Abstract
A strategy was developed for the high-throughput localization of unknown expressed proteins in Nicotiana benthamiana. Libraries of random, partial cDNAs fused to the 5′ or 3′ end of the gene for green fluorescent protein (GFP) were expressed in planta using a vector based on Tobacco mosaic virus. Viral populations were screened en masse on inoculated leaves using a confocal microscope fitted with water-dipping lenses. Each viral infection site expressed a unique cDNA-GFP fusion, allowing several hundred cDNA-GFP fusions to be screened in a single day. More than half of the members of the library carrying cDNA fusions to the 5′ end of gfp that expressed fluorescent fusion proteins displayed discrete, noncytosolic, subcellular localizations. Nucleotide sequence determination of recovered cDNA sequences and subsequent sequence searches showed that fusions of GFP to proteins that had a predicted subcellular “address” became localized with high fidelity. In a subsequent screen of >20,000 infection foci, 12 fusion proteins were identified that localized to plasmodesmata, a subcellular structure for which very few protein components have been identified. This virus-based system represents a method for high-throughput functional genomic study of plant cell organelles and allows the identification of unique proteins that associate with specific subcompartments within organelles.
INTRODUCTION
Plant genomic and cDNA sequencing programs have identified large numbers of genes, many of which can be ascribed putative functions through bioinformatic approaches. However, for many others, a function cannot be ascribed, and there is currently a need to relate genes to their functions. Because eukaryotic cells are highly compartmentalized and the subcellular localization of a protein is intrinsic to its function, insights into the functions of genes can be gained through the subcellular localization of their products.
The use of green fluorescent protein (GFP), which as a reporter allows the facile in vivo subcellular localization of fusion proteins, has greatly facilitated cell biological studies. Fusion of genomic or cDNA fragments to the 5′ or 3′ end of the gene for GFP has permitted the development of high-throughput cell-based methods for localizing the products of ectopically expressed yeast genes (Sawin and Nurse, 1996; Ding et al., 2000) and mammalian genes (Fujii et al., 1999; Rolls et al., 1999; Misawa et al., 2000). More recently, a protein trap strategy, in which the gfp gene is carried as a mobile, artificial exon by a transposable element, has been used to localize interrupted full-length proteins expressed from their endogenous locus in Drosophila (Morin et al., 2001). In plants, random cDNA fragments fused to the 3′ end of the gfp gene have been transformed en masse into Arabidopsis using Agrobacterium tumefaciens to localize plant gene products (Cutler et al., 2000). In that study, many useful subcellular markers were obtained, although a high proportion of out-of-frame fusions to gfp displaying artifactual localizations also were recovered. A major limitation to plant transformation approaches for screening random cDNA libraries is that they are labor intensive and time consuming.
The speed and ease with which plant virus-based vectors can be produced and used subsequently to express cDNA sequences in differentiated plant tissues has made them an attractive alternative to stable transformation in studies of plant gene function (Lacomme et al., 2001). These attributes make vectors based on RNA viruses, such as Tobacco mosaic virus (TMV) and Potato virus X (PVX), amenable to high-throughput screening for gain- and loss-of-function phenotypes by protein overexpression and virus-induced gene silencing, respectively (Pogue et al., 2002). However, foreign genes inserted into such vectors can be lost during in planta propagation through recombination between duplicated or homologous sequences (Rabindran and Dawson, 2001), and this instability increases with increasing foreign insert size.
Plant virus-based vectors have been used extensively to express free GFP to localize viruses to specific cells and tissues (Oparka et al., 1997) and to express GFP fusion proteins to determine the subcellular localization of both plant and viral proteins (Oparka et al., 1997; Boevink et al., 1998; Gillespie et al., 2002). We wished to determine whether such vectors could be used in a high-throughput manner, in preference to stable transformation, to determine the subcellular localization of proteins expressed from random cDNA-gfp fusions. Vectors based on TMV and PVX are particularly suitable for population screening, because each infection focus on a leaf is initiated essentially by a single RNA molecule (Haupt et al., 2001). Previously, TMV and PVX vector populations have been used to express and screen libraries of full-length tobacco and Cladosporium fulvum cDNAs for genes that elicit hypersensitive resistance responses (Karrer et al., 1998; Takken et al., 2000). More recently, we screened a population of TMV-based vectors that contained a library of shuffled viral movement protein (mp) genes, through the simple observation of fluorescent lesion size, for mutants enhanced in mp gene function (Toth et al., 2002).
The 30-kD MP of TMV, which has been shown to bind single-stranded nucleic acids in vitro (Citovsky et al., 1990), also accumulates in plasmodesmata, the intercellular cytoplasmic bridges between plant cells, during viral infection (Tomenius et al., 1987). Although these intercellular structures are believed to play a critical role in coordinating plant physiology and development, little is known about their protein composition, and to date, no plasmodesmata-specific genes have been identified (Aaziz et al., 2001; Blackman and Overall, 2001). The difficulty of isolating plasmodesmata without accompanying contaminants makes proteomics approaches based on the large-scale isolation of plasmodesmata difficult (Kotlizky et al., 1992; Turner et al., 1994; Blackman and Overall, 2001) compared with the isolation of plasma membranes, mitochondria, and chloroplasts (van Wijk, 2001). Therefore, we tested whether a virus-based library screen could be used to identify proteins associated with a subcellular structure that is refractory to fractionation. In addition, many plant proteins are thought to interact transiently with plasmodesmata (reviewed by Ding, 1998). Such proteins are unlikely to be major constituents of the plasmodesmata pore and consequently would not be identified from isolated plasmodesmata fractions. We reasoned that such interacting proteins might be identified more readily by their colocalization with plasmodesmata using a random virus-based screen.
Here, we describe the use of 30B.GFPc3, a previously described TMV-based vector in which GFP expression is driven from a duplicated viral subgenomic promoter (Shivprasad et al., 1999), to screen libraries of random cDNAs that were fused to both the 5′ and 3′ ends of gfp for high-throughput localization studies. Using this approach, we were able to screen several hundred cDNA-gfp fusions daily. Furthermore, we describe the use of this system to identify genes that encode specific proteins that associate with plasmodesmata and demonstrate the utility of this approach in ascribing proteins to subcellular “addresses” that are not amenable to fractionation and proteomics approaches.
RESULTS
Viral Library Construction
To allow the fusion of partial plant cDNAs to both the 5′ and 3′ ends of the gfp gene, derivatives were produced from the full-length cDNA hybrid tobamovirus vector 30B.GFPc3 (Shivprasad et al., 1999). The vector's gfp gene was modified in one of two ways. For fusions to the 5′ end of the gfp gene, a vector (pOL2-Asc) was produced in which the initiating Met codon of gfp was replaced with a polylinker containing unique AscI and NotI sites (Figure 1). For fusions to the 3′ end of gfp, a set of three vectors (pXNE0, pXNE1, and pXNE2) was produced in which the termination codon was replaced with a polylinker containing unique EcoRV and NotI sites, with the three vectors differing in the number of intervening nucleotides to allow insertions in all three frames. RNA extracted from Nicotiana benthamiana roots was used to synthesize both the 5′ and 3′ fusion libraries to avoid the prevalence of mRNAs encoding proteins involved in photosynthesis that might bias the libraries toward chloroplast localizations. For fusions to the 5′ end of gfp, cDNA was synthesized using a cap structure-dependent protocol and size selected. After digestion, this cDNA was ligated to the vector pOL2-Asc and transformed into Escherichia coli, producing a population of ∼6.5 × 104 transformants that is referred to hereafter as the 5′ fusion library. The cDNA synthesized for fusion to the 3′ end of gfp was subjected to normalization and size selection before ligation to an equipartite mix of the three receptor vectors described above and transformation of E. coli to produce a population of ∼3.5 × 103 transformants that is referred to hereafter as the 3′ fusion library. DNA prepared from these populations was used to produce in vitro transcripts, which were reassembled subsequently with purified TMV coat protein into virus particles to increase their infectivity.
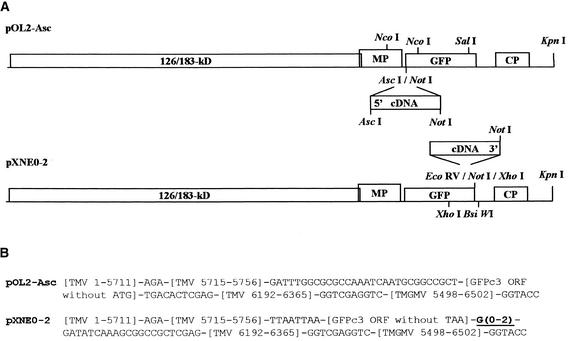
Figure 1.
Library Vectors.
(A) Schemes of the genome organizations of the four vectors (pOL2-Asc, pXNE0, pXNE1, and pXNE2) used for cDNA insertions. Restriction enzyme sites used in various constructions are indicated. CP, coat protein gene; GFP, green fluorescent protein gene; MP, movement protein gene; 126/183-kD, genes for viral components of RNA-dependent RNA polymerase.
(B) cDNAs sequences of TMV and Tobacco mild green mosaic virus (TMGMV) hybrid vectors. Sequence numbering is according to Shivprasad et al. (1999). The variable G stretch in the pXNE series of vectors is underlined.
