Abstract
The Drosophila melanogaster sex determination factor Tra2 positively regulates the splicing of both doublesex (dsx) and fruitless (fru) pre-mRNAs but negatively affects the splicing of the M1 intron in tra2 pre-mRNA. Retention of the M1 intron is known to be part of a negative-feedback mechanism wherein the Tra2 protein limits its own synthesis, but the mechanism responsible for accumulation of M1-containing RNA is unknown. Here we show that the recombinant Tra2 protein specifically represses M1 splicing in Drosophila nuclear extracts. We find that the Tra2 protein binds directly to several sites in and near the M1 intron and that, when Tra2 binding is competed with other RNAs, the splicing of M1 is restored. Mapping the RNA sequences functionally required for M1 repression identified both a 34-nucleotide (nt) A/C-rich sequence immediately upstream of the M1 5′ splice site and a region within the intron itself. The AC-rich sequence is largely composed of a repeated 4-nt sequence that also forms a subrepeat within the repeated 13-nt splicing enhancer elements of fru and dsx RNAs. Although required for repression, the element also enhances M1 splicing in the absence of Tra2. We propose that Tra2 represses M1 splicing by interacting with multiple sequences in the pre-mRNA and interfering with enhancer function.
The exons in many eukaryotic pre-mRNAs are spliced together in alternative patterns to produce multiple mRNAs. This not only allows a single DNA sequence to encode multiple proteins, increasing the size of the proteome, but also provides an important avenue for cell-specific control of gene function through the regulation of splicing patterns (26). A number of proteins that participate in the control of alternative splicing have been identified (16, 23). These factors affect the selection of specific combinations of splice sites through a variety of different mechanisms. A common theme of the mechanisms so far described is that most of these regulators act by binding initially to the targeted pre-mRNAs to control the assembly and activity of splicing complexes on them rather than by modulating the activity of the general splicing machinery. Bound regulatory proteins influence splice site selection by affecting the ability of prespliceosomal factors to recognize signals within the intron. Both positive control and negative control of splicing site recognition have been observed. For example, SR proteins are known to bind to exonic splicing enhancers (ESEs) and to recruit components of the general splicing machinery to nearby 5′ and 3′ splice sites that have relatively poor splicing signals and that would otherwise be ignored (16). On the other hand, polypyrimidine tract binding proteins are able to bind intronic sequences and antagonize functional interactions with general splicing factors or with positive regulators (2, 11, 27, 33, 45).
One of the best-studied systems in which the activities of splicing regulators are known derives from the Drosophila melanogaster sex determination regulatory pathway. Key genes in this pathway (Sex-lethal, transformer, and transformer-2) encode splicing regulators that direct several sex-specific splicing events determining the expression of critical downstream sex factors (3, 4, 32). For instance the Transformer-2 (Tra2) and Transformer (Tra) proteins participate directly in the regulation of sex-specific splicing of doublesex (dsx) and fruitless (fru) pre-mRNAs, leading to sex-specific expression of transcription factors encoded by these genes (17, 42). The mechanism of sex-specific dsx splicing has been well studied and involves the cooperative assembly of complexes containing Tra, Tra2, and one or more SR proteins at individual repeats of a 13-nucleotide (nt) sequence within an ESE in the dsx pre-mRNA (18, 24, 25, 43, 44). These complexes function to activate the upstream female-specific 3′ splice site, most likely by facilitating interactions of U2AF (14, 15, 48) and/or other general splicing factors (21, 22) with the RNA.
In contrast to its positive role in activating dsx and fru splicing, the Tra2 protein negatively affects the splicing of the M1 intron in the tra2 pre-mRNA itself (28). Transcripts in which this intron remains unspliced encode a nonfunctional protein isoform lacking one of the two RS domains (30). M1-containing RNAs are found almost exclusively in the male germ line, where their accumulation is dependent on the presence of the functional Tra2 protein isoform. In wild-type flies these partially spliced transcripts comprise about 50% of the germ line Tra2 mRNA. M1 is efficiently spliced in the absence of Tra2, and virtually no mRNA containing the intron is found. In previous studies we have shown that M1 retention is part of a normal negative-feedback mechanism by which the functional Tra2 isoform limits its own synthesis by promoting the accumulation of M1-containing transcripts (29). This limitation in Tra2 expression is necessary in the germ line, as transgenic males in which feedback has been circumvented have defects in spermatogenesis and reduced fertility (31).
The mechanism responsible for accumulation of M1-containing RNAs is unknown. Other introns in tra2 pre-mRNA are not retained, indicating that the mechanism responsible is specific for M1 (30). Mutations near the 5′ and 3′ ends of the intron that block splicing also block regulation of intron retention, arguing in favor of a splicing mechanism (28). However, direct evidence that Tra2 acts at the level of splicing rather than for instance, RNA export, has been lacking. Here we show that Drosophila Schneider 2 nuclear extracts can support effective and specific repression of M1 splicing by the recombinant Tra2 protein. Using this system we identify at least two elements in the RNA that interact with the Tra2 protein and that are necessary for normal levels of splicing repression. Based on the observation that one required element is also an exonic splicing enhancer, we propose that Tra2 represses M1 splicing in part by interfering with the function of this enhancer.
MATERIALS AND METHODS
In vitro transcription of RNA substrates and competitors.
For the ptra2X3-4 splicing substrate an ApaI-to-BglII fragment encompassing the male-specific portion of exon 3, the M1 intron, and all of exon 4 was cloned into the ApaI and BamHI sites of pBluscript II SK(+) (Stratagene). The vector was then linearized with XbaI for in vitro transcription. Similarly, for the M1/ftz 3′ splicing substrate the ApaI-to-BglII fragment from P[ftz3′], a tra2 construct containing a substitution of the polypyrimidine tract and 3′ splice site from the fushi-tarazu (ftz) gene and a 7-nt insertion in exon 4 (10), was cloned into pBluescript II SK(+) (Stratagene) to generate pM1/ftz3′. The positive-control splicing substrate derived from ftz was produced from the plasmid pG2V61 (34) and was a kind gift from Lisa Ryner. This plasmid consists of a SalI-to-BglII fragment encompassing the ftz intron inserted into pGEM (Promega). It was linearized with XhoI for in vitro transcription of splicing substrates. The tra2 intron 4 substrate was generated by linearization of the plasmid pINT4 with PstI for in vitro transcription. pINT4 was constructed by insertion of a tra2 genomic fragment including all sequences from the beginning of exon 4 to the PstI site in intron 5 into pBluescript II SK(+) (Stratagene). Additional splicing substrates were generated directly from PCR products generated with a T7 RNA polymerase recognition sequence at one end. The T7 promoter sequence 5′GGAATTCTAATACGACTCACTATAGGG3′ was included in sense orientation PCR primers and was followed immediately by tra2 sequences beginning at the 5′ end of each substrate as indicated below with nucleotides numbered as described earlier (30). The substrate Δ5′X3/ΔX4 extends from nt 1182 to 1496 and was amplified from pM1/ftz3′ with primers T7-1182-X3 and X4-1496as. ΔX3/ΔX4 extends from nt 1233 to 1496 and was amplified from the same plasmid with primers T7-1233-X3 and X4-1496as. Splicing substrate ΔX3/X4+50 was transcribed from a PCR amplimer derived from pSK1.5AΔ11F with a sense primer from the plasmid's T7 promoter and the X4-1496as antisense primer. It contains tra2 sequences from nt 1240 to 1496 and is preceded by 53 nt of plasmid-derived sequences. The plasmid sequences correspond to the region between the T7 transcription start site and the HindIII site of pBluescript SK(+). The ΔACE substrate contains sequences starting at nt 1182 and ending at nt 1496. The transcription template was amplified by PCR with T7-1182-X3 and X4-1496as by using the plasmid pM1/ftz3′ ΔACE, from which the 34-nt A/C-rich element (ACE; nt 1200 to 1233) was deleted. The ACE deletion was produced by primer-directed mutagenesis of the plasmid p[tra2+1] with the tra2-ΔGAR/ACE oligonucleotide 5′TTACATAGTGATAATGAAATGA3′. The mutagenized region was then substituted into pM1/ftz3′ by replacing the 256-nt ApaI/StyI fragment. The splicing substrate Δ1200 extends from nt 1200 to 1496 and was transcribed from a PCR product amplified with T7-1200-X3 and X4-1496as from pM1/ftz3′. A template for the splicing substrate BSEsub was created by a similar PCR amplification from the plasmid ptra2-BSE, in which the 18-nt bidirectional splicing enhancer (BSE) from simian virus 40 (SV40) (8) was substituted for the 34-nt ACE. To generate hybrid M1/ftz intron substrates, plasmids pBSE+29, pBSE+71, pBSE+126, and pBSE+208 were made from ptra2-BSE by substituting various PCR-amplified ftz intron sequences analogous to regions of the M1 intron. In addition 29-nt ftz exon sequences were included in these constructs to replace the tra2 exon 4. Splicing substrates were transcribed from PCR products amplified with a primer containing the T7 promoter followed immediately by BSE sequences.
