Abstract
A significant number of self-reactive T cell clones escape thymic negative selection and are released into the periphery, where some are potentially pathogenic. The clonal expansion of self-reactive T cells is known to be limited during initial antigen encounter by apoptotic or anergic mechanisms, regulatory CD4+ T cells, and cytokines. Here we report that superimposed on these mechanisms, during the evolution of autoimmunity in experimental autoimmune encephalomyelitis (EAE), CD8+ T cells are induced, which fine-tune the peripheral self-reactive T cell receptor (TCR) repertoire. We assayed the myelin basic protein-reactive TCR repertoire in naive, EAE-recovered mice as well as EAE-recovered mice depleted of CD8+ T cells by TCRVβ surface expression, complementarity-determining region 3 length distribution, and complementarity-determining region 3 sequencing analysis. In EAE-recovered mice, certain myelin basic protein-reactive CD4+Vβ8.2+ clones are significantly decreased and this decrease is not observed if CD8+ T cells were depleted from these mice. The clones that persist in CD8+ T cell-intact mice are highly diverse in contrast to the clones expanded in CD8+ T cell-depleted mice, which are dominated by the significant outgrowth of a few clones. Importantly, the T cell clones that expand in the absence of CD8+ T cell control are enriched in potentially pathogenic self-reactive T cell clones capable of inducing EAE in vivo.
Thymocytes expressing T cell receptors (TCRs) with high affinity for self-peptide/MHC complexes undergo apoptosis and are deleted centrally in the thymus. However, recent experiments have highlighted the fact that many self-reactive T cells with low to intermediate affinity for self-antigen escape thymic negative selection and are released into the periphery. Although these self-reactive T cells display relatively low avidity to self-peptide/MHC complexes, they are capable of self-peptide-driven proliferation and some may differentiate into potentially pathogenic effector cells (1–4). Thus, mechanisms that normally regulate the outgrowth or function of these self-reactive T cells may ultimately control the initiation and progression of autoimmune disease. One level of control resides at the initial clonal activation of the TCR itself by MHC/peptide complexes and, reminiscent of thymic negative selection, involves antigen-induced apoptosis. For example, a series of experiments by Anderton et al. (5) demonstrated that immunization of B10PL mice with the encephalitogenic NH2-terminal-acetylated nanopeptide from mouse myelin basic protein (MBP), 1–9Nac MBP, induces experimental autoimmune encephalomyelitis (EAE) and triggers the proliferation of a heterogeneous population of T cells that express diverse TCRs with varying affinities for this encephalitogenic peptide (5). Interestingly, mutated forms of 1–9Nac MBP, which bind to MBP-reactive T cells with very high affinity, trigger the T cells to undergo apoptosis and these peptides are incapable of inducing EAE. In contrast, the nonmutated, wild-type 1–9Nac MBP binds with lower affinity to MBP-reactive T cells, does not induce apoptosis, but readily induces EAE. Thus, the encephalitogenic MBP-reactive repertoire is comprised of T cells with a heterogeneous set of affinities, which contain the affinities high enough to induce EAE, but not sufficiently high enough to trigger apoptosis.
It is clear that homeostatic mechanisms other than apoptosis may be required to fine-tune the autoreactive TCR repertoire and limit the clonal expansion of the potentially pathogenic self-reactive clones with TCRs of too-low avidity to induce antigen-induced cell death. In this article we directly test whether one of these mechanisms involves CD8+ T cells, which are known to mediate resistance to EAE. Thus, it is known that B10PL mice immunized with the wild-type 1–9Nac MBP generate encephalitogenic CD4+ T cell clones that migrate to the CNS and induce EAE. Approximately 85% of encephalitogenic clones express the TCR Vβ8.2 (6). Mice typically fully recover from the first episode of disease. Interestingly, the first episode of EAE renders the mice highly resistant to the reinitiation of EAE by secondary immunization (7). This resistance to EAE is CD8+ T cell-dependent because B10PL mice recovered from EAE and then depleted of CD8+ T cells develop EAE on reimmunization with 1–9Nac MBP (7). Similarly, mice depleted of CD8+ T cells during the initial induction of EAE and allowed to recover normal levels of CD8+ T cells develop EAE again on rechallenge with 1–9Nac MBP (8). Thus, the CD8+ T cells require priming during the first episode of EAE to regulate CD4+ T cells triggered by secondary MBP stimulation in vivo. Interestingly, CD4+ T cells isolated from EAE-recovered mice do respond to 1–9 Nac MBP in vitro (8). Thus, there is an abundance of self-reactive T cell clones in the periphery of both EAE-recovered (EAE) and CD8+ T cell depleted EAE-recovered (CD8-/EAE) mice, despite the fact that EAE mice are resistant but CD8-/EAE mice are susceptible to EAE reinduction. This observation led us to propose the hypothesis that the clonal composition of the peripheral MBP-reactive CD4+ TCR repertoire is regulated by CD8+ T cells and thus will be different in EAE mice and CD8-/EAE mice.
To test this hypothesis in vivo, we compared the composition of the MBP-reactive TCR repertoire, after MBP immunization, in control, EAE, and CD8-/EAE mice by TCRVβ surface expression, PCR-based complementarity-determining region (CDR)3 length-distribution analysis, and direct CDR3 sequencing. We found, as expected, that the clonal composition of the peripheral MBP-reactive TCR repertoire changes during the evolution of EAE. In EAE mice, the major MBP-reactive CD4+Vβ8.2+ T cell population is significantly decreased. This decrease is accompanied by persistence of some MBP-reactive Vβ8.2 clones as well as other MBP-reactive clones expressing different Vβs. In contrast, this specific decrease is not observed in CD8-/EAE mice, where the TCR Vβ repertoire is characterized by the expansion of only a handful of distinct MBP-reactive CD4+ Vβ8.2+ cell clones expressing particular TCR CDR3 sequences. Importantly, the outgrowth MBP-reactive CD4+ Vβ8.2 T cell clones that are deleted from the TCR repertoire of EAE mice and observed only in CD8-/EAE mice are enriched in encephalitogenic CD4+ Vβ8.2 clones capable of inducing EAE in vivo. Taken together, these data provide evidence that during the evolution of EAE, regulatory CD8+ T cells are induced, which fine-tune the MBP-reactive CD4+ TCR repertoire in vivo, by selectively down-regulating the outgrowth of potentially pathogenic self-reactive T cell clones. The residual highly diverse nonpathogenic self-reactive TCR repertoire is preserved by this selective down-regulation.
