Abstract
Forebrain cholinergic neurons play important roles as striatal local circuit neurons and basal telencephalic projection neurons. The genetic mechanisms that control development of these neurons suggest that most of them are derived from the basal telencephalon where Lhx8, a LIM-homeobox gene, is expressed. Here we report that mice with a null mutation of Lhx8 are deficient in the development of forebrain cholinergic neurons. Lhx8 mutants lack the nucleus basalis, a major source of the cholinergic input to the cerebral cortex. In addition, the number of cholinergic neurons is reduced in several other areas of the subcortical forebrain in Lhx8 mutants, including the caudate-putamen, medial septal nucleus, nucleus of the diagonal band, and magnocellular preoptic nucleus. Although cholinergic neurons are not formed, initial steps in their specification appear to be preserved, as indicated by a presence of cells expressing a truncated Lhx8 mRNA and mRNA of the homeobox gene Gbx1. These results provide genetic evidence supporting an important role for Lhx8 in development of cholinergic neurons in the forebrain.
The mammalian forebrain contains two general types of cholinergic neurons. One group is composed of local circuit neurons, such as the cholinergic interneurons of the striatum (1). These neurons play an important role in the regulation of locomotor behavior through their modulation of γ-aminobutyric acid (GABA)ergic projection neurons in the striatum (2). The other group consists of projection neurons whose cell bodies can be found in a series of nuclei in the subcortical telencephalon, including the medial septum (also designated as Ch1 cholinergic group by Mesulam et al., ref. 3), the vertical and horizontal limbs of the nucleus of the diagonal band (designated as Ch2 and Ch3, respectively; ref. 3), and the basal magnocellular complex (designated as Ch4; ref. 3), which comprises cholinergic neurons scattered through the magnocellular preoptic nucleus, substantia innominata, ventral pallidum, and nucleus basalis. The axonal projections of these neurons provide the cerebral cortex and hippocampus with their principal cholinergic input (3–6), which has a critical role in cognitive functions (for reviews, see refs. 7 and 8). Accordingly, abnormalities of the cholinergic projection neurons in the basal forebrain are implicated in neurodegenerative disorders such as Alzheimer's disease (9, 10).
The genetic and developmental mechanisms that control the formation of forebrain cholinergic neurons are just beginning to be elucidated. The vast majority of forebrain cholinergic neurons derive from a region of the subcortical telencephalon that expresses the Nkx2-1 homeobox gene (11, 12). This region contains different progenitor zones, including the medial ganglionic eminence (MGE), anterior entopeduncular area and preoptic area (POa) (13). It has been proposed that these progenitor domains contribute projection neurons to the globus pallidus, ventral pallidum, nucleus of the diagonal band, and parts of the septum and amygdala, regions that generically are known as the basal telencephalon. In addition, a substantial fraction of striatal, cortical, and hippocampal interneurons originates from these progenitor zones and migrate tangentially to reach their final destinations (reviewed in ref. 14).
Nkx2-1 appears to specify the development of the basal telencephalon by repressing more dorsally expressed transcription factors (e.g., Pax6) and by positively regulating the expression of other transcription factors such as the LIM-homeodomain proteins Lhx6 and Lhx8 (11) [Lhx8 is also known as L3 (15) and Lhx7 (16)]. At birth, Nkx2-1 mutants lack all cholinergic neurons in the subpallium, including striatal interneurons and basal forebrain projection neurons, and such defects are associated with a complete loss of Lhx6 and Lhx8 expression in the developing telencephalon (11, 12). Interestingly, although Lhx6 expression appears to be associated with GABAergic neurons, Lhx8 expression is coupled to cholinergic neurons (12, 17).
To establish the role of Lhx8 in regulating the development of cholinergic neurons in the telencephalon, we have analyzed the development of the basal telencephalon in mutant mice that lack Lhx8 function (18). Our results demonstrate that Lhx8 is required for the formation of specific subsets of forebrain cholinergic neurons.
Materials and Methods
Animals and Tissue Preparation. As described in ref. 18, the Lhx8 mutant mice were generated by deleting exons 4–6 of the Lhx8 gene that encode part of the first LIM domain, the entire second LIM domain, and part of the homeodomain of the Lhx8 protein. About 60% of the Lhx8 homozygous mutants show a complete cleft of the secondary palate and die soon after birth, whereas the remaining 40% appear healthy and have a normal lifespan. For analysis, mouse embryos were isolated from pregnant females at various stages of development. Noon of the day when a vaginal plug was identified was designated as embryonic day (E) 0.5. The embryos were fixed by immersion in 4% paraformaldehyde/0.1 M phosphate buffer (pH 7.4) overnight at 4°C. After washes in PBS, the embryos were either dehydrated through a graded series of ethanol and embedded in paraffin, or cryoprotected in 30% sucrose and frozen in OCT compound (Sakura, Torrance, CA). To fix newborn (P0) pups or 6- to 7-week-old mice, animals were anesthetized and perfused transcardially with 4% paraformaldehyde/0.1 M phosphate buffer (pH 7.4). The brains were dissected out, postfixed in the same fixative overnight at 4°C, and processed with the same procedures used for the embryos.
