Abstract
We have defined a minimal Arabidopsis CATALASE 3 (CAT3) promoter sufficient to drive evening-specific circadian transcription of a LUCIFERASE reporter gene. Deletion analysis and site-directed mutagenesis reveal a circadian response element, the evening element (EE: AAAATATCT), that is necessary for evening-specific transcription. The EE differs only by a single base pair from the CIRCADIAN CLOCK ASSOCIATED 1-binding site (CBS: AAAAAATCT), which is important for morning-specific transcription. We tested the hypothesis that the EE and the CBS specify circadian phase by site-directed mutagenesis to convert the CAT3 EE into a CBS. Changing the CAT3 EE to a CBS changes the phase of peak transcription from the evening to the morning in continuous dark and in light-dark cycles, consistent with the specification of phase by the single base pair that distinguishes these elements. However, rhythmicity of the CBS-containing CAT3 promoter is dramatically compromised in continuous light. Thus, we conclude that additional information normally provided in the context of a morning-specific promoter is necessary for full circadian activity of the CBS.
The circadian clock enables an organism to specifically partition aspects of its biology to precise times over the day (Dunlap, 1999). Although the circadian clock is, by definition, endogenous and continues to run in the absence of external time cues, environmental stimuli such as light and temperature act to entrain the internal processes of an organism both to the exact external daily period and in a defined relationship, or phase angle, to the diurnal cycle. For example, in Arabidopsis, light and temperature information are integrated to partition physiological activities such as circadian-regulated leaf movement, stomatal opening, and gene expression to distinct times of day or phases (McClung et al., 2002).
A central theme that has emerged in circadian biology is that the core oscillator is composed of a negative feedback loop grounded in positive and negative transcriptional regulation (Dunlap, 1999). It has recently been demonstrated that the Arabidopsis circadian clock entails such a transcriptional feedback loop (Alabadí et al., 2001) that includes at least three components: TIMING OF CAB EXPRESSION 1 (TOC1; also called Arabidopsis PSEUDO-RESPONSE REGULATOR 1, APRR1; Millar et al., 1995; Makino et al., 2000), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1; Wang and Tobin, 1998), and LATE ELONGATED HYPOCOTYL (LHY; Schaffer et al., 1998). CCA1 and LHY are single-Myb domain transcription factors, and DNA-binding activity of CCA1 to a CCA1-binding site (CBS: AAAAATCT) has been characterized (Wang et al., 1997). The hypothesized role of TOC1 as a transcription factor is based on similarity to CONSTANS, although DNA binding by TOC1 has not been experimentally established (Strayer et al., 2000). However, TOC1 (APRR1) has been shown to bind to PHYTOCHROME-INTERACTING FACTOR 3 (PIF3), a Myc-related basic helix-loop-helix transcription factor, and to the related PIF3-LIKE 1 (PIL1; Makino et al., 2002). Expression of each of the three clock components, TOC1, CCA1, and LHY, is circadian regulated (Schaffer et al., 1998; Wang and Tobin, 1998; Matsushika et al., 2000; Strayer et al., 2000). TOC1 (APRR1) and CCA1/LHY make up a feedback loop in which TOC1 acts as a positive regulator of CCA1 and LHY, which in turn are negative regulators of TOC1 (Alabadí et al., 2001). CCA1 and LHY bind to the TOC1 promoter in vitro at a CBS-related motif called the evening element (EE: AAAATATCT), and overexpression of either LHY or CCA1 results in nonoscillating, low-level accumulation of TOC1 mRNA, indicating that both CCA1 and LHY are negative regulators of TOC1 (Alabadí et al., 2001; Matsushika et al., 2002). In plants homozygous for the strong loss-of-function toc1-2 allele, oscillations of LHY and CCA1 mRNA exhibit both the short period characteristic of toc1 mutations (Millar et al., 1995; Somers et al., 1998) and greatly reduced CCA1 and LHY mRNA abundance, consistent with a role of TOC1 as a positive regulator (Alabadí et al., 2001). TOC1 (APRR1) overexpression disrupts rhythmic expression of many genes, including CCA1 and LHY, but the results are not entirely consistent with the simple explanation of TOC1 (APRR1) acting directly as a positive regulator at the promoters of CCA1 and LHY (Makino et al., 2002).
It has been demonstrated in Arabidopsis, cyanobacteria, fruitfly (Drosophila melanogaster), and mammals and that the oscillations in the mRNA abundance of circadian-regulated transcripts peak at many unique phases that span the entire day (Liu et al., 1995; Harmer et al., 2000; Claridge-Chang et al., 2001; Grundschober et al., 2001; McDonald and Rosbash, 2001; Akhtar et al., 2002; Duffield et al., 2002). Included among these are genes encoding a number of key clock components, such as CCA1, LHY, and TOC1, that function within the circadian oscillator. Among other clock-controlled genes are a number of additional transcription factors, which leads to the simple and attractive hypothesis that the phasing of transcription of clock-controlled genes to specific times of day emerges through the interaction of a specific clock-controlled transcription factor with its cognate DNA target. Genes transcribed at a specific times of day share a promoter motif that binds a specific transcription factor whose activity peaks at that time of day, and genes transcribed at other times of day possess different promoter motifs that interact with distinct clock-regulated transcription factors.
Two elements implicated in circadian control of transcription, the EE and CBS (also called the lhc motif), were originally identified in the promoters of clock controlled genes (Carré and Kay, 1995; Wang et al., 1997; Harmer et al., 2000). The CBS and EE are closely related with a difference of only 1 bp (AAAaATCT versus AAAtATCT). The similarity of CBS and EE, coupled with their specific association with genes phased to morning and evening, respectively (Carré and Kay, 1995; Wang et al., 1997; Harmer et al., 2000), suggests that phase may be specified by the 1-bp difference that distinguishes the two motifs. To test this directly we used the promoter of the Arabidopsis CATALASE 3 (CAT3) gene, which oscillates with an evening-specific peak in circadian-regulated mRNA abundance (Zhong and McClung, 1996). Deletion analysis and site-directed mutagenesis of the CAT3 promoter reveals that the EE is necessary for evening-specific transcription. Converting the CAT3 EE to a CBS (aaaTatct to aaaAatct) renders the promoter substantially arrhythmic when examined in continuous light (LL), whereas in continuous dark (DD) conditions or in entraining conditions of 12 h light and 12 h dark (12/12 LD), this promoter confers morning-specific rhythmicity. These results reinforce the centrality of the CBS/EE in circadian transcription and demonstrate that the single base pair difference between these elements is sufficient to specify the time of day at which transcription occurs. However, our results also make it clear that additional promoter elements provide critical contextual information that is essential for complete circadian regulation.
