Abstract
The field of systems biology has attracted the attention of biologists, engineers, mathematicians, physicists, chemists and others in an endeavour to create systems-level understanding of complex biological networks. In particular, systems engineering methods are finding unique opportunities in characterizing the rich behaviour exhibited by biological systems. In the same manner, these new classes of biological problems are motivating novel developments in theoretical systems approaches. Hence, the interface between systems and biology is of mutual benefit to both disciplines.
Keywords: systems biology, identification, constraints, optimality, stochastics, robustness
1. Introduction
The term ‘complexity’ is often invoked in the description of biophysical networks that underlie gene regulation, protein interactions and metabolic networks in biological organisms. There are categorically two distinct characterizations of complexity: (i) the classical notion of behaviour associated with the mathematical properties of chaos and bifurcations, and (ii) the descriptive or topological notion of a large number of constitutive elements with non-trivial connectivity. In both biological and more general contexts, a key implication of complexity is that the underlying system is difficult to understand and verify (Wen et al. 1998). Simple low-order mathematical models can be constructed that yield chaotic behaviour, and yet rich complex biophysical networks may be designed to reinforce reliable execution of simple tasks or behaviours (Lauffenburger 2000).
A systematic approach for analysing complexity in biophysical networks was previously untenable owing to the lack of suitable measurements and the limitations imposed in simulating complex mathematical models. Advances in molecular biology over the past decade have made it possible to probe experimentally the causal relationships between microscopic processes initiated by individual molecules within a cell and their macroscopic phenotypic effects on cells and organisms. These studies provide increasingly detailed insights into the underlying networks, circuits and pathways responsible for the basic functionality and robustness of biological systems and create new and exciting opportunities for the development of quantitative and predictive modelling and simulation tools. Model development involves the translation of identified biological processes to coupled dynamical equations, which are amenable to numerical simulation and analysis. These equations describe the interactions between various constituents and the environment, and involve multiple feedback loops responsible for system regulation and noise attenuation and amplification.
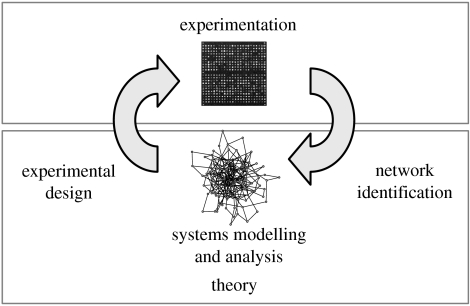
The discipline of Systems Biology has emerged in response to the challenges mentioned earlier (Kitano 2002b), and combines approaches and methods from systems engineering, computational biology, statistics, genomics, molecular biology, biophysics and other fields (Klipp et al. 2005; Palsson 2006; Szallasi et al. 2006). The recurring themes include: (i) integrative viewpoints towards unravelling complex dynamical systems, and (ii) tight iterations between experiments, modelling and hypothesis generation (figure 1).
Figure 1.
Systems biology cycle. Interactions between experimental analysis and theoretical approaches, and the main tasks for theory at the interfaces.
The central thesis of this paper is that systems engineering methods are finding unique opportunities in characterizing the rich behaviour exhibited by biological systems. In the same manner, these new classes of biological problems are motivating novel developments in theoretical systems approaches. Hence, the interface between systems and biology is of mutual benefit to both disciplines.
2. Elements of systems biology
2.1 Networks and motifs in gene regulation
Biophysical networks can be decomposed into modular components that recur across and within given organisms. One hierarchical classification is to label the top level as a network, which is comprised of interacting regulatory motifs consisting of groups of 2–4 genes (Lee et al. 2002; Shen-Orr et al. 2002; Zak et al. 2003). At the lowest level in this hierarchy is the module that describes transcriptional regulation, of which a nice example is given in Barkai & Leibler (2000).
At the motif level, one can use pattern searching techniques to determine the frequency of occurrence of these simple motifs (Shen-Orr et al. 2002), leading to the postulation that these are basic building blocks in biological networks. Of relevance to the present discussion is the fact that many of these components have direct analogues in system engineering architectures. Consider the three dominant network motifs found in Escherichia coli (Shen-Orr et al. 2002):
coherent feedforward loop: in this, one transcription factor regulates another factor, and in turn the pair jointly regulates a third transcription factor,
single input module (SIM): in systems terminology, a single-input multiple output block architecture and
densely overlapping regulons: in systems terminology, a multiple-input multiple output block architecture.
Similar studies in a completely different organism, Saccharomyces cerevisiae, yielded six-related or overlapping network motifs (Lee et al. 2002):
autoregulatory motif: in which, a regulator binds to the promotor region of its own gene,
feedforward loop: as described earlier,
multi-component loop. effectively, a closed-loop with two or more transcription factors,
regulator chain: a cascade of serial transcription factor interactions,
single input module: as described earlier (SIM) and
multi-input module: a natural extension of preceding motif.
In effect, these studies prove that, in both eukaryotic and prokaryotic systems, cell function is controlled by sophisticated networks of control loops, which are cascading onto and interconnected with, other (transcriptional) control loops. The noteworthy insight is that the complex networks, which underlie biological regulation, appear to be made of elementary systems components like a digital circuit. This lends credibility to the notion that analysis tools from systems engineering should find relevance in this problem domain.
As emphasized in the introduction, an important point in systems biology is the integrative perspective, that is to say, the analysis of the system considered as a whole and across the different levels (gene, protein, metabolite, etc.), and not the reductionist analysis of individual components. So while it is useful to categorize the elements and levels of a hierarchical regulatory scheme, it is more useful to analyse such schemes for behaviours that emerge from combinations of motifs. Some simple examples of canonical regulatory constructs that yield specific classes of behaviour in gene networks include (Smolen et al. 2000):
positive feedback: multistability, oscillations, state-dependent response,
integral feedback: robust adaptation,
negative feedback: steady-state (homeostasis, adaptation),
time delay: complex response, oscillations and
protein oligomerization: multistability, oscillations, resonant stimulus frequency response.
In addition, stochastic fluctuations can induce random response to stimuli, random outcomes, as well as stochastic focusing. Such properties are characteristic of general networks, including social networks, communication networks and biological networks (Committee on Network Science for Future Army Applications 2006).
2.2 Dynamic models
While the consideration of motifs and network topology is essential for unravelling design principles in complex biophysical networks, it is necessary to understand the role of dynamic behaviour in ascribing meaning to the rich hierarchies of regulation. Some of the intrinsically dynamic features of biophysical networks have been analysed in a recent paper that shows the close relationship between dynamic measures of robustness and the abundance of particular network motifs for a wide range of organisms (Prill et al. 2005).
Attempts to detail dynamic behaviour in these networks have fallen into three broad classes of modelling techniques: (i) first-principles approaches, (ii) empirical model identification and (iii) a hybrid approach that combines minimum metabolic network knowledge with an objective function to yield a predictive model. In this section, we outline some key results in the development of mechanistic models, and in the following sections, we will address the other two topics as they are related to network inference and constraints.
Given detailed knowledge of a biological architecture, mathematical models can be constructed to describe the behaviour of interconnected motifs or transcriptional units (TUs). A number of excellent review papers have been detailed in recent years (Smolen et al. 2000; Hasty et al. 2001). In the majority of these studies, gene expression is described as a continuous-time biochemical process, using combinations of algebraic and ordinary differential equations (ODEs; Goldbeter 1996; Cherry & Adler 2000; Smolen et al. 2000). In a similar manner, models at the signal transduction pathway level have been developed in a continuous-time framework, yielding ODEs (Kholodenko et al. 1999). At the TU level, a detailed mathematical treatment of transcriptional regulation is described in Barkai & Leibler (2000). Mechanistic models for a number of specific biological systems have been reported, including basic operons and regulons in E. coli (trp, lac and pho) and bacteriophage systems (T7 and λ; e.g. Gilman & Arkin 2002).
Systems theory has found an enabling role in the analysis of the complex mathematical structures that result from the previously described modelling approaches. The language of systems theory now dominates the quantitative characterization of biological regulation, as robustness, complexity, modularity, feedback and fragility are invoked to describe these systems. Even classical control theoretic results, such as the Bode sensitivity integral, are being applied to describe the inherent tradeoffs in sensitivity across frequency (Csete & Doyle 2002). Robustness has been introduced as both a biological system-specific attribute, as well as a measure of model validity (Ma & Iglesias 2002). In the sections that follow, brief accounts of systems-theoretic analysis of biological regulatory structures are given, emphasizing where new insights into biological regulation have been uncovered.
3. Interfaces: examples
3.1 Network identification
Currently, our knowledge of essentially all biological systems is incomplete. Despite genome projects that allow enumeration—and, to a certain extent, characterization—of all genes in a system, this does not imply knowledge about all network components (for instance, all protein variants that can be derived from a single gene), interactions, and properties thereof (Kitano 2002a). Hence, an important task in systems biology consists of specifying network interactions, which can concern qualitative or quantitative properties (existence and strength of couplings), or detailed reaction mechanisms, for genome-based inventories of components.
Essentially, this is a systems identification problem. Given a set of experimental data and prior knowledge, the network generating the data is to be determined (Ljung 1999). Alternatively termed ‘reverse engineering’ (Tegner et al. 2003), ‘network reconstruction’ (MacCarthy et al. 2005) or ‘network inference’ (Gardner et al. 2003), the general network identification problem provides a key interface between science and engineering. Several, qualitatively different approaches for biological systems have been proposed, which can be roughly classified into three categories: data-driven, approximative and mechanistic.
