In a tour de force in this issue of PNAS, Bourgeois et al. (1) have used 2.2-ns x-ray pulses to observe the motion of carbon monoxide (CO) through myoglobin (Mb) and the relaxation of the protein from 3.2 ns to 3 ms after photodissociation. This work follows the pioneering experiments of Moffat and collaborators (2). It demonstrates how far advances in x-ray sources and computers have moved the field of protein structure determination since the path-breaking work of Kendrew et al. and Perutz (3, 4). The recent breakthrough shows how careful studies of proteins, in particular of Mb, impact many different fields. Mb is a monomeric protein that gives muscle its red color. Thirty years ago the textbook function of Mb, storage of dioxygen at the heme iron, was considered to be simple, fully understood, and consequently boring. Mb was essentially written off as a topic of serious research. Since then, the situation has changed: Mb is no longer fully understood. It plays roles other than O2 storage, serves as a prototype for complex systems, and yields insight into the chemistry and physics of soft matter and of chemical reactions.
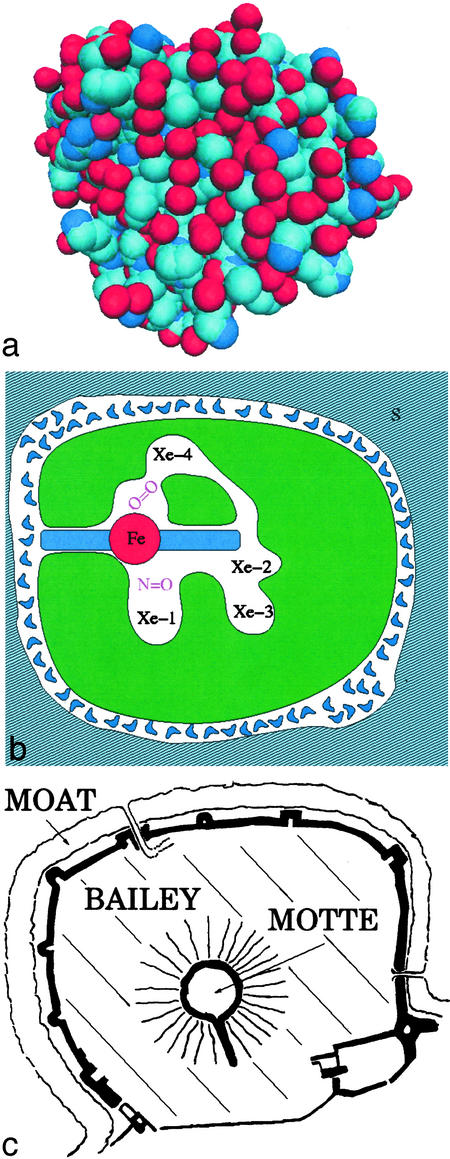
Mb consists of 153 amino acids that fold into a structure that is ≈3 nm in diameter, as depicted in Fig. 1a. Fig. 1b gives a schematic cross section through Mb that shows the active center: a heme group with a central iron atom. Surrounding the heme group are five cavities, the heme cavity and four cavities denoted by Xe1 to Xe4 (5). The amino acids lining the xenon cavities are much more conserved than other amino acids in mammalian Mb; they are thus likely to be important for function. A major part of Mb is taken up by amino acids that do not appear to have an obvious function; we denote this part as “bailey.” Surrounding the protein proper is the hydration shell, consisting of one to two layers of water molecules with properties that are different from bulk water. No openings are visible in Fig. 1a; entrance and exit of ligands must occur through gates. The entire structure is reminiscent of the Chateau Gisors, built in 1096 and sketched in Fig. 1c. The central part, called motte, resembles the active center, the moat is the equivalent of the hydration shell, and the bailey is the field between motte and moat, with no obvious function. Gates permit access to the castle.
Fig. 1.
(a) An atomistic view of myoglobin. The positions of the atoms are deduced from x-ray diffraction. The atoms are nearly close-packed, leaving no static path for the entry or exit of ligands such as CO and O2. Individual atoms are represented as van der Waals spheres of ≈2.6-Å diameter with oxygen shown in red, nitrogen shown in dark blue, and carbon shown in light blue. (b) A schematic cross section through Mb. The folded polypeptide chain (green) surrounds a heme group with a central iron atom. Cavities are clearly visible. The protein is surrounded by a hydration shell and the solvent S. (c) The Chateau Gisors (Normandy, France) as a “model” of myoglobin.