Materials and Methods
Preparation of CD4+ T Cells. The TCR repertoire was assessed in three groups of mice including (i) naive B10PL mice (control), (ii) mice recovered from EAE (EAE), and (iii) EAE-recovered mice that were depleted of CD8+ T cells (CD8-/EAE). EAE was induced in B10PL mice by using 1–9Nac MBP as described (7). The great majority of mice develop and subsequently recover from EAE. In the CD8-/EAE group, mice were depleted of CD8+ T cells 3 days before EAE induction by using anti-murine CD8 mAb 53-6.72. The mice in the EAE group were treated with rat Ig as described (7). Eight to 10 weeks after the induction of EAE, the three groups of mice were immunized with 1–9Nac MBP emulsified with complete Freund's adjuvant at 100 μg per mouse s.c. Seven days later, CD4+ T cells were purified from draining lymph nodes, using MACS beads (8), and restimulated in vitro with 1–9Nac MBP. One week after retriggering with 1–9Nac MBP in vitro, the CD4+ T cells were assayed for TCR Vβ surface expression by fluorescence-activated cell sorter (FACS) analysis, PCR-based TCR, CDR3 length analysis, and Vβ-chain CDR3 sequencing as described below.
Antibodies and Flow Cytometry. Antibodies to CD4 (GK1.5), CD8 (53-6.72), and a panel of 18 anti-murine TCR mAbs, which detect 85% of murine TCR families, were purchased from PharMingen. Analysis of stained cells was performed by using a FACScan flow cytometer and cellquest software (Becton Dickinson) as described (8).
PCR-Based TCR CDR3 Length-Distribution and CDR3-Sequence Analyses. Total RNA was extracted from ≈2 × 105 CD4+ T cells with TRI reagent, according to the manufacturer's instructions (Molecular Research Center, Cincinnati; refs. 9–11). RT-PCR was performed with oligo (dT) primers and a range of mouse TCR Vβ and Cβ primer sets. Briefly, cDNA was prepared from 200 ng of total RNA, and was added into a total volume of 20 μl containing 100 mM KCl, 20 mM Tris·HCl (pH 8.3), 2.5 mM MgCl2, 30 pM Vβ and Cβ primer, 250 mM dNTP, and 1.25 units of AmpliTaq gold DNA polymerase (Perkin–Elmer). The initial heat denaturation at 96°C for 12 min was followed by 40 cycles of 30 sec at 94°C, 50 sec at 60°C, 45 sec at 72°C, and a final elongation step at 72°C for 10 min. Using 2 μl of the PCR products as templates, the extension reaction was performed by using a single Hex-labeled Cβ or Jβ antisense primer. The extension PCR products were analyzed on a 4.5% denaturing polyacrylamide gel in an Applied Biosystems 373 or 377 automated sequencer with internal Rox-labeled size standards. While analyzed with genescan analysis 3.1 software (Perkin–Elmer Applied Biosystems), CDR3 length distribution was revealed by fluorescein of bands representing different CDR3 length in sequencing gels. Subsequently, the TCR Vβ8.2 cDNA was amplified again by using the specific TCR Vβ region and unlabeled Cβ region primer. Vβ8.2 GATCCATTATTCATATGGTGCTGGC with Cβ TTGCGAGGATTGTGCCAGAAGG, was subcloned and sequenced as described (11). Subcloning was done by using the Topo TA cloning kit (Invitrogen). Plasmid minipreparations were preformed according to the manufacturer's instructions (5 Prime → 3 Prime). Clones containing the insert were sequenced by using the BigDye Terminator cycle-sequencing ready-reaction kit (Perkin–Elmer, Applied Biosystems) and run on 4.5% denaturing polyacrylamide gel and analyzed by using sequencing analysis 3.3 software (Perkin–Elmer Applied Biosystems).
The EAE Model Induced by Adoptive Transfer of 1–9Nac MBP-Specific CD4+ T Cell Clones. The 1–9Nac MBP-specific CD4+ T cell clones were generated by limiting dilution from EAE and CD8-/EAE mice as described (8). To test the encephalitogenicity of 1–9Nac MBP-specific clones, CD4+Vβ8.2+ T cell clones isolated from EAE and CD8-/EAE mice were adoptively transferred into naive mice, and the pathogenicity of transferred cells was evaluated by their capacity to induce EAE in recipient mice. Briefly, different groups of mice were sublethally irradiated (350 rad) and after 24 h were injected intravenously with 8–10 × 106 cells of each 1–9Nac MBP-specific clones tested. These CD4+ T clones were activated with 1–9Nac MBP for 4 days before injection. Pertussis toxin was injected at 0.1 μg per mouse i.v. 24 and 72 h after transfer of T cells. Mice were clinically monitored daily for symptoms and EAE was evaluated as described (8).
Adoptive Transfer of CD8+ T Cells. CD8+ T cells were isolated from spleens of EAE-recovered mice (6–10 weeks after the induction of EAE) or naive mice by using MACS beads as described (8). Purified CD8+ T cells (2–5 × 106) were adoptively transferred into naive recipient B10PL mice by i.v. injection. Mice were immunized with 1–9Nac MBP 1 day after T cell transfer. Seven days later, CD4+ T cells isolated from draining nodes of recipient mice were stimulated with 1–9Nac MBP in vitro and used to perform PCR-based TCR CDR3 length analysis as described.