In Situ Hybridization. In situ hybridization was performed on 5-μm paraffin sections or 10-μm frozen sections as described (12, 19). [33P]- or [35S]UTP-labeled antisense probes were synthesized by in vitro transcription using vectors containing cDNAs of Gad67, Gbx2, Islet1 (Isl1), Lhx6, and Lhx8 (12, 18, 20). For analysis of Gbx1 expression, 20-μm frozen sections were hybridized with digoxigenin-labeled probes synthesized from a vector containing the Gbx1 coding sequence (M. Frohman, personal communication) as described (21).
Immunohistochemistry. Immunostaining was performed on 40-μm free-floating frozen sections. For immunoperoxidase labeling, sections were treated with ice-cold PBS containing 10% methanol and 0.1% hydrogen peroxide to inactivate endogenous peroxidase. Sections were incubated in PBS containing 2% BSA, 5% normal serum of the species from which the secondary antibody was derived, and 0.3% (for sections from P0 pups) or 0.4% (for sections from adult mice) Triton X-100. The sections were then incubated with primary antibodies diluted with PBS containing 2% normal serum and 0.3% (P0 sections) or 0.4% (adult sections) Triton X-100 for 40–60 h at 4°C. The antibodies used and their dilutions are as follows: goat anti-choline acetyltransferase (ChAT) (Chemicon), 1:250; rat anti-somatostatin (Chemicon), 1:100; rabbit anti-calretinin (Chemicon), 1:2,500. After primary antibody incubation, sections were washed in PBS/0.3% (for P0 sections) or 0.4% (for adult sections) Triton X-100, incubated with biotinylated secondary antibodies (KPL, Gaithersburg, MD or Vector Laboratories), and processed with a Vectastain ABC kit (Vector Laboratories). The sections were developed by using AEC as a chromogen (Zymed), counter-stained with hematoxylin, and mounted with Aqua PolyMount (Polysciences). For labeling of parvalbumin-positive neurons, sections were stained with a mouse anti-parvalbumin antibody (Sigma, 1:2,000) by using a histomouse kit (Zymed). TrkA-positive neurons were labeled with a rabbit anti-TrkA antibody (generously provided by L. Reichardt, University of California, San Francisco), as described (11).
Results
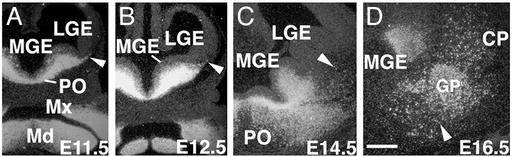
Expression of Lhx8 mRNA in the Developing Basal Forebrain. To provide a context for the phenotypic characterization of Lhx8 mutants, we analyzed the expression of Lhx8 mRNA in the telencephalon from E11.5 to E16.5 (Fig. 1). In agreement with previous reports (11–17), Lhx8 expression between E11.5 and E12.5 is largely restricted to the basal telencephalon, including the MGE, and POa (Fig. 1 A and B). Lhx8 mRNA was found in scattered cells in the ventricular zone and in most cells throughout the subventricular and mantle zones. In addition, a thin layer of expression was found to extend into the mantle zone of the lateral ganglionic eminence. By E14.5, strong expression remained throughout the subventricular zone of the basal telencephalon, particularly in the POa (Fig. 1C). Abundant cells were also found in the basal telencephalic mantle zone, and a few Lhx8-positive cells were present in the striatum (Fig. 1C). At E16.5, in addition to its expression in the basal telencephalic subventricular zone, strong expression was found in the globus pallidus as well as in scattered cells throughout the POa, ventral pallidum, and striatum (Fig. 1D). These observations, together with results from the previous studies (11, 12, 17), suggest that Lhx8 is broadly expressed in the developing basal telencephalon and therefore could potentially regulate the development of both GABAergic and cholinergic neurons derived from this region.
Fig. 1.
In situ analysis of Lhx8 mRNA expression in the developing mouse basal forebrain. (A and B) Lhx8 expression in the MGE and POa (PO) of E11.5 (A) and E12.5 (B) mouse embryos. Arrowheads point at labeled cells migrating from the MGE into the lateral ganglionic eminence (LGE). (C) Detection of Lhx8 mRNA in a dispersed population of cells in the striatum (pointed by an arrowhead) in addition to the MGE and POa at E14.5. (D) Expression of Lhx8 mRNA in the caudate-putamen (CP), globus pallidus (GP), and in cells dispersed ventral-medial to the globus pallidus (pointed by an arrowhead) at E16.5. Mx and Md, the maxillary and mandibular processes of the first branchial arch. (Scale bar in D represents 250 μm for all panels.)
