. 1997. AMoRe: an automated molecular replacement program package. Methods Enzymol. 276:581-594. [DOI] [PubMed] [Google Scholar]
- 37.Olry, A., S. Boschi-Muller, M. Marraud, S. Sanglier-Cianferani, A. Van Dorsselear, and G. Branlant. 2002. Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J. Biol. Chem. 277:12016-12022. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski, Z., and W. Minor. 2001. DENZO and SCALEPACK, p. 226-235. In M. G. Rossmann and E. Arnold (ed.), International tables for crystallography, 1st ed., vol. F, crystallography of biological macromolecules. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 39.Read, R. J. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A. 42:140-149. [Google Scholar]
- 40.Rossmann, M. G., and D. M. Blow. 1962. The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr. 15:24-31. [Google Scholar]
- 41.Sharov, V. S., D. A. Ferrington, T. C. Squier, and C. Schoneich. 1999. Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 455:247-250. [DOI] [PubMed] [Google Scholar]
- 42.Singh, V. K., J. Moskovitz, B. J. Wilkinson, and R. K. Jayaswal. 2001. Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by the cell-wall-active antibiotic oxacillin. Microbiology 147:3037-3045. [DOI] [PubMed] [Google Scholar]
- 43.Stadtman, E. R., and P. B. Chock. 1977. Superiority of interconvertible enzyme cascades in metabolic regulation: analysis of monocyclic systems. Proc. Natl. Acad. Sci. USA 74:2761-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadtman, E. R., and R. L. Levine. 2000. Protein oxidation. Ann. N. Y. Acad. Sci. 899:191-208. [DOI] [PubMed] [Google Scholar]
- 45.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tete-Favier, F., D. Cobessi, S. Boschi-Muller, S. Azza, G. Branlant, and A. Aubry. 2000. Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 Å resolution. Structure 8:1167-1178. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei, J., J. L. Dahl, J. W. Moulder, E. A. Roberts, P. O'Gaora, D. B. Young, and R. L. Friedman. 2000. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weissbach, H., F. Etienne, T. Hoshi, S. H. Heinemann, W. T. Lowther, B. Matthews, G. St. John, C. Nathan, and N. Brot. 2002. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 397:172-178. [DOI] [PubMed] [Google Scholar]
- 50.Wizemann, T. M., J. Moskovitz, B. J. Pearce, D. Cundell, C. G. Arvidson, M. So, H. Weissbach, N. Brot, and H. R. Masure. 1996. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc. Natl. Acad. Sci. USA 93:7985-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, Y., D. Yin, G. S. Jas, K. Kuczer, T. D. Williams, C. Schoneich, and T. C. Squier. 1996. Oxidative modification of a carboxyl-terminal vicinal methionine in calmodulin by hydrogen peroxide inhibits calmodulin-dependent activation of the plasma membrane Ca-ATPase. Biochemistry 35:2767-2787. [DOI] [PubMed] [Google Scholar]