Results
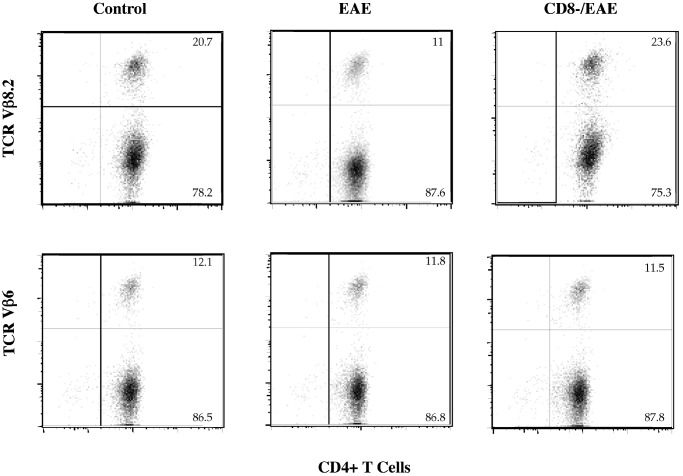
CD8+ T Cells Down-Regulate the Peripheral 1–9Nac MBP-Reactive CD4+Vβ8.2+ T Cell Population in EAE Mice. We first assayed CD4+ T cells derived from control, EAE, and CD8-/EAE mice for surface expression of TCR Vβ families by FACS analysis, using a panel of monoclonal anti-Vβ antibodies, which detect >85% of murine TCRs. The results, which are representative of four separate experiments (Fig. 1), show that compared with control mice, the EAE mice displayed a significant reduction in the proportion of CD4+ T cells expressing Vβ8.2. Expression of other TCR Vβ families, including Vβ6, remained unchanged or were slightly increased in EAE mice. In contrast, this specific reduction of CD4+ Vβ8.2+ T cells responding to 1–9Nac MBP was not observed in the CD8-/EAE mice. This finding is consistent with the hypothesis that during the evolution of EAE there is a change in the MBP-reactive TCR repertoire that is regulated by CD8+ T cells.
Fig. 1.
TCR Vβ8.2 and Vβ6 surface expression of MBP-reactive CD4+ T cells derived from control, EAE, and CD8-/EAE mice. The CD4+ T cells were prepared and assayed for TCR Vβ expression by two-color FACS analysis as described.
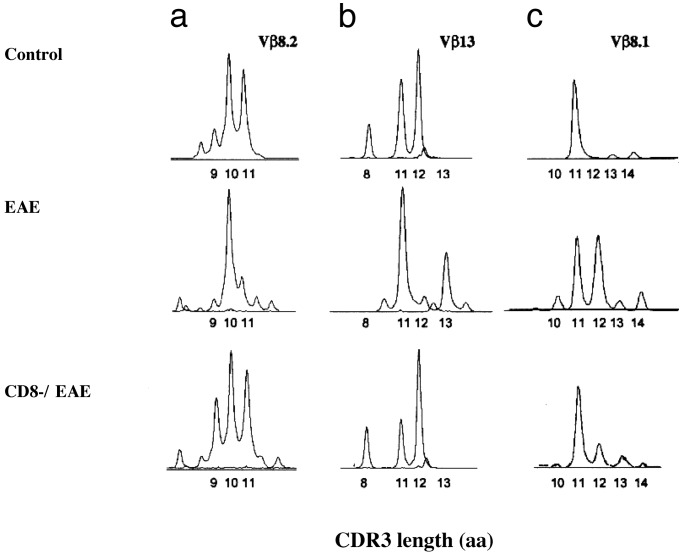
Analysis of the TCR Repertoire Among Control, EAE, and CD8-/EAE Mice by TCR CDR3 Length Distribution. We then directly analyzed the CDR3 length distributions of all of the TCR Vβ families by amplification of cDNA prepared from CD4+ T cells derived from control, EAE, and CD8-/EAE mice using the same cultures described above (9–11). In four separate experiments, this analysis confirmed a major effect of CD8+ T cell depletion on the TCR BV repertoire of both Vβ8.2 (BV8.2) and Vβ13 (BV13) families, but not other TCR Vβ families. Representative results are shown in Fig. 2. In control mice there was a distribution of Vβ8.2+ MBP-reactive oligoclonal expansions of various CDR3 lengths with major peaks observed at CDR3 lengths of 9, 10, and 11 aa (Fig. 2a). In contrast, in EAE mice there was a marked reduction of Vβ8.2 clonal expansions with CDR3 lengths of 9 and 11 aa, but with preservation of those with CDR3 length of 10 aa. Similarly, within the Vβ13 family of MBP-reactive clones in control mice, CDR3 length peaks were observed at 8, 11, and 12 aa (Fig. 2b). In contrast, in EAE mice there was a marked reduction observed in Vβ13+ clones with CDR3 lengths of 8 and 12 aa, with preservation of clones with CDR3 length of 11 aa. Importantly, these reductions of certain clonal expansions in both Vβ8.2 and Vβ13 families were not observed in CD8-/EAE mice. Although the particular dominant oligoclonal expansions observed differed somewhat in each experiment, the general pattern of a broader repertoire of outgrowing clones was evident in the CD8-/EAE mice, compared with EAE mice. Thus, our data show that CD8+ T cells, in vivo, selectively down-regulate certain, but not all, 1–9Nac MBP-reactive CD4+ clones within both TCR Vβ8.2 and Vβ13 families. Importantly, the Vβ8.2 and Vβ13 families of CD4+ T cells represent the two major T cell populations that respond to 1–9Nac MBP stimulation, and are largely responsible for clinical EAE (12, 13). This CD8+ T cell-mediated selective down-regulation of certain Vβ8.2+ and Vβ13+ 1–9Nac MBP-reactive CD4+ T cell clones in vivo may very well explain the abrogation of resistance to reinduction of EAE in CD8+ T cell-depleted mice, as well as the increased frequency of relapses in CD8-/- mice (14).
Fig. 2.
Distribution of the TCRβ-chain CDR3 length of CD4+ T cells isolated from control, EAE, and CD8-/EAE mice. The CD4+ T cells were prepared and the CDR3 length distribution of all Vβ families was assayed as described in Materials and Methods. The CDR3 length distributions for Vβ8.2, Vβ13, and Vβ8.1 families are shown. The data depict representative results of four independent experiments.
