Abstract
A “gene-island” sequencing strategy has been developed that expedites the targeted acquisition of orthologous gene sequences from related species for comparative genome analysis. A 152-kb bacterial artificial chromosome (BAC) clone from sorghum (Sorghum bicolor) encoding phytochrome A (PHYA) was fully sequenced, revealing 16 open reading frames with a gene density similar to many regions of the rice (Oryza sativa) genome. The sequences of genes in the orthologous region of the maize (Zea mays) and rice genomes were obtained using the gene-island sequencing method. BAC clones containing the orthologous maize and rice PHYA genes were identified, sheared, subcloned, and probed with the sorghum PHYA-containing BAC DNA. Sequence analysis revealed that approximately 75% of the cross-hybridizing subclones contained sequences orthologous to those within the sorghum PHYA BAC and less than 25% contained repetitive and/or BAC vector DNA sequences. The complete sequence of four genes, including up to 1 kb of their promoter regions, was identified in the maize PHYA BAC. Nine orthologous gene sequences were identified in the rice PHYA BAC. Sequence comparison of the orthologous sorghum and maize genes aided in the identification of exons and conserved regulatory sequences flanking each open reading frame. Within genomic regions where micro-colinearity of genes is absolutely conserved, gene-island sequencing is a particularly useful tool for comparative analysis of genomes between related species.
Many species within the grass family Poaceae provide staple grain and forage supplies for humans and animals and thus are of great economic and humanitarian importance. Members of the grass family are found in wide ranging areas of the world, demonstrating adaptability to diverse environmental conditions. Although genome sizes can vary greatly in the grasses (e.g. 420 Mb and 16,000 Mb for rice [Oryza sativa] and hexaploid wheat [Triticum aestivum], respectively [Arumuganathan and Earle, 1991]), recombinational mapping studies using common DNA markers indicate that gene order is generally conserved within long physical intervals between family members (Hulbert et al., 1990; Ahn and Tanksley, 1993; Van Deynze et al., 1998; Goff et al., 2002). This information has been used to construct comparative genetic maps among many grass species (for review, see Devos and Gale, 1997, 2000; Gale and Devos, 1998). The large variation in genome size observed in the grass family is attributable in part to differences in ploidy and to variation in the abundance of repetitive elements, primarily retrotransposons, located between low copy number genic regions in the genome (SanMiguel et al., 1996). In maize (Zea mays), retrotransposons are estimated to make up 50% to 80% of the genome (SanMiguel and Bennetzen, 1998). The repetitive sequences have little apparent sequence conservation among species (Hulbert et al., 1990; Bennetzen et al., 1994; Chen et al., 1998).
Because of their widespread economic importance and genetic resources, rice and maize have been focal points for studies of genome organization and evolution in the Poaceae (Gaut et al., 2000; Isawa and Shimamoto, 1996). The small size of the rice genome makes it a particularly attractive target for large-scale sequencing projects (Goff et al., 2002; Yu et al., 2002) and physical mapping studies (http://rgp.dna.affrc. go.jp). In contrast, maize presents several challenges because of its relatively large ancient allotetraploid genome (Gaut et al., 2000; Moore, 2000), approximately six times larger than rice, and high levels of retrotransposons and segmental duplications. The sorghum (Sorghum bicolor) genome is approximately one-third the size of the genome of maize, 750 Mb and 2,400 Mb, respectively (Arumuganathan and Earle, 1991). With its smaller and less complex genome, sorghum could serve as a useful complementary species to rice in the study of comparative genomics and evolution of the grasses.
Maize and sorghum are closely related C4 cereal species in the Andropogoneae tribe, separated by approximately 16 million years of evolution (Gaut and Doebley, 1997). Similar to others in the Poaceae family, early linkage studies established orthology in large segments of the maize and sorghum genomes, although numerous segmental duplications and deletions were noted (Hulbert et al., 1990; Binelli et al., 1992; Chen et al., 1997; Helentjaris et al., 1988; Peng et al., 1999). Genetic markers shared between rice and sorghum have been directly compared (Ventelon et al., 2001), and similar conclusions have been reached. In grasses, a limited number of direct sequence comparisons demonstrate that micro-colinearity of genes within long physical intervals is generally preserved among related species, although in some instances, duplications and deletions are present that are only observable at the nucleotide level (for review, see Bennetzen, 2000; Keller and Feuillet, 2000). The likely colinear relationship between large portions of the maize and sorghum genomes coupled with the smaller genome size of sorghum makes comparative map-based cloning studies especially attractive and feasible for this species. Dense genetic maps have been established for sorghum (Whitkus et al., 1992; Melake-Berhan et al., 1993; Chittenden et al., 1994; Dufour et al., 1997; Boivin et al., 1999; Peng et al., 1999; Bhattramakki et al., 2000; Menz et al., 2002). These sorghum maps contain DNA markers common to maize and other grass genetic maps, making cross-referencing between these species possible. Construction of an integrated physical and genetic map for sorghum has recently been initiated, using six-dimensional pooling of sorghum bacterial artificial chromosome (BAC) libraries, coupled with amplified fragment-length polymorphism technology (Klein et al., 2000; http://SorghumGenome.tamu.edu). This resource will facilitate map-based cloning of genes and the acquisition of sorghum gene sequences for comparative sequence analysis.
Two approaches are generally taken to identify a coding region within a given sequence. Computer algorithms have been developed that identify coding regions and exon/intron boundaries based on probability and current knowledge of the sequence composition of exons versus introns. Although indispensable for mass identification of open reading frames in large-scale sequencing projects, this approach is still not without errors in correctly assigning exons and introns (Pavy et al., 1999; Guigó et al., 2000; Pertea and Salzberg, 2002). Moreover, whole open reading frames are sometimes missed using this approach alone. Comparisons of genomic sequences with their cognate full-length cDNAs are useful in defining boundaries between exons and introns, transcription start sites, and regulatory sequences present in the untranslated portions of mRNAs. For most plants, deep collections of full-length cDNA sequences are unfortunately not available. Expressed sequence tag (EST) projects typically yield partial cDNA sequences, inadequate for direct comparison and assembly of entire genes. Identification of a gene by similarity searches through extant databases has a low probability for success for genes expressed at very low levels or for atypically large transcripts because of under representation in a database. Furthermore, no information on regulatory elements in non-transcribed regions can be obtained from the cDNA sequences. Comparative genome sequence analysis between related species is a useful alternative to identify important conserved regions in and around genes, providing clues to gene structure, function, and evolution (Stojanovic et al., 1999; Hardison, 2000; Levy et al., 2001; Ohler and Niemann, 2001; Blanchette and Tompa, 2002). Common developmental and biochemical processes in distantly related species could be identified by their common regulatory elements, functionally conserved throughout evolution, leading to a more refined understanding of signal transduction pathways and global controls over cellular functions.