Early Molecular Defects in the Development of the Basal Telencephalon in Lhx8 Mutants. To begin to assess the role of Lhx8 in the generation of specific neuronal populations in the basal telencephalon, we analyzed the expression of markers implicated in the development of cells derived from the basal telencephalon at E13.5. First, we studied expression from the mutant Lhx8 locus because the mutation did not delete the promoter and some of the exonic sequences of Lhx8 (18). Truncated Lhx8 transcripts were found in roughly a normal pattern in Lhx8 mutants, although their level of expression was reduced (Fig. 2 A and B). This observation suggests that the deletion did not remove an essential enhancer/silencer domain from the Lhx8 locus and that at least some of the cells specified to express Lhx8 are produced in the absence of Lhx8.
Fig. 2.
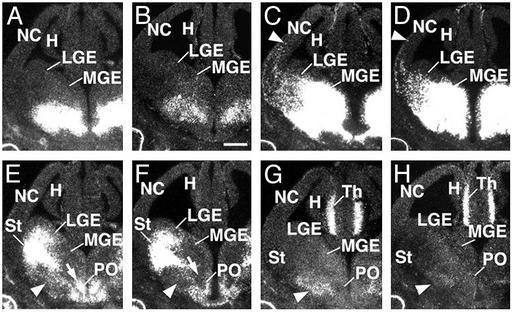
Embryonic defects in the organization of the basal forebrain of Lhx8 mutants. Adjacent coronal sections through the telencephalon of E13.5 embryos showing expression of Lhx8 (A and B), Lhx6 (C and D), Isl1 (E and F), and Gbx2 (G and H) mRNA in wild-type controls (A, C, E, and G) and Lhx8 mutants (B, D, F, and H). (C and D) Arrowheads point at labeled cells migrating from the basal telencephalon to the cortex. (E–H) Arrows and arrowheads indicate the proliferative and mantle regions of the preoptic area, respectively. Note that the expression of Isl1 and Gbx2 is reduced in these regions in Lhx8 mutants. H, hippocampus; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; NC, neocortex; PO, preoptic area; St, striatum; Th, thalamus. (Scale bar in B represents 500 μm for all panels.)
The initial expression of Lhx8 in the basal telencephalon is very similar to that of Lhx6 (11, 16) (Figs. 1 and 2 A and C). Despite this, loss of functional Lhx8 did not affect Lhx6 expression at E13.5. As in control mice, Lhx6 mRNA was expressed in cells throughout the developing basal telencephalon as well as in cells tangentially migrating to the cerebral cortex in Lhx8 mutants (Fig. 2 C and D). This observation suggests that Lhx8 is not required for the development of tangentially migrating GABAergic neurons. Accordingly, analysis of expression of Gad67 mRNA, a marker specific for GABAergic neurons, revealed no perceptible changes in Lhx8 mutants (data not shown), reinforcing the notion that Lhx8 does not play a major role in the early development of GABAergic neurons derived from the basal telencephalon.
We next assessed the expression of Isl1, a LIM-homeobox gene that regulates the development of cholinergic neurons in the spinal cord (22) and that is expressed in telencephalic cholinergic neurons at late embryonic stages (23). In addition to its prominent expression in the developing striatum, Isl1 mRNA was found in progenitor cells of the POa and in adjacent mantle areas (Fig. 2E). In Lhx8 mutants, expression of Isl1 was roughly normal in the striatal mantle, but it was reduced in the POa and in the superficial mantle zone of the MGE (i.e., the developing ventral pallidum and substantia innominata; Fig. 2F). The developing ventral pallidum also expresses Gbx1, a homeobox gene found in basal telencephalic cholinergic neurons at late embryonic stages (17). Expression of Gbx1, however, was roughly normal in Lhx8 mutants (data not shown). In contrast, expression of Gbx2 (20), a homeobox gene closely related to Gbx1, was reduced in Lhx8 mutants (Fig. 2 G and H). Thus, the reduction in Isl1 expression in the POa suggests a possible defect in the development of cholinergic neurons, whereas the normal expression of Lhx6 and Gad67 implies that GABAergic neurons are not affected by the Lhx8 mutation. These hypotheses were examined by analyzing Lhx8 mutants at P0 and in adulthood.
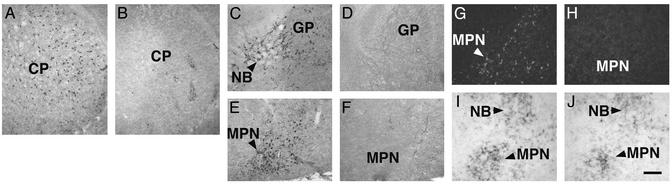
Reduction of Cholinergic Neurons in the Telencephalon of P0 Lhx8 Mutants. To establish whether Lhx8 regulates the development of cholinergic neurons in the telencephalon, we studied the expression of ChAT, a key enzyme in the synthesis of acetylcholine, at P0, when it is first detectable in telencephalic cholinergic neurons. Compared with wild-type controls, Lhx8 mutants contained far less ChAT-positive (ChAT+) cells in the striatum (Fig. 3 A and B) and in the basal magnocellular complex (Ch4), including the nucleus basalis (Fig. 3 C and D) and the magnocellular preoptic nucleus (Fig. 3 E and F). In addition, the number of cells positive for the nerve growth factor receptor TrkA, another marker of the basal forebrain cholinergic neurons, was also diminished in Lhx8 mutants (Fig. 3 G and H, and data not shown). In contrast, the expression of Gbx1 was roughly normal in the basal telencephalon of Lhx8 mutants (Fig. 3 I and J), suggesting that basal telencephalic cholinergic neurons are born but fail to differentiate properly in the mutants. To determine whether the reduction in expression of the cholinergic markers reflects a developmental delay or a definitive loss of cholinergic neurons, we next studied adult Lhx8 mutants.