Of interest is that some Vβ13+ oligoclonal expansions with CDR3 length of 13 aa emerge in the EAE mice but are not observed in either the control or CD8-/EAE mice (Fig. 2b). This emergence of oligoclonal expansions in the EAE mice is also observed in other Vβ families. For example, the CDR3 length-distribution analysis of the Vβ8.1 family did not show any reduction of CDR3 peaks in the EAE mice. In contrast, there was emergence of new oligoclonal expansions with the CDR3 length of 12 aa in EAE mice, but not in control and CD8-/EAE mice (Fig. 2c). The emergence of MBP-reactive clones in EAE mice but not in control and CD8-/EAE mice may simply reflect the growth of minor populations of self-reactive clones, with perhaps lower-affinity TCRs for MBP, which appear in the setting of the down-regulation of the major encephalitogenic Vβ8.2+ and Vβ13+ CD4+ T cell clones by the CD8+ T cells.
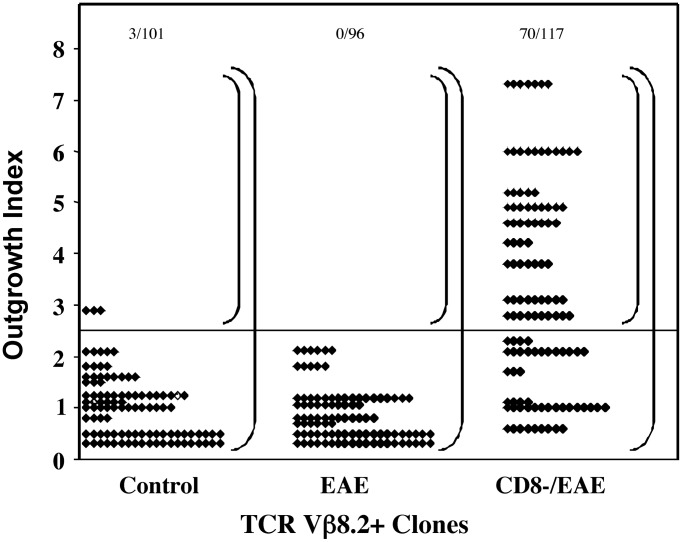
Comparison of the TCR Vβ8.2 Repertoire by Analysis of TCR CDR3 Sequences. The CDR3 length-distribution method is inherently limited by the fact that the individual amino acid differences in the CDR3 are not taken into account and that one or several T cell clones may be contained within a peak at a particular CDR3 length. To gain insight into the nature of the changes in the Vβ8.2 repertoire at a clonal level, in three separate experiments, we ligated the TCR Vβ8.2-specific PCR products of MBP-reactive CD4+ T cells derived from control, EAE, and CD8-/EAE mice into the pTOPO vector and obtained a total of 314 β-chain sequences. Among these Vβ sequences, some were represented only once, whereas others were found multiple times. Because T cell clones are defined by the expression of TCR CDR3 sequences, the frequency of a particular CDR3 sequence reflects the frequency of a distinct T cell clone. This enables us to calculate an outgrowth index to indicate the proportion of the repertoire occupied by a given T cell clone, defined by a β-chain sequence, and thus reflects the degree of expansion of that particular clone. To obtain the outgrowth index, the frequency of clones with same CDR3 sequence was first calculated as a fraction of total clones analyzed in each experimental group. This fraction was further normalized with respect to the actual percent of Vβ8.2+ T cells present in the total CD4+ T cell population assayed by FACS analysis.
The distribution of the outgrowth index of all of the clones analyzed from three experiments is shown in Fig. 3. The actual CDR3 amino acid sequences of MBP-reactive Vβ8.2+ clones with an outgrowth index of >2.5 are shown in Table 1. In the control group only 3 of 101 clones (≈3.0%) had an outgrowth index of >2.5. Among 96 clones obtained from the EAE mice, none of the clones had an outgrowth index of >2.5. In contrast, of 117 clones obtained from the CD8-/EAE mice, 70 clones had an outgrowth index of >2.5 (59.8%). Thus, in the absence of CD8+ T cell regulation both the clonal size and, importantly, the number of clones with outgrowth >2.5 is markedly increased over that found in EAE mice containing the regulatory CD8+ T cells. These results confirm in a more direct and quantitative manner the observations obtained by CDR3 length-distribution analysis that CD8+ T cells fine-tune the CD4+ TCR repertoire by controlling the expansion of certain, but not all, 1–9Nac MBP-reactive Vβ8.2+ clones. Moreover, these data are consistent with the idea that the CD4+ T cell clones with an outgrowth index of >2.5, which constitute ≈60% of peripheral MBP-reactive Vβ8.2 repertoire in CD8-/EAE mice, represent the clones that are subject to down-regulation by the CD8+ T cells in vivo.
Fig. 3.
CD8+ T cells control the clonal outgrowth of CD4+ Vβ8.2+ MBP-reactive T cells in EAE mice. Each point represents one Vβ8.2 clone with its outgrowth index. Outgrowth index = [(the number of Vβ8.2 clones with the same CDR3 sequence)/(total clones analyzed) × (CD4+ Vβ8.2+ T cells/total CD4+ T cells assayed by FACS)] × 100%. The cloning and sequencing were performed as described in Materials and Methods.