Library Characterization
Because each lesion is initiated by a single RNA molecule, the proportions of the two populations that expressed fluorescent fusion proteins were investigated through the inoculation of reassembled transcripts onto tobacco cv Xanthi NN, which carries the hypersensitivity resistance gene N. After propagation at a permissive high temperature and counting of the fluorescent infection foci, plants were shifted to a lower, nonpermissive temperature and the necrotic lesions that developed were counted. The means for the percentage of infection foci that expressed a fluorescent product were 20 and 98% for the libraries of fusions to the 5′ and 3′ ends of gfp, respectively. All members of the 3′ fusion library are expected to produce some form of GFP fusion protein; thus, the 2% of lesions that were not discernibly fluorescent could represent variants that expressed/accumulated low levels of fusion protein or in which the GFP portion of the fusion protein did not fold correctly. That 20% of the 5′ fusion library was fluorescent was encouraging, because for members of this population to produce a fluorescent protein, the cDNAs must have had an initiation codon that was in frame with an open reading frame (ORF) that was in frame with the gfp ORF and between which there were no intervening stop codons. Thus, if all aspects of the cloning procedure had been completely successful, only one-third of this population would have been expected to be fluorescent.
In an initial characterization of the localizations of the fusion proteins expressed by the library members, reassembled virus was inoculated onto the leaves of N. benthamiana. Between 2 days and 2 weeks after inoculation, the localizations of the GFP fusion protein in fluorescent infection foci were observed with a confocal laser scanning microscope. Because it is not always possible to unambiguously define subcellular compartments with the confocal laser scanning microscope, at least without a secondary marker, and to avoid being too prejudiced concerning the localizations observed, the members of the two libraries were categorized into the following broad classes (Table 1): cytosolic (the phenotype of the unfused GFP and that expected for members of the 3′ fusion library that carried no targeting information); nuclear (including homogeneous nuclear labeling, nucleolar labeling, nucleolar exclusion, and other subnuclear structures); chloroplastic; endoplasmic reticulum and associated structures; membrane (including the plasma membrane and tonoplast) and extracellular (including labeling of specific wall regions and the free space); blobs (a general descriptive term referring to small regular structures within the cytosol that probably included mitochondria, peroxisomes, and small vesicles); and granules (a general descriptive term for large irregular aggregates within the cytosol). Concerning the last category, many of these granular phenotypes were presumed to result from the very high level of expression from the plant virus vector, leading to saturation and aggregate formation. Inoculations of the libraries to tobacco, which is a less permissive host for the vector, resulted in a lower proportion of infection foci with the granular phenotype (data not shown), substantiating the notion that such irregular granules may have arisen from protein overexpression. Furthermore, the granular phenotype was discernible more frequently in mesophyll cells than in epidermal cells and occurred early in the infection process, both features that correlate with higher levels of fluorescent fusion protein expression.
Table 1.
Distributions of Phenotypic Classes for 5′ and 3′ Fusion Libraries
| Phenotype | 5′ Fusion Library | 3′ Fusion Library |
|---|---|---|
| Cytosol | 56.5% | 83.4% |
| Nucleus | 9.9% | 5.3% |
| ER | 1.2% | 0.2% |
| Membrane | 6.2% | 0.3% |
| Chloroplast | 5.0% | 0.2% |
| Blobs | 6.8% | 7.4% |
| Granules | 14.3% | 3.2% |
| Plasmodesmata | 0.0% | 0.05% |
The proportions of fluorescent infection foci in each phenotypic class are given as percentages. The number of random fluorescent infection foci inspected by confocal laser scanning microscopy was 4395 (3′ fusion library) or 161 (5′ fusion library).
In the case of the 3′ fusion library, only ∼17% of the fluorescent infection sites showed subcellular localizations other than cytosolic localizations, whereas for the 5′ fusion library, ∼43% of the fluorescent infection sites expressed GFP fusions with discrete subcellular localizations. In light of this result and the greater complexity of the 5′ fusion library, subsequent work focused mainly on the 5′ fusion library. In addition to clearly recognizable subcellular distributions, many of the expressed fusion proteins showed novel localizations. Some examples of the observed fusion protein localizations, for which the encoding cDNA sequences were determined, are described below.
Nucleus
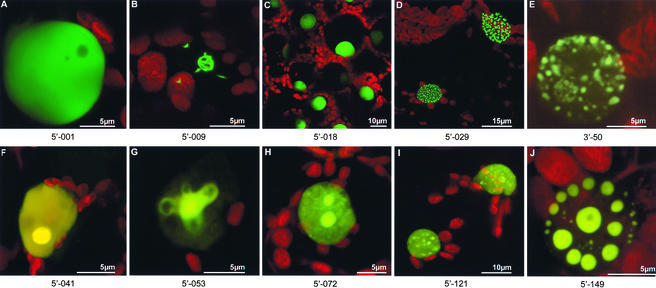
A high proportion of both libraries showed nuclear localizations. Subsequent nucleotide sequencing showed many of these localizations to be predictable based on sequence homology (Table 2). However, some unidentified protein fusions revealed unique structures associated with the nuclear envelope or nucleoplasm (Figure 2). Some protein fusions were excluded from the nucleolus (Figure 2A), whereas others showed discrete nucleolar localization (Figure 2F). One unknown protein decorated the nuclear envelope and possibly was associated with nuclear pore complexes (Figure 2D). Another unidentified protein fusion formed discrete, mobile, doughnut-shaped structures that sometimes were arranged helically within the nucleus (Figure 2J), whereas yet another unknown protein appeared to be wrapped around the nucleolus in the form of filamentous structures (Figure 2B).
Table 2.
Examples of In-Frame Fusions Homologous with Known and Unknown Proteins
| Phenotypic Class | Isolate | Homolog | Accession Number a |
|---|---|---|---|
| 3′ library | |||
| Nuclear | 3′-50 | Putative RSZp22 splicing factor (At) | AAD23894 |
| Nuclear | 3′-14 | Ribosomal protein L21 (Os) | T50602 |
| Nuclear | 3′-32 | Calmodulin (Bd) | AAA16320 |
| Nuclear | 3′-27 | Oxygen-evolving enhancer protein1, chloroplast precursor (Nt) | Q40459 |
| Membrane (plasma membrane) | 3′-06 | Pit1 protein, Pi starvation induced (Nt) | T03673 |
| Membrane (bulbs) | 3′-47 | Inorganic phosphate transporter (Nt) | AAF74025 |
| 5′ library | |||
| Chloroplast | 5′-012 | Thiazole biosynthetic enzyme, chloroplast precursor (Cs) | O23787 |
| Chloroplast | 5′-098 | Asp aminotransferase, chloroplast precursor (At) | NP_194927 |
| Chloroplast | 5′-083 | α-1,4-Glucan phosphorylase, chloroplast precursor (St) | P04045 |
| Chloroplast | 5′-111 | Biotin carboxyl carrier protein subunit (Gm) | AAG44776 |
| Chloroplast | 5′-014 | Ferritin (Gm) | BAB64536 |
| Chloroplast | 5′-131 | Probable lipoxygenase (St) | T07065 |
| Chloroplast | 5′-017 | ADP/ATP translocator (Le) | AAB49700 |
| Chloroplast | 5′-067 | ATP synthase (Gm) | S48643 |
| Nuclear | 5′-154 | Histone H1 (Nt) | BAA88671 |
| Nuclear | 5′-162 | Putative histone H2A (Os) | BAB44136 |
| Nuclear | 5′-053 | Putative histone H3 (At) | NP_189372 |
| Nuclear | 5′-072 | High-mobility-group protein2 (In) | AAC50019 |
| Nuclear | 5′-065 | Avr9/Cf-9 rapidly elicited protein 111B (Nt) | AAG43549 |
| Nuclear | 5′-041 | Putative TINY transcription factor (At) | NP_177301 |
| Nuclear | 5′-003 | Basic helix-loop-helix protein (At) | NP_563839 |
| Nuclear | 5′-151 | Nuclear protein Ytm1p (Mm) | AAF44683 |
| Nuclear | 5′-045 | 60S ribosomal protein L10A (Pc) | AAG17879 |
| Nuclear | 5′-124 | 60S ribosomal protein L23A (Nt) | Q07761 |
| Nuclear | 5′-166 | Elongation factor EF-1α (At) | Z35399 |
| Nuclear | 5′-133 | RNA binding protein AKIP1 (Vf) | AAM73765 |
| Nuclear | 5′-010 | RNA binding protein (At) | NP_188119 |
| Nuclear | 5′-167 | Putative RNA binding protein (Os) | BAA95888 |
| Nuclear | 5′-168 | Putative RNA binding protein LAH1 (At) | AAL07086 |
| ER | 5′-095 | Protein disulfide isomerase–like protein (At) | CAC81067 |
| ER | 5′-148 | Probable protein disulfide–isomerase precursor (Nt) | T03644 |
| Blobs (mitochondria) | 5′-024 | Threonyl-tRNA synthetase (At) | NP_198035 |
| Blobs | 5′-005 | Enoyl-CoA hydratase (At) | NP_193413 |
| Membrane (cell wall) | 5′-126 | Rapid alkalinization factor precursor (Nt) | AAL26478 |
| Membrane (plasma membrane) | 5′-016 | Plasma membrane protein (Nt) | T03680 |
| Nuclear | 5′-001 | Putative protein (At) | NP_190563 |
| Nuclear | 5′-009 | Hypothetical protein (At) | AAD08935 |
| Nuclear | 5′-121 | Expressed protein (At) | NP_566175 |
| Nuclear | 5′-149 | Unknown protein (At) | AAL06537 |
| Nuclear | 5′-159 | γ response I protein (At) | CAB41332 |
| Nuclear | 5′-165 | UDP glucose dehydrogenase–like protein (At) | CAC01748 |
| ER | 5′-147 | Putative integral membrane protein (At) | NP_187740 |
| ER | 5′-032 | Expressed protein (At) | NP_563744 |
| Blobs (mitochondria) | 5′-144 | Expressed protein (At) | NP_565389 |
| Blobs (mitochondria) | 5′-143 | Hypothetical protein (At) | AAG52341 |
| Blobs (mitochondria) | 5′-145 | Putative protein (At) | NP_196176 |
| Membrane (plasma membrane) | 5′-138 | Shock protein SRC2 (Gm) | T07080 |
| Membrane (plasma membrane) | 5′-074 | Ser/Thr/Tyr kinase (Ah) | AAK11734 |
| Membrane (cell wall) | 5′-127b | Glucosyltransferase-like protein (At) | BAB01433 |
Broad phenotypic classes are given, with more refined descriptions in some instance. Ah, Arachis hypogaea; At, Arabidopsis thaliana; Bd, Bryonia dioica; Cs, Citrus sinensis; Gm, Glycine max; In, Ipomoea nil; Le, Lycopersicon esculentum; Mm, Mus musculus; Nt, Nicotiana tabacum; Os, Oryza sativa; Pc, Phaseolus coccineus; St, Solanum tuberosum; Vf, Vicia faba.