For UV cross-linking experiments, a set of nested DNA templates that spanned various parts of the M1 intron and flanking exons were generated and transcribed by a PCR strategy similar to that described above. The regions amplified in each RNA were as follows: RNA 1, nt 1103 to 1213 (111 nt); RNA 2, nt 1163 to 1298 (136 nt); RNA 3, nt 1200 to 1328 (129 nt); RNA 4, nt 1245 to 1372 (128 nt); RNA 5, nt 1311 to 1436 (126 nt); RNA 6, nt 1367 to 1496 (130 nt). For the RNA2 ΔACE substrate PCR was performed on the p[tra2+1] ΔACE plasmid with oligonucleotides to amplify the region between nt 1163 and 1298. As a positive control in splicing and cross-linking experiments a 187-nt segment of the dsx splicing enhancer containing four 13-nt repeat elements and the purine-rich element (PRE) was transcribed from a PCR product generated with a sense primer that included the T7 promoter followed by the dsx sequence 5′TCAGCTTTCTTCAATCAAC3′. The antisense primer was 5′CGTTTACTACATTTTGTCC3′. A 207-nt negative-control RNA was similarly transcribed from DNA sequences at the 3′ end of dsx exon 4. These were amplified by using primers with the dsx sequences 5′TATCTTAGCAAGGCAAT3′ (sense) and 5′CTTAAGGTCGTAACAATA3′ (antisense). In vitro transcription reactions were carried out with either 5 μl of a 100-μl PCR mixture or 1 μg of linearized plasmid for standard T7 transcription reactions, as described in the Ambion Maxiscript (splicing substrates) or Megascript (unlabeled competitors) RNA transcription kits. The m7G cap analog (New England Biolabs) was included in the synthesis of all splicing substrates. All RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE) before being used as substrates or competitors in splicing and binding reactions. Quantities of unlabeled competitors were determined by UV absorbance after gel purification.
In vitro splicing assay.
Radiolabeled substrates were incubated at 22°C for 2 h in the presence of a mixture containing 50% S2 nuclear extract, 1.6 mM MgCl2, 20 mM phospho-l-arginine, 1.2 mM dithiothreitol, 1.2% polyethylene glycol, and 2 mM ATP. For Tra2 repression studies, 100, 50, or 25 ng of recombinant His-Tra2264 protein was added to the splicing reaction mixtures. To analyze the ability of specific RNAs to compete for Tra2 binding and repression of splicing, 0.5, 2.5, and 12.5 pmol of unlabeled competitor RNAs were added to reaction mixtures containing either 0 or 100 ng (3 pmol) of recombinant His-Tra2264 protein. The RNA products and intermediates were separated on 5% denaturing polyacrylamide gels and visualized by autoradiography.
RNA-protein UV cross-linking experiments.
After a 10-min room temperature incubation of labeled RNA substrates with 100 ng of Tra2 protein and S2 nuclear extracts under splicing conditions, mixtures were UV irradiated on ice for 10 min. Reaction products were digested with RNase A and RNase T1 for 30 min at 37°C. Labeled proteins were separated on sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gels and visualized by autoradiography. For competition experiments, 0.5, 2.5, or 12.5 pmol of the specified unlabeled RNAs was added to the reaction mixtures and mixtures were incubated at room temperature for 5 min prior to addition of RNA and cross-linking.
Recombinant proteins and nuclear extracts.
Recombinant Tra2264 with an amino-terminal six-His tag was expressed from a baculovirus strain kindly provided by K. Lynch and T. Maniatis and was purified as described by Tian and Maniatis (42). To check the purity of the recombinant Tra2 protein, Western blotting was performed using monoclonal antibody (MAb) 104. Specifically, SF9 cells lysed directly with Laemmli buffer and the recombinant Tra2 protein resuspended in Laemmli buffer were subjected to SDS-12.5% PAGE, electrophoretically transferred to a Hybond Plus membrane, blocked with Tris-buffered saline-0.1% Tween 20-7.5% nonfat dried milk, and incubated with the primary antibody diluted in Tris-buffered saline with 0.1% Tween 20 and 7.5% nonfat dried milk overnight at 4°C. Blots were washed with Tris-buffered saline-0.1% Tween 20 and incubated with the secondary antibody diluted in Tris-buffered saline with 0.1% Tween 20 and 7.5% nonfat dried milk for 1 h at 25°C. Detection was performed by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech). Nuclear extracts were prepared from plate cultures of Drosophila S2 cells essentially as described by Dignam et al. (13).
RESULTS
Regulated splicing of the M1 intron in Drosophila nuclear extracts.
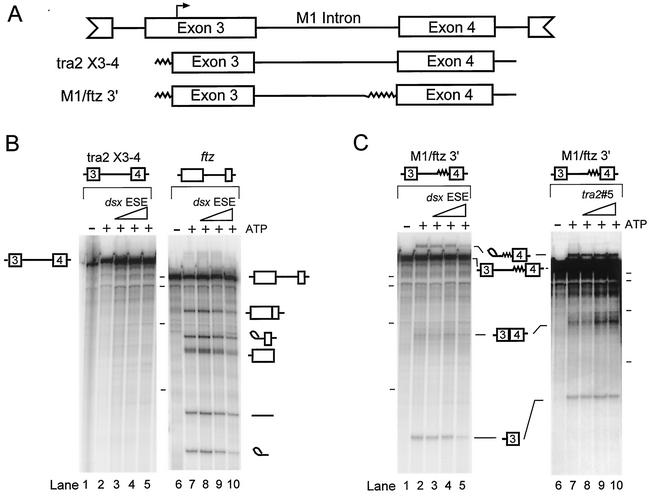
To investigate the regulation of M1 splicing, we initially sought to define a cell-free system in which Tra2-dependent processing of the intron could be carried out. For this purpose we introduced a labeled RNA substrate containing this intron and flanking exons (tra2 exons 3 and 4) into nuclear extracts prepared from Drosophila S2 cells (Fig. 1A). Reactions were carried out at different concentrations of nuclear extract, Mg2+, K+, ATP, and polyethylene glycol to identify optimal conditions for splicing (data not shown). Although parallel reactions with a positive-control RNA from the ftz gene resulted in efficient splicing under a variety of conditions, splicing products or intermediates were never observed from reactions carried out on substrates containing the native M1 intron (Fig. 1B, compare lanes 2 and 7). Because S2 cells have previously been shown to express Tra2 mRNA (35), we considered the possibility that the endogenous Tra2 protein was present in the extracts and was repressing M1 splicing. To test this, we added various amounts of an unlabeled 187-nt RNA containing the central portion of the dsx ESE, previously shown to efficiently bind Tra2, to splicing reactions carried out on both tra2 exons 3 and 4 and the ftz splicing substrates. Although we found the dsx RNA to be an effective competitor for Tra2 protein binding in other experiments (see below), its presence failed to activate the splicing of labeled tra2 exon 3 and 4 RNA (Fig. 1B, lanes 3 to 5 and 8 to 10), suggesting that the lack of M1 splicing in these extracts is not due to repression by the endogenous Tra2 protein.