RESULTS
Circadian Evening-Specific Transcription of the CAT3 Promoter
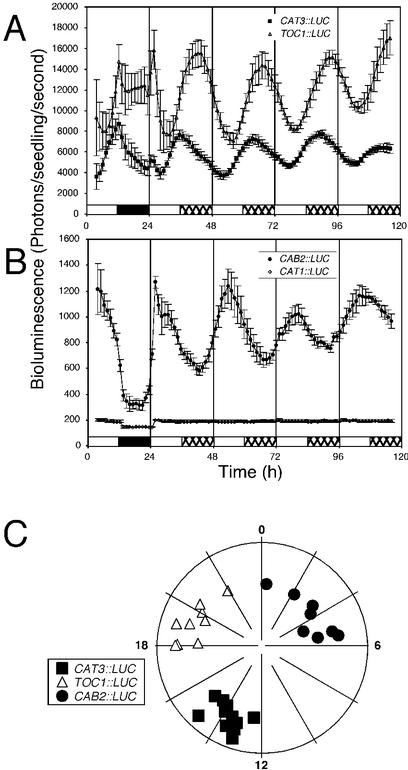
The circadian clock regulates CAT3 mRNA abundance with a peak at dusk and a trough at dawn (Zhong and McClung, 1996). CAT3 promoter:: LUCIFERASE fusions (CAT3::LUC) were constructed and transformed into ecotype Columbia (Col) plants to address whether circadian regulation is at the level of transcription. T2 plants containing CAT3::LUC were grown in entraining conditions of a 12/12 LD cycle at 22°C for 7 d. Seedlings were moved to a luminometer (TopCount, Packard, Meriden, CT), entrained in LD for 3 d, and then released into LL at 22°C. Figure 1A shows that, in LL, luciferase activity of CAT3::LUC seedlings oscillates with a period of about 24 h and with an evening-specific phase (period = 24.85 ± 0.19 h; phase = 13.78 ± 0.22 circadian time [CT] h; n = 12). In contrast, neither a CAT1::LUC fusion (Fig. 1B) nor the promoterless LUC gene alone (data not shown) demonstrated oscillations in luciferase activity in LL. Therefore, we conclude that circadian clock regulation of CAT3 transcription contributes to the circadian oscillation previously described for CAT3 mRNA abundance (Zhong and McClung, 1996). Similar period and phase results were obtained for the ecotypes Rschew (RLD), Wassilewskija (WS), Landsberg erecta (Ler), and Cape Verde Islands (Cvi; data not shown). When CAT3::LUC seedlings were entrained to different photoperiods (long days: 16/8 LD or short days: 8/16 LD), there was no significant difference in period or phase compared with plants that were entrained to 12/12 LD cycles (data not shown). The evening-specific phase of transcription of the maize (Zea maize) CAT3 ortholog similarly has been shown to be insensitive to photoperiod (Abler and Scandalios, 1994).
Figure 1.
Transcription of a CAT3::LUC transgene is regulated by the circadian clock. Plants were grown under 12-h light/12-h dark photoperiod at 22°C for 7 d. Plants were moved to a Packard TopCount luminometer and further entrained for 3 d in the LD cycle before being released into LL. The LD regime is indicated by the bars beneath the traces, with day (light) indicated by white bars, night (dark) indicated by black bars, and subjective night (dark of the entraining cycle) indicated by hatched bars. A, Traces present average values (± se, n = 12) from individual seedlings expressing CAT3::LUC (squares) or TOC1::LUC (triangles). B, Traces present average values (± se, n = 12) from individual seedlings expressing CAB2::LUC (circles) and CAT1::LUC (diamonds). C, Phase plot in which phases of individual seedlings are plotted against the strength of the rhythm. Phase is expressed in CT (phase/period × 24 h) around the circumference of a 24-h clock face. Strength of the rhythm is expressed as relative amplitude error (RAE), where a perfect sine wave is defined as 0 and a value of 1 defines the weakest rhythm considered to be statistically significant. The strength of the rhythm is plotted along the radius with the strongest rhythms (RAE = 0) at the outer edge of the circle and weakest rhythms (RAE = 1) at the center. CAT3::LUC, squares; CAB2::LUC, circles; TOC1::LUC, triangles. [−221/−103]2 CAT3::LUC seedlings are depicted because of their highly reproducible and accurate representation of endogenous CAT3 circadian-regulated transcription. Similar results have been obtained with all other rhythmic CAT3::LUC fusions tested.
In entraining LD conditions, clock-controlled reporters like CAB2::LUC, CAT3::LUC, and TOC1::LUC display sinusoid circadian rhythms with clear anticipation of dawn and dusk, respectively (Fig. 1). In addition, TOC1::LUC shows pronounced acute responses to the lights on signal at dawn and to the lights off signal at dusk. In contrast, during LD cycles both CAT1::LUC (Fig. 1B) and promoterless::LUC (data not shown) demonstrate driven rhythms as seen by “square waves,” in which LUC activity increases and decreases in direct response to lights on and lights off, with no evidence of anticipation of either dawn or dusk. This may reflect altered plant metabolism in light and dark affecting basal luciferase activity.
The phase of peak CAT3::LUC transcription is distinct from that of other clock-regulated genes. For example, CAB2::LUC, a well-documented clock-regulated gene fusion (Millar et al., 1992), cycles with a mid-day-specific phase (period = 24.61 ± 0.52 h; phase = 4.39 ± 0.75 CT h; n = 12) and TOC1::LUC cycles with a midnight-specific phase (period = 24.67 ± 0.32; phase = 18.89 ± 0.55 CT h; n = 12) in LL (Fig. 1). The TOC1::LUC phase lags by about 6 h that reported by Alabadí et al. (2001; phase approximately 12 CT h). One possible explanation is that Alabadí et al. (2001) describe a translational fusion in which the 5′-untranslated region of TOC1 is present (−834/+1 from the ATG), whereas the TOC1::LUC fusion described in this study is a transcriptional fusion that includes only promoter elements upstream of the transcriptional start (−890/−381). We suspect that the distinct phases of these two constructs results from different regulatory elements provided in the two fusion constructs. It is worth noting that TOC1 transcript abundance displays biphasic peaks, one at approximately CT12 and another at approximately CT18 (Makino et al., 2000; Strayer et al., 2000); possibly the transcriptional and translational TOC1::LUC fusions separate two bouts of transcriptional activity that contribute to this biphasic pattern of mRNA abundance. Others have shown that CCR2 and ELF3 promoters confer circadian transcription with afternoon- and late evening-specific phases (CT approximately 10 and approximately 16, respectively; Staiger and Apel, 1999; Strayer et al., 2000; Covington et al., 2001). To highlight phase differences between CAT3::LUC, TOC1::LUC and CAB2::LUC, phase was plotted against the strength of the rhythm (Fig. 1C). Strong rhythms are plotted close to the outer edge of the circle, whereas weaker rhythms are plotted near the center of the circle (see “Materials and Methods” for details).