Table 1. CDR3 amino acid sequences of MBP-reactive TCR Vβ8.2+ clones with an outgrowth index of >2.5.
| Experiment no. | Experimental condition | CDR3 amino acid sequence | Jβ sequences | Frequencies of identical sequences | Outgrowth index, % |
|---|---|---|---|---|---|
| 1 | Control | — | — | 0/32 | — |
| EAE | — | — | 0/35 | — | |
| CD8-/EAE | CASGDWGAYEQY | 2.7 | 11/36 | 6.0 | |
| CD8-/EAE | CASRDNNYAEQF | 2.1 | 9/36 | 4.9 | |
| CD8-/EAE | CASGSGGDYEQY | 2.7 | 7/36 | 3.8 | |
| 2 | Control | CASSDQNTLY | 2.4 | 3/21 | 2.9 |
| EAE | — | — | 0/19 | — | |
| CD8-/EAE | CASGAPRGTNSDYT | 1.2 | 8/29 | 4.5 | |
| CD8-/EAE | CASRGLGQDTQY | 2.5 | 5/29 | 2.8 | |
| CD8-/EAE | CASSPTGVNTGQLY | 2.2 | 5/29 | 2.8 | |
| 3 | Control | — | — | 0/48 | — |
| EAE | — | — | 0/42 | — | |
| CD8-/EAE | CASGEQGGQNTLY | 2.4 | 7/52 | 7.3 | |
| CD8-/EAE | CASGPTGYQDTQY | 2.5 | 5/52 | 5.2 | |
| CD8-/EAE | CASGEQGANGNTLY | 1.3 | 4/52 | 4.2 | |
| CD8-/EAE | CASASGTANTEVF | 1.1 | 3/52 | 3.1 | |
| CD8-/EAE | CASGDAPGQGNTGQLY | 2.2 | 3/52 | 3.1 | |
| CD8-/EAE | CASSDVRDWGDQDTQY | 2.5 | 3/52 | 3.1 |
We further analyzed the CDR3 amino acid sequences of MBP-reactive clones with respect to the degree of overall diversity of the TCR repertoire as well as the Jβ element used. We found that the degree of overall diversity in the primary and secondary MBP-reactive TCR repertoire as measured by the percent distinct CDR3 sequences in the control and EAE mice, respectively, remained basically the same (73.2% versus 64.8%). In contrast, the degree of overall diversity in the CD8-/EAE mice compared with the EAE mice was significantly decreased (36.8%, P < 0.001; Table 2). This decrease in diversity in the CD8-/EAE mice reflects, in part, the fact that 60% of the clones in this group have an outgrowth index of >2.5 and represent only 12 sequences (Table 1). Nevertheless, the outgrowth clones found in CD8-/EAE mice used a variety of distinct Jβ sequences in different mice, including Jβ2.1, Jβ2.2, Jβ2.4, Jβ2.5, and Jβ2.7, which may reflect the Jβ families used by pathogenic Vβ8.2 clones (Table 1).
Table 2. Analysis of the degree of the diversity of MBP-reactive TCR Vβ8.2 T cell repertoire among control, EAE, and CD8-/EAE mice.
| Experimental groups | Frequencies of distinct CDR3 sequences that only appear once, % | P | Degree of overall diversity (diversity index*), % | P |
|---|---|---|---|---|
| Control | 57.3 ± 20.1 | 73.2 ± 16.5 | ||
| EAE | 50.2 ± 4.2 | >0.1† | 64.8 ± 1.9 | >0.1† |
| CD8-/EAE | 18.6 ± 9.4 | <0.005‡ | 36.8 ± 3.6 | <0.001‡ |
Data and statistical analyses were obtained from three separate experiments
Diversity index = (the number of distinct CDR3 sequences/total CDR3 sequences obtained) × 100%
P value between control group and EAE group
P value between EAE group and CD8-/EAE group
CD8+ T Cells Control Expansion of Potentially Pathogenic CD4+Vβ8.2+ T Cell Clones in EAE Mice. To test the hypothesis that the outgrowth clones isolated from CD8-/EAE mice contain potentially pathogenic clones, CD4+Vβ8.2+ T cell clones were isolated from EAE or CD8-/EAE mice and adoptively transferred into irradiated naive mice. The pathogenicity of transferred cells was evaluated by their capacity to induce clinical EAE in recipient mice. As shown in Table 3, among four clones isolated from EAE mice, only one was capable of inducing EAE. Whereas among four clones isolated from CD8-/EAE mice, all induced EAE in vivo (P < 0.005). These data support the hypothesis that the MBP-reactive population down-regulated by CD8+ T cells contain the pathogenic self-reactive clones.
Table 3. Capacity of 1-9Nac MBP-specific CD4+Vβ8.2+ T cell clones to induce EAE in vivo by adoptive transfer.
|
Severity
|
||||
|---|---|---|---|---|
| Clones | Incidence | Mean | Maximum | Frequency of encephalitogenic clones, % |
| T cell clones from EAE mice | ||||
| 3EAC3 | 3/5 | 2.5 | 5.0 | |
| 5EB2 | 0/5 | 0 | 0 | |
| 6EB9 | 0/5 | 0 | 0 | |
| 3ED11 | 0/12 | 0 | 0 | 25 |
| T cell clones from CD8-/EAE mice* | ||||
| 4DC5 | 5/5 | 3.2 | 5.0 | |
| 4DH2 | 5/5 | 3.3 | 5.0 | |
| 5DC9 | 5/5 | 3.5 | 5.0 | |
| 5DH4 | 5/5 | 3.4 | 5.0 | 100 |
EAE was induced and evauluated as described in Materials and Methods
P < 0.05, value between EAE group and CD8-/EAE group
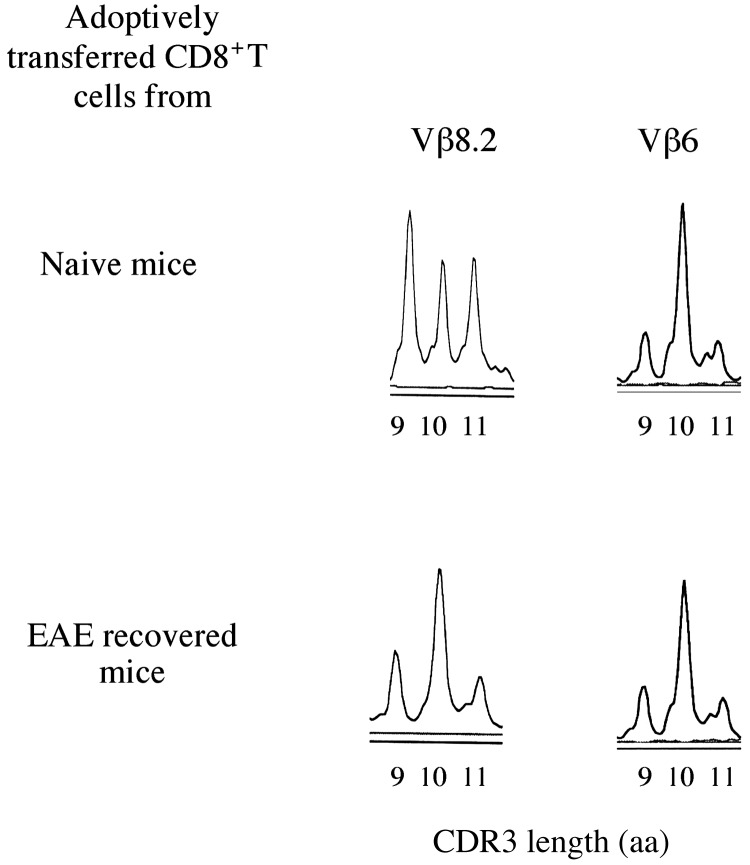