FIG. 1.
In vitro splicing of Tra2 substrates in Schneider 2 nuclear extracts. (A) Schematic representing the tra2 transcripts that were used in the in vitro splicing assays. Boxes, tra2 exons; lines, tra2 intron sequences; jagged line, heterologous sequences, including the ftz 3′ splice site, that were included in these splicing substrates. The top line shows the region of Tra2 pre-mRNA around M1. Arrow, position of the male germ line transcription start site. (B) Tra2 X3-4, an RNA substrate from the native tra2 gene, was labeled with 32P and used for in vitro splicing as shown in lanes 1 through 5. As a positive control a labeled RNA containing the intron and flanking exons from the Drosophila ftz gene was also subjected to splicing reactions under the same conditions (lanes 6 through 10). Lanes 1 and 6, control reactions without ATP showing no detectable splicing; lanes 2 and 7, reactions carried out with the native M1 intron only; lanes 3 to 5 and 8 to 10, reactions carried out in the presence of 0.5, 2.5, and 12.5 pmol, respectively (triangles), of unlabeled RNA from the dsx ESE. Products from the splicing reaction were resolved on denaturing acrylamide-urea gels. Reaction input pre-mRNAs, intermediates, and products are indicated at the side of each gel. The positions of radiolabeled in vitro-transcribed RNA molecular weight markers run on the same gel (dashes) correspond to 500, 400, 300, and 200 nt from top to bottom. (C) Similar splicing reactions were carried out on tra2 RNA substrates in which the native 3′ splice site region was replaced with the 3′ splice site of the ftz gene (M1/ftz 3′). Lanes 1 and 6, control reactions without ATP. Splicing was carried out in the presence of 0, 0.5, 2.5- and 12.5 pmol of unlabeled dsx ESE RNA (lanes 2 to 5, respectively) or an RNA from within the M1 intron (RNA 5 in Fig. 3A; lanes 7 to 10, respectively).
We inferred from the above experiments that the M1 intron is inefficiently recognized by the general splicing machinery in S2 nuclear extracts. In previous studies on M1 splicing we have shown that the intron contains a variant 3′ splice site and that suboptimal recognition of this site is required for Tra2-dependent repression of M1 splicing in vivo (10). We reasoned that this suboptimal site might preclude M1 splicing in vitro (even in the absence of Tra2) because reactions in extracts are inherently slower than those that occur in living cells. To “improve” the substrate, we substituted sequences from the polypyrimidine tract and 3′ splice site of the ftz intron, which closely match the Drosophila consensus splicing signals, for those of the M1 intron to generate M1/ftz 3′ RNA (Fig. 1A). As shown in Fig. 1C, lane 2, incubation of this hybrid substrate with ATP in S2 nuclear extracts yielded RNAs with mobilities expected for the lariat and 5′ exon splicing intermediates as well as a smaller amount of mature product (the lariat intron product comigrates with the pre-mRNA). Splicing efficiency of the hybrid substrate was not stimulated by addition of unlabeled competitor RNAs that bind Tra2 and that are derived from either the dsx ESE or the M1 intron itself (Fig. 1C, lanes 3 to 5 and 8 to 10). From these results we conclude that the M1/ftz 3′ substrate is spliced in S2 nuclear extracts and that the endogenous Tra2 protein, if present, is insufficient to repress M1 splicing.
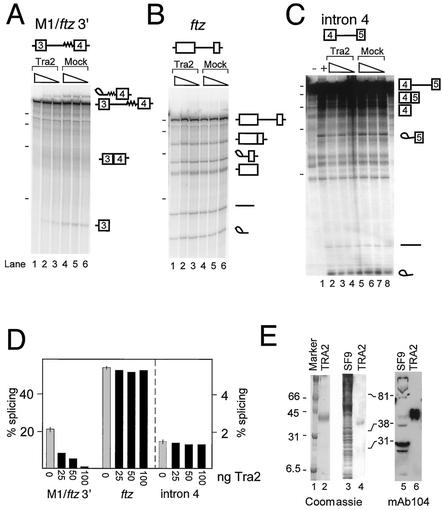
Further experiments next established that, although they lack a functional Tra2 protein, S2 extracts could be used to study the regulation of M1 splicing with added recombinant Tra2 protein. To determine if Tra2 is capable of repressing M1 splicing in vitro, we purified a His-tagged recombinant Tra2 protein using a baculovirus expression system. The purified protein was free of other SR proteins, as demonstrated by Western blotting using MAb 104, which reacts with a conserved phosphoepitope present in all SR proteins (Fig. 2E). When the recombinant protein was used to supplement splicing reactions, we found that accumulation of both lariat and 5′ exon intermediates from M1/ftz 3′ substrate were reduced in a manner dependent on the concentration of recombinant Tra2 added (Fig. 2A, lanes 1 to 6), indicating that Tra2 represses the first step of splicing. The splicing of the ftz intron, on the other hand, was unaffected by the same amounts of added Tra2 protein (Fig. 2B and D), suggesting that the observed repression of M1/ftz 3′ is not due to general inhibition of splicing activity. Quantitation of spliced products revealed over 30-fold repression of M1/ftz 3′ splicing but essentially no repression of the ftz intron (Fig. 2D). The ftz and M1/ftz 3′ RNAs also differed significantly in their basal splicing efficiencies. In a 2-h reaction without added Tra2 54% of the ftz substrate underwent at least one step of splicing whereas only 21% of M1/ftz 3′ RNA did so. We therefore considered the possibility that added Tra2 might have a general inhibitory effect on splicing that is only evident in reactions carried out on inefficient substrates. To test this we added the same amounts of recombinant Tra2 protein to splicing reactions carried out on an RNA containing intron 4 from the tra2 gene, which is not expected to be regulated by the Tra2 protein but which had previously been determined to have a low splicing rate in vitro (data not shown). As shown in Fig. 2C and D, although this intron is spliced less efficiently than M1/ftz 3′ in vitro (1.6% after 2 h), the addition of the recombinant Tra2 protein had little, if any, significant effect on the amount of splice products generated. We conclude that the recombinant Tra2 protein specifically represses M1/ftz 3′ splicing in vitro.
FIG. 2.
Repression of M1/ftz 3′ splicing by recombinant Tra2 protein. In vitro splicing assays were performed with S2 nuclear extracts on the 32P-labeled in vitro-transcribed RNAs depicted above each reaction set. In each set, splicing reactions were carried out in the presence of 100, 50, and 25 ng of recombinant Tra2 protein or in buffer controls without Tra2 (mock) (triangles indicate amounts of recombinant Tra2 and buffer). The positions of reaction intermediates and products are shown schematically at the right. The positions of radiolabeled in vitro-transcribed RNA molecular weight markers run on the same gel (dashes) correspond to 500, 400, 300, and 200 nt from top to bottom. In panel C, the position of the 100-nt band is also indicated. (A) Reactions carried out on M1/ftz 3′. The lariat products comigrate with the pre-mRNA on this gel. (B) Splicing reactions showing that the ftz intron is not repressed by recombinant Tra2. (C) Splicing reactions carried out on inefficiently spliced intron 4 of tra2, which is not regulated by the Tra2 protein in vivo. Lanes 1 and 2, control reactions without and with, respectively, ATP. (D) Quantitation of the splicing reactions shown in panels A to C. Grey bars, average percentages of splicing (products plus intermediates) for the three mock reactions (error bars, ranges of values); black bars, percentages of splicing reaction mixtures containing various amounts of Tra2 protein. Note that the scale for reactions with intron 4, shown to the right of the dashed line, is different from that for M1/ftz and ftz. (E) To demonstrate the degree of purity of the recombinant Tra2 protein used in the above assays, 3.5 μg of Tra2 was loaded onto a SDS-12.5% polyacrylamide gel and stained with Coomassie blue (lane 2). To further test if the purified Tra2 protein was not contaminated with SR proteins, whole-cell lysate from SF9 cells and 1 μg of Tra2 were run in duplicate on separate SDS-12.5% polyacrylamide gels and either stained with Coomassie blue (lanes 3 and 4) or electroblotted and probed with MAb 104 (lanes 5 and 6).