a Accession number in NCBI, PIR, or SWISS-Prot database.
b Clones recovered for isolate 5′-127 were out of frame with gfp.
Figure 2.
Nuclear Localizations of Virally Expressed cDNA-gfp Fusions.
Fusions of unknown proteins to GFP produced a variety of subnuclear distributions: nucleolar exclusion (A); filamentous structures wrapped around the nucleolus (B); uniform labeling of the nucleoplasm (C); association with the nuclear envelope, possibly at or within nuclear pore complexes (D); hollow spheres that were sometimes arranged helically within the nucleoplasm (J); and “speckle”-like structures (I) similar to a GFP fusion with a homolog of a splicing factor (E). Likewise, GFP fusions to homologs of known proteins showed various localizations, including both nucleoplasmic and nucleolar labeling ([F] to [H]): a fusion between GFP and a putative histone H3 homolog (G) labeled loop-like structures associated with the nucleolus.
Chloroplast
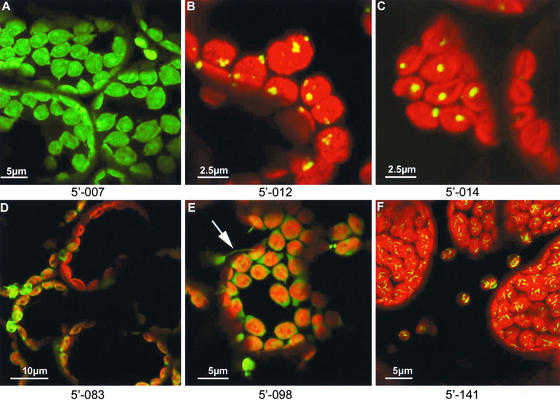
Despite the deliberate production of cDNA libraries from root mRNA, chloroplast-localizing fusion proteins were found when the libraries were inoculated onto leaves. For some, such plastidic localizations of the expressed fusion protein were predictable from sequence homology (Table 2), whereas others, labeling specific structures within chloroplasts, represented unknown proteins. Some of the expressed cDNA-gfp fusions labeled the stroma and were present in chloroplast protrusions known as “stromules” (Gray et al., 2001). One of these stromule-labeling sequences was similar to that of an aspartate aminotransferase (Figure 3E), whereas others represented unknown proteins (Figure 3A). During the course of screening, it was noted that some GFP fusion proteins (e.g., thiazole biosynthetic enzyme chloroplast precursor; Figure 3B) were associated with small vesicles that adhered to the outer chloroplast membrane. In these cases, it is possible that the partial protein sequences encoded by the cDNA-gfp fusions were sufficient to permit chloroplast targeting but insufficient to deliver the protein to its natural final address within the chloroplast. Furthermore, the filamentous structures observed in Figure 3F, which are composed of a GFP fusion to an unknown protein, may represent crystalline protein aggregates within the chloroplasts.
Figure 3.
Chloroplast Localizations.
Some GFP fusions were distributed throughout the stroma ([A], [D], and [E]), including stromules ([E], arrow; aspartate aminotransferase homolog fusion); others were associated with the chloroplast envelope ([B]; thiazole biosynthetic enzyme homolog fusion), single chloroplast inclusions ([C]; ferritin homolog fusion), or spindle-shaped structures within chloroplasts ([F]; unknown protein fusion).
Plasma Membrane and Tonoplast
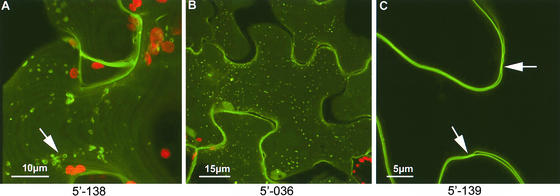
Although a large number of transporter proteins are known to be associated with the plasma membrane and the tonoplast, specific transporters from plants have been cloned only recently (Raghothama, 2000). During screening of the 5′ fusion library, a number of proteins were shown to associate with both the plasma membrane and the tonoplast. Plasma membrane–labeling proteins included GFP fusions to a Ser/Thr/Tyr kinase and a plasma membrane integral protein (Table 2, Figure 4C). Although some plasma membrane–associating proteins were distributed evenly across the plasma membrane, others showed specific punctate accumulations associated with the outer plasma membrane leaflet (Figure 4B) or associations with discrete vesicle populations (Figure 4A). One of the tonoplast-associated proteins identified from screening of the 3′ fusion library localized to motile vesicular structures that were present within the central vacuole (Figure 5A). These structures appeared to be anchored to the tonoplast and frequently fused and budded from it. Treatment of cells with latrunculin, an inhibitor of the actin cytoskeleton, completely inhibited the movement and fusion of the vesicular structures, allowing their reconstruction by optical sectioning (Figure 5B).
Figure 4.
Plasma Membrane Localizations.
(A) The inner plasma membrane surface is shown as part of an optical stack taken into a single epidermal cell. The fusion protein is concentrated in vesicular structures adhering to the inner surface of the plasma membrane.
(B) and (C) A GFP fusion to an unknown protein formed punctae associated with the outer plasma membrane surface (B), and a fusion to a plasma membrane–intrinsic protein homolog uniformly labeled the plasma membrane (C). The plasma membrane indents (arrows in [C]) represent primary pit fields.
Figure 5.
Tonoplast Localizations.
A GFP fusion to a protein homologous with a high-affinity phosphate transporter (NtPT1) was localized to vacuolar bulb-like structures attached to the tonoplast. Bulbs that were budding from the tonoplast are shown in (A). Incubation in the presence of 25 μM latrunculin, an inhibitor of the actin cytoskeleton, for 1 h allowed immobilization of the bulbs and their three-dimensional reconstruction (B).
Cell Wall
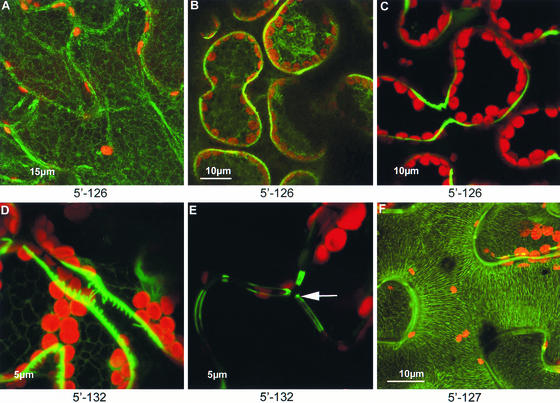
Some of the proteins identified by virus-based screening were detected in the apoplast. Interestingly, secreted GFP normally is not stable in the apoplast of N. benthamiana leaves (Boevink et al., 1999) and undergoes rapid degradation by proteases. Thus, some protein fusions appeared to confer apoplastic stability to GFP. One of these encoded a RALF (Rapid Alkalization Factor) peptide precursor (Figure 6C), a recently discovered peptide hormone involved in defense signaling (Pearce et al., 2001). Other apoplastic proteins remain to be identified. For example, the fusion protein depicted in Figure 6D was stable in the apoplast and formed discrete protein “bridges” between adjacent palisade and mesophyll cells. The same fusion protein, when expressed by the virus in epidermal cells, was present along the inner wall layers but was absent from the three-way junctions between adjoining cells (Figure 6E). Another isolate, containing a cDNA that is similar to a glucosyltransferase-like protein, expressed a fusion protein that associated stably with cell wall microfibrils (Figure 6F).
Figure 6.
Cell Wall Localizations.