Fig. 3.
Defects in development of telencephalic cholinergic neurons in P0 Lhx8 mutants (B, D, F, H, and J) as compared with wild-type controls (A, C, E, G, and I). (A–F) Anti-ChAT staining of the caudate-putamen (A and B), nucleus basalis (C and D), and magnocellular preoptic nucleus (E and F). (G and H) Anti-TrkA staining of the magnocellular preoptic nucleus. Note that the number of labeled cells is reduced in Lhx8 mutants. (I and J) In situ analysis showing that the expression of Gbx1 mRNA was maintained in the nucleus basalis and magnocellular preoptic nucleus in Lhx8 mutants. CP, caudate-putamen; GP, globus pallidus; MPN, magnocellular preoptic nucleus; NB, nucleus basalis. (Scale bar in J represents 180 μm for all panels.)
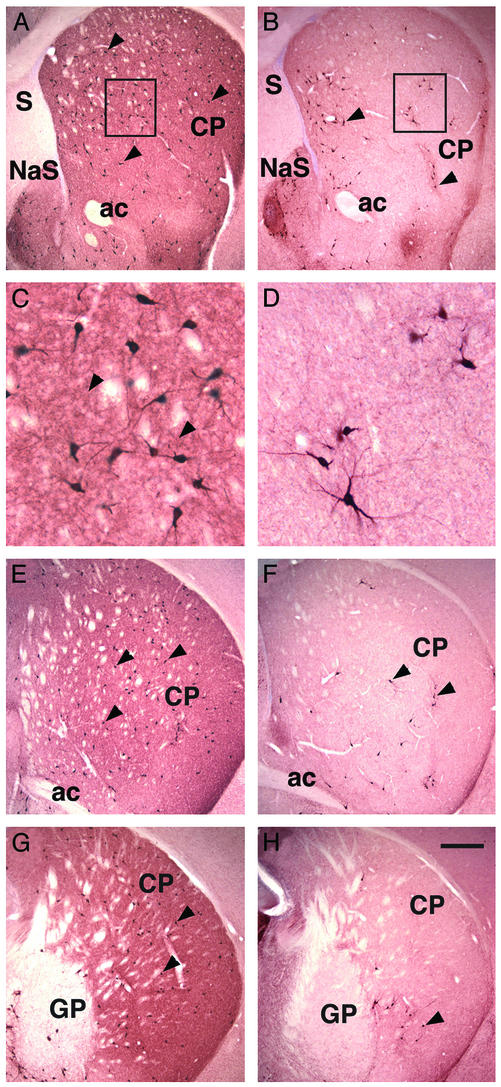
Reduction of Cholinergic Interneurons in the Telencephalon of Adult Lhx8 Mutants. Immunohistochemical analysis of the distribution of ChAT+ neurons in the brain of adult mice revealed that the number of cholinergic interneurons was reduced severely throughout the rostral-caudal axis of the caudate-putamen in Lhx8 mutants (Fig. 4 B, D, F, and H) as compared with wild-type controls (Fig. 4 A, C, E, and G). For quantification, the neurons labeled in the caudate-putamen were counted on three serial sections (with a distance of 200 μm from each other) in a region anterior to the crossover of the anterior commissure. Lhx8 mutants were found to contain only ≈25% of ChAT+ cells that were normally present in the controls (average number of neurons ± standard deviation: controls: 843 ± 151, n = 4; Lhx8 mutants: 218 ± 112, n = 4; t test, P = 0.0008). Consistent with the reduction in number of ChAT+ cell bodies, the density of the ChAT+ neuropil formed by the dendrites and axons of the cholinergic interneurons in the dorsal striatum was also reduced in the mutants (Fig. 4). In contrast to the dorsal striatum, the number of ChAT+ cells in the ventral striatum, including the nucleus accumbens and the olfactory tubercle, seemed not to be affected in Lhx8 mutants (Fig. 4 A and B and data not shown).
Fig. 4.
Anti-ChAT staining showing a reduction in the number of cholinergic interneurons in the caudate-putamen of adult Lhx8 mutants (B, D, F, and H)as compared with wild-type controls (A, C, E, and G). (A and B) Sections from a rostral level. (C and D) Enlargement of the outlined areas in A and B, respectively. (E and F) Sections from a middle level. (G and H) Sections from a caudal level. Arrowheads in A, B, and E–H indicate the somas of ChAT+ neurons. Arrowheads in C point at ChAT+ neuropil, which is reduced in Lhx8 mutants. ac, anterior commissure; CP, caudate-putamen; GP, globus pallidus; NaS, nucleus accumbens, shell; S, septum. (Scale bar in H represents 300 μm for A, B, and E–H, and 60 μm for C and D.)