To gain further understanding of gene density and the organization of genes in the sorghum genome, a BAC clone containing a 152-kb region encoding PHYA and other genes was sequenced. A direct hybridization procedure was developed that expedites the targeted acquisition and analysis of orthologous gene sequences between related species, such as sorghum, maize and rice. This technique has been used to identify the conserved sequences surrounding the orthologous PHYA genes of sorghum, maize, and rice. The “gene-island” sequencing technology can be applied to any study requiring targeted gene sequencing, such as map-based cloning projects.
RESULTS
Sequencing Sorghum BAC sbb3766
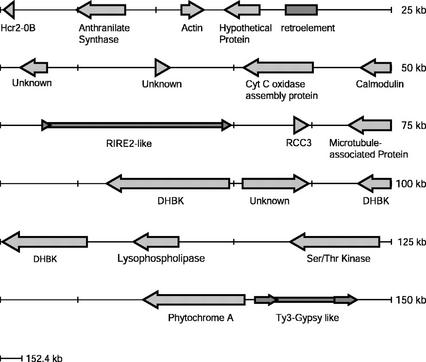
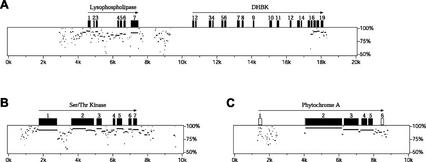
Sorghum BAC sbb3766 was identified by screening an ordered library of 13,440 BAC clones (Woo et al., 1994) with a sorghum PHYA cDNA clone (Childs et al., 1997). The sequence of sorghum BAC sbb3766 consists of 152,439 bp (accession no. AF369906). Not including the retrotransposon sequences, 16 separate open reading frames were identified within this region of DNA (Fig. 1; Table I). Twelve genes possess sequence similarity to previously identified genes from various species. Two of these genes, Phytochrome A (PHYA) and Actin, code for proteins with sequence identity to known proteins. The remaining 10 genes are putatively identified based on their closest protein homologs. Only a truncated portion of the putative Hcr2-0B gene is located on the end of BAC sbb3766. A gene coding for a hypothetical protein, identified by computer algorithms, has sequence similarity to a hypothetical protein in Arabidopsis. Three genes coding for unknown proteins were identified based on EST sequence similarity. One of the unknown proteins, identified by EST accession no. BG051875, has sequence similarity to the conceptual translation of a full-length cDNA coding for a protein of unknown function in Arabidopsis (accession no. AAL27510).
Figure 1.
Diagrammatic representation of genes and repetitive elements on sorghum BAC sbb3766 (accession no. AF369906). Genes are labeled according to putative identity. Arrows indicate the direction of transcription.
Table I.
Predicted genes on Sorghum bicolor BAC sbb3766 (accession no. AF369906)
| Predicted Genea | Position | Strand | Best BLASTX | Matchb | BLASTX E Value |
|---|---|---|---|---|---|
| Hcr2-0B protein, putative | 402–653 | − | BAB21161 | (Os) | 4e-26 |
| Anthranilate synthase, β-chain, putative | 4,902–8,041 | − | BAB08859 | (At) | 8e-64 |
| Actin | 11,596–13,035 | + | CAA34356 | (Os) | 0.0 |
| Hypothetical protein, conserved | 14,643–16,639 | − | CAB37558 | (At) | 5e-44 |
| Unknown protein | 26,253–28,029 | − | BG101726c | (Sb) | – |
| Unknown protein | 35,169–35,675 | + | BG051875c | (Sp) | – |
| Cytochrome c oxidase assembly protein, putative | 40,536–44,958 | − | AAG00893 | (At) | 1e-20 |
| Calmodulin, putative | 48,236–50,542 | − | P04464 | (Ta) | 9e-43 |
| Root-specific protein RCC3, putative | 69,137–69,337 | + | AAA65513 | (Os) | 3e-11 |
| Microtubule-associated protein, putative | 72,681–75,118 | − | CAC17794 | (Nt) | 7e-43 |
| 3,4-Dihydroxy-2-butanone kinase, putative | 81,679–89,546 | − | O04059 | (Le) | 9e-27 |
| Unknown protein | 89,962–94,688 | + | BI074510c | (Sb) | – |
| 3,4-Dihydroxy-2-butanone kinase, putative | 97,876–105,441 | − | O04059 | (Le) | 3e-25 |
| Lysophospholipase, putative | 108,545–111,366 | − | AAM47308 | (Os) | 2e-51 |
| Ser/Thr Protein kinase, putative | 118,584–124,256 | − | AAG30976 | (At) | 1e-88 |
| Phytochrome a | 134,066–140,636 | − | AAB41397 | (Sb) | 0.0 |
Predicted gene designation corresponds to closest protein homolog found in GenBank.
GenBank accession no. of closest protein homolog and species. At, Arabidopsis; Le, Lycopersicon esculentum; Nt, Nicotiana tabacum; Os, rice; Sb, sorghum; Sp, Sorghum propinquum; Ta, wheat. Dashes (–) indicate no protein homologs found in GenBank.
EST nucleotide sequence identified by BLASTN.
Cross-Species Gene-Island Sequencing
To acquire the sequence of maize PHYA and other nearby genes for comparison with the corresponding sorghum genes, an 80-kb maize BAC containing PHYA (ZmPHYA) was identified by screening of a maize BAC library with the sorghum PHYA cDNA probe. This BAC clone was initially shotgun sequenced at low coverage (192 sequences). Low-pass sequencing revealed the presence of four genes (PHYA, lysophospholipase, dihydroxybutanone kinase (DHBK), and Ser/Thr kinase) also located in the sorghum BAC sequence described above (Fig. 1; Table I). However, the majority (approximately 80%) of the DNA sequences initially acquired from maize BAC ZmPHYA corresponded to unknown or repeat sequences, indicating that random sequencing was not an efficient way to acquire gene sequences, at least in this region of the maize genome. On the basis of prior analysis by Bennetzen and colleagues (1994), we predicted that orthologous gene sequences in the sorghum and maize genome would be highly conserved and cross-hybridize but that repetitive sequences would not cross-hybridize. Therefore, sorghum BAC DNA could be used as a hybridization probe to identify the subset of maize BAC subclones derived from regions with high sequence conservation, thereby targeting sequence analysis to the gene-islands present in the maize BAC.