3.1.1 Data-driven methods
Empirical or data-driven methods rely on large-scale datasets that can be generated, for instance, through microarray analysis for gene regulatory networks. They include singular value decomposition analysis of microarray data (Alter et al. 2000; Holter et al. 2000), self-organizing maps (Tamayo et al. 1990), k-means clustering or hierarchical clustering (D'haeseleer et al. 2000), protein correlation and dynamic deviation factors (You & Yin 2000), and robust statistics approaches (Thomas et al. 2001; Zhao et al. 2001). For instance, clustering methods are routinely applied for identifying groups of co-regulated genes from microarray data. The interpretation of clustering results employs (implicit) models such as ‘co-expressed genes are likely to have a common regulator’. Data quality and algorithmic choices (for instance, of distance measures) critically influence the clustering results; in addition, validation of clustering results and techniques is an open issue (Datta & Datta 2003; Handl et al. 2005; Allison et al. 2006).
In contrast to the mechanistic approaches discussed later, most empirical approaches employ discrete-time grey box models (D'haeseleer et al. 1999; Weaver et al. 1999; Wessels et al. 2001; Hartemink et al. 2002). For instance, inference methods based on probabilistic graphical (e.g. Bayesian) models help to elucidate causal couplings between the network components (Friedman 2004). Their scalability for large systems and the ability to integrate heterogeneous datasets make them attractive (Lee et al. 2004; Klipp et al. 2005). Yet, these approaches deliver only qualitative descriptions of network function, and have inherent limitations. For instance, Bayesian models cannot cope with the ubiquitous feedback in cellular networks, since causal relationships have to be represented by directed acyclic graphs (Friedman 2004).
However, a number of challenges are present in treating experimental data for such problems: (i) the sampling rate is rarely uniform, and may be exponentially spaced by design, and (ii) data from multiple research groups are often combined (e.g. from WWW-posted data) to yield data records with inconsistent sampling, experimental bias, etc. From a systems engineering perspective, another critical point is the potentially divergent qualitative behaviour between continuous-time and discrete-time models of corresponding order (Pearson 1999). Recent work has shown the promise of continuous-time formulations of empirical models using modulating function approaches (Zak et al. 2003).
More generally, correctly identifying network topologies (corresponding to the model structure) clearly does not suffice for establishing predictive mathematical models. Experiences with engineered genetic circuits illustrate this point: with identical topology, qualitatively different behaviour can result and vice versa (Guet et al. 2002). Hence, quantitative characteristics, which are usually incorporated through model parameters in deterministic models, are also required. Corresponding identification methods are rooted in systems and information theory and, thereby, also provide the largest intersection among biology, other sciences and engineering.
3.1.2 Linear approximations
The identification of dynamically changing interactions requires corresponding dynamic models. In a first approximation, we can consider linear systems, i.e. systems with additive responses to perturbations. In systems engineering, a standard form for linear time-invariant (where the shape of the output does not change with a delay in the input) systems with n states and m inputs is given by
| (3.1) |
with n×1 state vector x(t), n×n system matrix A, n×m input matrix B and m×1 input vector u(t). Linearization of the general dynamic system dx(t)/dt=f(x, p, t) with parameter p provides first approximations to the network dynamics, even for highly nonlinear systems such as those encountered in biological networks. Linear models capture the local dynamics, for instance, in the vicinity of a steady state, instead of aiming at more complicated global behaviours.
Mathematically, most methods reconstruct the system matrix A, which corresponds to the Jacobian matrix J=∂f(x, p)/∂x, from the measured effects of (sufficiently small) perturbations. However, direct recovery of the system matrix A will be unreliable with noisy data and inputs. In one of the studies using linear models and perturbation experiments to identify the structure of genetic networks, Tegner et al. (2003) therefore proposed an iterative algorithm that uses rational choices of perturbations to improve the identification quality. For a developmental circuit, despite high nonlinearities in the system, the reverse engineering algorithm, which involves building and refining an ‘average’ connectivity matrix in successive steps, recovered all genetic interactions (Tegner et al. 2003). A related approach that uses linear models and multiple linear regression showed similar performance. The algorithm attempts to exploit the sparsity of systems matrices for biological networks owing to, for example, (estimated) upper bounds on the number of connections per node (Gardner et al. 2003; Bansal et al. 2006). Both algorithms are scalable—a central concept in engineering, but until recently considered of less importance in biology.
Newer approaches to systems identification aim at exploiting modularity in biological networks. For a modular system with one output per module, the method employs inversion of the global response matrix for identification of network connectivities and of local responses from perturbation experiments (Kholodenko et al. 2002). It requires a reduced number of measurements compared with other methods because only changes in so-called ‘communicating intermediates’ have to be recorded. Apparently, some simplifying assumptions have to be made; for example, modules are coupled by information flow only, and mass flow is negligible (Kholodenko et al. 2002). An important result of extending the modular identification to time-series data is that, for identifying all connections of a node, it is not necessary to perturb this node directly—inference can rely on detecting the network responses to remote perturbations (Sontag et al. 2004). Extensions to include the effects of uncertainties in experimental data and prior knowledge (Andrec et al. 2005), as well as the possibility of a unified mathematical framework (Cho et al. 2005) make modular identification methods particularly promising.
3.1.3 Mechanistic models, identifiability and experimental design
Mechanistic models, owing to effects such as saturation in enzymatic reactions, pose particular challenges because they involve identification of nonlinear systems. Depending on whether model structure and parameters, or only the parameters have to be identified, the problems fall into the classes of mixed-integer nonlinear programs or nonlinear programs, respectively. As a clear limitation, finding a unique global optimum in the estimation, or convergence of the algorithms cannot be guaranteed. In addition, model identification comes at high computational costs owing to numerous model simulations (Maria 2004).
In terms of parameter estimation, which is a common problem in different scientific domains (Ljung 1999), realistic modelling of complex, nonlinear dynamics of biological networks has given new impulses for the evaluation of existing methods and development of new methods. For instance, though stochastic algorithms show superior performance over deterministic methods for parameter optimization in these systems, they are computationally expensive (Moles et al. 2003). Novel hybrid methods try to exploit synergies between both approaches in order to increase robustness and efficiency (e.g. Rodriguez-Fernandez et al. 2006 and references therein).
More fundamentally, ‘identifiability’ and design of informative experiments need to be addressed. Unstructured approaches to model identification are completely ill-posed when faced with, for instance, modelling a yeast cell with 6200 genes and four possible states per gene; we obtain an overall expression state dimension in excess of 1015 (Lockhart & Winzler 2000)! Clearly a number of a priori constraints and correlations must be exploited. For discrete models, usage of the experimentally observed upper bound on the number of interactions per species brings the amount of data needed for identification into realistic dimensions (Selinger et al. 2003). However, mere extrapolation of current high-throughput technology will not solve these high dimensional data issues. Several recent studies have highlighted the importance of proper design of perturbations to reveal the logical connectivity of gene networks (Wagner 2004; MacCarthy et al. 2005). Systems engineering concepts of experimental design to provide ‘rich’ datasets can be exploited to develop predictive mechanistic models.
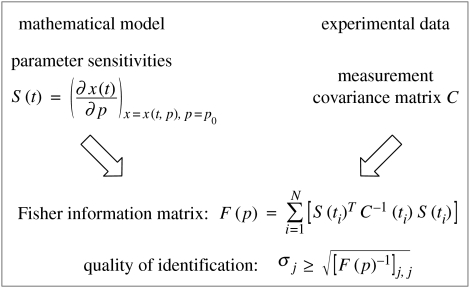
Parameter estimation accuracies are central to measuring identifiability of mechanistic models. Low accuracies mean that the corresponding parameters may be varied to a greater extent—and still describe the data—than it is possible for parameters with high estimation accuracy (low associated error). They combine information on model sensitivities with experimental data (figure 2). More specifically, the Fisher information matrix (FIM) F(p) (Emery & Nenarokomov 1998), for a point in parameter space p, links model and experiment via state sensitivities S(t)=∂x/∂p (see §3.4.1) and measurement covariance matrix for a discrete sampling time ti, C(ti). For an unbiased estimator, the Cramér–Rao theorem then gives a lower bound for the estimation error.
Figure 2.
FIM and identification quality. The lower bound for the estimation error of parameter i, σi, is derived from the inverse of the main diagonal of the FIM. See main text for details.
FIM-based approaches, for instance, yielded insight into the importance of suitable design of input perturbations for signalling networks (Zak et al. 2003), optimality criteria for the design of such inputs (Faller et al. 2003) and algorithms for the optimization of sampling times for dynamic experiments (Kutalik et al. 2004). New hybrid parameter estimation methods (Rodriguez-Fernandez et al. 2006) and closed-loop (i.e. integrating iterations between estimation, evaluation of the identifiability and experimental design) optimal identification procedures (Feng et al. 2004) rely on the FIM formalism. Note, however, that while these proof-of-concept studies with small models and synthetic data are valuable, the performance for real biological problems awaits assessment.
Information-rich datasets for integrative models will have to be derived from sources across all levels of biological regulation, such as the transcriptome, proteome and metabolic fluxes. Concomitantly, we need novel statistical frameworks for data integration (Hwang 2005). Systems identification would greatly benefit from the direct in vivo determination of kinetic parameters; the work by Ronen et al. (2002) for transcriptional control is a first step into this direction. As a complement, synthetic genetic circuits could provide means for controlled excitation of the system, for instance, by inducible genetic switches (Tegner et al. 2003), or through genetic oscillators to incorporate analysis methods in the frequency domain (Ljung 1999). Novel methods could also take known uncertainties associated with measurements—such as experimentally determined characteristics of stochastic noise (see §3.3)—explicitly into account. Finally, identification depends on adequate specification of the system and model (e.g. Kim & Tidor 2003). While models are currently either set up ad hoc, or through manual comparison of few alternative structures (including kinetic terms in the equations), uncertainties in biology pose a major challenge for systems sciences: deriving advanced approaches to model discrimination for the simultaneous identification of model structures and parameters.