In addition to cholinergic interneurons, the striatum contains three types of GABAergic interneurons (24) that can be distinguished by their expression of different markers such as parvalbumin, somatostatin and calretinin, respectively. Staining for these markers revealed that all these three types of GABAergic striatal interneurons were normally present in Lhx8 mutants (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). Thus, loss of Lhx8 causes a permanent deficit of cholinergic interneurons in the dorsal striatum, but does not affect the development of GABAergic interneurons.
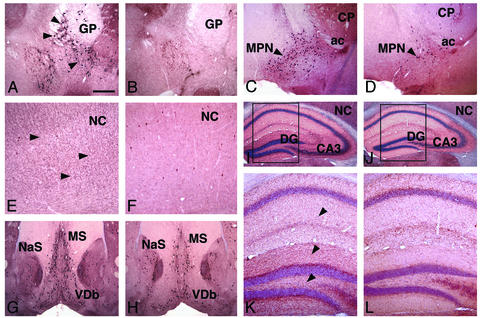
Reduction of Cholinergic Projection Neurons in the Telencephalon of Adult Lhx8 Mutants. ChAT immunostaining also revealed a severe reduction in the number of cholinergic projection neurons in the basal telencephalon of adult Lhx8 mutants (Fig. 5). The nucleus basalis (Ch4) was almost entirely devoid of ChAT+ neurons in Lhx8 mutants (Fig. 5 A and B). The number of ChAT+ neurons in the magnocellular preoptic nucleus (Ch4) was also drastically reduced in the mutants as compared with wild-type controls (Fig. 5 C and D). Because these cholinergic neurons provide major cholinergic input to the cortex, consequentially the cholinergic innervation of the cerebral cortex was diminished in the mutants (Fig. 5 E and F). The cholinergic input to the hippocampus is largely derived from the septum (Ch1) and the nucleus of the diagonal band (Ch2 and 3). In Lhx8 mutants, the number of ChAT+ neurons in some of those regions was also reduced as compared with that in wild-type controls (Fig. 5 G and H). By counting ChAT+ cells on three selected sections that represent the anterior, middle, and posterior levels of the septum, it was found that there was a 56% reduction in the number of these neurons in the medial septal nucleus (Ch1) and the vertical limb of the nucleus of the diagonal band (Ch2) (average number of neurons ± standard deviation: controls: 347 ± 92, n = 3; Lhx8 mutants, 151 ± 72, n = 4; t test, P = 0.04). Consistent with such a reduction in ChAT+ neurons, the cholinergic projection to the hippocampus was significantly reduced in the mutants (Fig. 5 I–L).
Fig. 5.
Anti-ChAT staining showing a reduction in number of the telencephalic cholinergic projection neurons in adult Lhx8 mutants (B, D, F, H, J, and L) as compared with wild-type controls (A, C, E, G, I, and K). (A and B) Absence of ChAT+ neurons in the nucleus basalis. Arrowheads in A point at ChAT+ neurons of the nucleus basalis in the control, which are missing in the mutant (B). (C and D) Reduction in number of ChAT+ neurons in the magnocellular preoptic nucleus (MPN). (E and F) Reduction of ChAT+ nerve fibers in the neocortex (NC). Arrowheads in E point at ChAT+ fibers in the control, which are largely missing in the mutant (F). (G and H) Reduction in number of ChAT+ neurons in the medial septal nucleus (MS) and the vertical limb of the nucleus of the diagonal band (VDb). (I–L) Reduction of ChAT+ neuropil in the hippocampus. K and L are enlargement of the areas outlined in I and J, respectively. Arrowheads in K point at ChAT+ fibers in the controls, which are reduced in the mutants. ac, anterior commissure; CA3, CA3 region of the Ammon's horn; CP, caudate-putamen; DG, dentate gyrus; GP, globus pallidus; NaS, nucleus accumbens, shell. (Scale bar in A represents 500 μm for A–D and G–J, and 200 μm for E, F, K, and L.)
Discussion
Herein we demonstrate that loss of Lhx8 function, via a targeted mutation in the mouse, blocks the formation of most cholinergic neurons in the mouse forebrain (see Fig. 6 for summary). Thus, LIM-homeobox genes have a central role in regulating cholinergic development throughout the central nervous system, as Isl1, Lhx1, Lhx3, and Lhx4 have been shown to function in controlling motor neuron development in the hindbrain and spinal cord (22, 25, 26). Given that spinal cord cholinergic neurons require multiple LIM-homeobox genes for their development, and that loss of Lhx8 does not prevent the development of all telencephalic cholinergic neurons, it is likely that Lhx8 is not the only LIM-homeobox gene that is required in generating telencephalic cholinergic neurons. Likely candidates include Lhx6 and Isl1, because they are also expressed in the basal telencephalon (11, 12, 16, 23). Presently, no information is available on Lhx6 function and, unfortunately, Isl1 mutants die before their ability to generate telencephalic cholinergic neurons can be established (22). On the other hand, it is likely that Isl1 cooperates with Lhx8 in the development of telencephalic cholinergic neurons. Isl1 is expressed in these neurons at birth (23). In addition, Isl1 is known to form complexes with products of other LIM-homeobox genes (e.g., Lhx3) that are implicated in controlling whether a differentiating progenitor cell in the spinal cord generates a motor neuron (cholinergic) or a V2 interneuron (GABAergic) (27). It remains to be determined whether Isl1 can complex with Lhx8.