The Tra2 protein binds to multiple sites in the tra2 pre-mRNA.
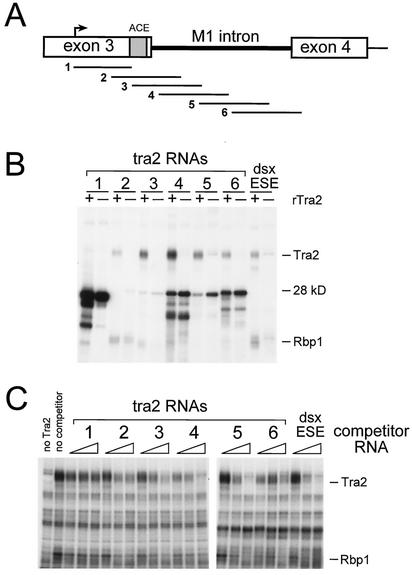
To determine if the Tra2 protein interacts with sequences in or near the M1 intron, we incubated the recombinant protein with a set of overlapping 32P-labeled RNA fragments (Fig. 3A) extending from the male germ line transcription start site (in exon 3) to 24 nt downstream of the native M1 3′ splice site. This region was chosen because fusion RNAs containing it have sufficient sequences to allow Tra2-dependent regulation of M1 splicing in vivo (31). The binding reaction products were then subjected to UV cross-linking, RNase digestion, and electrophoresis on SDS gels to determine whether Tra2 had become covalently attached to labeled nucleotides from the RNAs. As shown in Fig. 3B, in reactions supplemented with the recombinant protein, five out of the six tra2-derived RNA segments cross-linked with a 38-kDa band comigrating with Tra2. For comparison, similar reactions were carried out in parallel on the dsx ESE RNA and the reaction products cross-linked to both Tra2 and an endogenous band comigrating with the SR protein Rbp1 (another component of the dsx enhancer complex) in a manner dependent on the added Tra2 protein. The tra2-derived RNA fragments that cross-linked (RNAs 2, 3, 4, 5, and 6) spanned the region beginning 54 nt downstream of the male germ line transcription start site and ending in exon 4 (Fig. 3A). Because some of these fragments do not overlap (i.e., RNAs 2 and 5), more than one binding site must be present in the tra2 pre-mRNA. It is worth noting that several proteins deriving from the nuclear extracts also bound to tra2 RNAs in a sequence-specific manner. In some cases cross-linking of such proteins was dependent on the presence or absence of Tra2. For instance a 28-kDa protein that cross-linked with RNA 2 was displaced when recombinant Tra2 was added to the reactions (Fig. 3B, compare lanes + and − for RNA 2).
FIG. 3.
UV cross-linking of Tra2 with sequences near and within the M1 intron. Multiple RNAs spanning the M1 intron of tra2 were used in cross-linking experiments to determine the site of Tra2 protein binding. (A) The tra2 RNA segments used for cross-linking. The location of the ACE in exon 3 is shown. Arrow, position of the male germ line transcription start site. (B) UV cross-linking was performed with the radiolabeled tra2 RNAs depicted in panel A or a 188-nt RNA from the dsx ESE. Cross-linked proteins after label transfer and RNase digestion are resolved on an SDS-PAGE gel. The dsx RNA used in this experiment contains regulatory elements that have previously been shown to bind to Tra2. The RNAs used for cross-linking are indicated above each set of reactions. Cross-linking was performed in splicing-competent S2 nuclear extracts and in the presence (+) or absence (−) of recombinant Tra2 (100 ng). The position of the 28-kDa protein and the mobilities of Tra2 and endogenous Rbp1 proteins, as judged by Western blotting, are indicated. (C) Titration-competition cross-linking experiments performed using the radiolabeled dsx ESE RNA under splicing conditions in the presence of S2 nuclear extract supplemented with 100 ng (3 pmol) of Tra2 protein. Reactions in which recombinant Tra2 or competitor RNA was omitted are shown in the first and second lanes, respectively. In the remaining lanes, increasing amounts of various unlabeled competitor RNAs were added (0.5, 2.5, and 12.5 pmol), as indicated by the triangles.
To confirm the specificity of Tra2 binding, the same set of RNAs were also used as unlabeled competitors against 32P-labeled dsx ESE RNA in similar cross-linking experiments. Again it was observed that RNAs 3, 4, and 5 competed as well with labeled dsx RNA as did unlabeled dsx RNA itself in control reactions. RNAs 2 and 6 competed to a lesser extent. The labeled dsx RNA cross-linked with a number of extract-derived proteins in this experiment, but, significantly, only the binding of Tra2 and Rbp1 was competed by the M1-derived RNAs. Based on the results of these experiments we conclude that a minimum of three distinct sequences in tra2 pre-mRNA must be able to bind the Tra2 protein.
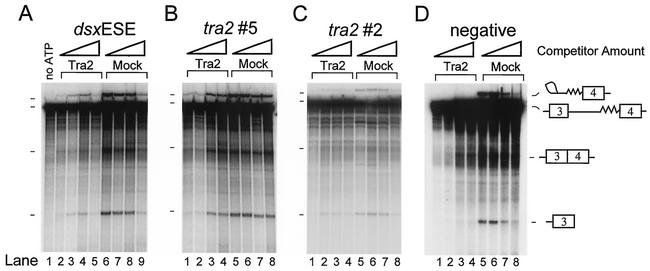
The observation that Tra2 can bind directly to sequences in and adjacent to the M1 intron suggests that its direct interaction with the RNA may be necessary for repression of splicing. To test this idea, we carried out in vitro splicing reactions on M1/ftz 3′ RNA in the presence of various unlabeled competitors to determine if they blocked the ability of recombinant Tra2 to repress splicing. The amount of Tra2 repressor used in these reactions was sufficient to completely repress splicing of M1/ftz 3′ RNA (Fig. 4A, lane 2 compared to lane 6). Addition of unlabeled dsx ESE RNA competitor restored splicing in these reactions in a concentration-dependent manner (Fig. 4A, lanes 2 to 5). At the highest concentration of dsx RNA used (Fig. 4A, lane 5) the level of splicing intermediates was reduced due to nonspecific effects on splicing that were also seen in controls in which no Tra2 protein was added (Fig. 4A, lanes 9). Similar stimulation of splicing was observed when the M1-derived RNAs 5 (Fig. 4B) and 2 (4C) were tested as competitors. A negative-control competitor containing sequences from dsx exon 4 outside the ESE did not stimulate splicing in parallel reactions (Fig. 4D). Together, the above data indicate that Tra2 binds to RNA sequences in and near the M1 intron and that the Tra2 protein can repress M1 splicing only when its RNA binding site is not otherwise occupied.
FIG. 4.