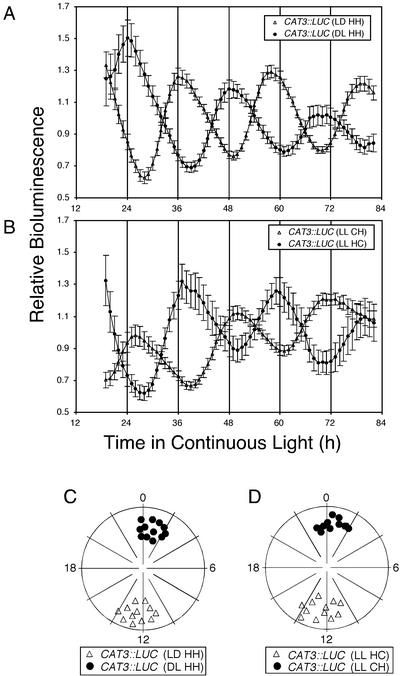
LD cycles entrain the circadian rhythm in CAT3::LUC activity (Fig. 2, A and C). Although light serves as a major external entrainment stimulus in plants, temperature cycles have also been shown to entrain the circadian clock (Heintzen et al., 1994; Somers et al., 1998). Consistent with this, CAT3::LUC expression is entrained by temperature cycles of 12-h hot (22°C) and 12-h cold (18°C) in LL (LL HC), where 22°C acts as a “day” signal and 18°C acts a “night” signal. After entrainment to LL HC, CAT3::LUC activity peaks at the beginning of the subjective cold period (Fig. 2, B and D), whereas CAB2::LUC activity has been shown to peak in the middle of the subjective hot period (Somers et al., 1998). Either light (Fig. 2, A and C) or temperature (Fig. 2, B and D) cycles provided 180° out of phase can be used to entrain two populations of seedlings antiphase to one another; CAT3::LUC expression is always phased to the beginning of the subjective dark or cold period. Both light and temperature cycles provide strong entraining stimuli that can override previous time-of-day information that the plant may have received.
Figure 2.
CAT3::LUC expression can be entrained by light or temperature cycles. A, Plants were grown at 22°C either under a 12/12 LD photoperiod (LD HH; triangles) or under a 12-h/12-h dark-light (DL HH; circles) photoperiod for 7 d before release into LL at T = 0. Traces present average values (± se, n = 12) from individual independent transgenic lines. Data are normalized to the average luciferase activity of the individual seedling and are presented as relative bioluminescence. B, Plants were grown in LL either under a 12-h hot (22°C)/12-h cold (18°C) thermoperiod (LL HC; circles) or under a 12-h cold (18°C)/12-h hot (22°C) thermoperiod (LL CH; triangles) for 7 d before release into constant temperature (22°C) and LL at T = 0. Traces present average values (± se, n = 12) from individual independent transgenic lines. C and D, Phase plots as described in the legend to Figure 1C for multiple seedlings from A and C, respectively. [−221/−103]2 CAT3::LUC seedlings are depicted, but similar results have been obtained with all other CAT3::LUC fusions tested, except those constructs that have lost rhythmicity.
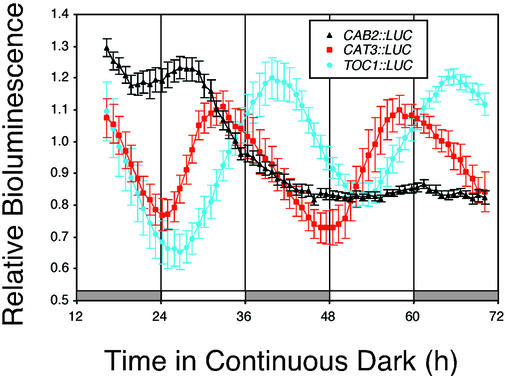
Rhythmic oscillation of CAT3::LUC (all fusions discussed in this study) persists in DD with evening-specific phase and 24-h period (Fig. 3; data not shown). This is interesting because CAT3 mRNA oscillations damp to constitutively high levels in DD (Zhong et al., 1997). That CAT3 mRNA abundance oscillations damp in DD while transcription continues to oscillate suggests posttranscriptional control in mRNA abundance; either CAT3 mRNA becomes stabilized in DD or CAT3 mRNA abundance is destabilized in the light. Of course, it is also possible that the CAT3::LUC fusions do not completely recapitulate endogenous CAT3 transcriptional activity. The persistence of robust circadian oscillations in CAT3 transcription in DD contrasts strikingly with the rapid damping seen in CAB2 transcription in DD (Fig. 3; Millar et al., 1992). However, transcription as measured with transcriptional LUC fusions has been shown to oscillate in DD for several genes in addition to CAT3, including CCR2 (Strayer et al., 2000), TOC1 (Strayer et al., 2000), EARLY FLOWERING 3 (Covington et al., 2001), PHYTOCHROME (PHY) A, PHYB, PHYD, PHYE, CRYPTOCHROME 1, and CRYPTOCHROME 2 (Hall et al., 2001; Tóth et al., 2001). Moreover, overexpression of tobacco (Nicotiana tabacum) ZGT allows sustained oscillation of CAB2 transcription in extended dark (Xu and Johnson, 2001).
Figure 3.
CAT3::LUC activity continues to oscillate in DD. Plants grown as described in the Figure 1 legend and released into DD conditions instead of LL. Traces present average values (± se, n = 12), normalized as described in the legend to Figure 2, from CAB2::LUC (black triangles), CAT3::LUC (red squares), and TOC1::LUC (blue circles) seedlings. The LD regime is indicated by the bars beneath the traces, with subjective day indicated by white bars and subjective night indicated by gray bars. As discussed in the Figure 1 legend, [−221/−103]2 CAT3::LUC seedlings are depicted.
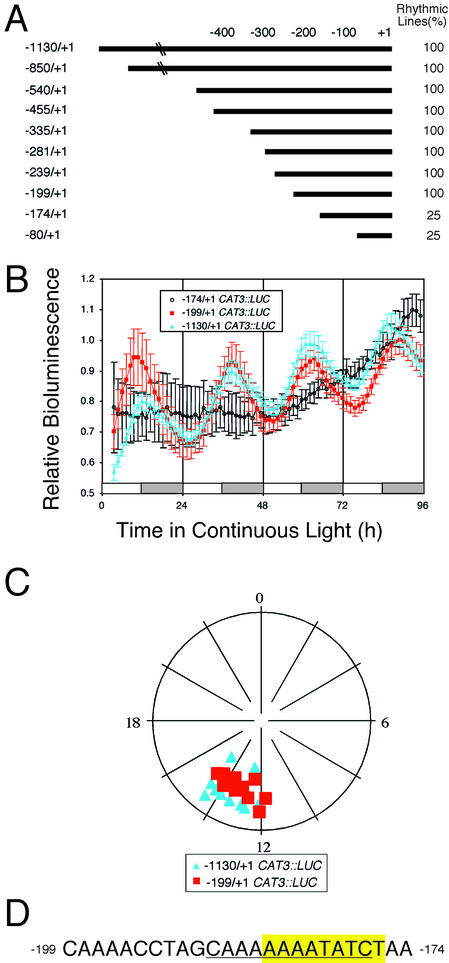
CAT3 Promoter Deletion Series Reveals That an EE Is Necessary for Evening-Specific Transcription
Progressive deletion of the CAT3 promoter from −1,130 to −199 yielded a series of eight promoter fragments that conferred similar evening-specific rhythmicity with a period of about 24 h (Fig. 4). The strength of the promoter fragment, as indicated by absolute LUC activity, was correlated with the size of the promoter fragment (data not shown), suggesting the presence multiple additive positive elements. At least nine independent lines of T2 seedlings were tested for each construct, and the vast majority (>85%) of the seedlings for any given line were rhythmic (Fig. 4A). In contrast, transgenic lines carrying the two shortest CAT3 promoter fragments tested, −174/+1 and −80/+1, were substantially arrhythmic (Fig. 4A). From these results, we conclude that an element necessary for evening-specific circadian transcription lies in the 25-bp region between −199 and −174 of the CAT3 promoter (Fig. 4D).