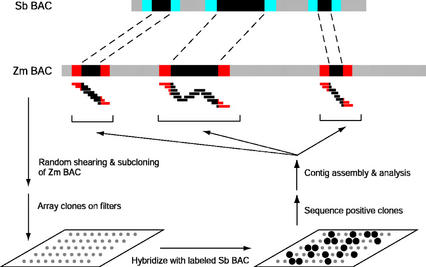
The gene-island sequencing approach was tested by shearing the maize PHYA-containing BAC DNA into 1- to 3-kb fragments to create a library of random subclones (Fig. 2). Approximately 1,200 ZmPHYA subclones were arrayed on nylon filters and probed with radiolabeled DNA from sorghum PHYA BAC sbb3766. The complete sorghum BAC was labeled without further separation of the sorghum DNA insert from the pBeloBAC11 vector. Several factors were employed to reduce the amount of background hybridization and increase the signal-to-noise ratio on the membranes. After isolation of relatively pure sbb3766 BAC DNA, the DNA preparation was further processed with an ATP-dependent DNase that selectively degrades linear DNA molecules. This minimized bacterial genomic DNA contamination in the subsequent random primer labeling reaction, which was necessary to reduce general background hybridization with the bacterial genomic DNA on the filters. A preblocking step with BAC vector sequences was carried out before the addition of radioactive probe to reduce hybridization of labeled sbb3766 vector sequences with the portions of the maize subclones containing the lacZ region of pBluescript and with subclones containing random fragments of the vector pBeloBAC11. Both measures were necessary to minimize the background hybridization so that positively hybridizing clones could be visualized (Fig. 3, compare A and B).
Figure 2.
Diagrammatic outline of the gene-island sequencing protocol. A maize (Zm) BAC is sheared and subcloned into a standard cloning vector. Zm subclones arrayed on filters are hybridized with an orthologous, radiolabeled sorghum (Sb) BAC. Only low-copy regions conserved between the Sb and Zm BACs will hybridize. Positive Zm clones are identified and sequenced. Contigs are assembled and analyzed. Black areas along hypothetical BACs indicate conserved open reading frames. Colored areas flanking open reading frames represent non-transcribed regions.
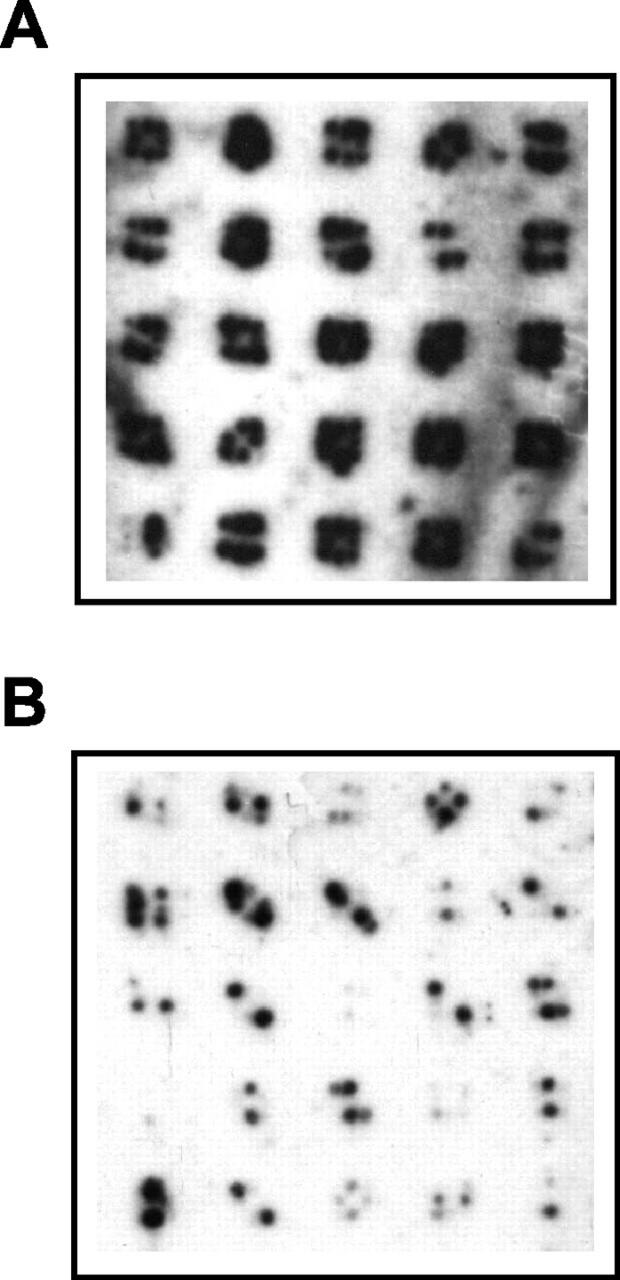
Figure 3.
Gene-island filter hybridization. Representative regions of duplicate filters arrayed with random maize ZmPHYA subclones either untreated (A) or treated (B) with specific blocking DNAs before hybridization with labeled sorghum BAC sbb3766. Clones were grown on filters in duplicate 3 × 3 arrays overnight and processed before hybridization.
Of the 1,152 ZmPHYA subclones screened, approximately 20% produced a positive hybridization signal, indicating that they had some sequence similarity to the sorghum BAC sbb3766 probe. Plasmids from these clones were isolated and sequenced bidirectionally. After sequence editing, four contigs were assembled, totaling about 26.5 kb with an average coverage within the contigs of approximately 6.7×. Approximately 25% of the plasmids sequenced corresponded to unknown sequences or repetitive elements, which did not form significant contigs. Only a small fraction of the sequenced clones contained fragments of the vector pBeloBAC11, indicating that the blocking step with pBSBeloBAC11 and pBSBeloBAC-NotI was quite efficient in suppressing hybridization with vector sequences. In addition, the radioactive signal derived from clones containing the unknown or vector sequences was generally much lower than that from the clones containing low copy number sequences (data not shown).
Sequence similarity searches carried out with the assembled maize sequence contigs identified four separate open reading frames, corresponding to DHBK (e = 8 × 10−20, accession no. O04059), lysophospholipase (8.7 kb; e = 2 × 10−57, accession no. AAG30967), Ser-Thr kinase-like protein (4.7 and 6.3 kb; e = 1 × 10−119, accession no. AC012396), and PHYA (6.7 kb; accession no. AAB41397; Fig. 4). The sequence of the contig containing the maize PHYA gene was identical to a previously published maize PHYA sequence, phyA1, localized on chromosome 1L (Christensen and Quail, 1989). Two of the assembled contigs, containing contiguous portions of the Ser/Thr kinase-like gene, are separated by a gap located within the intron between exons three and four of the sorghum gene. For all contigs, approximately 0.5 to 1.0 kb of sequence immediately flanking both ends of the putative coding regions was also obtained. The depth of sequence coverage was greatest over the putative coding regions within the contigs with sequence coverage tapering off closer to the ends of the genes.
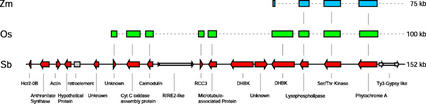
Figure 4.