3.2 Constraints and optimality
To understand complex biological systems, instead of starting from actual implementations and observations, one can reduce the problem by first separating the possible from the impossible, such as configurations and behaviours that would violate constraints. Systems approaches try to exploit three broad classes of constraints:
empirical: large-scale experimental analysis can provide constraints on possible network structures, such as the average or maximal number of interactions per component (see §3.1),
physico-chemical: laws of physics such as conservation of mass and thermodynamics impose constraints on cellular and network behaviours. These are used, in particular, for structural network analysis (SNA) with roots in the analysis of chemical reaction networks (Clarke 1988) and
functional: biological systems perform certain functions and their building blocks are confined to a large, yet finite set. Network structures and behaviours have to conform with both aspects.
Functional constraints constitute the main differences between complex physics and biology. In physics, they do not exist. Biological (as well as engineered) systems evolve to fulfil functions, and are constantly evaluated for their performance. Insufficient performance will lead to extinction, and better solutions are likely to survive. Hence, it is reasonable to assume some kind of optimality in biological systems. The immediate consequence of a purpose is a considerably smaller design space, in which effective and reliable network are rare and presumably highly structured. Understanding complexity in biology could, thus, employ a ‘calculus of purpose’—by asking teleological questions such as why cellular networks are organized as observed, given their known or assumed function (Lander 2004).
3.2.1 Physico-chemical constraints in metabolism
Essential constraints for the operation of metabolic networks are imposed by (i) reaction stoichiometries, (ii) thermodynamics that restrict flow directions through enzymatic reactions and (iii) maximal fluxes for individual reactions. For instance, metabolism usually involves fast reactions and high turnover of substances when compared with regulatory events. Therefore, on longer time-scales, it can be regarded as being in quasi-steady state. The metabolite balancing equation for a system of m internal metabolites and q reactions
| (3.2) |
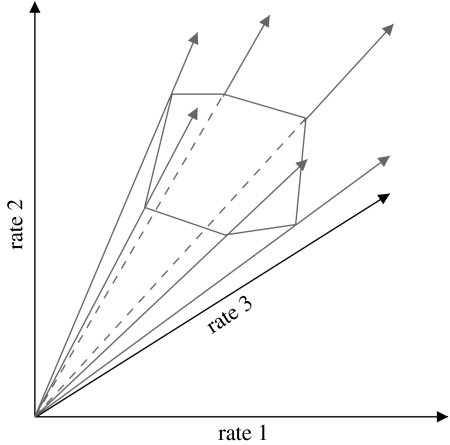
with the m×q stoichiometric matrix N and the q×1 vector of reaction rates (fluxes) r formalizes this main constraint in SNA. As for most real networks q≫m, the system of linear equation in equation (3.2) is underdetermined. However, all possible solutions are contained in a convex vector space, or flux cone (figure 3). Methods from convex analysis allow to investigate this space (Rockafellar 1970; Heinrich & Schuster 1996).
Figure 3.
Linear constraints specify a flux cone, with pathways as generating rays; projection on three-dimensional flux space.
Two broad classes of methods for SNA have been developed: metabolic pathway analysis (MPA) and flux balance analysis (FBA; see (Papin et al. 2004; Price et al. 2004; Borodina & Nielsen 2005) for recent reviews). MPA computes and uses the set of independent pathways—generating rays in figure 3—that uniquely describe the entire flux space; owing to the algorithmic complexity, it can currently only handle networks of moderate size. FBA, in contrast, determines a single flux solution through linear optimization (Varma & Palsson 1993c), often assuming that cells try to achieve optimal growth rates. The computational costs are modest, even for genome-scale models. The approach was successful, for instance, in predicting the effects of gene deletions and the outcomes of convergent evolution in micro-organisms (Fong et al. 2003; Fong & Palsson 2004; Price et al. 2004). FBA, however, has to reverse-engineer and operate with an essentially unknown objective function. While maximal growth proved a reasonable assumption for lower organisms, higher cells may tend to minimize overall fluxes in the network (Holzhütter 2004). In general, FBA has proven effective for simpler organisms, and when the steady-state assumption is valid. However, there are many situations where these conditions do not apply, many of which are biophysically meaningful, such as the dynamic diauxic shift in E. coli.
3.2.2 Extensions: dynamics and control
Stoichiometric constraints restrict the systems dynamics. Thus, the stoichiometric matrix N is fundamental, not only for SNA, but also for dynamic processes in reaction networks, in which the reaction rates r in equation (3.2) are time-dependent. For biological systems, the conservation of total amounts of certain molecular subgroups (‘conserved moieties’ such as ATP, ADP and AMP) is characteristic, and can be exploited for systems analysis. Classical work in chemical engineering addressed this topic for chemical reaction networks. For instance, Feinberg derived theorems to determine the possible dynamic regimes, such as multistability and oscillations, based on network structure alone (Feinberg 1987, 1988). Challenges posed by biological systems lead to renewed interest in these approaches and induced further theory development (Sontag 2001). Application areas in biology include stability analysis (Sontag 2001) and model discrimination by safely rejecting hypotheses on reaction mechanisms, thus, identifying crucial reaction steps (Conradi et al. 2005). Algorithms for the identification of dependent species in large biochemical systems—to be employed, for instance, in model reduction—have recently become available (Vallabhajosyula et al. 2006).
Enabling FBA to deal with dynamics and regulation proceeded by incorporating additional time-dependent constraints that reflect knowledge on the operation of cellular control circuits—an approach termed ‘regulatory FBA’ (Covert et al. 2001). For instance, using superimposed Boolean logic models to capture transcriptional regulatory events has extended the validity of the methodology for a number of complex dynamic system responses (Covert et al. 2001) and for data integration (Covert et al. 2004). Other dynamic extensions of the FBA algorithm have been proposed in Mahadevan et al. (2002). With these more detailed models, steady-state analysis suggested that the complex transcriptional control networks operate in a few dominant states, i.e. generate simple behaviour (Barrett et al. 2005). Finally, pathway analysis also allows to approach features of intrinsically dynamic systems: for instance, it helps to identify feedback loops in cellular signal processing (Klamt et al. 2006). Hence, SNA-related approaches are about to extend to non-classical domains, in particular, through theory development induced by new challenges in systems biology.
3.2.3 Functional constraints, optimality and design
In analysing living systems, one possibility is to start from the assumption that they have to fulfil certain functions, and that cells have been organized over evolutionary time-scales to optimize their operations in a manner consistent with mathematical principles of optimality. FBA demonstrates the utility of this assumption; note that its implicit functional constraint, i.e. steady-state operation of metabolic networks, is not self-explanatory. Similarly, other approaches invoking principles of optimal control theory have opened new avenues for systems analysis in biology.
The cybernetic approach developed by Ramkrishna & co-workers (Kompala et al. 1986; Varner & Ramkrishna 1998) is based on a simple principle: evolution has programmed or conditioned biological systems to optimally achieve physiological objectives. This straightforward concept can be translated into a set of optimal resource allocation problems that are solved at every time-step in parallel with the model mass balances (basic metabolic network model). Thus, at every instant in time, gene expression and enzyme activity are rationalized as choice between sets of competing alternatives, each with a relative cost and benefit for the organism. Mathematically, this can be translated into an instantaneous objective function. The researchers in this area have defined several postulates for specific pathway architectures, and the result is a computationally tractable (i.e. analytical) model structure. The potential shortcoming is a limited handling of more flexible objective functions that are commonly observed in biological systems (Savinell & Palsson 1992a,b; Varma & Palsson 1993a,b; Bonarious et al. 1997).
Instead of focusing on a single objective function, mathematical models and experimental data can be used to test hypotheses on optimality principles, given a specific cellular function to be fulfilled. For instance, extensions of FBA suggested that E. coli optimizes the tradeoff between achieving high growth rates and maintaining wild-type metabolic fluxes after gene deletions (Segre et al. 2002). MPA showed that the interplay between the metabolic network (the controlled plant) and gene regulation (the controller) in E. coli might be designed to achieve optimal tradeoffs between long-term objectives, such as metabolic flexibility, and short-term adjustment for metabolic efficiency (Stelling et al. 2002). Optimal production pipelines for biomass components, with fast responses to environmental changes and minimal additional efforts for enzyme synthesis, were predicted in detail to employ wave-like gene expression programs, which was later confirmed experimentally (Klipp et al. 2002; Zaslaver et al. 2004). Hence, at least certain cellular design principles can be revealed by evaluating assumptions on cellular optimality principles.
Finally, without assuming optimality, we can ask how functions in biological systems could be established in principle. Among others, drawing from analogies with engineered systems helps to understand more general design principles in biology. From nonlinear dynamics, for instance, it is well-known that functions such as oscillators and switches require some source of nonlinearity. Establishing such a function with biological building blocks, thus, allows only for certain circuit designs (Tyson et al. 2003; Kholodenko 2006). Similar ideas can prove powerful at different levels of abstraction. For instance, highly structured ‘bow-ties’ with multiple inputs, channelled through a core with standardized components and protocols to multiple outputs, could be the common organizational principles to establish complex production systems in engineering and biology (Csete & Doyle 2004). On the other hand, El-Samad & colleagues studied the bacterial heat-shock response, pointing out that the intertwined feedback and feedforward loops present can be assigned individual functions parallel to those loops in designed control circuits that have to yield fast responses in highly fluctuating environments (El-Samad et al. 2005). Notably, most of the examples discussed here involved new developments in theory to address challenges posed by biology; with respect to robustness as an important functional constraint, we will discuss these interfaces in more detail in §3.4.