Competition of Tra2 binding restores M1 splicing. In vitro splicing assays performed on 32P-labeled M1/ftz 3′ RNA in the presence (Tra2) or absence (Mock) of repressive amounts (3 pmol) of unlabeled recombinant Tra2 protein and RNA competitors are shown. Splicing precursors, intermediates, and products were resolved on denaturing polyacrylamide-urea gels, and their mobilities are indicated. The lariat intron product is not resolved from the pre-mRNA. (A) Competitions with the dsx ESE RNA in increasing amounts (0, 0.5, 2.5, and 12.5 pmol; triangles). Splicing is restored at moderate competitor concentrations (lanes 3 and 4), but the highest concentration of competitor RNA (lanes 5 and 9) caused nonspecific repression of splicing (compare control lanes 6 to 9). Lane 1 shows a reaction in which no ATP, Tra2, or competitors were added. (B) A similar set of reactions were carried out with RNA 5 (Fig. 3A) from within the M1 intron. Restoration of splicing is evident at the two highest concentrations of competitor (compare lane 1 to lanes 3 and 4). (C) A similar set of reactions carried out with RNA 2, which spans part of exon 3 and the 5′ end of the intron. Restoration of splicing can be seen by comparing lane 1 to lanes 2 to 4. Again, a nonspecific inhibition of splicing by high concentrations of the competitor was observed in lane 4 compared to lanes 5 to 8. (D) Competition with the same molar amounts of the unlabeled 207-nt negative-control RNA derived from sequences in dsx RNA outside of the enhancer (see Materials and Methods). No restoration of M1 splicing was observed (lanes 2 to 4).
Multiple sequences are needed for repression of M1 splicing in vitro.
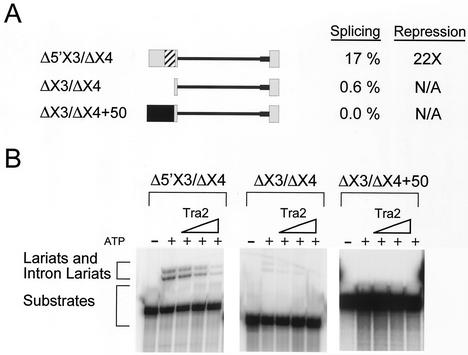
To determine what RNA sequences are necessary for regulation of M1 splicing, we truncated the flanking exons present in the M1/ftz 3′ RNA (Fig. 5A) and tested whether these shorter substrates were subject to Tra2-dependent repression in vitro. Substrates lacking the last 130 nt of exon 4 or the first 73 nt of exon 3 were effectively repressed by Tra2 (data not shown). We therefore tested a substrate lacking both of these regions (Δ5′X3/ΔX4). As shown in Fig. 5B this substrate was reduced only slightly in both the level of basal splicing (splicing in the absence of added Tra2 protein) and the amount of Tra2-dependent repression relative to that observed with more-extensive substrates (i.e., M1/ftz 3′; Fig. 2A). Deletion of 59 of the remaining 60 nt from exon 3 (ΔX3/ΔX4), however, resulted in a dramatic reduction of basal M1 splicing (from 17 to 0.6%; Fig. 5B, left and middle), suggesting that elements in the deleted region normally facilitate M1 splicing. To control for the possibility that the small size of the 5′ exon in ΔX3/ΔX4 (8 nt) is incompatible with efficient splicing, we also tested a substrate (ΔX3/ΔX4+50) in which a 50-nt plasmid-derived sequence is inserted upstream of the 5′ splice site creating an artificial 58-nt exon. The splicing of this construct was also minimal (Fig. 5B, right), indicating that exon length is not the cause of the reduced splicing and supporting the idea that sequences between 1 and 60 nt upstream of the 5′ splice site play an important role in facilitating M1 splicing.
FIG. 5.
Identification of sequences in exon 3 required for M1 splicing. (A) RNAs used for in vitro splicing assays. Boxes, exons; lines, M1 intron; thick line, substituted 3′ splice site region from ftz RNA; striped box in exon 3, position of the ACE. Δ5′X3/ΔX4 includes 58 nt from the promoter-distal end of exon 3, the M1 intron, and the first 24 nt of exon 4, while ΔX3/ΔX4 contains only the last 8 nt of exon 3, M1, and the first 24 nt of exon 4. ΔX3/ΔX4+50 contains an additional 50 nt of plasmid sequences (black box). The in vitro splicing efficiencies as a percentage of products and intermediates and the amounts of repression observed when maximum Tra2 protein is added (3 pmol) are indicated to the right. (B) Lariat products and intermediates from in vitro splicing reactions performed on the constructs in panel A in the presence of increasing amounts of Tra2 (0.5, 2.5, and 12.5 pmol). Control reactions without added Tra2 both with (+) and without ATP (−) are also shown for each substrate.
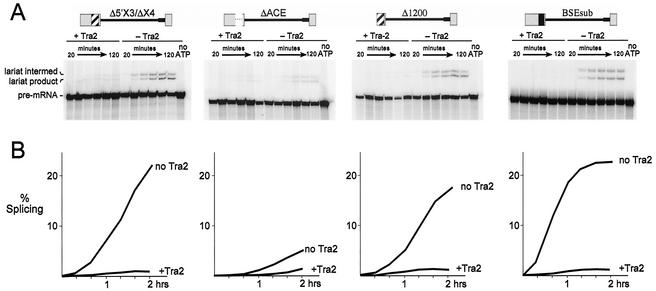
Inspection of the sequences in exon 3 identified above revealed the presence of an A/C-rich region of 34 nt that includes a fivefold direct repeat of the sequence CAAC located 16 nt upstream of the 5′ splice site. Several ACEs have been shown to have splicing enhancer activity (7, 12, 38), and a very similar sequence (CAATCAACA) is a nearly invariant feature of the nine 13-nt repeat elements found in the dsx and fru splicing enhancers (Fig. 6A) (9, 36). Also within the A/C-rich region, and adjacent to the CAAC repeat, is the sequence GAAGAA, which has been associated with the binding of the human Tra2 protein (19, 41, 46). We therefore tested whether deletion of this 34-nt region affected M1 splicing and/or repression. Parallel time course experiments were carried out with Δ5′X3-ΔX4 and analogous constructs with deletions of the ACE (ΔACE) and the region upstream of it (Δ1200) (Fig. 6). In the absence of the recombinant Tra2 protein, 22% of the Δ5′X3/ΔX4 RNA was converted to products and intermediates but only 3.9% of the ΔACE RNA was. Δ1200, on the other hand, was spliced to a similar level as Δ5′X3/ΔX4 (18%). Thus, the ACE appears to be the major sequence in exon 3 promoting M1 splicing. Interestingly this sequence also had a strong effect on the amount of Tra2-dependent repression observed. With ΔACE RNA, only fourfold repression was observed, compared to the 27-fold repression observed with Δ5′X3/ΔX4 RNA in a parallel reaction (Fig. 6B). These results suggested the possibility that the ACE acts to both enhance basal splicing of the M1 intron and facilitate splicing repression in response to Tra2.
FIG. 6.
A splicing enhancer is required for normal levels of basal splicing and repression. Two-hour time course splicing assays were performed on the various substrates with (+ Tra2) and without (− Tra2) repressor, and the results of these reactions were quantified. (A) The substrates used are schematized at the top. Striped box in exon 3, ACE. This 34-nt element (Fig. 7) is deleted in the ΔACE substrate, which is otherwise identical to Δ5′X3/ΔX4. The substitution of the BSE from SV40 is indicated by a black box at the same position in the substrate BSEsub. The Δ1200 RNA lacks all sequences upstream of the ACE. For the time course reactions shown, each lane corresponds to aliquots removed from the splicing reaction at 20-min intervals. Input pre-mRNA and lariats (intermediates and products) are denoted at the left. Control reactions without ATP showed no detectable splicing after 120 min. (B) The percentage of splicing indicated corresponds to the percentage of signal from all products and intermediates relative to the total signal.