Diagrammatic representation of genes present on sorghum BAC sbb3766 and their orthologs identified on maize BAC ZmPHYA and rice BAC OsPHYA. Regions conserved between sorghum BAC sbb3766 (Sb) and ZmPHYA (Zm) or OsPHYA (Os) were identified by direct hybridization of labeled sbb3766 to subclones of either BACs ZmPHYA or OsPHYA. Positively hybridizing subclones were sequenced. The resulting sequences were compared with the genes located on sorghum BAC sbb3766 (Sb). The order of genes on ZmPHYA and OsPHYA is assumed, based on the order of genes on sbb3766. Sizes reflect the size of the entire BAC insert.
The maize contigs were individually compared with the sorghum sbb3766 sequence using percent identity plots (PIPs; Schwartz et al., 2000; Fig. 5). A total physical interval of approximately 40 kb of the sorghum genome is covered by the maize contigs derived from BAC ZmPHYA (80 kb). Alignments between the conserved regions of sorghum and maize exhibited 80% to 95% similarity, closely corresponding to the predicted exons of the sorghum genes. High levels of similarity between sorghum and maize lysophospholipase are difficult to discern in the PIP plots because of the relatively short lengths of the exons (Fig. 5). The maize DHBK sequence is similar to the 3′ end of the sorghum ortholog, not to the 5′ end as would be expected if the gene arrangement were completely conserved, suggesting that a rearrangement of these genes has taken place. Sequence similarity significantly declines immediately flanking the exons, which may aid in exon detection and intron/exon boundary definition. Short, localized regions of high sequence conservation in the 5′-untranslated regions may represent conserved regulatory elements or transcription factor binding sites (see below and Fig. 6).
Figure 5.
PIPs comparing sorghum genes from BAC sbb3766 with orthologous maize sequences derived from cross-species hybridization. Regions shown in A, B, and C correspond to regions 96 to 116 kb, 116 to 126 kb, and 134 to 144 kb of the sbb3766 sequence, respectively. Presumptive sorghum exons and untranslated regions are represented as black boxes above the PIPs. Direction of transcription is indicated by the arrows.
Figure 6.
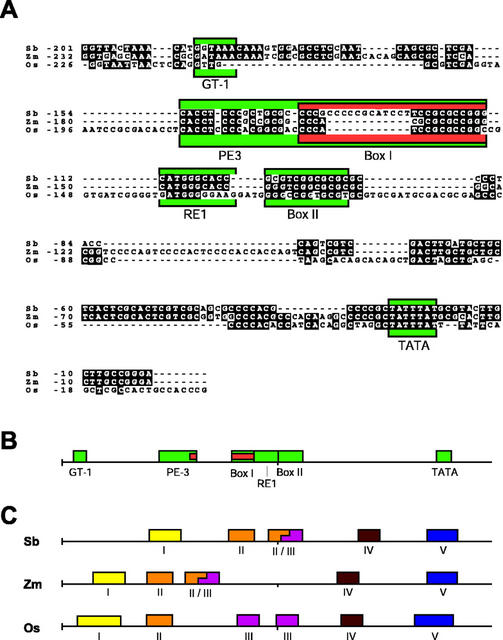
A, Alignment of sorghum, maize, and rice PHYA promoter regions. Numbers designate basepairs upstream from the presumed transcription initiation site. Black boxes denote identical nucleotides. Dashes indicate introduction of gaps for optimal alignment. Functionally conserved motifs within the promoters are outlined with colored boxes (see text for details). B, Linear representation of the sorghum PHYA promoter (−200 to −1 bp) and location of functionally conserved motifs denoted in A. C, Linear representation of highly conserved sequence motifs identified by FootPrinter along the PHYA promoters (−200 to −1 bp) of sorghum (Sb), maize (Zm), and rice (Os). Numbers correspond to related sequence motifs.
Comparative Analysis of the Sorghum and Maize PHYA Genes
The sorghum PHYA gene is highly similar to the PHYA gene of maize (this study; Christensen and Quail, 1989) and rice (Kay et al., 1989). The positions of the six exons and five introns are conserved among these species. Between sorghum and maize, the coding regions were 95% similar, whereas intron sequences were 85% similar. An 86% level of similarity in the coding regions was observed between either sorghum or maize and the PHYA gene from rice (data not shown). The first intron relative to the transcription initiation site is substantially shorter in maize than in sorghum or rice with relatively short regions of sequence similarity present in this intron. Comparison of the 5′-promoter regions of maize and sorghum reveal a relatively high level of sequence conservation from nucleotide positions −201 to −1, relative to the transcription initiation site in sorghum (Fig. 6A). Several motifs within the promoter, previously shown to control light-regulated expression, are present in sorghum (Fig. 6A). The motif RE1 is involved in the light-mediated transcriptional repression of PHYA (Bruce et al., 1991; Dehesh et al., 1994) and other negatively regulated genes (Neuhaus et al., 1997). PE3 promotes PHYA transcription under dark conditions (Bruce and Quail, 1990; Bruce et al., 1991). Conserved PE3 and RE1 sequences are also present in Arabidopsis and pea (Dehesh et al., 1994). Box I and II are sequence motifs conserved in monocot PHYA promoters (Christensen and Quail, 1989). No other conserved regions could be identified farther upstream from the transcription initiation site.
Conserved regulatory motifs in the PHYA promoter of sorghum, maize, and rice were identified using FootPrinter, a computer algorithm designed to identify short, conserved sequence motifs in orthologous genes from divergent species (Blanchette et al., 2002; Blanchette and Tompa, 2002). FootPrinter identified five highly conserved sequence motifs in the PHYA promoter within −200 bp of the transcriptional start site (Fig. 6C). A pair wise analysis between the sorghum and maize promoters yielded the same conserved motifs (data not shown). Three of the conserved motifs corresponded to sequences demonstrated to be functionally involved in the light-regulated expression of the PHYA gene in monocots (Fig. 6, compare B and C). One conserved motif identified by FootPrinter corresponded to the TATA-box region. No additional areas of sequence conservation beyond −200 bp of the PHYA transcription start site were identified. Three-way analyses of the Ser/Thr kinase and lysophospholipase promoters also revealed multiple, highly conserved motifs up to 1.0 kb upstream from the translational start sites of the genes (data not shown). In each case for the three species examined, the linear order of the conserved sequence motifs remained constant with respect to one another, although the spacing between the motifs varied to different degrees (Fig. 6C), further suggesting an evolutionarily conserved functional significance for these motifs.