3.3 Stochastic systems
Discrete stochastic modelling has recently gained popularity owing to its relevance in biological processes (McAdams & Arkin 1997; Arkin et al. 1998) that achieve their functions with low copy numbers of some key chemical species. Unlike the solutions to stochastic differential equations, the states/outputs of discrete stochastic systems evolve according to discrete jump Markov processes, which naturally lead to a probabilistic description of the system dynamics. A Markov process is a random process in which the future probabilities are dependent only on the present value, and not on past values. Such descriptions can find relevance in systems biology when the magnitude of the fluctuations in a stochastic system approaches the levels of the actual variables (e.g. protein concentrations). In addition, there are qualitative phenomena that are intrinsic to such descriptions that arise in biological systems, as mentioned later.
The idea that stochastic phenomena are essential for understanding complex transcriptional processes was nicely illustrated by Arkin & co-workers in the analysis of the phage λ lysis–lysogeny decision circuit (Arkin et al. 1998). The probabilistic division of the initially homogeneous cell population into subpopulations corresponding to the two possible fate outcomes was shown to require stochastic description (and could not be described with a continuous deterministic model). In particular, the coexistence of the two subpopulations necessitated such a formal characterization, and the relative sensitivity of the subpopulations to model parameters including external variables could be analysed with the resulting models. In a more recent work, Arkin & co-workers (Samoilov et al. 2005) have shown another example of a biological behaviour that is intrinsically stochastic in nature—namely the dynamic switching behaviour in a class of biochemical reactions (enzymatic futile cycles). In this case, the behaviour is more subtle than the lysis–lysogeny switch described earlier, where the existence of a bifurcation was at least evident in the continuous differential equation model. In the enzymatic futile cycle problem, the deterministic model gives no indication of multiplicity, yet the discrete stochastic model generates behaviours, including switching as well as oscillations, that indicate characteristics of bifurcation regimes. It is suggested that such noise-induced mechanisms may be responsible for control of switch and cycle behaviour in regulatory networks.
In the discrete stochastic setting, the states and outputs are random variables governed by a probability density function, which follows a chemical master equation (CME) (Gillespie 1976). The rate of reaction no longer describes the amount of chemical species being produced or consumed per unit time in a reaction, but rather the likelihood of a certain reaction to occur. Though analytical solution of the CME is rarely available, the density function can be constructed using the stochastic simulation algorithm (SSA; Gillespie 1976).
The discrete stochastic system of interest is described by a CME (Gillespie 1977)
| (3.3) |
where f(x, t|x0, t0) is the conditional probability of the system to be at state x and time t, given the initial condition x0 at time t0. Here, ak denotes the propensity functions, νk denotes the stoichiometric change in x when the kth reaction occurs and m is the total number of reactions. The propensity function ak(x, p)dt gives the probability of the kth reaction to occur between time t and t+dt, given the parameters p. As the state values are typically unbounded, the CME essentially consists of an infinite number of ODEs, whose analytical solution is rarely available except for a few simple problems. The SSA provides an efficient numerical algorithm for constructing the density function (Gillespie 1976). The algorithm follows a Monte Carlo approach based on the joint probability for the time to and the index of the next reaction, which is a function of the propensities. The SSA indirectly simulates the CME by generating many realizations of the states (typically of the order of 104) at specified time t, given the initial condition and model parameters, from which the distribution f(x, t|x0, t0) can be constructed.
This renewed interest in discrete stochastic simulation has motivated a number of systems engineering developments for the analysis of, and more efficient computation of, stochastic models. These include detailed analysis of the underlying assumptions invoked in using the SSA, with an emphasis on the distinction between separating fast and slow components as opposed to fast and slow reactions (Haseltine & Rawlings 2005). Gillespie has remained active in this area, and is currently collaborating with Linda Petzold to develop methods for accelerated tau-leaping methods, as well as effective numerical methods for step size selection (Cao et al. 2005, 2006). Kevrekidis & co-workers have introduced so-called ‘equation-free’ modelling approaches, which avoid the need for extensive Monte Carlo simulations. In the area of analysis, methods for formal sensitivity analysis of discrete stochastic equations enable the characterization of robustness properties of biological systems (Gunawan et al. 2005).
There has been simultaneous advancement in experimental methods for quantifying the characteristics of biological noise (Elowitz et al. 2002; Swain et al. 2002; Raser & O'Shea 2004) along with advances in computing and simulation. A number of groups have recently used dual reporter methods to track identical genes in the same cell to measure the impact of noise on expression. In the work of Elowitz & co-workers, the separate effects of stochastic behaviour in the transcriptional and translational processes in prokaryotes (so-called ‘intrinsic’ noise) are distinguished from noise effects arising from other cellular components that influence the rate of gene expression (so-called ‘extrinsic’ noise; Elowitz et al. 2002; Swain et al. 2002). O'Shea analyses eukaryotic systems with both cis- and trans-acting mutations to distinguish between the noise effects that are intrinsic to transcription as opposed to upstream processes that might ultimately influence expression (Raser & O'Shea 2004).
The interface of discrete stochastic systems and biology has clearly led to new insights into stochastic phenomena in biological systems, and has also spurred the development of more efficient computational methods for stochastic simulation, as well as analysis methods for these models. This interface will continue to motivate developments in systems engineering, with improved methods for imaging biological systems that include the ability to resolve spatial behaviours. Distributed stochastic models will require more sophisticated algorithmic developments, particularly as one builds models to truly address ‘systems-scale’ phenomena.
3.4 Robustness
In biology, as in engineering, robust performance refers to the attainment of a particular behaviour or response by a system in the presence of uncertainty. This appears to be a ubiquitous property of biological processes that are subject to constant uncertainty in the form of stochastic phenomena (McAdams & Arkin 1999), fluctuating environment and genetic variation (for a recent review on robustness in cellular functions, see Stelling et al. 2004b). Biology has adapted a number of approaches for coping with these sources of uncertainty, which include:
back-up systems (redundancy),
disturbance attenuation through feedback and feedforward control,
structuring of networked systems into semi-autonomous functional units (modularity) and
reliable coordination of network elements through hierarchies and protocols.
The robustness problems in systems biology have only begun to yield, in recent years, formal quantitative analyses, owing largely to their nonlinear (and non-stationary) nature. As with engineering systems, robust performance requires the precise specification of both a performance metric and the type/size of uncertainty. When both these elements are specified, it may be possible to analyse biological systems with the engineering tools, as will be shown in this paper. It is important to note that the performance metric is often difficult to be defined precisely in biology, as it is an implicit element of an evolved entity.
3.4.1 Parametric sensitivity approaches to robustness analysis
Parametric sensitivity has found widespread application in the analysis and design of both scientific and engineering systems (Varma et al. 1999). In the field of systems biology, sensitivity analysis has been employed in a number of applications, including optimized design of synthetic circuits (Feng et al. 2004), design of experiment for optimal parameter estimation (Zak et al. 2003; Gadkar et al. 2005) and robustness analysis to provide insights into design principles (Stelling et al. 2004a). The sensitivity operator describes the change in the system's outputs owing to variations in the parameters that affect the system dynamics. High sensitivity to a parameter suggests that the system's performance (e.g. growth, temperature, etc.) can drastically change with small variations in the parameter. Conversely, a small value of the sensitivity suggests that the system is not strongly affected by the parameter.
The classical parametric sensitivity analysis applies to continuous deterministic systems, e.g. systems described by differential (or differential-algebraic) equations. The first order sensitivity coefficients are given by Varma et al. (1999)
| (3.4) |
where yi denotes the ith output; t, the time; and pj, the jth parameter. Equation (3.4) follows directly from the definition of parametric sensitivity, and assumes implicitly that the output yi is continuous with respect to the parameter pj. Sensitivity analysis for stochastic systems can be applied to problems in which the stochastic effects enter as additive Gaussian white noise (e.g. Langevin-type problems; Costanza & Seinfeld 1981; Dacol & Rabitz 1984 or as uncertainty in the parameters Feng et al. 2004). Recent extensions allow the treatment of discrete stochastic systems (see §3.3).
Using the sensitivity operator, one can computer the FIM (§3.1.3), thus indicating robust elements (large variances) and fragile elements (tight variances). Corresponding to such a characterization are parametric sensitivities, which are high for fragile elements and low for robust elements. Previous work has shown the utility of this approach for analysing robustness in complex biophysical networks (Stelling et al. 2004a). The FIM allows flexibility in choosing the appropriate criterion for optimality depending on the goal of both robustness and model identification. D-optimal design aims to maximize the degree of informativeness in data by maximizing the determinant of the FIM, which corresponds to the area/volume of an information hyperellipsoid (Emery & Nenarokomov 1998). On the other hand, A-optimal design is equivalent to reducing the hyperellipsoid of uncertainty in parameter estimates.
One limitation to parametric sensitivity for the analysis of biological systems is the inherently nonlinear character of such systems, while the classical sensitivity methods yield linear (i.e. local) results. One can improve upon this by performing analyses in a neighbourhood of operating points, thus extending the region of validity of the method.