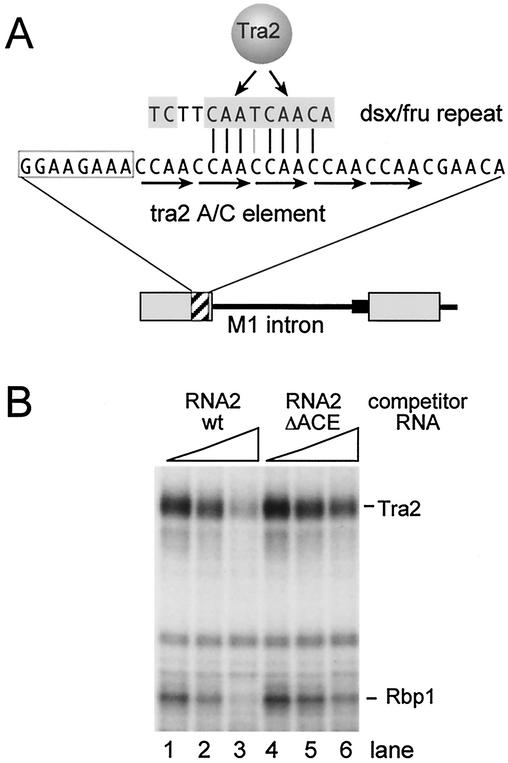
To determine if the sequences removed in the ΔACE mutation are required for Tra2 binding, we performed cross-linking competition experiments using wild-type and mutant versions of RNA 2 (Fig. 3A) as unlabeled competitors against labeled dsx ESE RNA. As shown in Fig. 7B, the deletion of the ACE (ΔACE) substantially reduced the ability of RNA to compete with dsx for binding. These results suggest that sequences within this region facilitate the binding of Tra2 to the RNA.
FIG. 7.
The ACE resembles sequences in the dsx and fru splicing enhancers, and its deletion reduces Tra2 binding. (A) Schematic showing the sequence and location of the tra2 ACE (striped box) and its relationship to the repeated enhancer elements found in dsx and fru RNA. The five CAAC repeats are underlined with arrows. Top arrows, sites in the 13-nt dsx repeat sequence of site-specific cross-linking to Tra2. (B) UV cross-linking was performed on the 188-nt dsx ESE fragment in the presence of splicing-competent Drosophila nuclear extract, 3 pmol of recombinant Tra2, and increasing amounts of RNA competitors (0.5, 2.5, and 12.5 pmol). The position of RNA 2 is shown in Fig. 3A. RNA 2 ΔACE contains a precise deletion of the 34-nt ACE but is otherwise identical. Rbp1 binding to the dsx ESE is dependent on Tra2 binding (Fig. 3B) and is reduced when Tra2 is competed (lane 3). Note that, although RNA 2 competition affects Tra2 and Rbp1, other cross-linking bands are unaffected. wt, wild type.
The above experiments do not distinguish whether the ACE plays a specific role in repression or whether other elements with enhancer function might have a similar effect. We therefore replaced the native 34-nt ACE with a purine-rich BSE from SV40 (8) to form the substrate BSEsub and tested its ability to both enhance basal splicing and support Tra2-dependent repression in S2 nuclear extracts. As shown in Fig. 6, this enhancer effectively replaced the native ACE both in promoting basal splicing in the absence of Tra2 and in stimulating Tra2-dependent repression. This observation supports the idea that high levels of repression in vitro require a splicing enhancer function and argues that this function is not a unique property of the native ACE in exon 3 (see Discussion).
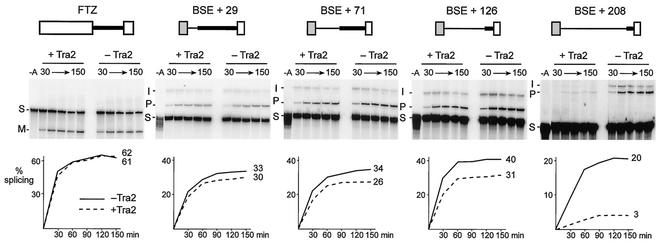
Further experiments indicated that the 34-nt ACE is not alone sufficient to confer responsiveness to Tra2 on other introns. When this enhancer was placed immediately upstream of either the ftz intron or intron 4, no significant Tra2-dependent repression was observed. This, taken together with the residual repression observed when the ACE is deleted (ΔACE; Fig. 6), suggests that additional sequences in the Tra2 RNA must play an important role in repression. To determine if sequences supporting repression are present in the M1 intron, we generated substrates in which various sequences of this intron are fused with the unregulated ftz intron. To facilitate basal splicing, the BSE from SV40 was used as an exogenous upstream enhancer in these substrates (Fig. 8).
FIG. 8.
Specific M1 intron sequences also promote Tra2-dependent repression. Schematic drawings of different splicing substrates are shown at the top. Grey boxes, BSE; open boxes, exon sequences derived from the ftz gene; thin lines, sequences derived from the M1 intron; thick lines, sequences derived from the ftz intron. Below each substrate are shown time courses of splicing reactions carried out with (+ Tra2) and without (− Tra2) the recombinant Tra2 protein. Quantitation of these reactions is shown in the graph below each gel. For simplicity not all products and intermediates are shown, but quantitation represents the total percentage of all products and intermediates detected. The positions of different RNAs are indicated beside the gels as splicing substrates (S), spliced products (M), lariat intermediates (I), and lariat intron products (P). Control lanes of 150-min reactions without ATP (lanes −A) are also shown for each substrate. The maximum percentages of products entering splicing are indicated to the right of each graph. No repression was observed with the ftz substrate, indicating that Tra2 does not have general repressive effects on splicing. Over sixfold repression (20 versus 3% splicing) was observed with BSE+208, but other substrates containing M1 sequences were repressed only slightly (BSE+71, BSE+126, and BSE+29).
A fusion containing only the first 29 nt of M1 (Fig. 8, BSE+29), was essentially unaffected by the added Tra2 protein, supporting the previous observation that the presence of an enhancer alone in the 5′ exon is not sufficient to support repression. In constructs with 71 and 126 nt of M1 sequences (BSE+71 and BSE+126), a small amount of repression was consistently observed. Repression was significantly increased (to sixfold) only when 208 nt of the M1 sequences were included (BSE+208). These results indicate that sequences in the M1 intron are sufficient to confer Tra2-dependent repression on a substrate in vitro and further indicate that sequences required for repression are located between 126 and 208 nt downstream of the 5′ splice site. We conclude that sequences both in exon 3 and within the M1 intron play important roles in the repression of splicing.
DISCUSSION
The results presented here indicate that Tra2 participates directly in the repression of M1 splicing. In Drosophila nuclear extracts recombinant Tra2 both reduces M1 splicing and cross-links to the RNA at several different positions. When the binding of the protein is competed by other RNAs, the splicing of M1 is enhanced, arguing that the availability of the protein's RNA binding domain is necessary for Tra2-dependent repression. The addition of recombinant Tra2 did not affect the general splicing activity of nuclear extracts but instead acted specifically on the M1 intron, indicating that splicing is repressed through specific interactions with the tra2 pre-mRNA. Given that it has already been established that Tra2 promotes accumulation of M1-containing transcripts in adult flies (28), it seems likely Tra2 also directly represses M1 splicing in vivo.