Identification of the Coding Regions Near the PHYA Gene in Rice
The utility of the gene-island sequencing strategy was further investigated using a more distantly related species. A 100-kb rice BAC clone (OsPHYA) containing the PHYA gene was identified in a rice BAC library (Zhang et al., 1996) by filter hybridization with a sorghum PHYA cDNA probe. To identify the DNA sequences conserved between sorghum and the PHYA-containing rice BAC, cross-species hybridization was carried out between labeled sbb3766 and random subclones of OsPHYA. Positive clones were isolated and sequenced. After assembly of the resulting sequences from the positive subclones, nine open reading frames in common with sbb3766 were identified within the OsPHYA BAC (Fig. 4). The region corresponded to a physical distance of about 100 kb in the sorghum BAC sbb3766, revealing similar gene densities within this region of the genomes of the two species. The rice orthologs of two genes, DHBK and an unknown open reading frame, arranged in tandem on sbb3766 were not identified among the rice sequences, suggesting that a deletion, duplication, or translocation has taken place in this region after the evolutionary split between rice and sorghum. A fully sequenced rice BAC (accession no. AF377946) partially overlaps with the sequence of sorghum BAC sbb3766. Within the 35-kb overlapping segment, micro-colinearity between four genes, PhyA, protein kinase, lysophospholipase, and DHBK, is observed (data not shown).
DISCUSSION
Sorghum is a close relative of maize, but with a substantially smaller genome, creating an ideal situation for comparative genomic studies. Genetic mapping studies with common DNA markers have shown that physical order has been generally preserved along relatively large intervals of these genomes. The conservation of gene order at the micro-colinear level has more recently been demonstrated in smaller regions of the two genomes. To further our understanding of the evolutionary relationship between the genomes of sorghum and maize, we have sequenced a region of the sorghum genome flanking the PHYA locus and compared genes encoded in this region to genes by the orthologous region of the maize genome.
Gene Density in Sorghum
Determining the distribution of genes and repetitive elements in whole genomes is an important problem to be addressed by comparative genomics and has broad implications for identification of genes in species with large genomes. Genes may be randomly dispersed throughout the genome or exist in more localized gene dense regions, separated from blocks of repetitive sequences. Because total DNA content for various grasses varies 40-fold, it would be expected that if genes were dispersed randomly throughout the genome, gene density in any given region would directly reflect the total DNA content of the organism. As an alternative, if genes were segregated from the repetitive sequences, gene density in these regions among species would be more comparable. Gene localization studies using DNA from different grass species fractionated on CsCl density gradients indicate that the majority of genes are clustered within small portions of the nDNA (Barakat et al., 1997). These gene dense regions are separated by large stretches of DNA largely lacking in gene sequences. Substantial differences in genome size among plant species were concluded to be attributable to the amount of DNA lacking coding sequences. For Arabidopsis, using similar separation techniques, it was determined that genes are more widely dispersed throughout the genome because of the relatively low level of repetitive sequences (Barakat et al., 1998). Gene density values determined by direct sequencing around orthologous loci in grass species with small and large genomes vary only 2- to 5-fold, a value relatively small considering the disparate sizes of grass genomes (e.g. Feuillet and Keller, 1999; Tikhonov et al., 1999; Tarchini et al., 2000). The data presented in this study further substantiate the notion that genes are concentrated in particular regions of the sorghum genome with other regions being relatively gene poor.
Within the 152.4-kb region near sorghum PHYA, 16 open reading frames were identified by sequence analysis and similarity searches, indicating a gene density of one gene per 9.5 kb. This value is much higher than the expected average gene density of about one gene per 20 kb if 35,000 genes are present in the 750-Mb genome of sorghum. The gene density value for BAC sbb3766 closely corresponds to the calculated gene density within a 339.5-kb region of the rice genome (Tarchini et al., 2000) despite the estimated 1.6X larger genome of sorghum. It has similarly been shown that the sh2 and a1 loci are separated by approximately the same physical distance in rice and sorghum (Chen et al., 1997), further indicating a similar gene density in some regions of the two genomes. The gene density around the PHYA locus of sorghum is approximately one-half the overall gene density observed in Arabidopsis (Arabidopsis Genome Initiative, 2000) despite a genome approximately one-fifth the size of sorghum. The data also indicate that the gene density is approximately two times greater in this portion of the sorghum genome, when compared with the orthologous region in maize. A density within the maize ZmPHYA BAC of one gene per 20 kb is in approximate agreement with the value for the 225-kb region surrounding the Adh1 locus of maize (Tikhonov et al., 1999).
Comparative Sequence Analysis by Gene-Island Sequencing
Comparative sequence analysis has become a major resource for the functional analysis of evolutionarily conserved regions among genomes. Large-scale sequencing efforts can provide comparative information on two levels: (a) conservation of gene order (co-linearity) between species; and (b) conservation of nucleotide sequence from two or more species in important areas of the genome. Comparisons of large genomic segments have demonstrated that regulatory elements and protein coding regions in genomic DNA tend to remain conserved, even between distantly related species, whereas the similarities in intergenic and intron regions are reduced (Koop, 1995; Chen et al., 1997; Hardison et al., 1997). Comparative genomics in animal model systems have relied on complete sequencing of large regions surrounding orthologous loci, followed by computer analysis to identify common sequences. Although this approach is useful, a great deal of effort is required to obtain the raw sequence data.
For systems in which large-scale sequencing is less tractable, alternative methods are required to extract useful comparative information within a locus of interest. In a previous study, the conserved coding regions in a relatively large physical interval were identified by local genomic cross-referencing, in which a sorghum BAC containing the Adh1 locus was used to identify conserved low copy number sequences within a contig of λ-subclones derived from a YAC, containing a 280-kb region surrounding the maize Adh1 locus (Avramova et al., 1996). Sequence information from the low-copy number λ-clones, identified by this approach, required further subcloning of the λ-inserts, because each insert contained 15 to 25 kb of DNA. The cross-species hybridization method described in this report has the advantage that with an insert size of 1 to 3 kb the sequence of each of the subclones was easily determined without further manipulation of the inserts. By assembly of overlapping sequences, entire low copy regions were determined in one step. For the BAC clones tested, the method yielded nearly the same amount of information as a full-scale BAC sequencing project, but with a greatly reduced level of time and effort. In the case presented here, about one-fifth the cost was expended to sequence the entire low-copy regions of the maize and rice BACs. The regions flanking the conserved domains were also identified, providing an expeditious means to obtain the sequence of promoter regions. Because the conditions used for hybridization in this study were sufficient to identify orthologous sequences between sorghum and rice, two relatively divergent species, it would be expected that the technique is applicable to most grass family members without a wholesale change inhybridization conditions.