Figure 4.
Deletion analysis of the CAT3::LUC promoter reveals an EE that is necessary for evening-specific circadian transcription. A, Summary of the CAT3::LUC promoter resection indicating the proportion of independent transgenic lines expressing evening-specific circadian LUC activity in LL. B, Plants were grown as described in the Figure 1 legend and released into LL. The LD regime is indicated by the bars beneath the traces, with subjective day indicated by white bars and subjective night indicated by gray bars. Traces present average values (± se, n = 12), normalized as described in the legend to Figure 2, from −1,130/+1 CAT3::LUC (blue triangles), −199/+1 CAT3::LUC (red squares), and −174/+1 CAT3::LUC (black circles) seedlings. C, Phase plots of 12 seedlings from single transgenic lines carrying either the −1,130/+1 CAT3::LUC (blue triangles) or the −199/+1 CAT3::LUC (red squares) constructs. D, Nucleotide sequence of the 25-bp CAT3 promoter region between −199 and −174, which is required for rhythmicity and contains the EE, AAAATATCT (highlighted), and the lhc motif, CAN2–4ATC (underlined; Piechulla et al., 1998).
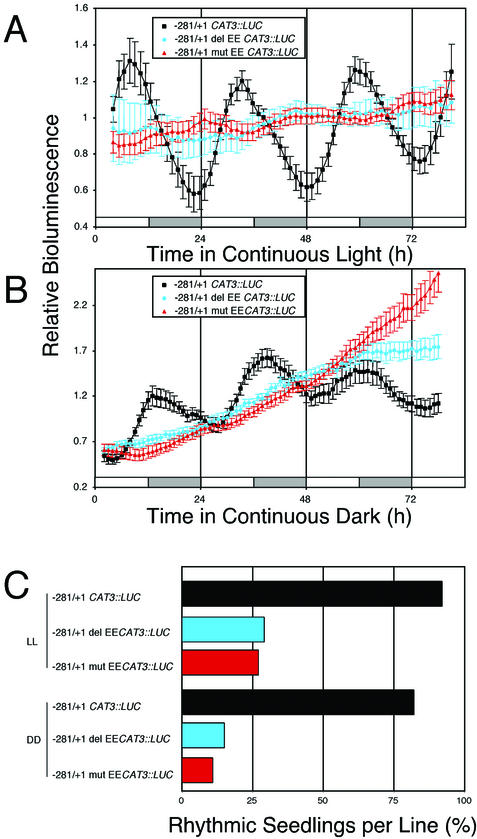
Located between −199 and −174 of the CAT3 promoter is an EE (AAATATCT; Fig. 4D; see Harmer et al., 2000) that is similar to the CBS (AAAAATCT, Wang et al., 1997) or the closely related lhc motif (Piechulla et al., 1998). To determine whether this EE is necessary for evening-specific circadian LUC activity, we performed two loss-of-function experiments. In the context of the −281/+1 CAT3::LUC construct, deletion of a 40-bp region from −194 to −153 that contains the EE (−281/+1 delEE CAT3::LUC) or mutation of three positions (AAATATCT to AtATAgCg; −281/+1 mutEE CAT3::LUC) previously shown by to be important for CCA1 binding to the CBS (Wang et al., 1997) rendered LUC activity substantially arrhythmic (<25% rhythmic seedlings) in both LL and DD conditions (Fig. 5, A–C). Therefore, we conclude that the EE is necessary for evening-specific circadian transcription of the minimal CAT3 promoter, as has been previously demonstrated for the CCR2 and TOC1 promoters (Harmer et al., 2000; Alabadí et al., 2001).
Figure 5.
Deletion and site-directed mutagenesis show that the EE is necessary for circadian-regulated transcription of CAT3::LUC. Plants were grown as described in the Figure 1 legend and released into LL (A) or DD (B). The LD regime is indicated by the bars beneath the traces, with subjective day indicated by white bars and subjective night indicated by gray bars. A, Traces present average values (± se, n = 12), normalized as described in the legend to Figure 2, from −281/+1 CAT3::LUC (black squares), −281/+1 delEE CAT3::LUC (blue circles), and −281/+1 mutEE CAT3::LUC (red triangles) seedlings assayed in LL. B, Traces present average values (± se, n = 12), normalized as described in the legend to Figure 2, from −281/+1 CAT3::LUC (black squares), −281/+1 delEE CAT3::LUC (blue circles), and −281/+1 mutEE CAT3::LUC (red triangles) seedlings assayed in DD. C, Average proportion (%) of seedlings per each independent transgenic line exhibiting circadian rhythmicity in LL and DD.
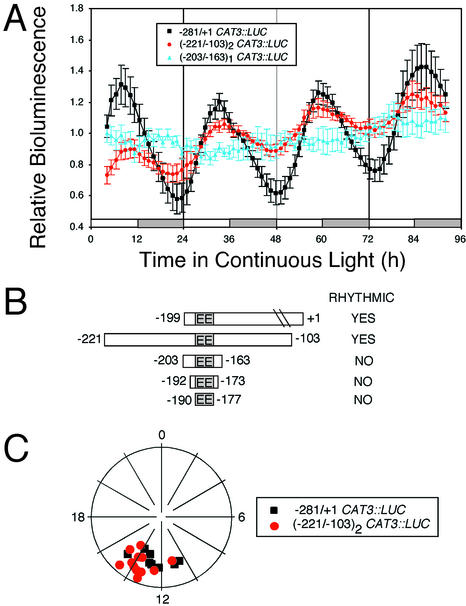
Is the EE sufficient to confer evening-specific circadian transcription? A dimerized 118-bp fragment of the CAT3 promoter encompassing the EE ([−221/−103]2 CAT3::LUC) is sufficient to confer robust evening-specific circadian rhythmicity on the LUC reporter, consistent with the other CAT3 fusions (Fig. 6, A and B). However, monomers of 41 (−203/−163) or 20 bp (−192/−173), or a dimer of 14 bp (−190/−177), each centered on the EE, failed to confer rhythmic LUC transcription (Fig. 6; data not shown).
Figure 6.