Adoptive Transfer of Regulatory CD8+ T Cells from EAE-Recovered Mice Fine-Tune the MBP-Reactive TCR Vβ Repertoire. To further confirm that the regulatory CD8+ T cells, which are induced and function in EAE mice fine-tune the MBP-reactive TCR Vβ repertoire, CD8+ T cells were isolated from EAE mice and adoptively transferred into naive B10PL mice. CD8+ T cells isolated from naive B10PL mice were adoptively transferred as control. Both groups of recipient mice were immunized with 1–9Nac MBP 1 day after the adoptive transfer of the CD8+ T cells. MBP-reactive CD4+ TCR repertoire was assessed as described in Materials and Methods. We emphasize that the secondary MBP repertoire in EAE mice is already modified by the regulatory CD8+ T cells as shown above but naive mice possess an unmodified MBP repertoire. Thus, in the adoptive transfer experiments, the primed CD8+ T cells from EAE mice were transferred into naive mice, which were subsequently immunized with 1–9Nac MBP, to reveal the effect of transferred CD8+ T cells in fine-tuning the MBP-reactive repertoire.
As shown in Fig. 4, in mice adoptively transferred with regulatory CD8+ T cells isolated from EAE mice, the major peaks of 9 and 11 in TCR Vβ8.2 family are significantly decreased, compared with mice adoptively transferred with unprimed CD8+ T cells isolated from naive mice. In contrast, there is no difference in the CDR3 length distribution of the TCR Vβ6 family found between two types of recipient mice. Thus, the MBP-reactive Vβ8.2, but not Vβ6 repertoire, were significantly affected by the adoptive transfer of regulatory CD8+ T cells from EAE mice.
Fig. 4.
Regulatory CD8+ T cells adoptively transferred into naive mice fine-tune the MBP-reactive TCR Vβ repertoire. CD8+ T cells isolated and adoptively transferred into naive B10PL mice as described and recipient mice were immunized with 1–9Nac MBP 1 day after the T cell transfer. CD4+ T cells were isolated from draining lymph nodes from recipient mice, and CDR3 length distribution was performed as described.
Discussion
Because some potentially pathogenic self-reactive T cells escape thymic negative selection and are released into the periphery (2–4), mechanisms that normally regulate the outgrowth of these self-reactive T cells ultimately control the initiation and progression of autoimmune disease. Here, employing the murine EAE model of autoimmunity, we report that one mechanism for limiting the outgrowth of potentially pathogenic self-reactive clones is the selective down-regulation of these clones by CD8+ T cells. Evidence that the control of CD4+ Vβ8.2+ MBP-reactive T cells was selective was revealed by PCR-based CDR3 length-distribution analysis of the TCR repertoire and by direct sequence analysis of the CDR3s. In CD8-/EAE mice, ≈60% of the MBP-reactive Vβ8.2 repertoire is dominated by the significant outgrowth of only a few clones. The significant outgrowth of these dominant clones, each representing >2.5% of the repertoire, was not observed in EAE mice with intact CD8+ T cells. In contrast, the TCR repertoire of MBP-reactive CD4+ T cells in the EAE mice was comprised predominantly of a highly diverse set of self-reactive clones with limited outgrowth. Because the CD8-/EAE mice are highly susceptible to clinical EAE, whereas CD8+ T cell-intact EAE mice are not. It is likely that the dominant clones that emerge after secondary MBP immunization in CD8-/EAE mice contain the potentially encephalitogenic CD4+ T cells. This idea was further supported by our observation that MBP-reactive CD4+Vβ8.2+ T cell clones derived from CD8-/EAE mice were more likely to induce EAE after adoptive transfer into naive mice than clones derived from EAE mice with intact CD8+ T cells. Moreover, adoptive transfer of the regulatory CD8+ T cells isolated from EAE-recovered mice into naive mice provided further evidence that the CD8+ T cells fine-tune MBP-reactive TCR Vβ repertoire. Taken together, these studies provide evidence that in addition to their TCR Vβ specificity CD8+ T cells selectively down-regulate certain but not all self-reactive T cells within TCR Vβ8.2 family. Thus, regulatory CD8+ T cells play a key role in controlling self-reactive TCR repertoire by selectively down-regulating the potentially pathogenic self-reactive T cells in the periphery.
In this regard, it is of interest to consider the potential relationship between the CD4+ T cell clones that are pathogenic and when adoptively transferred, induce EAE with the outgrowth clones we identified by Vβ8.2 TCR CDR3 length distribution and sequencing analyses. It is highly likely that in addition to TCR CDR3 sequence, that other factors including the expression of costimulatory molecules, chemokine receptors, and TCR receptor affinity (see below), independently dictate either pathogenicity or susceptibility to down-regulation. We have not yet determined which of these factors are most important in the relationship between pathogenicity of individual CD4+ clones and the 1–9Nac MBP-reactive CD4+ Vβ8.2 TCR repertoire. However, the important point of this article is that both pathogenicity and TCR repertoire are controlled by CD8+ T cells, as is the clinical susceptibility of EAE mice to reinduction of disease.