Studies of proteins play different roles in biology and biochemistry, biophysics, chemistry, and biological physics. In biology and biochemistry, protein functions and their interactions within networks are central. In biophysics, one goal is to explore in detail how proteins function as nanomachines with unusual mechanical and dielectric properties. In chemistry, proteins can help elucidate solvent properties and catalysis mechanisms. For biological physicists, proteins combine properties of crystals, glasses, and liquids with probes for all types of spectroscopies. Their study can lead to a better understanding of complex systems. Mb is a nearly ideal system for all these different endeavors. It is simple enough so that it can be studied in detail, yet complex enough that the research is not trivial. We will now present a few examples that show how Mb has already yielded rich results.
In this issue of PNAS, Bourgeois et al. (1) provide an example that touches biology, chemistry, and physics. One result can be summarized with the help of Fig. 1b. CO is initially bound to the heme iron. After a laser flash, the photodissociated CO moves rapidly to the cavity Xe4 and more slowly to Xe1 and then out into the solvent. These kinetic results are useful and are in general agreement with flash photolysis experiments that follow the progress of the CO spectroscopically (6, 7). After photodissociation below 150 K, the CO remains in the heme pocket and rebinds from there (8). Rebinding is nonexponential in time and must be described by a distribution of energy barriers. This fact was explained by assuming that Mb can exist in various conformations that are frozen at cryogenic temperatures (6). It was, however, a long time before the temperature-dependent kinetic data were explained in terms of the structural data (9). Clear structural and energetic data on the ligand migration process in Mb should lead to better understanding of how proteins position substrates for reaction.
Even more important, however, is the second result: The structures of MbCO and deoxy Mb differ. After photodissociation, the protein thus changes its structure in a quake-like motion (10, 11). The changes are not large but are important for the function. Bourgeois et al. follow these structural changes and show that they extend over a considerable range of times, in agreement with spectroscopic data of Anfinrud and collaborators (12). This observation leads directly to a central problem in understanding protein function, the existence of an energy (or conformation) landscape. A protein does not exist in a unique conformation but can assume a very large number of somewhat different conformations or conformational substates. A particular substate is characterized by the coordinates of all atoms, including the hydration shell (6, 10, 13). The conformational motions observed in the work of Bourgeois et al. are transitions between substates. If a protein had just a single conformation, it could not function and would be dead like a stone.
Experiments show that the protein motions fall, crudely speaking, into two classes, slaved and nonslaved (14). Nonslaved motions are nearly independent of the motions in the solvent. Slaved motions have the same temperature dependence as the configurational dielectric fluctuations in the solvent, but are slower. This observation has consequences, both for the function of proteins and for the understanding of the energy landscape. Consider first the function. Exit and entrance of ligands such as CO and O2 are slaved, they are controlled by the environment. It is as if the drawbridges in Fig. 1c were controlled from the outside of the castle! Slaving also changes the interpretation of the barriers between protein substates. Initially they were assumed to be given by enthalpy barriers intrinsic to the protein, but because the solvent determines the temperature dependence of the slaved transition rates, the internal barriers must be entropic. To open a gate, the protein must make a random walk in the energy landscape and the number of steps must be very large. Such a random walk is only possible if the protein has a sufficient number of substates or, in other words, has sufficient entropy. The logical place for the entropy is the bailey, the part of the protein away from the active center. This model could explain the size of proteins.
Myoglobin may have evolved in conjunction with life's ability to control basic oxygen chemistry.
A second extension of standard x-ray crystallography also shows great promise, probing the protein structure under high pressure (15). Pressure is at least as important as temperature for understanding biological phenomena, but it is far less used. Just as time- and temperature-dependent studies explore the energetics of protein motion, pressure studies will elucidate the effect of volume fluctuations and shape in controlling protein function. It is to be hoped that the development of new techniques will lead to more studies of proteins under pressure.