Gain of function experiments show that a 118-bp region from the CAT3 promoter is sufficient to confer evening-specific circadian LUC activity. Plants were grown as described in the Figure 1 legend and released into LL. A, Traces present average values (± se, n = 12), normalized as described in the legend to Figure 2, from (−221/−118)2 CAT3::LUC (red circles), −281/+1 CAT3::LUC (black squares), and (−203/−163)1 CAT3::LUC (blue triangles) seedlings. The LD regime is indicated by the bars beneath the traces, with subjective day indicated by white bars and subjective night indicated by gray bars. B, Cartoon comparing CAT3 promoter fragments used in gain-of-function experiments. C, Phase plots of 12 seedlings from one transgenic line for each of the two rhythmic constructs shown in A.
The T to A Difference between CBS and EE Determines Circadian Phase in DD and LD
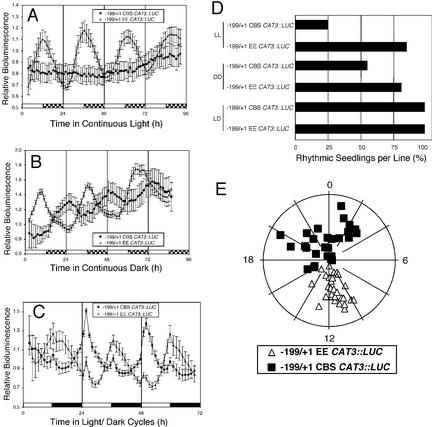
The EE (AAATATCT) is related to the CBS (AAAAATCT; Wang et al., 1997), and both have been shown in vitro to be the targets of the single MYB domain transcription factors CCA1 and LHY (Wang et al., 1997; Alabadí et al., 2001). Functional studies indicate that the EE is important for evening-specific transcription of CCR2 (Harmer et al., 2000) and that the CBS is important for mid-morning-specific transcription of the CAB2 (Carré and Kay, 1995). Because of the difference of only 1 bp between the EE and the CBS, we hypothesized that it is the difference at this single position that is responsible for the distinct phase properties of promoters carrying the two elements. To test this hypothesis, we changed the EE into a CBS (AAATATCT to AAAAATCT) in the −199/+1 CAT3::LUC context (−199/+1 CBS CAT3::LUC). In LL, >85% of seedlings carrying the intact EE (−199/+1 CAT3::LUC) expressed robust evening-specific circadian oscillations, whereas plants carrying the EE to CBS mutation (−199/+1 CBS CAT3::LUC) were substantially arrhythmic (<25% of the plants rhythmic; Fig. 7, A and D). Similar results were obtained when we changed the T to an A in the −333/+1 CAT3::LUC and −281/+1 CAT3::LUC constructs (data not shown). In contrast, the circadian dysfunction resulting from the T to A substitution was less pronounced in DD conditions; 55% of the −199/+1 CBS CAT3::LUC seedlings were rhythmic. Moreover, it is important to note that these rhythmic seedlings displayed the morning-specific phase characteristic of the CBS (Fig. 7, B and E). This is in contrast to the −281/+1 del EE CAT3::LUC or the −281/+1 mut EE CAT3::LUC seedlings, which were arrhythmic in DD conditions (Fig. 5C). Therefore, the morning-specific expression of the −199/+1 CBS CAT3::LUC in DD cannot be simply attributed to loss of EE function. These results suggest that the CBS cannot function properly in the context of the CAT3 promoter in LL but exhibits morning-specific activity in DD.
Figure 7.
The CBS and EE are phase-specific motifs. Plants were grown under a 12/12 LD photoperiod at 22°C for 7 d. Plants were grown as described in the Figure 1 legend and released into LL (A) or into DD (B) or retained in LD cycles (C). Traces present average values (± se, n = 12), normalized as described in the legend to Figure 2, from −199/+1 EE CAT3::LUC (triangles) and −199/+1 CBS CAT3::LUC (squares) seedlings. The LD regime is indicated by the bars beneath the traces, with subjective day indicated by white bars and subjective night indicated by hatched bars. C, The entraining LD cycle is indicated with white and black bars, respectively. D, Average proportion (%) of seedlings per each independent transgenic line exhibiting circadian rhythmicity in LL, DD, and LD cycles. E, Phase plots of all rhythmic seedlings, assayed in DD, from five transgenic lines of −199/+1 EE CAT3::LUC (triangles) and for eight transgenic lines of −199/+1 CBS CAT3::LUC (squares).
Furthermore, we hypothesized that if the clock confers morning-specific activity to the −199/+1 CBS CAT3::LUC in DD, then the circadian clock should drive morning-specific transcription during LD cycles also. In LD, >90% of the −199/+1 EE CAT3::LUC plants display driven circadian rhythms with dusk anticipation. That is, LUC activity increases throughout the light period, peaks at dusk, and declines throughout the dark period, as expected for an evening-specific promoter (Fig. 7C). A small acute response at both dawn and dusk is observed in all promoter luciferase fusions assayed in LD regardless of the promoter used (data not shown). In contrast, >90% of the −199/+1 CBS CAT3::LUC plants exhibit dawn anticipation where LUC activity increases throughout the dark period, peaks at dawn, and declines throughout the light period (Fig. 7C). This demonstrates that the −199/+1 CBS CAT3::LUC plants are responding to circadian clock control.
DISCUSSION
Resection of the CAT3 promoter has revealed that the EE is required for evening-specific circadian clock-regulated CAT3 transcription. Deletion of 40 bp centered on the CAT3 EE or mutation of 3 bp in the CAT3 EE renders CAT3::LUC expression substantially arrhythmic. The EE had been implicated previously in evening-specific clock-regulated transcription of AtGER3 (Staiger et al., 1999) and AtGRP7 (also called CCR2; Staiger and Apel, 1999). The EE was also identified through sequence analysis of the promoters of 31 genes that exhibited circadian oscillations in mRNA abundance that peaked in the evening (Harmer et al., 2000). Our study confirms the necessity of this element through site-directed mutagenesis and is consistent with the results of loss of function mutation of the EEs in the TOC1 and CCR2 minimal promoters (Harmer et al., 2000; Alabadí et al., 2001).
The available data strongly suggests that the EE is a phase-specific circadian clock response element that is necessary to confer not only circadian-regulated transcription but also time-of-day (phase) information. It is quite striking that the EE is closely related to the CBS, which has been identified in morning-specific promoters (Carré and Kay, 1995; Liu et al., 1996; Piechulla et al., 1998; Kellmann et al., 1999) and which differs from the EE by one base (CBS, AAAAATCT, and EE, AAAATATCT). Conversion of the CAT3 EE into a CBS within the context of a CAT3 minimal promoter dramatically reduces rhythmicity in LL; the low frequency of rhythmic plants (<25%) is similar to that seen when EE activity is eliminated by deletion or by site-directed mutation at three positions. Thus, we conclude that the EE to CBS mutation results in a loss of circadian promoter activity in LL. However, conversion of the CAT3 EE into a CBS shifts the phase of transcription from evening to morning in LD and DD. This represents the first attempt to define the mechanism by which the circadian clock imparts time-of-day-specific information to the transcriptional apparatus. Our results reinforce the centrality of the CBS/EE in circadian transcription in Arabidopsis and clearly establish that phase may be modulated through the 1-bp difference between the CBS and EE.