It is important to emphasize that CD8+ T cells require priming during the first episode of EAE to regulate the outgrowth of potentially pathogenic CD4+ T cells triggered by secondary MBP challenge in vivo. The evidence that regulatory CD8+ T cells require priming during the first episode of EAE is simply that B10PL mice depleted of CD8+ T cells during the initial induction of EAE recover from EAE normally, but are not resistant to rechallenge with 1–9Nac MBP. In contrast, EAE mice with CD8+ T cells primed during the first episode are resistant to reinduction of EAE unless they are depleted of CD8+ cells before reinduction of EAE (7, 8). In this study, CD8+ T cells were depleted during the induction of EAE (first episode). These mice developed EAE and clinically recovered. Newly generated CD8+ T cells reappear during the recovery and are present at the time of secondary MBP immunization in vivo. When the MBP-reactive TCR Vβ repertoire of these mice was compared with EAE mice with primed CD8+ T cells, the profound effect of the primed CD8+ T cells on the CD4+ MBP-reactive TCR Vβ repertoire was observed. This sequence of events is reminiscent of the general biology of CD8+ T cells involved in the response to viruses. During the initial infection with most viruses CD8+ T cells are not involved in the recovery that may occur within the first week or so (this initial recovery is mediated in part by the innate immune response), but they are primed during the initial infection period and are clearly involved in resistance to reinfection or persistent virus infection.
We envision that the regulatory CD8+ T cells function in concert with other homeostatic mechanisms, including those mediated by cytokines or by regulatory CD4+ T cells. It has been shown that a subset of regulatory CD4+ T cells reactive with pathogenic CD4+ T cells control the clinical development of EAE and their function depends on CD8+ T cells (15). Thus, regulatory CD4+ and CD8+ T cells interact with one another to control autoimmunity in EAE. Furthermore, there is growing evidence that regulatory CD4+ T cells can down-regulate self-reactive CD4+ T cells, independent of other T cells. These include the CD4+CD25+ (16–19) and CD4+CD25- cells (20) involved in the down-regulation of a number of autoimmune diseases including EAE. However, the specificity or mechanisms of the regulatory CD4+ T cell-mediated suppression is currently not completely understood, but they have been shown to express a diverse set of αβ TCR and to secrete immunosuppressive cytokines including transforming growth factor type-β, IL-4, or IL-10 (21–26). It is likely that immunoregulation mediated by CD4+ or natural killer T(NKT) cells may be most important during the initial stages of self-antigen-induced responses and may be highly effective in regulating the innate immune response and/or the early phases of antigen-driven T cell activation (18). Our data show that the regulatory CD8+ T cells are primed during the first episode of EAE and differentiate into effector cells, which fine-tune the CD4+ TCR Vβ repertoire at later stages of self-antigen-driven immune responses.
The precise mechanisms by which CD8+ T cells selectively recognize and down-regulate only certain, but not all, clones of T cells within Vβ8.2 family triggered by MBP in vivo are not known. However, in previous studies (8, 27) we showed that TCR Vβ-specific, Qa-1 restricted regulatory CD8+ T cells are induced in T cell-vaccinated mice, which are protected from EAE. It is of interest that the Qa-1 restricted CD8+ T cells have also been observed in the regulation of antibody synthesis in which Qa-1-expressing B cells were shown to induce regulatory CD8+ T cells (28). The effector mechanisms used by the regulatory CD8+ T cells to down-regulate target T cells may involve lysis of target T cells (8, 27, 29).
If the Vβ-specific, Qa-1-restricted CD8+ T cells are involved in the control of the TCR Vβ repertoire in EAE as shown here, it is of interest to consider why only some, but not all, 1–9Nac MBP-reactive TCR Vβ8.2+ clones are down-regulated. The differential recognition of TH1 versus TH2 cells by the regulatory CD8+ T cells we previously described may contribute to the selective down-regulation of certain, but not, all antigen-activated CD4+ T cells by the CD8+ T cells (8). Nevertheless, we have shown that at least some MBP-reactive TH1 clones in EAE mice escape CD8+ T cell regulation (8). Thus, differential recognition of TH1 versus TH2 cells by the CD8+ T cells alone cannot account for why only some, but not all, antigen-activated clones are down-regulated by the CD8+ T cells.
In this regard it is known that Qa-1 is only minimally expressed on resting lymphoid cells, and that unlike classical MHC class Ia molecules, its expression depends on activation. Although the precise Qa-1-binding peptide(s) in this system have not been identified yet, distinct self-peptide(s) binding to Qa-1, which may be determined by the affinity/avidity of the CD4+ T cells, may influence the susceptibility of activated CD4+ T cells to the down-regulation by the CD8+ T cells (30).
In addition, it is known that a significant number of murine CD8+ T cells express receptors, which bind to Qa-1-associated with Qdm peptide, the predominant self-peptide presented by Qa-1 (31), and modify the function of CD8+ T cells. These include the NKCD94/NKG2 receptors, which are known to either positively or negatively regulate cytotoxic T lymphocyte (CTL) function (32, 33), and the more recently reported Qa-1/Qdm binding receptor present on virtually all activated CD8+ T cells, which triggers IFN-γ secretion (34). The clones of the regulatory CD8+ T cells may coexpress these two distinct types of Qa-1 receptors. One type is the clonally distributed αβ TCR, which recognizes Qa-1/self-peptide complexes we discussed above, and another type of nonclonal receptors that recognizes Qa-1/Qdm. These two types of receptors could be differentially expressed on the surface of regulatory CD8+ T cells and functionally regulate one another. Thus, we hypothesize that the balance between different forms of Qa-1/peptide complexes expressed on antigen-activated CD4+ T cells binding the two types of Qa-1 receptors on the regulatory CD8+ T cells may also contribute to the susceptibility of CD4+ T cells to the down-regulation by the regulatory CD8+ T cells (30).
Acknowledgments
This work was supported by National Institutes of Health Grants AI39630 and AI44927, National Multiple Sclerosis Society Grant RG2938A (to H.J.), and National Institutes of Health Grant U19AI46132 (to L.C.).
Abbreviations: MBP, myelin basic protein; EAE, experimental autoimmune encephalomyelitis; TCR, T cell receptor; 1–9Nac MBP, NH2-terminal-acetylated nano peptide from mouse MBP; FACS, fluorescence-activated cell sorter; CDR, complementarity-determining region.