3.4.2 Systems engineering approaches to robustness
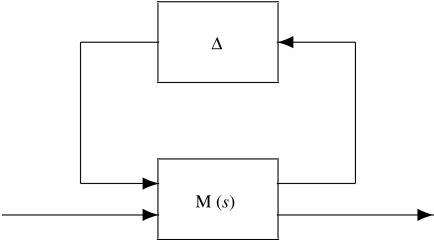
In control engineering, a standard tool for robustness analysis is the structured singular value (SSV), which allows to determine whether a particular dynamical system, subject to a specified (structured) uncertainty, is able to remain stable or to achieve a particular performance metric (e.g. Doyle 1982; Skogestad & Postlethwaite 1996; Zhou 1998). The two problems are known as robust stability and robust performance, respectively, and there are standard software packages available to facilitate this computation (e.g. Balas et al. 1995). The key idea is to transform the perturbed system into a new closed-loop operator, and then to test the stability of the operator. The basic idea is illustrated in figure 4, where the M operator denotes a nominal process system, and the Δ operator denotes the uncertainty in the system. Stability of the depicted system is equivalent to robust stability of the original problem, and if a feedback loop between suitably transformed input and output signals is closed, then an operator whose stability characteristics coincide with the attainment of robust performance in the original problem is obtained.
Figure 4.
Standard M–Δ diagram for robustness analysis.
There are straightforward computational algorithms for determining the solution to the corresponding linear time-invariant stability problem in figure 4, for example, MATLAB's μ-Analysis and Synthesis Toolbox (Balas et al. 1995). Note, however, that extensions for time-varying and nonlinear uncertainty (Doyle et al. 1989) are necessary before applying the SSV analysis on nonlinear systems. The SSV-based methods have begun to find application in biological systems, with recent papers on ‘robustness’ properties of models (Ma & Iglesias 2002) and robust performance analysis of a signal transduction cascade (Doyle & Stelling 2005). One of the more important messages from the engineering robust control literature is the notion of ‘performance’, which requires a precise description in order to calculate the so-called ‘robust performance’. This idea has important consequences in biology, as robustness in one performance attribute may be quite different from the behaviour of another attribute (Bagheri et al.). The unique attributes of problems encountered in biology are also motivating the development of new algorithms for formal theoretic analysis (Sontag 2004).
Although the aforementioned engineering methods outline a formal framework for robustness analysis, with potential application for simple biological systems, there are intrinsic scaling problems with such methods. In particular, in the case of larger and more complex nonlinear systems, computational tractability of the corresponding analysis problem is a limiting concern, as is the fact that the methods are inherently conservative for nonlinear systems. This suggests that scalability of methods for robustness analysis is a major opportunity for future research.
3.4.3 Robust yet fragile systems
There is a coexistence of fragility along with robustness in complex networked systems. This property is observed in a wide variety of systems that include forest fires, the Internet, metabolic networks and protein regulatory networks (Carlson & Doyle 1999, 2000, 2002). A nice treatment of the theoretical issues and algorithms in analysing robustness in biological networks is provided in the review paper (El-Samad et al. 2006). The concept of ‘Highly Optimized Tolerance’ has emerged to describe the optimal design of such networks in which robustness is maintained across a wide range of expected perturbations, while fragility (often catastrophic) is observed for the very rare and unlikely perturbations. This is accomplished through hierarchies of regulation that are built to withstand some expected class of disturbances. As an engineering system ‘evolves’ to incorporate ancillary layers of stabilizing control, these layers often expose the system to novel points of fragility, thus leading to so-called ‘spiralling complexity’.
In simple (linear) engineering systems, such tradeoffs are analysed using theory such as the Bode sensitivity integral to calculate the ‘conservation’ of robustness (captured by the sensitivity operator) across the frequency domain. This reveals that reduced sensitivity in some frequency range is exactly balanced by a heightened sensitivity in another range (i.e. ‘robust yet fragile’; Csete & Doyle 2004; Doyle & Csete 2005). The significance of the Bode sensitivity integral for biological (nonlinear) dynamics needs to be clarified, as the result is traditionally applied to linear systems. Yet it is widely understood that biological networks exhibit the same robust yet fragile tradeoffs. As an example, consider the exquisite timekeeping of circadian rhythm in neuronal cells. These clocks are well known to be robust to large fluctuations in temperature (Ruoff et al. 2005), and yet recent evidence has shown that timekeeping in cellular networks is fragile to the blocking of key receptors for intracellular synchronization (Aton et al. 2005). The former is an expected disturbance for which the system is designed to be robust, while the latter represents a highly unusual and unexpected disturbance for which the system is ill-equipped to handle. Furthermore, quantitative studies in systems such as the Drosophila circadian gene network have revealed similar tradeoffs between global (core cellular machinery) and local (circadian specific) functions (Stelling et al. 2004a).
3.4.4 Two biological examples of robustness
The signal transduction system that mediates chemotaxis exhibits a type of adaptation in which the response to a persistent stimulus is reset to the pre-stimulus value, thereby enabling an enhanced sensitivity. For a number of years, researchers speculated a mechanistic explanation for this robust behaviour, and two hypotheses had emerged: (i) precise fine-tuning of several parameters to yield a consistent (robust) response under varied conditions, and (ii) structural organization that yielded this robust behaviour intrinsically (Barkai & Leibler 1997). The assumptions in this work require a specific mechanism of fine-tuning of the network structure so as to produce integral feedback, which is sufficient to make ‘adaptation’ perfectly robust to all remaining network parameters. John Doyle & co-workers at Caltech were able to use the internal model principle to demonstrate that the regulatory system was exploiting integral feedback control to achieve the robust level of adaptation exhibited in chemotaxis, and more generally in systems with such behaviour (Yi et al. 2000; Lander 2004). In other words, they showed that integral control is a necessary condition for robust perfect adaptation, and if the mechanism described in Barkai & Leibler (1997) is incorrect in some aspects, then some other fine-tuned structure must be present. This understanding suggests that many seemingly complex biological networks may employ redundancy and other structural motifs or modules (enumerated in an earlier section) to achieve relatively simple overall system behaviour (Lauffenburger 2000).
The gene network underlying circadian rhythm in flies and mammals has been the focus of detailed analysis in recent years (Goldbeter 1996; Reppert 2000; Winfree 2001; Young & Kay 2001; Goldbeter 2002). The biological details are coming into sharper focus, as new experiments yield clues to the detailed (and somewhat overlapping) molecular circuitry of both flies and mammals (Panda et al. 2002). Building upon the evolving biological knowledge, there have been many postulated mathematical models (Leloup & Goldbeter 1998; Scheper et al. 1999; Tyson et al. 1999; Lema et al. 2000; Smolen et al. 2001) that range in complexity from simple two-state oscillators to more biophysically detailed transcriptional feedback schemes. As with adaptation in chemotaxis, robustness is the dominant characteristic often associated with the circadian rhythm regulatory loop (e.g. Vilar et al. 2002), although formal systems-theoretic treatment of this behaviour is a notable absence among the published reports. In recent work, we have shown that systems engineering tools, notably robustness analysis, shed light on the underlying design principles in the gene regulatory architectures (Stelling et al. 2004a). In particular, the organization of fragility and robustness between global cellular components and circadian-specific components enables precision in circadian clock function.
4. Summary
Biological regulation has been reviewed and analysed from the perspective of systems engineering. Mathematical modelling approaches, both empirical and fundamental, have yielded descriptions of many complex systems, and systems-theoretic tools have been employed to provide hypotheses for biological behaviour, such as system robustness. Open challenges were described in the areas of network identification, constraints and optimality, stochastic systems modelling and robustness analysis. Synthetic biology represents one of the more promising future directions in this field (Arkin 2001; Benner & Sismour 2005), which requires a fusion of methods from both engineering and molecular biology in the design of biological circuits in order to achieve the aims at the interface of these disciplines.
Acknowledgments
We acknowledge the financial support to F.J.D. from the Institute for Collaborative Biotechnologies through grant DAAD19-03-D-0004 from the US Army Research Office.