A GFP fusion to a RALF peptide homolog showed temporal changes in distribution after viral inoculation of leaves. The RALF-GFP fusion was first detected within the ER (A) at 4 days after inoculation. By 5 days after inoculation, the fusion protein was observed in both the ER and the cell wall (B), and by 7 days after inoculation, it was observed solely in the cell wall (C). A fusion between GFP and an unknown protein formed narrow bridges between adjacent palisade and mesophyll cell walls (D), and in epidermal cell, it was aligned along the inner wall region but absent from the three-way wall junctions ([E], arrow). A GFP fusion to a protein homologous with a putative glucosyltransferase was associated with wall microfibrils when viewed in optical sections of the wall face (F).
Endomembrane System
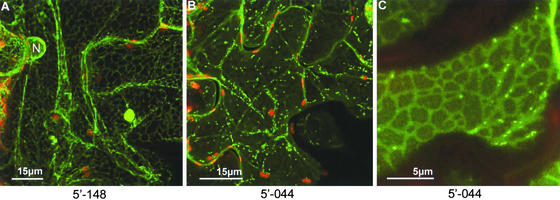
Several proteins from the 5′ fusion library were shown to associate stably with the endoplasmic reticulum (ER), including the cortical ER and the nuclear envelope (Figure 7A). In addition, some proteins labeled a motile population of structures that moved along the ER network (Figures 7B and 7C). We have tentatively identified these structures as Golgi bodies (Boevink et al., 1998; Nebenfuhr et al., 1999), although the association of novel proteins with this organelle requires future confirmation.
Figure 7.
Endomembrane Localizations.
Several fusion proteins stably labeled the cortical ER and nuclear envelope (A). Others labeled a small, motile population of structures, tentatively identified as Golgi bodies, that moved along the ER network ([B] and [C]).
Mitochondria
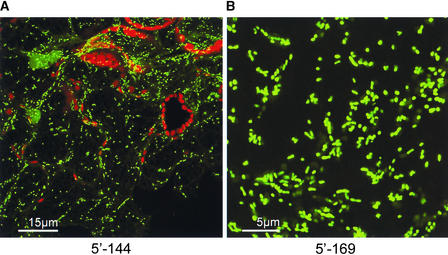
The small size of mitochondria made them difficult to distinguish from other small subcellular organelles, including motile vesicle populations. However, some proteins, such as a threonyl-tRNA synthetase mitochondrial precursor (Table 2), were associated with small, elongated structures that were likely to have been mitochondria (Figure 8).
Figure 8.
Putative Mitochondrial Localizations.
Low-magnification (A) and high-magnification (B) views of small, elongate structures labeled by GFP fusions.
Strategy Verification
To check the veracity of the approach for ascribing subcellular localizations to uncharacterized genes, 50 infection foci from the 3′ fusion library and 161 infection foci from the 5′ fusion library that showed localizations other than to the cytosol were selected for analysis. Individual infection foci were excised, and homogenates were passaged to separate N. benthamiana leaves to confirm the phenotypes. For the 5′ fusion library, 12% of the passaged viral subpopulations produced either no fluorescent infection foci or foci showing altered subcellular localizations for the fusion proteins by confocal laser scanning microscopy, whereas for the 3′ fusion library, 35% of the passaged viral subpopulations gave these results. There are a number of explanations for the lack of apparent infection foci: (1) simple infection failures, the insertion of foreign sequences correlating with reduced infectivity in a size-dependent manner; (2) for samples derived from the 5′ fusion library, a contaminant subpopulation originating from an infection initiated by a viral RNA that does not express a functional GFP fusion may have predominated; (3) during in planta propagation, recombination events may have resulted in a significant proportion of the subpopulation containing deletions so that they no longer could produce a functional GFP fusion. Such deletion events in the inserted cDNA sequences also can explain the observed alterations in sucellular localizations, although these alterations could merely reflect cross-contamination by other fluorescent infection foci.
During the rescreening procedure, it was noted that some library members displayed different subcellular localizations depending on the time of inspection. For instance, the RALF precursor described above initially showed localization to the ER (Figure 6A) but subsequently showed both ER and cell wall localization (Figure 6B), and by 7 days after inoculation, it showed localization only to the cell wall (Figure 6C). This temporal change in distribution for the GFP fusion to an apoplastically secreted protein is assumed to reflect the maturation process of the protein, with the protein initially being translated on the ER and subsequently being trafficked to its final destination in the apoplast.
As described in Methods, RNA was extracted from the passaged leaves from the recovered isolates that reproduced the original phenotype. This RNA was used in reverse transcriptase–mediated (RT) PCR with flanking primers specific to TMV sequences and gfp to recover the cDNA sequences. For both libraries, ∼40% of the amplification reactions produced single major products larger than the insert-free vector controls. The other reactions produced more than one major product, a product of the same size as the insert-free vector control, or failed completely. As discussed above, the extra bands are proposed to represent either contaminants or recombinant progeny arising from in planta deletion events. Major amplification products larger than those from the controls were cloned, and nucleotide sequences of the inserts were determined. The sequences obtained were analyzed using Basic Local Alignment Search Tool (BLASTX and BLASTN) searches. Sixty-four percent of the 44 sequences obtained from clones isolated from the 3′ fusion library had significant similarity to known or predicted genes, whereas for the 150 sequences from the 5′ fusion library, a higher proportion (74%) were similar.
3′ Library
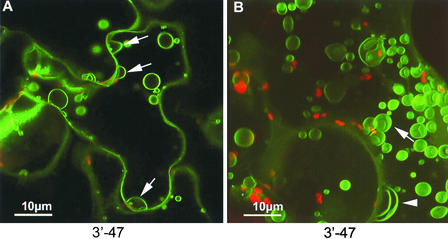
Of the 28 homologous sequences from the 3′ fusion library, only 11 were in frame; the others were in the wrong frame or orientation. Thus, only 39% of the identified sequences were in frame, which is only slightly more than the 33% expected on a purely random basis. Similarly, in the transgenic study by Cutler et al. (2000), a low proportion (52%) of the sequences identified were in frame, indicating the potential for many sequences that are naturally not translated to cause targeting. Of the identified sequences with predictable localizations, two (3′-14 and 3′-50) gave expected localizations (Table 2) and two (3′-27 and 3′-47) gave unpredicted localizations. That the GFP fusion to the oxygen-evolving enhancer protein homolog (Table 2) did not localize to chloroplasts as expected is unsurprising given the absence of the N-terminal chloroplast transit peptide. As mentioned previously, the fusion protein produced by 3′-47, which contained a sequence homologous with the tobacco high-affinity phosphate transporter PT1, localized to tonoplast-associated vesicles rather than demonstrating the predicted plasma membrane distribution.
5′ Library
Sixty-six percent of the clones from the 5′ fusion library that were homologous with known or predicted sequences were found to be in frame with gfp. The other one-third of the sequences were excluded from the analysis because they could not be responsible for the observed patterns of fluorescence and represented either nonfluorescent contaminants or recombinant progeny that had undergone in planta deletions. All of the genes whose fusion products had a predictable localization fell within the expected phenotypic classes, with three exceptions (Table 2). These three exceptions (5′-017, 5′-067, and 5′-165) were homologous with an adenine nucleotide translocator, an ATP synthase subunit, and a UDP-glucose dehydrogenase–like protein, respectively. The fusion proteins expressed from 5′-017 and 5′-067 were predicted from their homology to localize to mitochondria, but instead they localized to chloroplasts. Notably, the product encoded by 5′-017 was extended at its N terminus by 21 amino acids relative to its potato homolog and also showed little similarity to the extreme N terminus of the homolog (data not shown).
Plasmodesmata
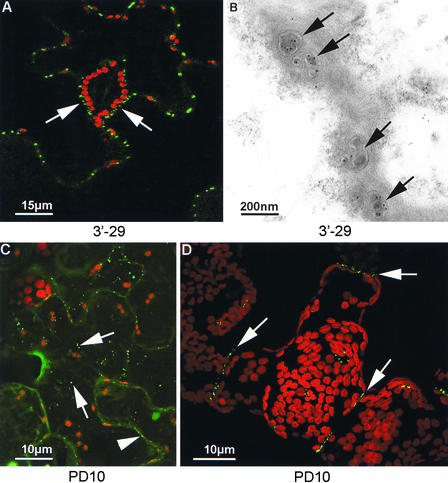
Our original rationale for developing virus-based cDNA-gfp screens was to identify plasmodesmata-interacting proteins. During our initial screening of the 3′ fusion library, two infection foci showing localization of the expressed fusion protein to plasmodesmata (Figure 9A) were isolated (3′-29 and 3′-42, alias plasmodesmata01 and plasmodesmata02), and the cDNA-derived sequences were recovered. However, foci with such a phenotype occurred with very low frequency (Table 1). Because of our interest in plasmodesmata proteins, large-scale screening of both the 5′ and 3′ fusion libraries was performed to obtain additional such isolates. Through screening of ∼20,000 fluorescent infection foci, another 10 isolates that displayed plasmodesmata localization were obtained: two from the 3′ fusion library (plasmodesmata03 and plasmodesmata04) and eight from the 5′ fusion library (plasmodesmata05 to plasmodesmata12). The isolates were passaged onto fresh leaves and RNA extracted from single infection sites showing the desired phenotype. Despite RNA preparation from single infection sites, three of the isolates, plasmodesmata07, plasmodesmata10, and plasmodesmata12, produced more than one major amplification product in RT-PCR. The major amplification products were cloned back into the progenitor, insert-free vectors, and the nucleotide sequences of the inserts were determined (Table 3). Clones derived from one of the isolates, plasmodesmata05, contained a deletion that resulted in the formation of an in-frame fusion between the TMV mp gene and gfp. Such MP-GFP fusion proteins have been shown previously to accumulate in plasmodesmata (Oparka et al., 1996; Crawford and Zambryski, 2000; Gillespie et al., 2002); thus, this isolate represented a false positive.