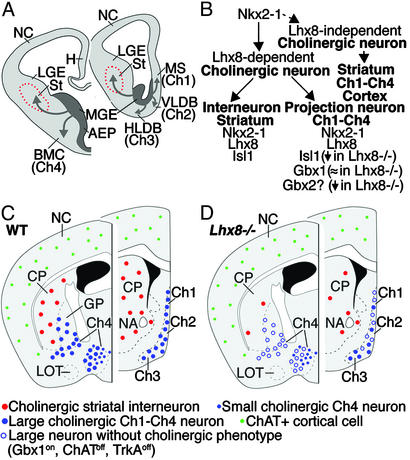
Fig. 6.
Schemas illustrating the origin and migration of cholinergic neurons derived from the basal telencephalon and the genes that are involved in their specification and differentiation. (A) The schematic of transversal hemisections through the embryonic telencephalon summarizes the expression of Lhx8. Lhx8+ neurons differentiate as interneurons in the striatum or as projection neurons in the basal telencephalon (Ch1–Ch4 cholinergic groups). (B) Model describing some of the genes that regulate production of cholinergic neurons in the telencephalon. Nkx2-1 regulates production of cells that express Lhx8 (11), which is necessary for the differentiation of most cholinergic neurons in the basal telencephalon (this study). Nkx2-1 is also required for the specification of GABAergic neurons (11), and some cholinergic neurons in the telencephalon develop in the absence of Lhx8 function (dashed line). In the striatum, cholinergic interneurons express Nkx2-1, Lhx8, and Isl1. In addition to Nkx2-1 and Lhx8, cholinergic projection neurons in the basal telencephalon express Isl1 and Gbx1. Whereas the Gbx1 expression is maintained, the Isl1 expression is reduced in Lhx8 mutants. The expression of Gbx2 is also reduced in the mantle zone of the MGE in Lhx8 mutants. Although Gbx2 is not expressed in the mature cholinergic neurons (17), its expression in the MGE suggests that it may be expressed in the progenitors of the cholinergic neurons. (C and D) Schemas illustrating the distribution of ChAT+ cells in the adult telencephalon of control and Lhx8 mutant mice. Lhx8 mutation preferentially blocks the differentiation of cholinergic interneurons in the striatum and large projection neurons in the basal forebrain. In Lhx8 mutants, small ChAT+ cells are still present in the cortex and basal forebrain. Expression of Gbx1 in the basal forebrain of Lhx8 mutants suggests that cholinergic cells are born in Lhx8 mutants, but they fail to differentiate properly (i.e., lack ChAT and TrkA expression). AEP, anterior entopeduncular region; BMC, basal magnocellular complex; Ch1–Ch4, Ch1–Ch4 cholinergic cell groups; CP, caudateputamen; GP, globus pallidus; H, hippocampus; HLDB, horizontal limb of diagonal band; LGE, lateral ganglionic eminence; LOT, lateral olfactory tract; MGE, medial ganglionic eminence; MS, medial septal nucleus; NA, nucleus accumbens; NC, neocortex; St, striatum; VLDB, vertical limb of diagonal band.
Lhx8 is required to generate multiple types of telencephalic cholinergic neurons, including hippocampal and cortical projection neurons in the septum and nucleus basalis, respectively, and local circuit neurons in the striatum. Thus, Lhx8 is proximal in the genetic hierarchy that controls telencephalic cholinergic development. Although Lhx8 is required to generate most of the cholinergic neurons in this region, at least some subsets are produced in its absence. For example, the cholinergic interneurons in the ventral striatum (e.g., the nucleus accumbens) (Fig. 4) and the parvocellular component of the cholinergic magnocellular preoptic nucleus (Fig. 5) are still present in Lhx8 mutants. Future studies are needed to identify the genes that specify subtypes of cholinergic projection and local circuit neurons.
Lhx8 is expressed in progenitor cells in the subventricular zone, and is maintained in postmitotic neurons (Fig. 1) (11, 12, 17). Thus, Lhx8 may function at multiple developmental stages and perhaps in mature cholinergic neurons. Lack of Lhx8 protein does not prevent the generation of cells that express Lhx8 mRNA (Fig. 2), although these cells fail to differentiate into cholinergic neurons. The fact that expression of Gbx1, which is normally found in cholinergic neurons in the nucleus basalis (17), is maintained in Lhx8 mutants reinforces the notion that Lhx8 is required for the acquisition of the cholinergic phenotype (e.g., ChAT and TrkA expression) rather than for the initial specification of these neurons.