We found that the splicing of the native M1 intron is extremely inefficient in vitro but that measurable regulated splicing could be obtained when the intron's suboptimal 3′ splicing signals were replaced with consensus-like sequences from the efficiently spliced ftz intron. In a previous study we tested the role of the 3′ splice site in M1 repression and found that the entire region upstream of this site including the polypyrimidine tract could be replaced with that of an unrelated intron without effect on Tra2-dependent repression. These studies also revealed the importance of splice site strength in M1 repression. A strong splice site (e.g., ftz) gave constitutive splicing in vivo that could not be repressed by M1, while mutations causing further deviations of the native site from the consensus increased the level of M1 repression. (10). We believe that the observation that the ftz 3′ splice is compatible with regulation in vitro but not in vivo is readily explained by the difference in splicing rates in these two contexts. In the cell, splicing occurs rapidly and cotranscriptionally on nascent pre-mRNAs (5, 6). As M1 is the first intron of male germ line transcripts and follows the transcription start site by only 133 nt, the Tra2 protein must interact rapidly with regulatory sequences in the RNA after their transcription and before the intron is spliced. We hypothesize that the suboptimal 3′ splice site is likely required to reduce the in vivo rate of splicing to allow the Tra2 protein a greater opportunity to interact with the RNA. Under in vitro conditions, splicing reactions are typically much slower, and the limitation in rate imposed by the suboptimal 3′ splice site is not required. For the M1 intron our data suggest that the presence of the weak 3′ splice site reduces the splicing rate to the point where products cannot be detected in a standard reaction. Thus, improvement of the splice site merely allows the detection of these products and does not present a kinetic barrier for repression.
Although the tra2 gene is transcribed in a variety of tissues in the adult fly, repression of M1 splicing is observed only in growing spermatocytes (1, 30). Previously it has been suggested that this cell type specificity might indicate a requirement for spermatocyte-specific factors that participate in M1 repression (29). However, in both the in vitro splicing experiments described here and in transfections of cultured cells with tra2 reporters (J. Qi and W. Mattox, unpublished data) we have found that the S2 cell line supports Tra2-dependent repression. As these cells are of somatic origin (39), this finding raises the question of how the observed germ line tissue specificity in adult male flies comes about. One possibility is that differences in the level of tra2 transcription in these cell types determine whether repression occurs. In adult flies significantly less Tra2 is expressed in somatic cells than in spermatocytes (1, 30), where, gene dosage studies suggest, M1 repression is part of a highly concentration-sensitive negative-feedback mechanism for limiting Tra2 expression below deleterious levels (31). Thus, somatic cells might have the ability to repress M1 splicing, but Tra2 expression never reaches a level high enough to necessitate activation of negative feedback. An alternative possibility is that S2 cells either fortuitously express a germ line factor required for repression of M1 or lack a somatic factor that would block repression.
The mechanism by which Tra2 affects alternative splicing has been best studied in the case of dsx pre-mRNA, where it binds cooperatively with Tra and other SR proteins at several different sites in the female-specific exon (18, 20, 24, 25, 43). Such complexes are known to form on two different types of elements in the dsx enhancer, a PRE and a 13-nt repeated element, nearly six identical copies of which are dispersed in the female-specific exon of dsx RNA. We found that, in tra2 RNA, the Tra2 protein interacts at several sites within and flanking the M1 intron; however the interacting regions contain no exact matches to the PRE or 13-nt repeat elements. Functional mapping of sequences required for repression of M1 splicing performed with the in vitro repression assay described here revealed the importance of a 34-nt A/C-rich region located immediately upstream of the intron's 5′ splice site. When this sequence is deleted, the ability of Tra2 to repress M1 splicing is substantially reduced. Interestingly, we found that in the absence of the Tra2 protein the same element plays an important role in facilitating M1 splicing, indicating that it plays a role in both activation and repression of M1 splicing. Several observations suggest that Tra2 binds directly to the 34-nt ACE. First Tra2 cross-links with two overlapping RNAs that encompass this sequence. Second, we have observed that an RNA lacking this element acts as a significantly less efficient competitor for Tra2 binding than an identical RNA containing the element. Third, the 34-nt sequence contains a fivefold direct repeat of the sequence CAAC. This sequence is closely related to a subrepeat that is perfectly conserved in eight of the nine copies of the 13-nt repeat elements found in dsx and fru RNAs (TCTTCAATCAACA) (9, 36). The subrepeat (underlined) is of particular interest because site-specific cross-linking experiments identify it as the portion of the 13-nt repeat element most closely associated with the bound Tra2 protein (24). Thus the subrepeat is likely to be a natural target for Tra2 binding.
Our observation that repression in vitro is empirically reduced when the ACE is deleted and that this sequence binds Tra2 suggests mechanisms in which this enhancer plays both positive and negative roles. One possibility is that the intron is dependent on the enhancer for splicing and that in the presence of bound Tra2 enhancer function is disrupted, reducing M1 splicing. However, there are other potential explanations for the observation that enhancer deletions reduce repression. For instance, the effect of Tra2 in our in vitro reactions may be limited by a threshold below which it cannot further repress. If so, then the differences in the magnitude of repression observed with and without the enhancer might be related to the efficiency of splicing in the absence of Tra2. Another possibility raised by our results is that the enhancer plays a generic role in repression. Consistent with this idea, we found that the 34-nt A/C-rich sequence could be functionally replaced by a BSE from SV40. Thus, functionally significant binding of Tra2 outside the upstream enhancer may interfere with enhancer-dependent splicing, possibly through interactions with SR proteins or other enhancer binding factors. Note in this regard that, although the BSE is known to bind SRp40, the major human Tra2 proteins comigrate with this protein, and the possibility they bind to BSE sequences has not been rigorously excluded (8). Ideally the issue of whether Tra2-enhancer binding is required for repression could be resolved by substitution of a known Tra2-independent enhancer, but few enhancers are known to be unambiguously Tra2 independent, and from among those, we have been unable to identify one able to promote M1 splicing in S2 extracts (J. Qi and W. Mattox, unpublished data).
Evidence from other systems supports the idea that A/C-rich sequences such as those found here in exon 3 have intrinsic enhancer function. For instance, in HeLa nuclear extracts it has been observed that the dsx 13-nt repeats act as constitutive splicing enhancers that bind SR proteins independently of Tra and Tra2 when moved to a position close to the dsx 3′ splice site (44). A/C-rich sequences with enhancer activity have also been identified in both in vivo and in vitro selections of random sequences for those promoting exon inclusion in vertebrate cells (7, 12, 37). More recently it has been shown that activation of exon inclusion in the CD44 gene depends largely on an A/C-rich splicing enhancer that binds the Y box protein YB-1 (40). Interestingly, in this case human Tra2 orthologs increase exon inclusion and facilitate the binding of YB-1 with the A/C-rich sequences (E. Stickeler, A. Honig, A. L. Chen, M. M. Rojas, A. J. McCullough, S. D. Fraser, T. A. Cooper, W. Mattox, and S. M. Berget, submitted for publication). Finally, we note that another sequence (GAAGAA) located within the 34-nt element exactly matches an element found to enhance splicing in a manner dependent on human Tra2 proteins (41). However, preliminary experiments indicate that repression is not reduced when this element is removed so its presence is not required (D. S. Chandler and W. Mattox, unpublished data).