The gene-island sequencing method described herein is easily scaleable to accommodate larger physical intervals or contigs of BACs. A labeled BAC clone from one species may be used to probe an arrayed BAC library from a related species. Given sufficient similarity and overlap within conserved low-copy regions, orthologous BAC clones may be easily distinguished. In regions of the maize genome with lower gene density, comparative gene-island sequencing would be even more efficient. High-throughput sequencing strategies are currently under development that selectively target gene-rich, low-copy, hypomethylated regions of the genome (Rabinowicz et al., 1999; Peterson et al., 2002). Because the gene-island sequencing method can be targeted to genomic regions of special interest or low representation, it should be a useful complementary approach to genome-wide sequencing projects.
We have compared the orthologous region near the PHYA genes of three members of the Poaceae family by direct cross-species hybridization and sequencing. The gene-island sequencing method applied in this study was very efficient in selectively identifying genic regions for sequence analysis. The results show that the gene composition surrounding the PHYA locus has been generally preserved between sorghum, maize, and rice (Fig. 4). Although the gene-island sequencing approach is useful to identify orthologous sequences, no conclusions can be drawn regarding the overall preservation of gene order within the BAC subjected to this type of sequence analysis. Furthermore, the method will not identify genes present on the maize or rice BACs, if the ortholog is absent from the sorghum BAC. However, single-pass sequencing of subclones derived from the maize and rice BACs did not reveal additional open reading frames (data not shown), suggesting that the full complement of genes on the BACs was uncovered by cross-species hybridization. Gene duplications or rearrangements within a BAC sequence will not affect the analysis of orthologous sequences using the gene-island sequencing approach. For example, one maize contig contained orthologs to both sorghum DHBK and lysophospholipase. The maize DHBK gene is in the opposite orientation to the sorghum ortholog, indicating that a rearrangement has taken place. If the genes were completely colinear, it would be expected that similarity to the 5′ end of the sorghum DHBK gene would be observed, in accordance with its orientation with respect to the lysophospholipase gene (Fig. 4). Only a portion of the maize DHBK gene is found on BAC ZmPHYA, so it is not possible to further deduce the nature of the rearrangement. A rearrangement in this portion of the genome is further substantiated by the apparent absence of two genes corresponding to genes coding for a second DHBK and a protein of unknown function in the rice BAC (Fig. 4).
Identification of Conserved Regulatory Elements in Promoters
Comparative DNA sequence analysis between divergent species can be used to identify conserved sequences in both coding and non-coding regions of genomic DNA (Stojanovic et al., 1999; Hardison, 2000; Blanchette and Tompa, 2002). With sequence comparisons of this nature, the phylogenetic relationship between the species used in the study is an important variable to consider. Regulatory elements are generally short (10–20 nucleotides), buried within longer stretches of DNA sequence. During speciation, functionally important motifs will be conserved because of selective pressures. Closely related species with less time to accumulate genetic mutations would be expected to have a higher overall level of sequence similarity than highly divergent species, potentially masking true functional domains. Cross-species comparison of the PHYA promoters of sorghum and maize (15–20 million years divergence) reveals conserved sequence motifs within about 200 bp upstream of the transcriptional start site (Fig. 6). Many of these elements have been demonstrated to confer biological activity. Further analysis incorporating the PHYA promoter sequence from rice, separated from maize and sorghum by about 50 million years, reveals the same conserved sequence domains (Fig. 6). These data suggest that the sequence divergence between maize and sorghum is sufficient to identify biologically relevant promoter motifs by cross-species analysis. Despite their relatively close phylogenetic relationship, sufficient evolutionary time has elapsed so that the functionally important sequences were conserved. Comparison of promoter elements carried out between Arabidopsis and cauliflower (separated by 14–20 million years; Yang et al., 1999) similarly revealed significant blocks of conservation in the promoter regions (Colinas et al., 2002). Together, these data indicate that the comparative analysis of promoter sequences between even closely related species provides unique sequence information that can be used for defining conserved DNA regulatory motifs.
MATERIALS AND METHODS
DNA Constructs and Subcloning
A BAC clone (sbb3766) containing an insert of 152 kb was identified from a sorghum (Sorghum bicolor) BTx623 BAC library (Woo et al., 1994) by hybridization with a sorghum PHYA cDNA clone (accession no. U56729; Childs et al., 1997). Large-scale isolation of sbb3766 BAC DNA was carried out using the Maxi-prep kit (Qiagen USA, Valencia, CA), according to the instructions provided by the manufacturer for low copy number plasmids. Purified BAC DNA was further treated with Plasmid-Safe DNase (Epicenter Technologies, Madison, WI) overnight at 37°C, according to the instructions provided by the manufacturer, to remove residual Escherichia coli genomic DNA. BAC DNA was randomly sheared by passage through a nebulizer (Aeromist, catalog no. 4207, IPI Medical Products, Chicago) at 10 p.s.i. for 2.5 min at 4°C (Roe et al., 1996). The sheared DNA was blunt-ended with T4 and Klenow DNA polymerases. The DNA fragments were size-fractionated using a 1% (w/v) low-gelling point agarose gel (FMC Bioproducts, Rockland, ME). Size fractions were excised from the gel and used directly for blunt-end cloning into the dephosphorylated EcoRV site of pBluescript (SK−) (Stratagene, La Jolla, CA). Plasmid was isolated from insert-containing clones using the 5Prime/3Prime plasmid isolation kit.
For probe synthesis sorghum BAC sbb3766 DNA was isolated using the Qiagen Mini Plasmid prep kit, according to the BAC DNA isolation protocol provided by the manufacturer. BAC DNA was subsequently treated with Plasmid-Safe DNase (Epicenter Technologies) overnight at 37°C.
A BAC clone (ZmPHYA) containing an 80-kb region of the maize (Zea mays) genome was identified by hybridization with a sorghum PHYA cDNA clone (Childs et al., 1997). A 100-kb BAC clone (OsPHYA) containing the rice (Oryza sativa) PHYA ortholog was similarly identified by screening a rice cv Teqing BAC library (Zhang et al., 1996) with the sorghum PHYA cDNA clone. Subcloning of 1- to 3-kb fragments of OsPHYA and ZmPHYA were carried out as for subcloning of sorghum BAC sbb3766. Clones containing inserts were randomly selected and stored in 96-well plates at −80°C before further use.
The plasmid vectors pBeloBAC11 and pBluescript share regions of significant sequence similarity around their respective multiple cloning regions. To reduce the level of potential background hybridization between radiolabeled pBeloBAC11 vector DNA and the arrayed subclones derived from pBluescript and also with subclones containing random fragments of the pBeloBAC11 vector, two plasmids were constructed and used in the prehybridization blocking step (see below). A 630-bp NotI fragment from pBeloBAC11, containing regions of high sequence similarity to pBluescript, including the lacZ gene, was cloned into pBluescript (SK−) to form pBSBeloBAC-NotI. To increase pBeloBAC11 DNA yields, pBSBeloBAC11 was constructed by ligating pBluescript (SK−) into the BamHI/HindIII sites of pBeloBAC11, thereby providing a high copy number origin of replication in the pBeloBAC11 vector. To reduce the potential for recombination of large inserts, pBeloBAC11 had originally been designed to be maintained in E. coli at a level of one to two copies per cell (Shizuya et al., 1992). Yields of pBSBeloBAC11 were increased five to 10 times over standard pBeloBAC11 yields (data not shown). pBSBeloBAC-NotI and pBSBeloBAC11 plasmid DNAs were isolated using the Qiagen Giga kit following the protocol provided by the manufacturer.