Figure 9.
Plasmodesmata Localizations.
A search of 20,000 infection sites identified 12 isolates that expressed fusion proteins that targeted plasmodesmata. The fusion protein expressed by isolate 3′-29 (alias plasmodesmata01) localized strongly to epidermal plasmodesmata, including the truncated plasmodesmata between epidermal cells and guard cells ([A], arrows). Immunogold labeling of thin sections with antiserum against the GFP portion of the expressed protein revealed gold particles associated with plasmodesmata pores ([B], arrows). The fusion protein expressed by isolate plasmodesmata10, which was homologous with a Rab protein, was associated first with a small, motile vesicle population ([C], arrows; 5 days after inoculation) and then associated stably with plasmodesmata ([C], arrowhead). By 10 days after inoculation, the fusion protein expressed by plasmodesmata10 was associated only with the plasmodesmata ([D], arrows).
Table 3.
Homologs of cDNAs Recovered from Isolates Expressing Plasmodesmata-Localizing Fusion Proteins
| Isolate | Homolog | Accession Number |
E Value | Comment |
|---|---|---|---|---|
| plasmodesmata01 | Hypothetical protein MMB12.12 (At) | T52393 | 9e-10 | |
| plasmodesmata02 | No significant similarity | GenBank accession number CB968091 |
||
| plasmodesmata03 | Unknown protein (At) | AAK93625 | 1e-26 | |
| plasmodesmata04 | No significant similarity | GenBank accession number CB968092 |
||
| plasmodesmata05 | Deletion mutant giving MP-GFP fusion protein |
|||
| plasmodesmata06 | Probable reticuline oxidase (Ps) | T07969 | 1e-38 | Product N-terminally truncated relative to homolog |
| plasmodesmata07 | Putrescine N-methyltransferase (Nt) | AAF14879 | 6e-73 | |
| plasmodesmata08 | Unknown protein (At) | AAK76647 | 6e-69 | Product N-terminally truncated relative to homolog |
| plasmodesmata09 | Monodehydroascorbate reductase (Le) | T06407 | e-114 | |
| plasmodesmata10 | Ras-related protein Rab11b (Nt) | Q40521 | 5e-43 | |
| plasmodesmata11 | Protein T1N15.3 (At) | D96524 | 9e-60 | Homology with 1-aminocyclopropane-1- carboxylate deaminases |
| plasmodesmata12 | Predicted NADH dehydrogenase 24-kD subunit (At) | AAK96519 | 1e-84 |
At, Arabidopsis thaliana; Le, Lycopersicon esculentum; Nt, Nicotiana tabacum; Ps, Papaver somniferum.
To confirm which sequences were responsible for the observed phenotypes, reassembled transcripts from the regenerated full-length viral clones were inoculated onto N. benthamiana plants. For the three isolates that produced more than one amplification product, only clones derived from one of the amplification products resulted in fluorescent infection foci in each case. Furthermore, the fusion protein in these fluorescent foci showed plasmodesmata localization, indicating that the clones derived from the other amplification products represented contaminants that did not express a functional GFP fusion. However, one of the isolates, plasmodesmata06, which produced a single amplification product, yielded no clones that produced fluorescent infection foci. Such clones could not be obtained directly in spite of repeated attempts: the ORF in the cDNA-derived sequence was always out of frame with gfp. Therefore, to generate clones with in-frame fusions, PCR was performed with mutagenic primers to adjust the reading frame. Inoculation of transcripts from the resulting modified, full-length clones produced fluorescent infection foci on plants, and the expressed fusion protein accumulated in plasmodesmata.
There was variation in the phenotypes produced by the regenerated plasmodesmata clones, with many of the clones initially producing fluorescent aggregates that were presumed to result from excess viral expression of the fusion proteins. Further, inoculations performed with plasmodesmata03-derived clones resulted in fluorescent infection foci that displayed two phenotypes on the same leaf: infection foci showed either cytosolic GFP fluorescence or plasmodesmata labeling. Passaging of those infection foci that showed cytosolic GFP localization resulted solely in infection foci showing the same phenotype. By contrast, passaging of infection foci that showed the plasmodesmata phenotype resulted in infection foci showing both phenotypes. This result suggested that the inserted sequence was unstable in the viral vector and was lost during in planta propagation, resulting in the cytoplasmic distribution. This hypothesis was confirmed by extracting RNA from the two lesion types followed by RT-PCR and nucleotide sequencing of clones. Although foci showing plasmodesmata phenotypes retained the insert sequence, those showing cytoplasmic phenotypes had undergone deletions within the inserted sequence (data not shown).
All of the clones, with the exception of those derived from isolate plasmodesmata08, eventually showed a low cytosolic background and specific labeling of plasmodesmata. Plasmodesmata labeling was confirmed by immunogold electron microscopy of infected tissue using GFP-specific antisera. Gold labeling was concentrated in plasmodesmata but not in adjacent wall regions (Figure 9B). In contrast to the other clones, those derived from isolate plasmodesmata08 produced labeling of the plasma membrane in addition to plasmodesmata localization. The phenotype displayed by clones based on isolate plasmodesmata10, which contained sequence homologous with that of a Rab protein (Table 3), was unusual in that fluorescence was observed in a motile vesicle population (Figure 9C) before the accumulation of the fusion proteins in plasmodesmata (Figure 9D). Such a phenotype is not incongruous with the known involvement of Rab GTPases in vesicular shuttling between donor and acceptor membranes (Rutherford and Moore, 2002).
DISCUSSION
In the postgenomics era, a variety of methods have been used to help ascribe functions to unknown genes. Because a protein's localization is intrinsic to its function, proteomics approaches have been used to elucidate plant gene function. These approaches have proven effective for subcellular compartments, such as the chloroplast and the plasma membrane, because they can be fractionated easily. Although more information may be extracted in the future from these studies, through correlation with known subcellular markers, the use of GFP fusions for in vivo localization offers the prospect of a much finer subcellular resolution and may be especially useful for gene products found in compartments that cannot be fractionated readily. These benefits make this strategy worthwhile, despite the fact that the expressed proteins are not completely native. Previously, transgenic Arabidopsis plants were used to screen a library of cDNAs fused to the 3′ end of gfp (Cutler et al., 2000). Because of the requirement to regenerate transgenic plants, this method is time consuming; therefore, we wished to develop a higher throughput method. We have been successful in this objective and have developed a plant virus vector-based method that allows the daily screening of several hundred clones. This method relies on the observation that each infection focus is initiated by an individual RNA transcribed from the library (Haupt et al., 2001; Toth et al., 2002).
In addition to the speed advantages, the use of a viral system also results in advantageous high-level expression of the fusion proteins. For the 3′ fusion library, this meant that fluorescence could be detected from virtually all of the fusions. This occurred despite the fact that the form of GFP used in this study is not expected to be fluorescent in some compartments, such as the apoplast (Boevink et al., 1999). In some respects, this high-level expression might be considered a disadvantage, because it could lead to the saturation of targeting pathways and to aggregate formation. The latter problem could be circumvented for isolates that show nonspecific aggregate formation by rescreening on a less susceptible host (e.g., tobacco) in which the “granular” phenotype is less prevalent. In future studies, appropriate development of RNA-based viral vectors for use in Arabidopsis should allow the direct screening of Arabidopsis cDNA libraries on this species. Because vector-based expression in differentiated leaf tissue is transient, it may permit the localization of fusion proteins whose expression would cause lethality in transgenic plants. Furthermore, the transient nature of the system allows the observation of temporal changes in fusion protein localization, exemplified by the progressive localization of the fusion proteins expressed by isolates 5′-126 and plasmodesmata10 from the ER to the apoplast and from vesicles to plasmodesmata, respectively.
As in the earlier work of Cutler et al. (2000), random cDNAs were fused to the 3′ end of gfp; additionally, random cDNAs, synthesized using a cap structure-dependent protocol, were fused to the 5′ end of gfp. Unsurprisingly, given the N-terminal location of most signal peptides, a much higher proportion of fluorescent infection foci showed specific localizations in the 5′ fusion library than in the 3′ fusion library. Therefore, we concentrated our efforts on the former.
3′ Library
For the 3′ fusion library, ∼17% of infection foci showed a phenotype that differed from the simple cytosolic phenotype of the control, whereas in the study by Cutler et al. (2000), only 2% of the transgenic lines obtained showed a distribution of fusion protein that differed from cytosolic GFP. This difference could reflect lower stringency in ascribing specific phenotypes during the screening of our 3′ fusion library or differences in the amounts of encoded targeting information. That the former might be the case is suggested by the fact that only 39% of the clones recovered from the 3′ fusion library were in the correct frame, whereas a figure of 52% was obtained by Cutler et al. (2000) and this was increased to 69% when one phenotypic class was excluded. Even so, these results demonstrate the potential of out-of-frame fusions to contain cryptic targeting information.