The CBS in the context of the CAT3 promoter functions as a morning-specific element in DD and LD, but fails to impart circadian control in LL. It is reasonable to suppose that the complement of proteins recruited to the promoter differs in light versus dark. For example, mRNA accumulation of CAB2 and CCA1 damps dramatically in the dark, which has been attributed to the depletion of phytochrome in the Pfr form (Kay and Millar, 1993). In contrast, the core clock components LHY and TOC1 robustly oscillate in DD conditions. Because there exist significant differences in the abundance and activity of transcription factors between light and dark (Terzaghi and Cashmore, 1995), it should not be surprising that the activity of the CBS or EE may differ in either condition, reflecting the altered milieu at the promoter environment surrounding the EE/CBS.
Although the EE is necessary for transcription of CAT3, a number of lines of evidence have established that the presence of an EE is insufficient to confer circadian-regulated transcription. For example, the 500-bp CAT1 promoter fragment contains one consensus EE (−124 AAAATATCT −132), yet transcription of the CAT1::LUC construct displays no circadian rhythm. Monomers of 41 or 20 bp, and a 14-bp dimer centered on the CAT3 EE are insufficient to confer robust circadian regulation. Furthermore, the −687/+1 TOC1::LUC retains an EE (−25/−39), yet is substantially arrhythmic (Alabadí et al., 2001). These findings collectively suggest that the EE and the CBS require additional contextual information to confer circadian-regulated transcription. Although a 41-bp fragment of the CAT3 promoter, centered on the EE, is insufficient to drive circadian-regulated transcription of the LUC reporter gene, a 118-bp CAT3 dimer is sufficient to confer robust circadian transcription with wild-type period and evening-specific phase. The implication is that additional information is contained in the additional 78 bp of this larger construct that is essential for the circadian activity of the EE. A minimal promoter consisting of the −199/+1 region of the CAT3 promoter similarly retains rhythmicity, as do minimal CCR2 and TOC1 promoters of 130 and 190 bp, respectively (Harmer et al., 2000; Alabadí et al., 2001). It seems reasonable to hypothesize that there are additional binding activities associated with these promoters that are necessary for circadian transcription. These activities are, themselves, insufficient for circadian transcription because deletion or mutation of the EE eliminates circadian activity. Rather, they provide a permissive context within which the EE can function.
Similar conclusions have been reached regarding circadian transcription in fruitfly. A 69-bp circadian regulatory sequence (CRS) from the period (per) promoter was initially identified as sufficient to confer circadian-regulated transcription (Hao et al., 1997). The CRS is sufficient to confer normal spatial and temporal expression on a per transgene and to drive per expression sufficient to restore normal behavioral rhythms to a per-null mutant (Hao et al., 1999). At the heart of the CRS is the E-box (CACGTG), which binds the dCLOCK-CYCLE heterodimer to drive rhythmic transcription (Darlington et al., 1998; Gekakis et al., 1998; Jin et al., 1999). In mammals, the E box plays a similar role and is bound by heterodimers of the mammalian orthologs, CLOCK and BMAL (Darlington et al., 1998; Gekakis et al., 1998; Jin et al., 1999). However, mutation of the core E-box of either the per or timeless (tim) genes, allows the retention of rhythmic transcription, although transcript levels are reduced (Hao et al., 1997; McDonald et al., 2001). Mutation of other per CRS sequences outside the E-box affects spatial and temporal expression and impairs the restoration of behavioral rhythms to per-null mutants by the driven per transgene (Lyons et al., 2000). Thus, the context of the E-box within the CRS is critical for fully functional spatial and temporal per transcription. The most parsimonious interpretation is that the interaction of other binding activities with dCLOCK-CYCLE bound to the E-box is necessary for wild-type per expression (Darlington et al., 2000; Kyriacou and Rosato, 2000; Lyons et al., 2000). Analysis of the tim promoter identified two non-canonical E-boxes as well as other elements, at least one of which is also found in the per promoter, that each contribute to robust rhythmic transcription (McDonald et al., 2001).
Although elements other than the canonical E-box contribute to rhythmic transcription of both per and tim, a tetramer of an 18-mer centered on the per E-box (and including 6 bp on either side) drives reduced rhythmic per-like LUC expression that displays partial spatial overlap with the pattern conferred by the intact CRS (Darlington et al., 2000). It is thought that multimerization enhances the strength of the element, compensating for the lack of the flanking elements provided in the context of the full CRS (Darlington et al., 2000; Kyriacou and Rosato, 2000). Although the multimerized E-box will drive rhythmic per-like LUC expression, it is not known whether this construct will rescue per-null flies when driving per expression (Darlington et al., 2000; Kyriacou and Rosato, 2000). Moreover, a single E-box is insufficient to drive transcription (Lyons et al., 2000), consistent with our observations that monomers up to 41 bp centered on the CAT3 EE are insufficient to confer circadian-regulated transcription. As with the per E-box, a tetramer of a 36-bp sequence including the CAB2 CBS is sufficient to drive robust morning-specific circadian transcription (Carré and Kay, 1995). It is worth noting that this 36-bp sequence binds at least four distinct factors that do not exhibit circadian oscillation in binding activity (Carré and Kay, 1995; Wang et al., 1997) but that may be providing contextual information.
Thus, we conclude that the EE/CBS are cis-acting elements central to the generation of rhythmic transcription in Arabidopsis and may be analogous to the E-box of fruitfly and mammals. Of course, we would not preclude the possibility of other motif/transcription factors interactions imparting clock regulation to other genes. Like the E-box, the EE and CBS are found in promoters both of clock component genes and of clock-controlled genes that function purely on circadian output loops. Although the EE and CBS have been defined as critical to evening- and morning-specific transcription of some genes, it is clear that the Arabidopsis circadian clock transcribes clock-controlled genes at multiple phases that span the entire day-night cycle (Harmer et al., 2000; Schaffer et al., 2001). There might be a DNA element and a cognate-binding factor for each distinct phase, but it seems more likely that additional information provided by the promoter context modulates activity at the CBS and EE. Combinatorial regulation of promoter activity is well established in light-regulated gene expression (Menkens et al., 1995; Puente et al., 1996; Chattopadhyay et al., 1998) and combinatorial interactions might contribute to the specification of circadian phase-specific promoter activity.