Hybridization
Randomly selected OsPHYA and ZmPHYA BAC subclones in 96-well plates were arrayed on Hybond-N+ membranes (Amersham Biosciences AB, Uppsala), overlaying LB plates supplemented with 100 μg mL−1 ampicillin. Plates were incubated at 37°C for 12 to 14 h. Colonies were processed according to the protocol provided by the manufacturer. Membranes were prehybridized in 1.0 m NaCl, 1% (w/v) SDS, 2× Denhardt's solution, 10% (w/v) dextran sulfate, and 100 μg mL−1 sheared and denatured salmon sperm DNA (Sigma-Aldrich, St. Louis) for 6 to 8 h at 65°C. After the nonspecific prehybridization step, heat-denatured pBSBeloBAC-NotI and pBSBeloBAC11 plasmid were added to the prehybridization buffer, so that each had a final concentration of 20 μg mL−1 buffer. Prehybridization continued for an additional 6 to 8 h. sbb3766 BAC DNA was labeled by random priming. Labeled BAC DNA was denatured by boiling and was added to the hybridization bottles to a final concentration of 1 × 106 cpm mL−1. Additional heat-denatured pBSBeloBAC-NotI and pBSBeloBAC11 plasmid were added with the radioactive probe to a final concentration of 5 μg DNA mL−1 hybridization buffer. Hybridizations were carried out at 65°C for 14 to 16 h. Membranes were washed at 65°C, twice with 2× SSC/0.5% (w/v) SDS and twice with 0.1× SSC/0.5% (w/v) SDS (1× SSC contains 150 mm NaCl and 15 mm sodium citrate). Filters were placed under photographic film with an intensifying screen at −80°C.
Sequencing and Contig Assembly
For sequencing of sorghum BAC, plasmids of random sbb3766 subclones were isolated using 5Prime/3Prime plasmid isolation kits. Plasmids were sequenced in either direction using SK (5′-CGCTCTAGAACTAGTGGATC-3′) or KS (5′-CTCGAGGTCGACGGTATCG-3′) primers. Sequencing was carried out using ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kits with FS AmpliTaq DNA polymerase. Standard sequencing reactions included 2 μL of plasmid DNA (typically 150–300 ng), 10 pmol of primer, 1 μL of FS AmpliTaq reaction mix (Applied Biosystems, Foster City, CA), 1 μL of 5× reaction buffer (5× = 400 mm Tris, pH 9.0, and 10 mm MgCl2), and 5 μL of dH2O. Sequencing reactions were carried out using an ABI 9600 thermocycler (Applied Biosystems) with the following cycling parameters: an initial denaturation at 95°C for 2 min, followed by 50 cycles of 95°C, 10 s; 50°C, 5 s; and 60°C, 4 min. Extension products were purified by G-50 Sephadex spin columns (Sigma-Aldrich) and dried in a vacuum evaporator (Savant Instruments, Holbrook, NY). Extension products were separated on an ABI 377 sequencer (Applied Biosystems). Some clones were sequenced using BigDye Terminator mix v.2.0 and an ABI 3700 DNA sequencer. To fill gaps in the sequence, clones bridging underrepresented areas of the working sbb3766 sequence were identified and used to probe of additional sbb3766 subclones arrayed on nylon filters. The corresponding clones were sequenced, and the information was used to supplement sequence data in underrepresented areas.
For sequencing positive maize and rice clones, plasmid templates for sequencing were isolated from positive clones by an automated plasmid preparation system using Wizard SV 96 DNA-binding plates (Promega, Madison, WI). Plasmids were sequenced in either direction using SK or KS primers. Standard sequencing reactions included 2 μL of plasmid DNA, 10 pmol of primer, 1 μL of BigDye Terminator mix v2.0 (Applied Biosystems), 1 μL of 5× reaction buffer (5× = 400 mm Tris, pH 9.0, and 10 mm MgCl2), and 5 μL of dH2O. Sequencing reactions were carried out using an ABI 9700 thermocycler with the following cycling parameters: an initial denaturation at 95°C for 2 min, followed by 99 cycles of 95°C, 10 s; 50°C, 5 s; and 60°C, 4 min. Extension products were purified by isopropanol precipitation. Products were separated on an ABI 3700 DNA sequencer.
Sequence Analysis
Sequences were edited and assembled using Sequencher (v3.1, Gene Codes Corp., Ann Arbor, MI). Sequence analysis was performed using the NIX program package (http://www.hgmp.mrc.ac.uk/NIX/), a World Wide Web tool to view the results from several different DNA analysis programs. The sequences were analyzed using the settings for grasses. Open reading frames were further analyzed by BLASTN and BLASTX sequence similarity searches (Altschul et al., 1997). PIP comparisons between sorghum and maize sequences were carried out using PIPmaker (Schwartz et al., 2000), using the default settings. Low complexity regions of the sbb3766 sequence were masked using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; A.F.A. Smit and P. Green, unpublished data). Promoters were aligned using FootPrinter (http://bio.cs.washington.edu/software.html; Blanchette et al., 2002; Blanchette and Tompa, 2002), a computer algorithm designed to identify conserved motifs in non-coding regions of orthologous genes from multiple species. Motifs were identified using a window size of 10 to 12 with maximum mutations of zero to two.
ACKNOWLEDGMENTS
We thank Drs. Patricia E. Klein and Robert R. Klein for their fruitful suggestions and critical reading of the manuscript. We gratefully acknowledge Julie McCollum for her expert assistance with automated plasmid preps and sequencing.
Footnotes
This material is based on work supported by the National Science Foundation (grant no. 0077713), by the Texas Agricultural Experiment Station, and by the Perry Adkisson Chair in Agricultural Biology.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.012567.