The use of a secondary marker could facilitate unambiguous subcellular localizations. For example, in the case of small subcellular organelles, such as mitochondria and Golgi bodies, inoculation of viral libraries expressing cDNA-gfp fusions, or passage of selected isolates onto transgenic plants expressing a second fluorescent protein (e.g., yellow fluorescent protein or red fluorescent protein) fused to a known subcellular targeting sequence, would provide unambiguous confirmation of the unknown fusion protein's localization.
Because out-of-frame fusions can be excluded from analysis, the authenticity with which in-frame fusions localize is perhaps more important. For the 3′ fusion library, only four isolates containing in-frame fusions of cDNAs with predictable subcellular localizations were obtained: two, 3′-50 (a splicing factor homolog) and 3′-14 (a ribosomal protein homolog), localized as expected, and one, 3′-27 (an oxygen-evolving enhancer protein homolog), clearly was mistargeted, probably as a result of the absence of its natural N-terminal transit peptide. This isolate, which was recovered independently from the 3′ fusion library twice, also is of interest because it was reported recently that silencing of this gene enhances TMV replication (Abbink et al., 2002), suggesting that the presence of this sequence within the TMV-based vector may confer some selective advantage to its propagation. The sequence found in the fourth isolate, 3′-47, was homologous with the tobacco high-affinity phosphate transporter NtPT1, and on this basis, it was expected to localize to the plasma membrane (Raghothama, 2000). Instead, the expressed fusion protein was found specifically in recently described vacuolar structures termed “bulbs” that were shown to be enriched in the aquaporin γ-TIP (Saito et al., 2002). Recently, high-affinity phosphate transporters were implicated in phosphate remobilization (Karthikeyan et al., 2002), and it is possible that the expression of PT1 on bulb structures may be associated with the remobilization of phosphate from the central vacuole. Recent GFP fusion and proteomic studies have localized the Arabidopsis phosphate transporter PHT2;1 and its spinach ortholog IEP60 to the chloroplast (Ferro et al., 2002; Versaw and Harrison, 2002), showing the spatial regulation of phosphate transporters within cells. In contrast to an earlier report regarding the vacuolar bulbs, in which they were found only in the vacuoles of cotyledon and hypocotyl cells (Saito et al., 2002), we found the bulbs in all cells in which the fusion protein was expressed.
5′ Library
Compared with the 3′ fusion library, the 5′ fusion library produced more specific localizations, and a greater proportion of the recovered cDNAs were in frame with gfp. Those clones that contained upstream ORFs that were out of frame with gfp were excluded from the subsequent analysis on the basis that they were either contaminants or recombinant progeny that had sequences deleted from the RNA molecule that initiated the infection focus. This level of contamination could have been reduced by diluting the initial inoculum to give fewer infection foci per leaf, by extracting RNA for cloning purposes only from single lesions rather than from passaged leaves, or by additional rounds of viral passage. That the out-of-frame exclusion criterion might have resulted in false negatives is demonstrated by the case of isolate plasmodesmata06, for which the upstream ORF was never recovered in frame with gfp, despite repeated attempts. That such false negatives might have occurred during library screening is illustrated by isolate 5′-127 (Figure 6F), which encoded a homolog to a glucosyltransferase-like protein and showed labeling of cell wall fibers, although the ORF for this homolog was out of frame with gfp.
Based on the library verification procedure, there was little evidence of false positives, because all of the recovered in-frame fusions with predictable localizations gave phenotypes consistent with their known properties, with three exceptions. Two of these exceptions, 5′-017 and 5′-067, were expected based on homology to be found in mitochondria; instead, they were localized to chloroplasts. Chloroplast localization in itself might be considered a mislocalization because the libraries were prepared from root tissue lacking such organelles. However, this is presumed to reflect the generality of plastid targeting information. Although missorting of chloroplast proteins to mitochondria using in vitro systems has been reported, the mistargeting of mitochondrial proteins has not (Rudhe et al., 2002). Some proteins are targeted to both of these organelles (Chabregas et al., 2001), and a recent study has shown that the use of multiple transcription start sites allows the expression of mitochondrial and chloroplast isozymes from the same gene (Obara et al., 2002). Previous studies have shown that the 76–amino acid presequence of the potato ADP/ATP translocator is neither necessary nor sufficient for mitochondrial targeting (Mozo et al., 1995). Thus, because the sequence encoded by 5′-017 was N-terminally extended over its potato homolog, the observed chloroplast localization might not be completely artifactual.
Some of the other chloroplast-localizing isolates showed a phenotype in which the GFP fusion protein accumulated in vesicles associated with the surface of the chloroplasts. This observation suggests that for these isolates, there may have been a block in the targeting pathway, resulting from either the fusion of GFP to the C terminus or the omission of sequence from the partial cDNAs necessary for the completion of targeting. The selection of larger cDNAs for insertion might result in more authentic end-point targeting. However, the consequences of the inclusion of larger foreign inserts are increased viral genomic instability and reduced levels of fusion protein expression. The latter phenomenon may not be a problem because it still may result in detectable levels of fluorescence and reduced aggregate formation. Genomic instability, or rather selection pressure for the recombinant progeny that arise from it, could be reduced by using more robust vectors that have been developed through DNA shuffling (Toth et al., 2002).
Putative Plasmodesmata Proteins
The major rationale for undertaking a high-throughput screen of cDNA libraries was to identify plant proteins that interact with plasmodesmata. To date, we have identified a number of protein sequences that consistently localize to plasmodesmata, and a future aim will be to characterize the functional significance of these interactions. The identification of the false-positive plasmodesmata05, which expresses a fusion protein with known plasmodesmata-localizing properties, indicates the veracity of this approach for identifying such proteins. However, further studies are being conducted to prove that the native plant proteins also localize to this subcellular organelle. That some others might be false positives is suggested by the isolation of plasmodesmata06, whose cDNA sequence showed similarity to a berberine bridge enzyme. The sequence recovered from this isolate encoded a protein that was N-terminally truncated relative to its homolog and that lacked the sequences shown previously to be necessary to direct GFP to the ER and subsequently to the vacuole (Bird and Facchini, 2001). The phenotype of plasmodesmata10, which contains a Rab sequence, suggests that the native protein may associate only transiently with plasmodesmata and that vesicular transport of the fusion protein is blocked at the acceptor membrane-targeting stage. Not surprisingly, given the paucity of known plasmodesmatal proteins, almost half of the sequences that localize to plasmodesmata exhibited no significant similarity or similarity to unknown proteins in BLASTX searches.
Of the other homologous sequences recovered from the plasmodesmata isolates, one encoded an enzyme related to the ethylene biosynthetic pathway (plasmodesmata11), one encoded the enzyme for the first committed step in the nicotine biosynthetic pathway (plasmodesmata07), and two (plasmodesmata09 and plasmodesmata12) encoded proteins involved in “redox”-based signaling reactions (Pastori and Foyer, 2002). However, to date, only partial cDNA sequences have been expressed, and these sequences have been expressed only from a TMV-based vector. It is possible that some of the sequences that we isolated associate with plasmodesmata in response to viral infection. For example, TMV infection is signaled by increased levels of monodehydroascorbate radicals (Fodor et al., 2001), suggesting that plasmodesmata09, which encodes a monohydroascorbate reductase homolog, might be localized to plasmodesmata in response to infection. This observation may be significant in light of the fact that the highly charged ascorbate radical can traffic significant distances away from the site of fungal infection (Muckenschnabel et al., 2001). Future studies in which the same plasmodesmata-localizing sequences and full-length cDNA sequences are expressed from both viral and nonviral expression systems should distinguish proteins that interact with plasmodesmata in response to pathogenesis from those that are normal constituents of plant signaling pathways and elucidate which localizations are truly significant.
Although the system we have developed is not easily applicable to genes that encode products with both N- and C-terminal targeting information, it will allow the fine subcellular localization of many gene products. For genes whose products have targeting sequences at both termini, it may be necessary to use transgenic plants and employ a strategy analogous to the mobile artificial gfp exon approach used with Drosophila (Morin et al., 2001). However, the virus-based system offers many advantages over transgenic plants; the most obvious of these is speed and the potential of the system to be truly high throughput, making it feasible to screen very large numbers to identify rare subcellular localizations, as exemplified by the isolation of plasmodesmata-localizing sequences. Second, because screening is performed on mature plants, sequences that might be lethal during transgenic plant regeneration will not be lost. Third, because there is a burst of protein expression, changes in subcellular localization during progression through a targeting pathway can be observed, rather than merely the final localization. Fourth, because viral isolates can be passaged to transgenic plants expressing known, fluorescent, subcellular marker proteins, more precise subcellular localizations can be achieved than by screening of transgenic plants by confocal laser scanning microscopy.
The development of “mammalian cell microarrays” (Wu et al., 2002) should facilitate studies to relate genes and functions. However, to date, no such systems exist for plant cells. Despite the various caveats we have highlighted, we believe that the high-throughput, virus vector–based system described here should provide a useful addition to the armory of techniques available to help relate plant genes to their functions.