References
- Allison D.B, Cui X, Page G.P, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. doi:10.1038/nrg1749 [DOI] [PubMed] [Google Scholar]
- Alter O, Brown P.O, Botstein B. Singular value decomposition for genome wide expression data processing and modeling. Proc. Natl Acad. Sci. USA. 2000;97:10 101–10 116. doi: 10.1073/pnas.97.18.10101. doi:10.1073/pnas.97.18.10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrec M, Kholodenko B.N, Levy R.M, Sontag E. Inference of signaling and gene regulatory networks by steady-state perturbation experiments: structure and accuracy. J. Theor. Biol. 2005;232:427–441. doi: 10.1016/j.jtbi.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Arkin A.P. Synthetic cell biology. Curr. Opin. Biotechnol. 2001;12:638–644. doi: 10.1016/s0958-1669(01)00273-7. doi:10.1016/S0958-1669(01)00273-7 [DOI] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams H.H. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton S.J, Colwell C.S, Harmar A.J, Waschek J, Herzog E.D. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri, N., Stelling, J., Doyle III, F. J. Submitted. Quantitative performance metrics for robustness in circadian rhythms. Bioinformatics [DOI] [PubMed]
- Balas G.J, Doyle J.C, Glover K, Packard A, Smith R. The Mathworks; Natick, MA: 1995. μ-Analysis and synthesis toolbox user's guide. [Google Scholar]
- Bansal M, Della Gatta G, di Bernardo D. Inference of gene regulatory networks and compound mode of action from time course gene expression profiles. Bioinformatics. 2006;22:815–822. doi: 10.1093/bioinformatics/btl003. doi:10.1093/bioinformatics/btl003 [DOI] [PubMed] [Google Scholar]
- Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. doi:10.1038/43199 [DOI] [PubMed] [Google Scholar]
- Barkai N, Leibler S. Circadian clocks limited by noise. Nature. 2000;403:267–268. doi: 10.1038/35002258. [DOI] [PubMed] [Google Scholar]
- Barrett C.L, Herring C.D, Reed J.L, Palsson B.O. The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states. Proc. Natl Acad. Sci. USA. 2005;102:19 103–19 108. doi: 10.1073/pnas.0505231102. doi:10.1073/pnas.0505231102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner S.A, Sismour A.M. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. doi:10.1038/nrg1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonarious H.P.J, Schmid G, Tramper J. Flux analysis of underdetermined metabolic: the quest for the missing constraints. Trends Biotech. 1997;15:308–314. doi:10.1016/S0167-7799(97)01067-6 [Google Scholar]
- Borodina I, Nielsen J. From genomes to in silico cells via metabolic networks. Curr. Opin. Biotechnol. 2005;16:350–355. doi: 10.1016/j.copbio.2005.04.008. doi:10.1016/j.copbio.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Cao Y, Gillespie D.T, Petzold L.R. Accelerated stochastic simulation of the stiff enzyme-substrate reaction. J. Chem. Phys. 2005;123:144 917. doi: 10.1063/1.2052596. doi:10.1063/1.2052596 [DOI] [PubMed] [Google Scholar]
- Cao Y, Gillespie D.T, Petzold L.R. Efficient step size selection for the tau-leaping simulation method. J. Chem. Phys. 2006;124:044 109. doi: 10.1063/1.2159468. doi:10.1063/1.2159468 [DOI] [PubMed] [Google Scholar]
- Carlson J.M, Doyle J. Highly optimized tolerance: a mechanism for power laws in designed systems. Phys. Rev. E. 1999;60:1412–1427. doi: 10.1103/physreve.60.1412. doi:10.1103/PhysRevE.60.1412 [DOI] [PubMed] [Google Scholar]
- Carlson J.M, Doyle J. Highly optimized tolerance: robustness and design in complex systems. Phys. Rev. Lett. 2000;84:2529–2532. doi: 10.1103/PhysRevLett.84.2529. doi:10.1103/PhysRevLett.84.2529 [DOI] [PubMed] [Google Scholar]
- Carlson J.M, Doyle J. Complexity and robustness. Proc. Natl Acad. Sci. USA. 2002;99:2538–2545. doi: 10.1073/pnas.012582499. doi:10.1073/pnas.012582499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.L, Adler F.R. How to make a biological switch. J. Theor. Biol. 2000;203:117–133. doi: 10.1006/jtbi.2000.1068. doi:10.1006/jtbi.2000.1068 [DOI] [PubMed] [Google Scholar]
- Cho K.-H, Choo S.-M, Wellstead P, Wolkenhauer O. A unified framework for unraveling the functional interaction structure of a biomolecular network based on stimulus–response experimental data. FEBS Lett. 2005;579:4520–4528. doi: 10.1016/j.febslet.2005.07.025. doi:10.1016/j.febslet.2005.07.025 [DOI] [PubMed] [Google Scholar]
- Clarke B.L. Stoichiometric network analysis. Cell Biophys. 1988;12:237–253. doi: 10.1007/BF02918360. [DOI] [PubMed] [Google Scholar]
- Committee on Network Science for Future Army Applications 2006 Network Science National Research Council, Washington, DC.
- Conradi C, Saez-Rodriguez J, Gilles E.-D, Raisch J. Using chemical reaction network theory to discard a kinetic mechanism hypothesis. IEE Proc. Syst. Biol. 2005;152:243–248. doi: 10.1049/ip-syb:20050045. doi:10.1049/ip-syb:20050045 [DOI] [PubMed] [Google Scholar]
- Costanza V, Seinfeld J.H. Stochastic sensitivity analysis in chemical kinetics. J. Chem. Phys. 1981;74:3852–3858. doi:10.1063/1.441615 [Google Scholar]
- Covert M.W, Schilling C.H, Palsson B. Regulation of gene expression in flux balance models of metabolism. J. Theor. Biol. 2001;213:73–88. doi: 10.1006/jtbi.2001.2405. doi:10.1006/jtbi.2001.2405 [DOI] [PubMed] [Google Scholar]
- Covert M.W, Knight E.M, Reed J.L, Herrgard M.J, Palsson B.O. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. doi:10.1038/nature02456 [DOI] [PubMed] [Google Scholar]
- Csete M.E, Doyle J.C. Reverse engineering of biological complexity. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. doi:10.1126/science.1069981 [DOI] [PubMed] [Google Scholar]
- Csete M, Doyle J. Bow ties, metabolism and disease. Trends Biotechnol. 2004;22:446–450. doi: 10.1016/j.tibtech.2004.07.007. doi:10.1016/j.tibtech.2004.07.007 [DOI] [PubMed] [Google Scholar]
- D'haeseleer, P., Wen, X., Fuhrman, S. & Somogyi R.1999 Linear modeling of mRNA expression levels during CNS development and injury. In Pac. Symp. Biocomput, 4, pp. 41–52. [DOI] [PubMed]
- D'haeseleer P, Liang S, Somogyi R. Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics. 2000;16:707–726. doi: 10.1093/bioinformatics/16.8.707. doi:10.1093/bioinformatics/16.8.707 [DOI] [PubMed] [Google Scholar]
- Dacol D.K, Rabitz H. Sensitivity analysis of stochastic kinetic models. J. Math. Phys. 1984;25:2716–2727. doi:10.1063/1.526478 [Google Scholar]
- Datta S, Datta S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics. 2003;19:459–466. doi: 10.1093/bioinformatics/btg025. doi:10.1093/bioinformatics/btg025 [DOI] [PubMed] [Google Scholar]
- Doyle J.C. Analysis of feedback systems with structured uncertainties. IEE Proc., Part D. 1982;129:242–250. [Google Scholar]
- Doyle J, Csete M. Motifs, control, stability. PLoS Biol. 2005;3:1868–1872. doi: 10.1371/journal.pbio.0030392. doi:10.1371/journal.pbio.0030392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle III, F. J. & Stelling, J. 2005 Robust performance in biophysical networks. In Proc. IFAC World Congress, 849–854.
- Doyle F.J, III, Packard A.K, Morari M. Robust controller design for a nonlinear CSTR. Chem. Eng. Sci. 1989;44:1929–1947. doi:10.1016/0009-2509(89)85133-4 [Google Scholar]
- El-Samad H, Kurata H, Doyle J.C, Gross C.A, Khammash M. Surviving heat shock: control strategies for robustness and performance. Proc. Natl Acad. Sci. USA. 2005;102:2736–2741. doi: 10.1073/pnas.0403510102. doi:10.1073/pnas.0403510102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Samad H, Prajna S, Papachristodoulou A, Doyle J, Khammash M. Advanced methods and algorithms for biological networks analysis. Proc. IEEE. 2006;94:832–853. doi:10.1109/JPROC.2006.871776 [Google Scholar]
- Elowitz M.B, Levine A.J, Siggia E.D, Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. doi:10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Emery A.F, Nenarokomov A.V. Optimal experiment design. Meas. Sci. Technol. 1998;9:864–876. doi:10.1088/0957-0233/9/6/003 [Google Scholar]
- Faller D, Klingmüller U, Timmer J. Simulation methods for optimal experimental design in systems biology. Simulation. 2003;79:717–725. doi:10.1177/0037549703040937 [Google Scholar]
- Feinberg M. Chemical reaction network structure and the stability of complex isothermal reactors-i. The deficiency zero and deficiency one theorems. Chem. Eng. Sci. 1987;42:2229–2268. doi:10.1016/0009-2509(87)80099-4 [Google Scholar]
- Feinberg M. Chemical reaction network structure and the stability of complex isothermal reactors-ii. Multiple steady states for networks of deficiency one. Chem. Eng. Sci. 1988;43:1–25. doi:10.1016/0009-2509(88)87122-7 [Google Scholar]
- Feng X.-J, Hooshangi S, Chen D, Li G, Weiss R, Rabitz H. Optimizing genetic circuits by global sensitivity analysis. Biophys. J. 2004;87:2195–2202. doi: 10.1529/biophysj.104.044131. doi:10.1529/biophysj.104.044131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S.S, Marciniak J.Y, Palsson B.O. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol. 2003;185:6400–6408. doi: 10.1128/JB.185.21.6400-6408.2003. doi:10.1128/JB.185.21.6400-6408.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S.S, Palsson B.O. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat. Genet. 2004;36:1056–1108. doi: 10.1038/ng1432. doi:10.1038/ng1432 [DOI] [PubMed] [Google Scholar]
- Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. doi:10.1126/science.1094068 [DOI] [PubMed] [Google Scholar]
- Gadkar K.G, Gunawan R, Doyle F.J., III Iterative approach to model identification of biological networks. BMC Bioinform. 2005;6:155. doi: 10.1186/1471-2105-6-155. doi:10.1186/1471-2105-6-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner T.S, di Bernardo D, Lorenz D, Collins J.J. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. doi:10.1126/science.1081900 [DOI] [PubMed] [Google Scholar]