References
- 1.Harrington, C. J., Paez, A., Hunkapiller, T., Mannikko, V., Brabb, T., Ahearn, M., Beeson, C. & Goverman, J. (1998) Immunity 8 571-580. [DOI] [PubMed] [Google Scholar]
- 2.Goldrath, A. W. & Bevan, M. J. (1999) Nature 402 255-262. [DOI] [PubMed] [Google Scholar]
- 3.Bouneaud, C., Kourilsky, P. & Bousso, P. (2000) Immunity 13 829-840. [DOI] [PubMed] [Google Scholar]
- 4.Kuchroo, V. K., Anderson, A. C., Waldner, H., Munder, M., Bettelli, E. & Nicholson, L. B. (2002) Annu. Rev. Immunol. 20 101-123. [DOI] [PubMed] [Google Scholar]
- 5.Anderton, S. M., Radu, C. G., Lowrey, P. A., Ward, E. S. & Wraith, D. C. (2001) J. Exp. Med. 193 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acha-Orbea, H., Mitchell, D. J., Timmermann, L., Wraith, D. C., Tausch, G. S., Waldor, M. K., Zamvil, S. S., McDevitt, H. O. & Steinman, L. (1988) Cell 54 263-273. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, H., Zhang, S. I. & Pernis, B. (1992) Science 256 1213-1215. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, H., Braunstein, N., Yu, B., Winchester, R. & Chess, L. (2001) Proc. Natl. Acad. Sci. USA 98 6301-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pannetier, C., Even, J. & Kourilsky, P. (1995) Immunol. Today 16 176-181. [DOI] [PubMed] [Google Scholar]
- 10.Kim, G., Tanuma, N., Kojima, T., Kohyama, K., Suzuki, Y., Kawazoe, Y. & Matsumoto, Y. (1998) J. Immunol. 160 509-513. [PubMed] [Google Scholar]
- 11.Costello, P. J., Winchester, R. J., Curran, S. A., Peterson, K. S., Kane, D. J., Bresnihan, B. & FitzGerald, O. M. (2001) J. Immunol. 166 2878-2886. [DOI] [PubMed] [Google Scholar]
- 12.Zamvil, S., Nelson, P., Trotter, J., Mitchell, D., Knobler, R., Fritz, R. & Steinman, L. (1985) Nature 317 355-358. [DOI] [PubMed] [Google Scholar]
- 13.Urban, J. L., Kumar, V., Kono, D. H., Gomez, C., Horvath, S. J., Clayton, J., Ando, D. G., Sercarz, E. E. & Hood, L. (1988) Cell 54 577-592. [DOI] [PubMed] [Google Scholar]
- 14.Koh, D.-R., Fung-Leung, W.-P., Ho, A., Gray, D., Acha-Orbea, H. & Mak, T.-W. (1992) Science 256 1210-1213. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, V. & Sercarz, E. E. (1993) J. Exp. Med. 178 909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shevach, E. M. (2000) Annu. Rev. Immunol. 18 423-449. [DOI] [PubMed] [Google Scholar]
- 17.Olivares-Villagomez, D., Wang, Y. & Lafaille, J. J. (1998) J. Exp. Med. 188 1883-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furtado, G. C., Olivares-Villagomez, D., Curotto de Lafaille, M. A., Wensky, A. K., Latkowski, J. A. & Lafaille, J. J. (2001) Immunol. Rev. 182 122-134. [DOI] [PubMed] [Google Scholar]
- 19.Hori, S., Haury, M., Coutinho, A. & Demengeot, J. (2002) Proc. Natl. Acad. Sci. USA 99 8213-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafaille, J. J., Nagashima, K., Katsuki, M. & Tonegawa, S. (1994) Cell 78 399-408. [DOI] [PubMed] [Google Scholar]
- 21.Powrie, F., Carlino, J., Leach, M. W., Mauze, S. & Coffman, R. L. (1996) J. Exp. Med. 183 2669-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpus, W. J. & Swanborg, R. H. (1989) J. Immunol. 143 3492-3497. [PubMed] [Google Scholar]
- 23.Karpus, W. J. & Swanborg, R. H. (1991) J. Immunol. 146 1163-1168. [PubMed] [Google Scholar]
- 24.Groux, H., O'Garra, A., Bigler, M., Rouleau, M., Antonenko, S., de Vries, J. E. & Roncarolo, M. G. (1997) Nature 389 737-742. [DOI] [PubMed] [Google Scholar]
- 25.Lafaille, J. J. (1998) Cytokine Growth Factor Rev. 9 139-151. [DOI] [PubMed] [Google Scholar]
- 26.Chen, Y., Kuchroo, V. K., Inobe, J., Hafler, D. A. & Weiner, H. L. (1994) Science 265 1237-1240. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, H., Kashleva, H., Xu, L. X., Forman, J., Flaherty, L., Pernis, B., Braunstein, N. S. & Chess, L. (1998) Proc. Natl. Acad. Sci. USA 95 4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble, A., Zhao, Z. S. & Cantor, H. (1998) J. Immunol. 160 566-571. [PubMed] [Google Scholar]
- 29.Jiang, H., Ware, R., Stall, A., Flaherty, L., Chess, L. & Pernis, B. (1995) Immunity 2 185-194. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, H. & Chess, L. (2000) Annu. Rev. Immunol. 18 185-216. [DOI] [PubMed] [Google Scholar]
- 31.Aldrich, C. J., Rodgers, J. R. & Rich, R. R. (1988) Immunogenetics 28 334-344. [DOI] [PubMed] [Google Scholar]
- 32.Lanier, L. L., Corliss, B. C., Wu, J., Leong, C. & Phillips, J. H. (1998) Nature 391 703-707. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Botet, M. & Bellon, T. (1999) Curr. Opin. Immunol. 11 301-307. [DOI] [PubMed] [Google Scholar]
- 34.Wang, R., Ramaswamy, S., Hu, D. & Cantor, H. (2001) Eur. J. Immunol. 31 87-93. [DOI] [PubMed] [Google Scholar]