Insights into some of the genetic pathways that regulate telencephalic cholinergic development are now available. Early steps in the program begin with specification of the anterior telencephalon at neural plate stages; this region includes the anlage of the basal telencephalon (30), and appears to require Six3 and Vax1 (31, 32). Sonic hedgehog and nodal type transforming growth factor (TGF)-β signaling are important for induction of Nkx2-1 expression in the telencephalon (33), which begins soon after neural tube closure and defines the basal telencephalic anlage (34, 35). Nkx2–1 is required for telencephalic expression of Sonic hedgehog, Lhx6, and Lhx8 (11, 12). Lhx8 controls cholinergic development perhaps through regulating Isl1 and Gbx2 expression (Fig. 2). Dlx1/2 and Mash1, though not directly regulating Lhx8, participate in controlling the number of cholinergic neurons that are formed in the telencephalon by regulating the differentiation of late and early-born neurons, respectively (12). Because cholinergic neurons are among the earliest born neurons in the telencephalon (36, 37), loss of Mash1 affects the differentiation of cholinergic neurons more severely than mutation of Dlx1/2. In addition, signaling molecules of the bone morphogenetic protein family also regulate the differentiation of the telencephalic cholinergic neurons (38). Finally, later steps in cholinergic differentiation and survival are regulated by galanin (39, 40) and nerve growth factor (41, 42). Future studies are required to establish whether Lhx8 controls the maturation and function of telencephalic cholinergic neurons, as these cells have critical roles in regulating cognitive and motor functions, and they are among the first neurons to degenerate in Alzheimer's disease.
Supplementary Material
Acknowledgments
We are grateful to L. Reichardt (University of California, San Francisco) for providing the Anti-TrkA antibody. This work was supported in part by grants from Nina Ireland, National Institute on Drug Abuse Grant R01DA12462, and National Institute of Mental Health Grant K02MH01046-01 (to J.L.R.R.). O.M. was a National Alliance for Research on Schizophrenia and Depression Young Investigator and a Medical Investigation of Neurodevelopmental Disorders Institute Scholar. N.F. is the recipient of a Predoctoral Fellowship from the Ministry of Education and Culture of Spain.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MGE, medial ganglionic eminence; P0, newborn; GABA, γ-aminobutyric acid; POa, preoptic area; En, embryonic day n; ChAT, choline acetyltransferase.
References
- 1.Woolf, N. J. (1991) Prog. Neurobiol. 37, 475-524. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko, S., Hikida, T., Watanabe, D., Ichinose, H., Nagatsu, T., Kreitman, R. J., Pastan, I. & Nakanishi, S. (2000) Science 289, 633-637. [DOI] [PubMed] [Google Scholar]
- 3.Mesulam, M. M., Mufson, E. J., Wainer, B. H. & Levey, A. I. (1983) Neuroscience 10, 1185-1201. [DOI] [PubMed] [Google Scholar]
- 4.Bigl, V., Woolf, N. J. & Butcher, L. L. (1982) Brain Res. Bull. 8, 727-749. [DOI] [PubMed] [Google Scholar]
- 5.Woolf, N. J., Eckenstein, F. & Butcher, L. L. (1983) Neurosci. Lett. 40, 93-98. [DOI] [PubMed] [Google Scholar]
- 6.Woolf, N. J., Eckenstein, F. & Butcher, L. L. (1984) Brain Res. Bull. 13, 751-784. [DOI] [PubMed] [Google Scholar]
- 7.Everitt, B. J. & Robbins, T. W. (1997) Annu. Rev. Psychol. 48, 649-684. [DOI] [PubMed] [Google Scholar]
- 8.Baxter, M. G. & Chiba, A. A. (1999) Curr. Opin. Neurobiol. 9, 178-183. [DOI] [PubMed] [Google Scholar]
- 9.Whitehouse, P. J., Price, D. L., Clark, A. W., Coyle, J. T. & DeLong, M. R. (1981) Ann. Neurol. 10, 122-126. [DOI] [PubMed] [Google Scholar]
- 10.Whitehouse, P. J., Price, D. L., Struble, R. G., Clark, A. W., Coyle, J. T. & Delon, M. R. (1982) Science 215, 1237-1239. [DOI] [PubMed] [Google Scholar]
- 11.Sussel, L., Marín, O., Kimura, S. & Rubenstein, J. L. R. (1999) Development (Cambridge, U.K.) 126, 3359-3370. [DOI] [PubMed] [Google Scholar]
- 12.Marín, O., Anderson, S. A. & Rubenstein, J. L. R. (2000) J. Neurosci. 20, 6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marín, O. & Rubenstein, J. L. R. (2002) in Mouse Development: Patterning, Morphogenesis, and Organogenesis, eds. Rossant, J. & Tam, P. P. L. (Academic, San Diego), pp. 75-106.