- Gillespie D.T. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 1976;22:403–434. doi:10.1016/0021-9991(76)90041-3 [Google Scholar]
- Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. doi:10.1021/j100540a008 [Google Scholar]
- Gilman A, Arkin A. Genetic ‘code’: representations and dynamical models of genetic components and networks. Annu. Rev. Genomics Hum. Genet. 2002;3:341–369. doi: 10.1146/annurev.genom.3.030502.111004. doi:10.1146/annurev.genom.3.030502.111004 [DOI] [PubMed] [Google Scholar]
- Goldbeter A. 2nd edn. Cambridge University Press; Cambridge, UK: 1996. Biochemical oscillations and cellular rhythms: the molecular bases of periodic and chaotic behaviour. [Google Scholar]
- Goldbeter A. Computational approaches to cellular rhythms. Nature. 2002;420:238–245. doi: 10.1038/nature01259. doi:10.1038/nature01259 [DOI] [PubMed] [Google Scholar]
- Guet C.C, Elowitz M.B, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. doi:10.1126/science.1067407 [DOI] [PubMed] [Google Scholar]
- Gunawan R, Cao Y, Petzold L, Doyle F.J., III Sensitivity analysis of discrete stochastic systems. Biophys. J. 2005;88:2530–2540. doi: 10.1529/biophysj.104.053405. doi:10.1529/biophysj.104.053405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handl J, Knowles J, Kell D.B. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21:3201–3212. doi: 10.1093/bioinformatics/bti517. doi:10.1093/bioinformatics/bti517 [DOI] [PubMed] [Google Scholar]
- Hartemink, A. J., Gifford, D. K., Jaakola, T. S. & Young, R. A. 2002 Combining location and expression data for principled discovery of genetic regulatory network models. In Proc. Pac. Symp. Biocomput., 7, 437–449. [PubMed]
- Haseltine E.L, Rawlings J.B. On the origins of approximations for stochastic chemical kinetics. J. Chem. Phys. 2005;123:164 115. doi: 10.1063/1.2062048. doi:10.1063/1.2062048 [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Isaacs F, Collins J.J. Computational studies of gene regulatory networks: in numero molecular biology. Nat. Rev. Gen. 2001;2:268–279. doi: 10.1038/35066056. doi:10.1038/35066056 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Schuster S. Chapman & Hall; New York, NY: 1996. The regulation of cellular systems. [Google Scholar]
- Holter N.S, Mitra M, Maritan A, Cieplak M, Banavar J.R, Fedoroff N.V. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc. Natl Acad. Sci. USA. 2000;97:8409–8414. doi: 10.1073/pnas.150242097. doi:10.1073/pnas.150242097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzhütter H.-G. The principle of flux minimization and its application to estimate stationary fluxes in metabolic networks. Eur. J. Biochem. 2004;271:2905–2922. doi: 10.1111/j.1432-1033.2004.04213.x. doi:10.1111/j.1432-1033.2004.04213.x [DOI] [PubMed] [Google Scholar]
- Hwang D, et al. A data integration methodology for systems biology. Proc. Natl Acad. Sci. USA. 2005;102:17 296–17 301. doi: 10.1073/pnas.0508647102. doi:10.1073/pnas.0508647102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko B.N. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell. Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko B.N, Demin O.V, Moehren G, Hoek J.B. Quantification of short term signaling by the epidermal growth factor receptor. J. Biol. Chem. 1999;274:30 169–30 181. doi: 10.1074/jbc.274.42.30169. doi:10.1074/jbc.274.42.30169 [DOI] [PubMed] [Google Scholar]
- Kholodenko B.N, Kiyatkin A, Bruggeman F.J, Sontag E, Westerhoff H.V, Hoek J.B. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc. Natl Acad. Sci. USA. 2002;99:12 841–12 846. doi: 10.1073/pnas.192442699. doi:10.1073/pnas.192442699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.M, Tidor B. Limitations of quantitative gene regulation models: a case study. Genome Res. 2003;13:2391–2395. doi: 10.1101/gr.1207003. doi:10.1101/gr.1207003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Computational systems biology. Nature. 2002a;420:206–210. doi: 10.1038/nature01254. doi:10.1038/nature01254 [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002b;295:1662–1664. doi: 10.1126/science.1069492. doi:10.1126/science.1069492 [DOI] [PubMed] [Google Scholar]
- Klamt S, Saez-Rodriguez J, Lindquist J, Simeoni L, Gilles E.D. A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinform. 2006;7:56. doi: 10.1186/1471-2105-7-56. doi:10.1186/1471-2105-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp E, Heinrich R, Holzhütter H.-G. Prediction of temporal gene expression. Metabolic opimization by re-distribution of enzyme activities. Eur. J. Biochem. 2002;269:5406–5413. doi: 10.1046/j.1432-1033.2002.03223.x. doi:10.1046/j.1432-1033.2002.03223.x [DOI] [PubMed] [Google Scholar]
- Klipp E, Herwig R, Kowald A, Wierling C, Lehrach H. Wiley; Weinheim, Germany: 2005. Systems biology in practice: concepts, implementation and application. [Google Scholar]
- Kompala D.S, Ramkrishna D, Jansen N.B, Tsao G.T. Investigation of bacterial growth on mixed substrates. Experimental evaluation of cybernetic models. Biotech. Bioeng. 1986;28:1044–1056. doi: 10.1002/bit.260280715. doi:10.1002/bit.260280715 [DOI] [PubMed] [Google Scholar]
- Kutalik Z, Cho K.-H, Wolkenhauer O. Optimal sampling time selection for parameter estimation in dynamic pathway modeling. Biosystems. 2004;75:43–55. doi: 10.1016/j.biosystems.2004.03.007. doi:10.1016/j.biosystems.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Lander A.D. A calculus of purpose. PLoS Biol. 2004;2:0712. doi: 10.1371/journal.pbio.0020164. doi:10.1371/journal.pbio.0020164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D.A. Cell signaling pathways as control modules: complexity for simplicity? Proc. Natl Acad. Sci. USA. 2000;97:5031–5033. doi: 10.1073/pnas.97.10.5031. doi:10.1073/pnas.97.10.5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. doi:10.1126/science.1075090 [DOI] [PubMed] [Google Scholar]
- Lee I, Date S.V, Adai A.T, Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. doi:10.1126/science.1099511 [DOI] [PubMed] [Google Scholar]
- Leloup J.-C, Goldbeter A. A model for circadian rhythms in Drosophila incorporting the formation of a complex between the PER and TIM proteins. J. Biol. Rhythms. 1998;13:70–87. doi: 10.1177/074873098128999934. doi:10.1177/074873098128999934 [DOI] [PubMed] [Google Scholar]
- Lema M.A, Golombek D.A, Echave J. Delay model of the circadian pacemaker. J. Theor. Biol. 2000;204:565–573. doi: 10.1006/jtbi.2000.2038. doi:10.1006/jtbi.2000.2038 [DOI] [PubMed] [Google Scholar]
- Ljung L. 2nd edn. Prentice Hall PTR; Upper Saddle River, NJ: 1999. System identification: theory for the user. [Google Scholar]
- Lockhart D.J, Winzler E.A. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. doi:10.1038/35015701 [DOI] [PubMed] [Google Scholar]
- Ma L, Iglesias P.A. Quantifying robustness of biochemical network models. BMC Bioinform. 2002;3:38–50. doi: 10.1186/1471-2105-3-38. doi:10.1186/1471-2105-3-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCarthy T, Pomiankowski A, Seymour R. Using large-scale perturbations in gene network reconstruction. BMC Bioinform. 2005;6:11. doi: 10.1186/1471-2105-6-11. doi:10.1186/1471-2105-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan K, Edwards J, Doyle F.J., III Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys. J. 2002;83:1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria G. A review of algorithms and trends in kinetic model identification for chemical and biochemical systems. Chem. Biochem. Eng. Q. 2004;18:195–222. [Google Scholar]
- McAdams H.H, Arkin A. Stochastic mechanisms in gene expression. Proc. Natl Acad. Sci. USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. doi:10.1073/pnas.94.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams H.H, Arkin A. Its a noisy business: genetic regulation at the nanomolar scale. Trends Genet. 1999;15:65–69. doi: 10.1016/s0168-9525(98)01659-x. doi:10.1016/S0168-9525(98)01659-X [DOI] [PubMed] [Google Scholar]
- Moles C.G, Mendes P, Banga J.R. Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res. 2003;13:2467–2474. doi: 10.1101/gr.1262503. doi:10.1101/gr.1262503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson B.O. Cambridge University Press; Cambridge, UK: 2006. Systems biology: properties of reconstructed networks. [Google Scholar]
- Panda S, Hogenesch J.B, Kay S.A. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. doi:10.1038/417329a [DOI] [PubMed] [Google Scholar]
- Papin J.A, Stelling J, Price N.D, Klamt S, Schuster S, Palsson B.O. Comparison of network-based pathway analysis methods. Trends Biotechnol. 2004;22:400–405. doi: 10.1016/j.tibtech.2004.06.010. doi:10.1016/j.tibtech.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Pearson R.K. Oxford University Press; Oxford, UK: 1999. Discrete-time dynamic models. [Google Scholar]
- Price N.D, Reed J.L, Palsson B.O. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. doi:10.1038/nrmicro1023 [DOI] [PubMed] [Google Scholar]
- Prill R.J, Iglesias P.A, Levchenko A. Dynamic properties of network motifs contribute to biological network organization. PLoS Biol. 2005;3:e343. doi: 10.1371/journal.pbio.0030343. doi:10.1371/journal.pbio.0030343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser J.M, O'Shea E.K. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. doi:10.1126/science.1098641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S.M. Comparing clockworks: mouse versus fly. J. Biol. Rhythms. 2000;15:357–364. doi: 10.1177/074873000129001459. doi:10.1177/074873000129001459 [DOI] [PubMed] [Google Scholar]
- Rockafellar R.T. Princeton University Press; Princeton, NJ: 1970. Convex analysis. [Google Scholar]