We suggest that one role of the contextual information provided by sequences surrounding the EE/CBS may be to modulate the phase at which the EE/CBS is transcribed. For example, we note that the CAT3 and TOC1 promoter elements described in this study each contain a single EE, yet drive transcription at distinct phases (CT14 versus CT19, respectively). Sequences flanking the CAT3 and TOC1 EEs apparently include an element or elements that function as “phase modifiers.” Alone, these phase modifiers are insufficient to confer rhythmicity but, instead, modulate activity of the EE/CBS to confer distinct phases seen with the two promoters. These phase modifiers might function constitutively to establish a stable phase that is distinct from that inherent in the interaction of the element with its clock-controlled-binding factor (e.g. CAT3 versus TOC1), but also might provide targets to integrate other environmental or developmental information with clock regulation. For example, the phase of both CAB2 and TOC1 transcription is modulated by daylength (Millar and Kay, 1996; Matsushika et al., 2000), which suggests that activities of the CBS and EE in the CAB2 and TOC1 promoters, respectively, are modulated by light- and/or photoperiod-sensitive phase modifiers. The CAB2 and TOC1 promoters both contain the Hexamer (Hex) element (TGACGTGG), a relative of the light-mediated motif, the G-box (CACGTG, curiously identical to the E-box of flies and mammals) that binds G-box-binding factor 1 (Schindler et al., 1992; Menkens et al., 1995). Both the Hex element and G-box are candidates for light-specific phase modifiers. It may be pertinent that casein kinase 2 phosphorylates G-box-binding factor 1 (Klimczak et al., 1992, 1995) in addition to CCA1 and LHY (Sugano et al., 1998, 1999).
Interestingly, a motif related to both the Hex motif and the E-box, the cAMP response element (CRE: TGACGTCA), has also been implicated in circadian transcription of mammalian c-fos and Arg vasopression genes (Robertson et al., 1995; Iwasaki et al., 1997). Multimers of the CRE confer circadian-regulated transcription in both the mouse SCN (Obrietan et al., 1999) and fruitfly (Belvin et al., 1999). CRE elements are present in both the per and tim promoters, although their contribution to circadian-regulated transcription remains unclear (Kyriacou and Rosato, 2000). It is possible that the CRE acts as a phase modifier, or perhaps modulates promoter activity in response to environmental or developmental cues.
The circadian transcriptional machinery must be responsive to environmental and developmental change. Combinatorial regulation in which the activity of core clock components is modulated through interaction with other factors recruited to clock-controlled promoters provides an important mechanism to integrate circadian control of gene expression with other levels of control (Kyriacou and Rosato, 2000). It is thought that interlocked feedback loops contribute to the robustness and stability of the circadian oscillator itself (Glossop et al., 1999; Lee et al., 2000; Shearman et al., 2000). It seems equally reasonable to posit that combinatorial control of rhythmic transcription is also likely to add to the stability of circadian transcription both of core oscillator components and of clock output circuits.
MATERIALS AND METHODS
CAT3::LUCIFERASE (CAT3::LUC) Constructs
CAT3 promoter fragments (−1,130, −850, −540, −455, −335, −281, −239, −199, −174, −80 to +1, where +1 denotes the transcriptional start site of CAT3 [Zhong and McClung, 1996]) were isolated from BAC T10F14 and subcloned into pZPXomegaLUC+ (Schultz et al., 2001). The [−221/−103]2 CAT3::LUC construct was created by digesting the 118-bp fragment from the CAT3 promoter and ligating into pZPXomegaLUC+; the resultant clone carried two tandem copies of the 118-bp fragment inserted in the reverse orientation. −281/+1 delEE CAT3::LUC was created by removing bases −194/−153 by restriction digestion, and religating the resulting CAT3 promoter fragments. The −281/+1 mutEE CAT3::LUC was created using the site-directed mutagenesis primer 5′-GCCCCCACTTCGCTATTATTTTGCTAGGTTTTG-3′ (where the mutated EE is underlined and shown in the inverse orientation). The CAT3::LUC, −335/+1 CBS CAT3::LUC, −281/+1 CBS CAT3::LUC, and −199/+1 CBS CAT3::LUC were made with overlapping primers containing the mutated base. The CAT1::LUC and TOC1::LUC transcriptional fusions contained 500 bp (starting 78 bp upstream of the ATG) and 509 bp (starting 381 bp upstream of the ATG) of their promoter regions, respectively. All constructs were sequenced to confirm fidelity and to check for mutations and/or unwanted DNA fragments introduced by the subcloning process.
Arabidopsis Transformation
Floral dip transformation was performed on different ecotypes (Col, COL CS933; Rschew, RLD CS913; WS, WS CS915; Ler, LER CS20; and Cvi, Cvi CS902) with slight modifications (Clough and Bent, 1998). Agrobacterium tumefaciens strain GV3101 was used in all transformations. T0 seeds were collected, and resistant seeds were selected on 1% (w/v) agar Murashige and Skoog (1962) plates with 70 μL mL−1 gentamicin and 150 μL mL−1 carbenicillin. T1 seedlings were collected and allowed to self, and T2 seeds were collected and analyzed for luciferase activity.
Luciferase Assays
T2 plants containing CAT3::LUC constructs were analyzed using a Packard TopCount luminometer and scintillation counter (Packard) as described (Carré and Kay, 1995). Seeds were vapor-phase sterilized (Clough and Bent, 1998) and plated on 1% (w/v) agar Murashige and Skoog media containing 70 μL mL−1 gentamicin. Seeds were stratified 3 d in the dark at 4°C and then transferred into 12-h white light (70 μmol m−2 s−1)/12-h dark (LD) cycle for 7 d at 22°C. For temperature experiments plants were grown in 12-h 18°C/12-h 22°C in constant white light (70 μmol m−2 s−1). Seedlings were transferred to black microtiter plates (Dynex Technologies, Chantilly, VA) containing, per well, 200 μL of 0.8% (w/v) agar Murashige and Skoog medium plus 2% (w/v) Suc and 35 μL of 0.5 mm luciferin (Biosynth AG, Staad, Switzerland). Microtiter plates were covered with clear plastic TopSeal (Packard) in which holes were placed above each well for seedling gas exchange. Plates were moved to the Packard TopCount and interleaved with four clear plates to allow light diffusion to the seedlings. Seedlings were entrained in white light (15–25 μmol m−2 s−1) for 3 d with 12/12 LD cycles. Luciferase activity was measured every 1 h by integrating photons emitted by seedlings during a 10-s sampling period. DD experiments were conducted as above with the exception that they received DD after they were entrained on the Packard TopCount.