- 14.Marín, O. & Rubenstein, J. L. R. (2001) Nat. Rev. Neurosci. 2, 780-790. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, K., Tanaka, T., Furuyama, T., Kashihara, Y., Mori, T., Ishii, N., Kitanaka, J., Takemura, M., Tohyama, M. & Wanaka, A. (1996) Neurosci. Lett. 204, 113-116. [DOI] [PubMed] [Google Scholar]
- 16.Grigoriou, M., Tucker, A. S., Sharpe, P. T. & Pachnis, V. (1998) Development (Cambridge, U.K.) 125, 2063-2074. [DOI] [PubMed] [Google Scholar]
- 17.Asbreuk, C. H., van Schaick, H. S., Cox, J. J., Kromkamp, M., Smidt, M. P. & Burbach, J. P. (2002) Neuroscience 109, 287-298. [DOI] [PubMed] [Google Scholar]
- 18.Zhao, Y., Guo, Y. J., Tomac, A. C., Taylor, N. R., Grinberg, A., Lee, E. J., Huang, S. & Westphal, H. (1999) Proc. Natl. Acad. Sci. USA 96, 15002-15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson, G. W., Wray, S. & Mahon, K. A. (1991) New Biol. 3, 1183-1194. [PubMed] [Google Scholar]
- 20.Bulfone, A., Puelles, L., Porteus, M. H., Frohman, M. A., Martin, G. R. & Rubenstein, J. L. (1993) Neuroscience 13, 3155-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaeren-Wiemers, N. & Gerfin-Moser, A. (1993) Histochemistry 100, 431-440. [DOI] [PubMed] [Google Scholar]
- 22.Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. & Jessell, T. M. (1996) Cell 84, 309-320. [DOI] [PubMed] [Google Scholar]
- 23.Wang, H.-F. & Liu, F.-C. (2001) Neuroscience 103, 999-1016. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., Wilson, C. J., Augood, S. J. & Emson, P. C. (1995) Trends Neurosci. 18, 527-535. [DOI] [PubMed] [Google Scholar]
- 25.Sharma, K., Sheng, H. Z., Lettieri, K., Li, H., Karavanov, A., Potter, S., Westphal, H. & Pfaff, S. L. (1998) Cell 95, 817-828. [DOI] [PubMed] [Google Scholar]
- 26.Kania, A., Johnson, R. L. & Jessell, T. M. (2000) Cell 102, 161-173. [DOI] [PubMed] [Google Scholar]
- 27.Thaler, J. P., Lee, S.-K., Jurata, L. W., Gill, G. & Pfaff, S. L. (2002) Cell 110, 237-249. [DOI] [PubMed] [Google Scholar]
- 28.Lavdas, A. A., Grigoriou, M., Pachnis, V. & Parnavelas, J. G. (1999) J. Neurosci. 19, 7881-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson, S. A., Marín, O., Horn, C., Jennings, K. & Rubenstein, J. L. R. (2001) Development (Cambridge, U.K.) 128, 353-363. [DOI] [PubMed] [Google Scholar]
- 30.Cobos, I., Shimamura, K., Rubenstein, J. L., Martinez, S. & Puelles, L. (2001) Dev. Biol. 239, 46-67. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, D., Kobayashi, M., Matsumoto, K., Ogura, T., Nakafuku, M. & Shimamura, K. (2002) Development (Cambridge, U.K.) 129, 83-93. [DOI] [PubMed] [Google Scholar]
- 32.Hallonet, M., Hollemann, T., Pieler, T. & Gruss, P. (1999) Genes Dev. 13, 3106-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohr, K. B., Barth, K. A., Varga, Z. M. & Wilson, S. W. (2001) Neuron 29, 341-351. [DOI] [PubMed] [Google Scholar]
- 34.Shimamura, K. & Rubenstein, J. L. (1997) Development (Cambridge, U.K.) 124, 2709-2718. [DOI] [PubMed] [Google Scholar]
- 35.Crossley, P. H., Martinez, S., Ohkubo, Y. & Rubenstein, J. L. (2001) Neuroscience 108, 183-206. [DOI] [PubMed] [Google Scholar]
- 36.Semba, K., Vincent, S. R. & Fibiger, H. C. (1988) J. Neurosci. 8, 3937-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brady, D. R., Phelps, P. E. & Vaughn, J. E. (1989) Dev. Brain Res. 47, 81-92. [DOI] [PubMed] [Google Scholar]
- 38.López-Coviella, I., Berse, B., Krauss, R., Thies, R. S. & Blusztajn, J. K. (2000) Science 289, 313-316. [DOI] [PubMed] [Google Scholar]
- 39.O'Meara, G., Coumis, U., Ma, S. Y., Kehr, J., Mahoney, S., Bacon, A., Allen, S. J., Holmes, F., Kahl, U., Wang, F. H., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 11569-11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steiner, R. A., Hohmann, J. G., Holmes, A., Wrenn, C. C., Cadd, G., Jureus, A., Clifton, D. K., Luo, M., Gutshall, M., Ma, S. Y., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4184-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, Y., Holtzman, D. M., Kromer, L. F., Kaplan, D. R., Chua-Couzens, J., Clary, D. O., Knusel, B. & Mobley, W. C. (1995) J. Neurosci. 15, 2888-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naumann, T., Casademunt, E., Hollerbach, E., Hofmann, J., Dechant, G., Frotscher, M. & Barde, Y. A. (2002) J. Neurosci. 22, 2409-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.