Although there are several elements in the 34-nt A/C-rich sequence that may have regulatory activity, it is unlikely that this element acts alone to mediate regulation of M1 splicing. While deletion of this sequence reduced Tra2-dependent repression, repression was not eliminated. Nor did its deletion reduce basal splicing of M1 to the same degree as did more-extensive deletions from exon 3. Thus other elements in the M1 intron or upstream flanking sequences must also be able to mediate activation and Tra2-dependent repression. We identified sequences in the M1 intron necessary to support repression located between 126 and 208 nt downstream of the 5′ splice site. This region immediately precedes the branch point and corresponds to RNA fragments that were observed to strongly cross-link with Tra2. Our analysis also established that a 208-nt region of the intron is sufficient to support significant M1 repression in the presence of an exogenous splicing enhancer (the BSE). Notably subsegments of this region supported a small amount of repression in a manner correlated with the extent of the M1 sequences present. Taken together with our binding data, these results, we believe, support the idea that Tra2 acts at multiple sites in the M1 intron and the upstream enhancer. The idea that a repressor binds to multiple sites in an RNA in a cooperative manner to “coat” or sequester has been suggested previously; such binding was recently demonstrated in the regulation of human immunodeficiency virus tat splicing (47), and it is possible that Tra2 acts in a similar way. Another attractive possibility for the mechanism by which Tra2 regulates M1 splicing is that Tra2 alters the function of splicing enhancer complexes that form on sequences near or within the M1 intron. Proteins that might bind at or near the A/C-rich sequence upstream of M1 and activate splicing in the absence of Tra2 have yet to be identified. In UV cross-linking experiments with an RNA spanning this region, several Drosophila proteins from S2 nuclear extracts were labeled. One of these, a 28-kDa cross-linked species, was bound to an RNA containing the ACE and was displaced by the addition of the recombinant Tra2 protein. Thus its binding correlated with conditions permissive for M1 splicing. The identification of proteins binding specifically to sequences with enhancer activity in the ACE and an understanding of their functional interactions with Tra2 are likely to lead to further insights into the mechanism responsible for repression of M1 splicing.
Acknowledgments
We thank Tom Cooper, Brigitte Dauwalder, and Tomoo Iwakuma for helpful comments on the manuscript. We are grateful to Kristen Lynch, Mitzi Kuroda, Tom Cooper, Brigitte Dauwalder, Michelle Wilson, Kathy Kennedy, Sue Berget, and members of the Houston RNA group for providing useful materials and advice.
This work was supported by an NIH grant (GM 58625) to W.M.; DNA sequencing was supported by Cancer Center Support (Core) grant CA16672; D.S.C. received support from an NIH training grant (CA 09299).
REFERENCES
- 1.Amrein, H., T. Maniatis, and R. Nothiger. 1990. Alternatively spliced transcripts of the sex-determining gene tra-2 of Drosophila encode functional proteins of different size. EMBO J. 9:3619-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashiya, M., and P. J. Grabowski. 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3:996-1015. [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, B. S. 1989. Sex in flies: the splice of life. Nature 340:521-524. [DOI] [PubMed] [Google Scholar]
- 4.Bell, L. R., J. I. Horabin, P. Schedl, and T. W. Cline. 1991. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell 65:229-239. [DOI] [PubMed] [Google Scholar]
- 5.Beyer, A. L., and Y. N. Osheim. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2:754-765. [DOI] [PubMed] [Google Scholar]
- 6.Beyer, A. L., and Y. N. Osheim. 1991. Visualization of RNA transcription and processing. Semin. Cell Biol. 2:131-140. [PubMed] [Google Scholar]
- 7.Boukis, L. A., and J. P. Bruzik. 2001. Functional selection of splicing enhancers that stimulate trans-splicing in vitro. RNA 7:793-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourgeois, C. F., M. Popielarz, G. Hildwein, and J. Stevenin. 1999. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol. 19:7347-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burtis, K. C., and B. S. Baker. 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56:997-1010. [DOI] [PubMed] [Google Scholar]
- 10.Chandler, D. S., M. E. McGuffin, and W. Mattox. 2001. Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing. Nucleic Acids Res. 29:3012-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlet, B. N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 12.Coulter, L. R., M. A. Landree, and T. A. Cooper. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell Biol. 17:2143-2150. (Erratum, 17:3468.) [DOI] [PMC free article] [PubMed]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley, B. R., K. J. Hertel, and T. Maniatis. 1999. SR proteins are ‘locators' of the RNA splicing machinery. Curr. Biol. 9:R6-R7. [DOI] [PubMed] [Google Scholar]
- 15.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1:765-771. [DOI] [PubMed] [Google Scholar]
- 16.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 17.Heinrichs, V., L. C. Ryner, and B. S. Baker. 1998. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell Biol. 18:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertel, K. J., and T. Maniatis. 1998. The function of multisite splicing enhancers. Mol. Cell 1:449-455. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, Y., C. L. Lorson, S. Stamm, E. J. Androphy, and B. Wirth. 2000. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc. Natl. Acad. Sci. USA 97:9618-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue, K., K. Hoshijima, I. Higuchi, H. Sakamoto, and Y. Shimura. 1992. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc. Natl. Acad. Sci. USA 89:8092-8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kan, J. L., and M. R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., and B. J. Blencowe. 1999. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 274:35074-35079. [DOI] [PubMed] [Google Scholar]
- 23.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 24.Lynch, K. W., and T. Maniatis. 1996. Assembly Of specific SR protein complexes on distinct regulatory elements of the Drosophila Doublesex splicing enhancer. Genes Dev. 10:2089-2101. [DOI] [PubMed] [Google Scholar]
- 25.Lynch, K. W., and T. Maniatis. 1995. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 9:284-293. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 27.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattox, W., and B. S. Baker. 1991. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 5:786-796. [DOI] [PubMed] [Google Scholar]
- 29.Mattox, W., M. E. McGuffin, and B. S. Baker. 1996. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator Transformer-2. Genetics 143:303-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattox, W., M. J. Palmer, and B. S. Baker. 1990. Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 4:789-805. [DOI] [PubMed] [Google Scholar]
- 31.McGuffin, M. E., D. Chandler, D. Somaiya, B. Dauwalder, and W. Mattox. 1998. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics 149:1477-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagoshi, R. N., M. McKeown, K. C. Burtis, J. M. Belote, and B. S. Baker. 1988. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 53:229-236. [DOI] [PubMed] [Google Scholar]
- 33.Polydorides, A. D., H. J. Okano, Y. Y. Yang, G. Stefani, and R. B. Darnell. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 97:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rio, D. C. 1988. Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc. Natl. Acad. Sci. USA 85:2904-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryner, L. C., and B. S. Baker. 1991. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 5:2071-2085. [DOI] [PubMed] [Google Scholar]
- 36.Ryner, L. C., S. F. Goodwin, D. H. Castrillon, A. Anand, A. Villella, B. S. Baker, J. C. Hall, B. J. Taylor, and S. A. Wasserman. 1996. Control of male sexual behavior and sexual orientation in Drosophila by the Fruitless gene. Cell 87:1079-1089. [DOI] [PubMed] [Google Scholar]
- 37.Schaal, T. D., and T. Maniatis. 1999. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaal, T. D., and T. Maniatis. 1999. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19:1705-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 40.Stickeler, E., S. D. Fraser, A. Honig, A. L. Chen, S. M. Berget, and T. A. Cooper. 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tacke, R., M. Tohyama, S. Ogawa, and J. L. Manley. 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93:139-148. [DOI] [PubMed] [Google Scholar]
- 42.Tian, M., and T. Maniatis. 1992. Positive control of pre-mRNA splicing in vitro. Science 256:237-240. [DOI] [PubMed] [Google Scholar]
- 43.Tian, M., and T. Maniatis. 1993. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74:105-114. [DOI] [PubMed] [Google Scholar]
- 44.Tian, M., and T. Maniatis. 1994. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 8:1703-1712. [DOI] [PubMed] [Google Scholar]
- 45.Valcarcel, J., and F. Gebauer. 1997. Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 7:R705-R708. [DOI] [PubMed] [Google Scholar]
- 46.Yeakley, J. M., J. P. Morfin, M. G. Rosenfeld, and X. D. Fu. 1996. A complex of nuclear proteins mediates Sr protein binding to a purine-rich splicing enhancer. Proc. Natl. Acad. Sci. USA 93:7582-7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 48.Zuo, P., and T. Maniatis. 1996. The splicing factor U2af(35) mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10:1356-1368. [DOI] [PubMed] [Google Scholar]