METHODS
Vector Construction
Four vectors (Figure 1), from which GFP fusions could be expressed after the insertion of partial cDNAs, were produced from a previously described plasmid, p30B.GFPc3 (Shivprasad et al., 1999) using standard techniques (Sambrook et al., 1989). An intermediate plasmid, pOL2, in which the initiating Met codon of the gfp gene was replaced with unique EcoRV and NotI sites, was produced through overlap extension PCR with mutagenic oligonucleotides (Higuchi, 1990). The mutagenized DNA was recloned as an NcoI fragment. The plasmid pOL2 was used to create the vector pOL2.Asc by digestion with EcoRV and insertion of an AscI linker (New England Biolabs, Beverly, MA). The three vectors pXNE0, pXNE1, and pXNE2, in which the stop codon of the gfp gene was replaced with unique EcoRV and NotI sites, were produced by PCR with three different mutagenic oligonucleotides to allow fusions in each of the three frames. The mutagenized DNAs were recloned as XhoI fragments.
Library Construction
To create random cDNA fusions to the 3′ end of gfp, 4 g of root tissue from 5-week-old Nicotiana benthamiana plants was ground in liquid nitrogen and RNA extracted using Tri Reagent (Sigma) according to the manufacturer's instructions. mRNA was purified from the total RNA preparation using Oligo(dT)25 Dynabeads (Dynal, Oslo, Norway) according to the manufacturer's instructions. The immobilized mRNA was used to synthesize first-strand cDNA with a Superscript cDNA Synthesis Kit (Invitrogen, Carlsbad, CA), and after heat inactivation of the enzyme, the first-strand reaction products were used for second-strand synthesis. After collection of the magnetic beads, second-strand products were eluted in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and a half volume of 3× hybridization buffer (12× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate], 15× Denhardt's solution [1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA], and 1.5% SDS) was added. After denaturation by boiling for 5 min, the second-strand products were hybridized with the immobilized first-strand products at 65°C for 8 h. The partially normalized second-strand products were collected, and bound products were eluted in water before discarding. This hybridization process was repeated three times before purification of the normalized second-strand products with a QIAquick PCR Purification Kit (Qiagen, Valencia, CA).
Double-stranded cDNA was regenerated from the normalized second-strand products by priming with NotI-dT oligonucleotide [(T)6GCGGCCGC(T)20] and extension with cloned T7 DNA polymerase (Amersham Biosciences) at 37°C for 1 h. The products were purified by phenol:chloroform extraction and, after T4 DNA polymerase treatment, phosphorylation with T4 polynucleotide kinase and digestion with NotI separated by agarose gel electrophoresis. Fragments of >400 bp were recovered with a QIAEX II Gel Extraction Kit (Qiagen), ligated to an equipartite mixture of the three vectors, pXNE0, pXNE1, and pXNE2, that had been digested previously with EcoRV and NotI, and dephosphorylated with calf intestinal phosphatase. The ligated populations were transformed into Escherichia coli, and after plating of aliquots to determine the library complexity, the library was grown for DNA preparation.
To create random cDNA fusions to the 5′ end of gfp, total RNA was prepared from root tissue as described above. mRNA was purified from total RNA using a Micro Poly(A)Pure Kit (Ambion, Austin, TX) according to the manufacturer's instructions, and the eluted mRNA was precipitated with ethanol. The mRNA was resuspended in water and used for first-strand cDNA synthesis with Superscript II in the presence of 0.6 M trehalose in a thermocycler (Carninci et al., 1998). The mRNA was incubated initially at 65°C for 10 min with N6-NotI primer (5′-AAGCAGTGGTAACAAAGCAGAGTACGCGGCCGCNNNNNN-3′) before cooling to 45°C and the addition of other reagents. The reaction then was subjected to the following thermal profile: incubation at 45°C for 10 min; ramp to 35°C at 0.2°C/s; incubation at 35°C for 5 min; incubation at 45°C for 5 min; ramp to 55°C at 0.2°C/s; incubation at 55°C for 1 min, followed by an additional nine 1-min incubations at 0.5°C incremental temperatures; and 10 cycles of 2-min incubations at 60°C followed by 2-min incubations at 55°C. At the end of this process, the reaction was cooled, proteinase K was added to a final concentration of 0.4 mg/mL, and the reaction was incubated at 45°C for 15 min.
After purification by ethanol precipitation, the heteroduplex was biotinylated as described by Carninci et al. (1996). After isopropanol precipitation, the resuspended, biotinylated heteroduplex was treated with RNase I (Ambion) at 37°C for 15 min and immobilized on M-280 Streptavidin Dynabeads (Dynal) according to the manufacturer's instructions before second-strand synthesis with a Superscript cDNA Synthesis Kit (Invitrogen). The double-stranded cDNA products in the supernatant were collected and purified by phenol:chloroform extraction and ethanol precipitation. The purified products were ligated to AscI linkers and digested with AscI and NotI. The digested products were passed through cDNA Size Fractionation Columns (Invitrogen) according to the manufacturer's instructions, and pooled fractions were concentrated by ethanol precipitation. The resuspended products were ligated to pOL2.Asc, which had been digested with AscI and NotI, and dephosphorylated. The ligated populations were transformed into E. coli and, after plating of aliquots to determine the library complexity, the library was grown for DNA preparation. The sizes of the inserts present in the library were characterized by PCR analysis of individual clones from the library and sizing of the amplified products after gel electrophoresis. The mean insert size was 956 bp, and the standard error of the mean was 67.
Library Screening
Unlinearized, plasmid DNA prepared from the libraries was transcribed with a T7 mMessage mMachine kit (Ambion), reassembled with purified Tobacco mosaic virus (TMV) coat protein, and inoculated to plants as described by Toth et al. (2002). After the development of fluorescent infection foci, inoculated leaves were detached, stuck to microscope slides with double-sided adhesive tape, and imaged with a spectral confocal laser scanning microscope fitted with water-dipping lens as described by Gillespie et al. (2002). Infection foci that displayed GFP distributions of interest were excised, homogenized in 10 mM sodium phosphate buffer, and passaged as described by Toth et al. (2002).
Cloning of Library Isolates
Leaf tissue was homogenized in liquid nitrogen, and total RNA was isolated using Tri Reagent (Sigma-Aldrich) according to the manufacturer's instructions. RNA was reverse transcribed with Superscript II (Invitrogen). For isolates from the 3′ fusion library, first-strand cDNA synthesis was primed with oligonucleotide E (5′-ATCCAAGACACAACCCTTC-3′), which is complementary to TMV sequence downstream of the cDNA inserts, whereas for isolates from the 5′ fusion library, first-strand cDNA synthesis was primed with oligonucleotide C (5′-ACCTTCACCCTCTCCACT-3′), which is complementary to the downstream gfp sequence. Regions encompassing the cDNA inserts were amplified by PCR from the first-strand cDNA products with Taq DNA polymerase (Invitrogen) with primer pairs flanking the cDNA inserts. For isolates from the 5′ fusion library, primers A and B (5′-AAAAAAGTCGACGCAAAGGGAAAAATAGTAGTAGT-3′ and 5′-GAATTGGGACAACTCCAGTG-3′) were used, and for isolates from the 3′ fusion library, primers D (5′-CCTGTCGACACAATCTGC-3′) and E were used. The amplification products were subjected to agarose gel electrophoresis, and fragments of interest were cloned into the vector pGEM-T Easy (Promega). Insert sequences were determined with an ABI PRISM Big-Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) and analyzed using Basic Local Alignment Search Tool (BLASTN and BLASTX) searches (Altschul et al., 1990; Gish and States, 1993).
Reconstitution of Plasmodesmata-Localizing Clones
To regenerate full-length clones of isolates from the 3′ fusion library, reverse transcriptase–mediated (RT) PCR–derived products were digested with SalI and NotI or SalI and BsiWI where the NotI site was not intact and inserted into plasmid pXNE0 digested with the same pairs of restriction enzymes. Similarly, to regenerate full-length clones of isolates from the 5′ fusion library, RT-PCR–derived products were digested with AscI and NotI and inserted into pOL2-Asc that had been digested with the same pair of restriction enzymes. For the isolate plasmodesmata9, in which the NotI site was not intact, RT-PCR was repeated with primers A and E, the amplification product was digested with AscI and SalI, and the released fragment was inserted between the same sites of pOL2-Asc. To correct the frame in the full-length clone produced from isolate plasmodesmata6, the cDNA insert was reamplified by PCR with a mutagenic antisense primer (5′-TTGCTAGCGGCCGCACCACCTGAAATATGA-3′), which introduced two extra nucleotides between the cDNA sequence and the downstream NotI site on one side, and primer A on the other. The reamplified product was digested with AscI and NotI and inserted into pOL2-Asc. DNA produced from the regenerated full-length clones was used for transcription before reassembly of in vitro transcripts and plant inoculations as described above. Inoculated tissue was inspected with a confocal laser scanning microscope as described above. Immunogold labeling of ultrathin sections from infected tissue with GFP-specific antiserum was performed as described by Santa Cruz et al. (1996).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Chai-Lin Wei of Large Scale Biology Corporation for providing library construction methods and Linda Cardle for assistance with sequence searches. This work was supported by Biosource Genetics Corporation and by the Scottish Executive Environment and Rural Affairs Department.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013284.