Mb is also an excellent laboratory for physics and chemistry. Below 40 K, CO binds through quantum-mechanical tunneling, an unexpected phenomenon (16). The binding of CO and O2 at cryogenic temperatures also led to a reexamination of the role of friction and of the Kramers theory in chemical reactions (17). The electronic properties of the heme group have long been a model system for studying electronic structures (18–20). The well characterized nano-structure of Mb may provide structural insight and specific experimental probes into the nature of motions that remain when glassy systems are frozen (21). Mb is a benchmark system for developing femtosecond optical absorption and photon echoes (21, 22). Site-directed mutagenesis of valine 68 to aspartic acid was a first use of this technique to probe the dielectric properties of the protein interior (23), as is the recent use of the iron carbonyl to probe vibrational Stark shift (24).
Mb was initially considered to have just one function, and thus be
nonallosteric. It turns out, however, that Mb is actually allosteric and has
at least two functions (25,
26). The allostery is
explained through the hierarchical organization of the energy landscape. At
the top of the landscape hierarchy are three, or possibly four, taxonomic
substates. They have distinct properties, and two, denoted by A0
and A1, have different functions. In A1, dominant at
high pH, the distal histidine is inside the heme pocket; in A0,
dominant at low pH, the distal histidine has swung out into the solvent.
A1 stores O2, A0 catalyzes the conversion of
NO to 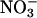 . The important observation is
that Mb, a simple monomeric protein, is allosteric. This observation leads to
the speculation that most proteins may have more than one function.
. The important observation is
that Mb, a simple monomeric protein, is allosteric. This observation leads to
the speculation that most proteins may have more than one function.
The identification of Mb in numerous bacteria (27) and the human brain (28), together with the recent appreciation of the importance of small-molecule chemistry, such as NO and CO, in biology, suggest that Mb evolved in conjunction with life's ability to control the most basic oxygen chemistry. Most of our metabolic energy flows through a more complex heme protein, cytochrome c oxidase, whereas more than half of all drugs are covalently modified by a group of heme proteins, known collectively as P-450s. Neuroglobin, a myoglobin that has been found in the brain in micromolar quantities (28), has ligand migration and binding kinetics that are very similar to that of ordinary Mb (29).
The biased sample selected here in this commentary already shows that Mb can indeed be considered the hydrogen atom of biology. Mb studies have yielded concepts such as the hierarchical energy landscape and the importance of the Kramers theory that most likely are valid for many other proteins. The large number of substates and their organization and importance for function make Mb a paradigm of complexity. Although Mb may be the best-studied protein, the work is not over yet. Many fundamental problems remain to be solved, such as the detailed connections among structure, energy landscape, dynamics, and function and the role of protein–protein interactions. The work of Bourgeois et al. adds valuable information about protein conformational motions during function, but it is a beginning, not an end.
See companion article on page 8704.
References
- 1.Bourgeois, D., Vallone, B., Schotte, F., Arcovito, A., Miele, A. E., Sciara, G., Wulff, M., Anfinrud, P. & Brunori, M. (2003) Proc. Natl. Acad. Sci. USA 100, 8704–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srajer, V., Ren, Z., Teng, T.-Y., Schmidt, M., Ursby, T., Bourgeois, D., Pradervand, C., Schildkamp, W., Wulff, M. & Moffat, K. (2001) Biochemistry 40, 13802–13815. [DOI] [PubMed] [Google Scholar]