Data Analysis
Data were formatted using Import and Analysis Excel software (Plautz et al., 1997; Strayer et al., 1999). Rhythms were analyzed by fast Fourier transform-nonlinear least squares analysis (Plautz et al., 1997; Zhong et al., 1997). Except in Figure 1, all data were normalized to the average luciferase activity of the individual seedling and are presented as relative bioluminescence. Seedlings were determined to be rhythmic if their period was between 20 and 28 h, the peak signal strength exceeded 100 photons seedling−1 s−1, and the RAE, a measure of the strength of the rhythm, was <1.0. A perfect noise-free cosine wave would return an RAE = 0, because the analytical estimate of rhythmic amplitude would be determined with practically no error. A rhythmic component assessed to have an RAE approaching 1 is contrarily approaching the limit of statistical significance (i.e. RAE = 1 is the limit of statistical significance for any given rhythmic amplitude). For all experiments, between nine and 24 independent T2 lines were tested in a minimum of two independent experiments. All lines, except −174/+1 and −80/+1 CAT3::LUC, contained a proportion of plants that were rhythmic. Because lines varied in the proportion of seedlings that were rhythmic, we established the cutoff that 50% of the seedlings in a given line must be rhythmic for that line to be called “substantially rhythmic.” If fewer than 50% of the seedlings in that line were rhythmic, that line was considered to be “substantially arrhythmic.” All values are presented as mean ± se. CT (phase × 24-h period) allows the normalization of rhythms with different period to ascertain how phase compares in constant conditions. To compare phase of different genes or constructs, phases of individual seedlings are plotted against the strength of the rhythm. Phase (CT) is plotted around the circumference of a 24-h clock face. The strength of the rhythm is plotted along the radius with the strongest rhythms (RAE = 0) at the outer edge of the circle and weakest rhythms (RAE = 1) at the center.
ACKNOWLEDGMENTS
We thank Marty Straume and Carl Strayer for advice on data analysis. pZPXomegaLUC+ and seed of Col carrying CAB2::LUC were generous gifts from Steve Kay. The −281/+1 mutEE CAT3::LUC construct is a generous gift from Patrice Salomé. We thank Jay Dunlap, Allan Froehlich, Mary Lou Guerinot, Kwangwon Lee, and Patrice Salomé for helpful discussions. We thank the Arabidopsis Biological Resource Center for all Arabidopsis accessions used in this study.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN 9817603 to C.R.M.), by the U.S. Department of Agriculture (National Research Initiative-Competitive Grants Program grant no. 9602632 to C.R.M.), and by an institutional grant from the American Cancer Society to the Norris Cotton Cancer Center at Dartmouth College.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004929.
LITERATURE CITED
- Abler ML, Scandalios JG. Regulation of the cyclic repression of the Cat3 catalase gene in maize leaves and roots occurs via the dark/light transition. Maydica. 1994;39:83–88. [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JCP. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré IA, Kay SA. Multiple DNA-protein complexes at a circadian-regulated promoter element. Plant Cell. 1995;7:2039–2051. doi: 10.1105/tpc.7.12.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng X-W, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13:1305–1316. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Lyons LC, Hardin PE, Kay SA. The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J Biol Rhythms. 2000;15:462–471. doi: 10.1177/074873040001500603. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the clock protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Glossop NRJ, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Grundschober C, Delaunay F, Pühlhofer A, Triqueneaux G, Laudet V, Bartfai T, Nef P. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J Biol Chem. 2001;276:46751–46758. doi: 10.1074/jbc.M107499200. [DOI] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognár L, Tóth R, Nagy F, Millar AJ. Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol. 2001;127:1808–1818. [PMC free article] [PubMed] [Google Scholar]
- Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Glossop NRJ, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Förster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 1994;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Oiso Y, Saito H, Majzoub JA. Positive and negative regulation of the rat arginine vasopressin gene promoter. Endocrinology. 1997;138:5266–5274. doi: 10.1210/endo.138.12.5639. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, De Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kay SA, Millar AJ. Circadian-regulated cab gene transcription in higher plants. In: Young MW, editor. The Molecular Genetics of Biological Rhythms. New York: Marcel Dekker; 1993. pp. 73–89. [Google Scholar]
- Kellmann J-W, Hoffrogge R, Piechulla B. Transcriptional regulation of oscillating steady-state Lhc mRNA levels: characterization of two Lhca promoter fragments in transgenic tobacco plants. Biol Rhythm Res. 1999;30:264–271. [Google Scholar]
- Klimczak LJ, Collinge MA, Farini D, Giuliano G, Walker JC, Cashmore AR. Reconstitution of Arabidopsis casein kinase II from recombinant subunits and phosphorylation of transcription factor GBF1. Plant Cell. 1995;4:87–98. doi: 10.1105/tpc.7.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Schindler U, Cashmore AR. DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell. 1992;4:87–98. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou CP, Rosato E. Squaring up the E-box. J Biol Rhythms. 2000;15:483–490. doi: 10.1177/074873040001500605. [DOI] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- Liu Z, Taub CC, McClung CR. Identification of an Arabidopsis Rubisco activase (RCA) minimal promoter regulated by phytochrome and the circadian clock. Plant Physiol. 1996;112:43–51. doi: 10.1104/pp.112.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons LC, Darlington TK, Hao H, Houl J, Kay SA, Hardin PE. Specific sequences outside the E-box are required for proper per expression and behavioral rescue. J Biol Rhythms. 2000;15:472–482. doi: 10.1177/074873040001500604. [DOI] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/pcp/41.6.791. [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002;43:58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 2002;43:118–122. doi: 10.1093/pcp/pcf006. [DOI] [PubMed] [Google Scholar]
- McClung CR, Salomé PA, Michael TP. The Arabidopsis circadian system. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. http://www.aspb.org/publications/arabidopsis . DOI 10.1199/tab.0044 http://www.aspb.org/publications/arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua N-H, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua N-H, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige TR, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Obrietan K, Impe YS, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- Piechulla B, Merforth N, Rudolph B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol. 1998;38:655–662. doi: 10.1023/a:1006094015513. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Robertson LM, Kerpolla TK, Vendrell M, Luk D, Smeyne RJ, Bocchiaro C, Morgan JI, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcriptional control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. LATE ELONGATED HYPOCOTYL, an Arabidopsis gene encoding a MYB transcription factor, regulates circadian rhythmicity and photoperiodic responses. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore AR. DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF-1. EMBO J. 1992;11:1275–1289. doi: 10.1002/j.1460-2075.1992.tb05171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GTJ, Hastings MH et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Staiger D, Apel K. Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol Gen Genet. 1999;261:811–819. doi: 10.1007/s004380050025. [DOI] [PubMed] [Google Scholar]
- Staiger D, Apel K, Trepp G. The Atger3 promoter confers circadian clock-regulated transcription with peak expression at the beginning of night. Plant Mol Biol. 1999;40:873–882. doi: 10.1023/a:1006278030024. [DOI] [PubMed] [Google Scholar]
- Strayer C, Orozco C, Plautz JD, Kay SA (1999) IandA 99.8. http://www.scripps.edu/cb/kay
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang Z-Y, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Tóth R, Kevei É, Hall A, Millar AJ, Nagy F, Kozma-Bognár L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Xu Y, Johnson CH. A clock- and light-regulated gene that links the circadian oscillator to LHCB gene expression. Plant Cell. 2001;13:1411–1426. doi: 10.1105/tpc.13.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, McClung CR. The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol Gen Genet. 1996;251:196–203. doi: 10.1007/BF02172918. [DOI] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, McClung CR. Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell. 1997;9:947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]