References
- Aaziz, R., Dinant, S., and Epel, B.L. (2001). Plasmodesmata and plant cytoskeleton. Trends Plant Sci. 6, 326–330. [DOI] [PubMed] [Google Scholar]
- Abbink, T.E.M., Peart, J.R., Mos, T.N.M., Baulcombe, D.C., Bol, J.F., and Linthorst, H.J.M. (2002). Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 295, 307–319. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bird, D.A., and Facchini, P.J. (2001). Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213, 888–897. [DOI] [PubMed] [Google Scholar]
- Blackman, L.M., and Overall, R.L. (2001). Structure and function of plasmodesmata. Aust. J. Plant Physiol. 28, 709–727. [Google Scholar]
- Boevink, P., Martin, B., Oparka, K., Santa Cruz, S., and Hawes, C. (1999). Transport of virally expressed green fluorescent protein through the secretory pathway in tobacco leaves is inhibited by cold shock and brefeldin A. Planta 208, 392–400. [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Carninci, P., et al. (1996). High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37, 327–336. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Nishiyama, Y., Westover, A., Itoh, M., Nagaoka, S., Sasaki, N., Okazaki, Y., Muramatsu, M., and Hayashizaki, Y. (1998). Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc. Natl. Acad. Sci. USA 95, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabregas, S.M., Luche, D.D., Farias, L.P., Ribeiro, A.F., van Sluys, M.-A., Menck, C.F.M., and Silva-Filho, M.C. (2001). Dual targeting of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Mol. Biol. 46, 639–650. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Knorr, D., Schuster, G., and Zambryski, P. (1990). The P30 movement protein of tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell 60, 637–647. [DOI] [PubMed] [Google Scholar]
- Crawford, K.M., and Zambryski, P. (2000). Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10, 1032–1040. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B. (1998). Intercellular protein trafficking through plasmodesmata. Plant Mol. Biol. 38, 279–310. [PubMed] [Google Scholar]
- Ding, D.-Q., Tomita, Y., Yamamoto, A., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (2000). Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5, 169–190. [DOI] [PubMed] [Google Scholar]
- Ferro, M., Salvi, D., Riviere-Rolland, H., Vermat, T., Seigneurin-Berny, D., Grunwald, D., Garin, J., Joyard, J., and Rolland, N. (2002). Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. USA 99, 11487–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor, J., Hideg, E., Kecskes, A., and Kiraly, Z. (2001). In vivo detection of tobacco mosaic virus-induced local and systemic oxidative burst by electron paramagnetic resonance spectroscopy. Plant Cell Physiol. 42, 775–779. [DOI] [PubMed] [Google Scholar]
- Fujii, G., Tsuchiya, R., Ezoe, E., and Hirohashi, S. (1999). Analysis of nuclear localization signals using a green fluorescent protein-fusion protein library. Exp. Cell Res. 251, 299–306. [DOI] [PubMed] [Google Scholar]
- Gillespie, T., Boevink, P., Haupt, S., Roberts, A.G., Toth, R., Valentine, T., Chapman, S., and Oparka, K.J. (2002). Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of Tobacco mosaic virus. Plant Cell 14, 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish, W., and States, D.J. (1993). Identification of protein coding regions by database similarity search. Nat. Genet. 3, 266–272. [DOI] [PubMed] [Google Scholar]
- Gray, J.C., Sullivan, J.A., Hibberd, J.M., and Hansen, M.R. (2001). Stromules: mobile protrusions and interconnections between plastids. Plant Biol. 3, 223–233. [Google Scholar]
- Haupt, S., Santa Cruz, S., Chapman, S., Smolenska, L., and Oparka, K. (2001). Analysis of mixed infections of TMV and PVX expressing different fluorescent reporter genes. In Plasmodesma 2001 Abstracts, Fourth International Conference. (Capetown, South Africa: University of Capetown), pp. 69–71.
- Higuchi, R. (1990). Recombinant PCR. In PCR Protocols, M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, eds (San Diego, CA: Academic Press), pp. 177–183.
- Karrer, E.E., Beachy, R.N., and Holt, C.A. (1998). Cloning of tobacco genes that elicit the hypersensitive response. Plant Mol. Biol. 36, 681–690. [DOI] [PubMed] [Google Scholar]
- Karthikeyan, A.S., Varadarajan, D.K., Mukatira, U.T., D'Urzo, M.P., Damsz, B., and Raghothama, K.G. (2002). Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 130, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlizky, G., Shurtz, S., Yahalom, A., Malik, Z., Traub, O., and Epel, B.L. (1992). An improved procedure for the isolation of plasmodesmata embedded in clean maize cell-walls. Plant J. 2, 623–630. [Google Scholar]
- Lacomme, C., Pogue, G.P., Wilson, T.M.A., and Santa Cruz, S. (2001). Plant viruses. In Genetically Engineered Viruses, C.J.A. Ring and E.D. Blair, eds (Oxford, UK: BIOS Scientific Publishers), pp. 59–105.
- Misawa, K., Nosaka, T., Morita, S., Kaneko, A., Nakahata, T., Asano, S., and Kitamura, T. (2000). A method to identify cDNAs based on localization of green fluorescent protein fusion products. Proc. Natl. Acad. Sci. USA 97, 3062–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, X., Daneman, R., Zavortink, M., and Chia, W. (2001). A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozo, T., Fischer, K., Flugge, U.I., and Schmitz, U.K. (1995). The N-terminal extension of the ADP/ATP translocator is not involved in targeting to plant mitochondria in vivo. Plant J. 7, 1015–1020. [DOI] [PubMed] [Google Scholar]
- Muckenschnabel, I., Goodman, B.A., Deighton, N., Lyon, G.D., and Williamson, B. (2001). Botrytis cinerea induces the formation of free radicals in fruits of Capsicum annuum at positions remote from the site of infection. Protoplasma 218, 112–116. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, K., Sumi, K., and Fukuda, H. (2002). The use of multiple transcription starts causes the dual targeting of Arabidopsis putative monodehydroascorbate reductase to both mitochondria and chloroplasts. Plant Cell Physiol. 43, 697–705. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., Boevink, P., and Santa Cruz, S. (1996). Studying the movement of plant viruses using green fluorescent protein. Trends Plant Sci. 1, 412–418. [Google Scholar]
- Oparka, K.J., Roberts, A.G., Santa Cruz, S., Boevink, P., Prior, D.A.M., and Smallcombe, A. (1997). Using GFP to study virus invasion and spread in plant tissues. Nature 388, 401–402. [Google Scholar]
- Pastori, G.M., and Foyer, C.H. (2002). Common components, networks, and pathways of cross-tolerance to stress: The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 129, 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, G., Moura, D.S., Stratman, J., and Ryan, C.A. (2001). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue, G.P., Lindbo, J.A., Garger, S.J., and Fitzmaurice, W.P. (2002). Making an ally from the enemy: Plant virology and the new agriculture. Annu. Rev. Phytopathol. 40, 45–74. [DOI] [PubMed] [Google Scholar]
- Rabindran, S., and Dawson, W.O. (2001). Assessment of recombinants that arise from the use of a TMV-based transient expression vector. Virology 284, 182–189. [DOI] [PubMed] [Google Scholar]
- Raghothama, K.G. (2000). Phosphate transport and signalling. Curr. Opin. Plant Biol. 3, 182–187. [PubMed] [Google Scholar]
- Rolls, M.M., Stein, P.A., Taylor, S.S., Ha, E., McKeon, F., and Rapoport, T.A. (1999). A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J. Cell Biol. 146, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudhe, C., Chew, O., Whelan, J., and Glaser, E. (2002). A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J. 30, 213–220. [DOI] [PubMed] [Google Scholar]
- Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5, 518–528. [DOI] [PubMed] [Google Scholar]
- Saito, C., Ueda, T., Abe, H., Wada, Y., Kuroiwa, T., Hisada, A., Furuya, M., and Nakano, A. (2002). A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 29, 245–255. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Santa Cruz, S., Chapman, S., Roberts, A.G., Roberts, I.R., Prior, D.A.M., and Oparka, K.J. (1996). Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. USA 93, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K.E., and Nurse, P. (1996). Identification of fission yeast nuclear markers using random polypeptide fusions with green fluorescent protein. Proc. Natl. Acad. Sci. USA 94, 15146–15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivprasad, S., Pogue, G.P., Lewandowski, D.J., Hidalgo, J., Donson, J., Grill, L.K., and Dawson, W.O. (1999). Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255, 312–323. [DOI] [PubMed] [Google Scholar]
- Takken, F.L.W., Luderer, R., Gabriels, S.H.E.J., Westerink, N., Lu, R., de Wit, P.J.G.M., and Joosten, M.H.A.J. (2000). A functional cloning strategy, based on a binary PVX-expression vector, to isolate HR inducing cDNAs of plant pathogens. Plant J. 24, 275–283. [DOI] [PubMed] [Google Scholar]
- Tomenius, K., Clapham, D., and Meshi, T. (1987). Localization, by immunogold cytochemistry, of the virus-coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 160, 363–371. [DOI] [PubMed] [Google Scholar]
- Toth, R.L., Pogue, G.P., and Chapman, S. (2002). Improvement of the movement and host range properties of a plant virus vector through DNA shuffling. Plant J. 30, 593–600. [DOI] [PubMed] [Google Scholar]
- Turner, A., Wells, B., and Roberts, K. (1994). Plasmodesmata of maize root tips: Structure and composition. J. Cell Sci. 107, 3351–3361. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J. (2001). Challenges and prospects of plant proteomics. Plant Physiol. 126, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw, W.K., and Harrison, M.J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate starvation responses. Plant Cell 14, 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R.Z., Bailey, S.N., and Sabatini, D.M. (2002). Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 12, 485–488. [DOI] [PubMed] [Google Scholar]