- 3.Kendrew, J. C., Dickerson, R. E., Strandberg, B. E., Hart, R. G., Davies, D., Phillips, D. C. & Shore, V. C. (1960) Nature 185, 422–427. [DOI] [PubMed] [Google Scholar]
- 4.Perutz, M. F. (1979) Annu. Rev. Biochem. 48, 327–386. [DOI] [PubMed] [Google Scholar]
- 5.Tilton, R. F., Kuntz, I. D., Jr., & Petsko, G. A. (1984) Biochemistry 23, 2849–2857. [DOI] [PubMed] [Google Scholar]
- 6.Austin, R. H., Beeson, K. W., Eisenstein, L., Frauenfelder, H. & Gunsalus, I. C. (1975) Biochemistry 14, 5355–5373. [DOI] [PubMed] [Google Scholar]
- 7.Ansari, A., Jones, C. M., Henry, E. R., Hofrichter, J. & Eaton, W. A. (1994) Biochemistry 33, 5128–5145. [DOI] [PubMed] [Google Scholar]
- 8.Schlichting, I., Berendzen, J., Phillips, G. N., Jr., & Sweet, R. M. (1994) Nature 371, 808–812. [DOI] [PubMed] [Google Scholar]
- 9.McMahon, B. H., Stojkovic, B. P., Hay, P. J., Martin, R. L. & Garcia, A. E. (2000) J. Chem. Phys. 113, 6831–6850. [Google Scholar]
- 10.Ansari, A., Berendzen, J., Bowne, S. F., Frauenfelder, H., Iben, I. E. T., Sauke, T. B., Shyamsunder, E. & Young, R. D. (1985) Proc. Natl. Acad. Sci. USA 82, 5000–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dadusc, G., Ogilvie, J. P., Schulenberg, P., Marvet, U. & Miller, R. J. D. (2001) Proc. Natl. Acad. Sci. USA 98, 6110–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson, T. A., Lim, M. & Anfinrud, P. A. (1994) Chem. Phys. 180, 131–140. [Google Scholar]
- 13.Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. (1991) Science 254, 1598–1603. [DOI] [PubMed] [Google Scholar]
- 14.Fenimore, P. W., Frauenfelder, H., McMahon, B. H. & Parak, F. G. (2002) Proc. Natl. Acad. Sci. USA 99, 16047–16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urayama, P., Phillips, G. N., Jr., & Gruner, S. M. (2002) Structure (London) 10, 51–60. [DOI] [PubMed] [Google Scholar]
- 16.Alben, J. O., Beece, D., Bowne, S. F., Eisenstein, L., Frauenfelder, H., Good, D., Marden, M. C., Moh, P. P., Reinisch, L., Reynolds, A. H. & Yue, K. T. (1980) Phys. Rev. Lett. 44, 1157–1160. [Google Scholar]
- 17.Frauenfelder, H. & Wolynes, P. G. (1985) Science 229, 337–344. [DOI] [PubMed] [Google Scholar]
- 18.Gouterman, M. (1959) J. Chem. Phys. 30, 1139–1161. [Google Scholar]
- 19.Kirchner, R. F. & Loew, G. H. (1977) J. Am. Chem. Soc. 99, 4639–4647. [DOI] [PubMed] [Google Scholar]
- 20.Edwards, W. D., Weiner, B. & Zerner, M. C. (1986) J. Am. Chem. Soc. 108, 2196–2204. [DOI] [PubMed] [Google Scholar]
- 21.Thorn Leeson, D., Wiersma, D. A., Fritsch, K. & Friedrich, J. (1997) J. Phys. Chem. B 101, 6331–6340. [Google Scholar]
- 22.Petrich, J. W., Poyart, C. & Martin, J. L. (1988) Biochemistry 27, 4049–4060. [DOI] [PubMed] [Google Scholar]
- 23.Varadarajan, R., Lambright, D. J. & Boxer, S. J. (1989) Biochemistry 28, 3771–3781. [DOI] [PubMed] [Google Scholar]
- 24.Park, E. S., Andrews, S. S., Hu, R. B. & Boxer, S. G. (1999) J. Phys. Chem. B 103, 9813–9817. [Google Scholar]
- 25.Frauenfelder, H., McMahon, B. H., Austin, R. H., Chu, K. & Groves, J. T. (2001) Proc. Natl. Acad. Sci. USA 98, 2370–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourassa, J. L., Ives, E. P., Marqueling, A. L., Shimanovich, R. & Groves, J. T. (2001) J. Am. Chem. Soc. 123, 5142–5143. [DOI] [PubMed] [Google Scholar]
- 27.Hou, S., Freitas, T., Larsen, R. W., Piatibratov, M., Sivozhelezov, V., Yamamoto, A., Meleshkevitch, E. A., Zimmer, M., Ordal, G. W. & Alam, M. (2001) Proc. Natl. Acad. Sci. USA 98, 9353–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burmester, T., Weich, B., Reinhardt, S. & Hankeln, T. (2000) Nature 407, 520–523. [DOI] [PubMed] [Google Scholar]
- 29.Kriegl, J. M., Bhattacharyya, A. J., Nienhaus, K., Deng, P. C., Minkov, O. & Nienhaus, G. U. (2002) Proc. Natl. Acad. Sci. USA 99, 7992–7997. [DOI] [PMC free article] [PubMed] [Google Scholar]