- Rodriguez-Fernandez M, Mendes P, Banga J.R. A hybrid approach for efficient and robust parameter estimation in biochemical pathways. Biosystems. 2006;83:248–265. doi: 10.1016/j.biosystems.2005.06.016. doi:10.1016/j.biosystems.2005.06.016 [DOI] [PubMed] [Google Scholar]
- Ronen M, Rosenberg R, Shraiman B, Alon U. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc. Natl Acad. Sci. USA. 2002;99:10 555–10 560. doi: 10.1073/pnas.152046799. doi:10.1073/pnas.152046799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff P, Loros J, Dunlap J.C. The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc. Natl Acad. Sci. USA. 2005;102:17 681–17 686. doi: 10.1073/pnas.0505137102. doi:10.1073/pnas.0505137102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoilov M, Plyasunov S, Arkin A.P. Stochastic amplification and signaling in enzymatic futile cycles through noise-induced bistability with oscillations. Proc. Natl Acad. Sci. USA. 2005;102:2310–2315. doi: 10.1073/pnas.0406841102. doi:10.1073/pnas.0406841102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinell J.M, Palsson B.O. Network analysis of intermediary metabolism using linear optimization. Development of mathematical formalism. J. Theor. Biol. 1992a;154:421–454. doi: 10.1016/s0022-5193(05)80161-4. [DOI] [PubMed] [Google Scholar]
- Savinell J.M, Palsson B.O. Network analysis of intermediary metabolism using linear optimization. Interpretation of hybridoma cell metabolism. J. Theor. Biol. 1992b;154:455–473. doi: 10.1016/s0022-5193(05)80162-6. [DOI] [PubMed] [Google Scholar]
- Scheper T, Klinkenberg D, Pennartz C, van Pelt J. A mathematical model for the intracellular rhythm generator. J. Neurosci. 1999;19:40–47. doi: 10.1523/JNEUROSCI.19-01-00040.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre D, Vitkup D, Church G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl Acad. Sci. USA. 2002;99:15 112–15 117. doi: 10.1073/pnas.232349399. doi:10.1073/pnas.232349399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D.W, Wright M.A, Church G.M. On the complete determination of biological systems. Trends Biotechnol. 2003;21:251–254. doi: 10.1016/S0167-7799(03)00113-6. doi:10.1016/S0167-7799(03)00113-6 [DOI] [PubMed] [Google Scholar]
- Shen-Orr S.S, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. doi:10.1038/ng881 [DOI] [PubMed] [Google Scholar]
- Skogestad S, Postlethwaite I. Wiley; New York, NY: 1996. Multivariable feedback control. [Google Scholar]
- Smolen P, Baxter D.A, Byrne J.H. Mathematical modeling of gene networks. Neuron. 2000;26:567–580. doi: 10.1016/s0896-6273(00)81194-0. doi:10.1016/S0896-6273(00)81194-0 [DOI] [PubMed] [Google Scholar]
- Smolen P, Baxter D.A, Byrne J.H. Modeling circadian oscillations with interlocking positive and negative feedback loops. J. Neurosc. 2001;21:6644–6656. doi: 10.1523/JNEUROSCI.21-17-06644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E. Structure and stability of certain chemical networks and applications to the kinetic proofreading model of t-cell receptor signal transduction. IEEE Trans. Autom. Control. 2001;46:1028–1047. doi:10.1109/9.935056 [Google Scholar]
- Sontag E.D. Some new directions in control theory inspired by systems biology. IEE Syst. Biol. 2004;1:9–18. doi: 10.1049/sb:20045006. doi:10.1049/sb:20045006 [DOI] [PubMed] [Google Scholar]
- Sontag E, Kiyatkin A, Kholodenko B.N. Inferring dynamic architecture of cellular networks using time series of gene expression, protein and metabolite data. Bioinformatics. 2004;20:1877–1886. doi: 10.1093/bioinformatics/bth173. doi:10.1093/bioinformatics/bth173 [DOI] [PubMed] [Google Scholar]
- Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles E.D. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. doi:10.1038/nature01166 [DOI] [PubMed] [Google Scholar]
- Stelling J, Gilles E.D, Doyle F.J., III Robustness properties of circadian clock architectures. Proc. Natl Acad. Sci. USA. 2004a;101:13 210–13 215. doi: 10.1073/pnas.0401463101. doi:10.1073/pnas.0401463101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle F.J, III, Doyle J. Robustness of cellular functions. Cell. 2004b;118:675–685. doi: 10.1016/j.cell.2004.09.008. doi:10.1016/j.cell.2004.09.008 [DOI] [PubMed] [Google Scholar]
- Swain P.S, Elowitz M.B, Siggia E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA. 2002;99:12 795–12 800. doi: 10.1073/pnas.162041399. doi:10.1073/pnas.162041399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szallasi Z, Stelling J, Periwal V, editors. System modeling in cellular biology: from concepts to nuts and bolts. MIT Press; Cambridge, MA: 2006. [Google Scholar]
- Tamayo P, Slonium D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E, Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA. 1990;44:129–131. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegner J, Yeung M.K.S, Hasty J, Collins J.J. Reverse engineering gene networks: integrating genetic perturbations with dynamical modeling. Proc. Natl Acad. Sci. USA. 2003;100:5944–5949. doi: 10.1073/pnas.0933416100. doi:10.1073/pnas.0933416100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.G, Olson J.M, Tapscott S.J, Zhao L.P. An efficient and robust statistical modeling approach to discover differentially expressed genes using genomic expression profiles. Genome Res. 2001;11:1227–1236. doi: 10.1101/gr.165101. doi:10.1101/gr.165101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.J, Hong C.I, Thron C.D, Novak B. A simple model of circadian rhythms based on dimerization and proteolysis of per and tim. Biophys. J. 1999;77:2411–2417. doi: 10.1016/S0006-3495(99)77078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.J, Chen K.C, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell. Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. doi:10.1016/S0955-0674(03)00017-6 [DOI] [PubMed] [Google Scholar]
- Vallabhajosyula R.R, Chickarmane V, Sauro H.M. Conservation analysis of large biochemical networks. Bioinformatics. 2006;22:346–353. doi: 10.1093/bioinformatics/bti800. doi:10.1093/bioinformatics/bti800 [DOI] [PubMed] [Google Scholar]
- Varma A, Morbidelli M, Wu H. Oxford University Press; New York, NY: 1999. Parametric sensitivity in chemical systems. [Google Scholar]
- Varma A, Palsson B.O. Metabolic capabilities of Escherichia coli: I. Synthesis of biosynthetic precursors and cofactors. J. Theor. Biol. 1993a;165:477–502. doi: 10.1006/jtbi.1993.1202. doi:10.1006/jtbi.1993.1202 [DOI] [PubMed] [Google Scholar]
- Varma A, Palsson B.O. Metabolic capabilities of Escherichia coli: II. Optimal growth patterns. J. Theor. Biol. 1993b;165:503–522. doi: 10.1006/jtbi.1993.1202. doi:10.1006/jtbi.1993.1203 [DOI] [PubMed] [Google Scholar]
- Varma A, Palsson B.O. Metabolic flux balancing: basic concepts, scientific and practical use. Biotechnol. Bioeng. 1993c;12:994–998. [Google Scholar]
- Varner J, Ramkrishna D. Application of cybernetic models to metabolic engineering: investigation of storage pathways. Biotech. Bioeng. 1998;58:282–291. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<282::aid-bit24>3.0.co;2-d. doi:10.1002/(SICI)1097-0290(19980420)58:2/3<282::AID-BIT24>3.0.CO;2-D [DOI] [PubMed] [Google Scholar]
- Vilar J.M, Kueh H.Y, Barkai N, Leibler S. Mechanisms of noise-resistance in genetic oscillators. Proc. Natl Acad. Sci. USA. 2002;30:5988–5992. doi: 10.1073/pnas.092133899. doi:10.1073/pnas.092133899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Reconstructing pathways in large genetic networks from genetic perturbations. J. Comput. Biol. 2004;11:53–60. doi: 10.1089/106652704773416885. doi:10.1089/106652704773416885 [DOI] [PubMed] [Google Scholar]
- Weaver, D. C., Workman, C. T. & Stormo, G. D. 1999 Modeling regulatory networks with weight matrices. In Pac. Symp. Biocomput., 4, pp. 102–111. [DOI] [PubMed]
- Wen X, Fuhrman S, Michaels G.S, Carr D.B, Smith S, Barker J.L, Somogyi R. Large-scale temporal gene expression mapping of central nervous system development. Proc. Natl Acad. Sci. USA. 1998;95:334–339. doi: 10.1073/pnas.95.1.334. doi:10.1073/pnas.95.1.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, L. F. A., Van Someren, E. P. & Reinders, M. J. T. 2001 A comparison of genetic network models. In Pac. Symp. Biocomput., 6, pp. 508–519 [PubMed]
- Winfree A.T. 2nd edn. Springer; New York, NY: 2001. The geometry of biological time. [Google Scholar]
- Yi T.M, Huang Y, Simon M.I, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl Acad. Sci. USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. doi:10.1073/pnas.97.9.4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Yin J. Patterns of regulation from mRNA and protein time series. Metabolic Eng. 2000;2:210–217. doi: 10.1006/mben.1999.0139. doi:10.1006/mben.1999.0139 [DOI] [PubMed] [Google Scholar]
- Young M.W, Kay S.A. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. doi:10.1038/35088576 [DOI] [PubMed] [Google Scholar]
- Zak D.E, Gonye G.E, Schwaber J.S, Doyle F.J., III Importance of input perturbations and stochastic gene expression in the reverse engineering of genetic regulatory networks: insights from an identifiability analysis of an in silico network. Genome Res. 2003;13:2396–2405. doi: 10.1101/gr.1198103. doi:10.1101/gr.1198103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Mayo A.E, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette M.G, Alon U. Just-in-time transcription program in metabolic pathways. Nat. Genet. 2004;36:486–491. doi: 10.1038/ng1348. doi:10.1038/ng1348 [DOI] [PubMed] [Google Scholar]
- Zhao L.P, Prentice R, Breeden L. Statistical modeling of large microarray data sets to identify stimulus–response profiles. Proc. Nat. Acad. Sci. USA. 2001;98:5631–5636. doi: 10.1073/pnas.101013198. doi:10.1073/pnas.101013198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. Prentice-Hall; Englewood Cliffs, NJ: 1998. Essentials of robust control. [Google Scholar]