LITERATURE CITED
- Ahn S, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Avramova Z, Tikhonov A, SanMiguel P, Jin Y-K, Liu C, Bennetzen JL. Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- Barakat A, Carels N, Bernardi G. The distribution of genes in the genomes of Gramineae. Proc Natl Acad Sci USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Matassi G, Bernardi G. Distribution of genes in the genome of Arabidopsis thalianaand its implications for the genome organization of plants. Proc Natl Acad Sci USA. 1998;95:10044–10049. doi: 10.1073/pnas.95.17.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schrick K, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- Bhattramakki D, Dong J, Chhabra AK, Hart G. An integrated SSR and RFLP linkage map of Sorghum bicolor(L.) Moench. Genome. 2000;43:988–1002. [PubMed] [Google Scholar]
- Binelli G, Gianfranceschi L, Pe ME, Taramino G, Busso C, Stenhouse J, Ottaviano E. Similarity of maize and sorghum genomes as revealed by maize RFLP probes. Theor Appl Genet. 1992;84:10–16. doi: 10.1007/BF00223975. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Schwikowski B, Tompa M. Algorithms for phylogenetic footprinting. J Comput Biol. 2002;9:211–223. doi: 10.1089/10665270252935421. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Tompa M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 2002;12:739–748. doi: 10.1101/gr.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin K, Deu M, Rami J-F, Trouche G, Hamon P. Towards a saturated sorghum map using RFLP and AFLP markers. Theor Appl Genet. 1999;98:320–328. [Google Scholar]
- Bruce WB, Deng X-W, Quail PH. A negatively acting DNA sequence element mediates phytochrome-directed repression of phyAgene transcription. EMBO J. 1991;10:3015–3024. doi: 10.1002/j.1460-2075.1991.tb07852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce WB, Quail PH. cis-Acting elements involved in photoregulation of an oat phytochrome promoter in rice. Plant Cell. 1990;2:1081–1089. doi: 10.1105/tpc.2.11.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, SanMiguel P, Bennetzen JL. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, SanMiguel P, DeOliveira AC, Woo S-S, Zhang H, Wing RA, Bennetzen JL. Microcolinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Miller FR, Cordonnier-Pratt M-M, Pratt LH, Morgan PW, Mullet JE. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 1997;113:611–619. doi: 10.1104/pp.113.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden LM, Schertz KF, Lin YR, Wing RA, Paterson AH. A detailed RFLP map of Sorghum bicolor × S. propinquum, suitable for high-density mapping, suggests ancestral duplication of sorghum chromosomes or chromosomal segments. Theor Appl Genet. 1994;87:925–933. doi: 10.1007/BF00225786. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Structure and expression of a maize phytochrome-encoding gene. Gene. 1989;85:381–399. doi: 10.1016/0378-1119(89)90431-9. [DOI] [PubMed] [Google Scholar]
- Colinas J, Birnbaum K, Benfey PN. Using cauliflower to find conserved non-coding regions in Arabidopsis. Plant Physiol. 2002;129:451–454. doi: 10.1104/pp.002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K, Franci C, Sharrock RA, Somers DE, Welsch JA, Quail PH. The Arabidopsisphytochrome A gene has multiple transcription start sites and a promoter sequence motif homologous to the repressor elements of monocot phytochrome A genes. Photochem Photobiol. 1994;59:379–384. doi: 10.1111/j.1751-1097.1994.tb05051.x. [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Comparative genetics in the grasses. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour P, Deu M, Grivet L, D'Hont A, Paulet F, Bouet A, Lanaud C, Glaszmann JC, Hamon P. Construction of a composite sorghum genome map and comparison with sugarcane, a related complex polyploid. Theor Appl Genet. 1997;94:409–418. [Google Scholar]
- Feuillet C, Keller B. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, d'Ennequin ML, Peek AS, Sawkins MC. Maize as a model for the evolution of plant nuclear genomes. Proc Natl Acad Sci USA. 2000;97:7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Guigó R, Agarwal P, Abril JF, Burset M, Fickett JW. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 2000;10:1631–1642. doi: 10.1101/gr.122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: a reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Helentjaris T, Weber DL, Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988;118:353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Richter TE, Axtell JD, Bennetzen JL. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc Natl Acad Sci USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa T, Shimamoto K. Becoming a model plant: the importance of rice to plant science. Trends Plant Sci. 1996;1:95–99. [Google Scholar]
- Kay SA, Keith B, Shinozaki K, Chua N-H. The sequence of the rice phytochrome gene. Nucleic Acids Res. 1989;17:2865–2866. doi: 10.1093/nar/17.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B, Feuillet C. Colinearity and gene density in grass genomes. Trends Plant Sci. 2000;5:246–251. doi: 10.1016/s1360-1385(00)01629-0. [DOI] [PubMed] [Google Scholar]
- Klein PE, Klein RR, Cartinhour SW, Ulanch PE, Dong J, Obert JA, Morishige DT, Schlueter SD, Childs KL, Ale M et al. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res. 2000;10:789–807. doi: 10.1101/gr.10.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop BF. Human and rodent DNA sequence comparisons: a mosaic model of genomic evolution. Trends Genet. 1995;11:367–371. doi: 10.1016/s0168-9525(00)89108-8. [DOI] [PubMed] [Google Scholar]
- Levy S, Hannenhalli S, Workman C. Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics. 2001;17:871–877. doi: 10.1093/bioinformatics/17.10.871. [DOI] [PubMed] [Google Scholar]
- Melake-Berhan A, Hulbert SH, Butler LG, Bennetzen JL. Structure and evolution of the genomes of Sorghum bicolor and Zea mays. Theor Appl Genet. 1993;86:598–604. doi: 10.1007/BF00838715. [DOI] [PubMed] [Google Scholar]
- Menz MA, Klein RR, Mullet JE, Obert JA, Unruh NC, Klein PE. A high-density genetic map of Sorghum bicolor(L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Mol Biol. 2002;48:483–499. doi: 10.1023/a:1014831302392. [DOI] [PubMed] [Google Scholar]
- Moore G. Cereal chromosome structure, evolution and pairing. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:195–222. doi: 10.1146/annurev.arplant.51.1.195. [DOI] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Hiratsuka K, Yamagata H, Chua N-H. Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 1997;16:2554–2564. doi: 10.1093/emboj/16.10.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Niemann H. Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet. 2001;17:56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- Pavy N, Rombauts S, Dehais P, Mathe C, Ramana DV, Leroy P, Rouze P. Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thalianasequences. Bioinformatics. 1999;15:887–899. doi: 10.1093/bioinformatics/15.11.887. [DOI] [PubMed] [Google Scholar]
- Peng Y, Schertz KF, Cartinhour S, Hart GE. Comparative genome mapping of Sorghum bicolor(L.) Moench using an RFLP map constructed in a population of recombinant inbred lines. Plant Breed. 1999;118:225–235. [Google Scholar]
- Pertea M, Salzberg SL. Computational gene finding in plants. Plant Mol Biol. 2002;48:39–48. [PubMed] [Google Scholar]
- Peterson DG, Schulze SR, Sciara EB, Lee SA, Bowers JE, Nagel A, Jiang N, Tibbitts DC, Wessler SR, Paterson AH. Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res. 2002;12:795–807. doi: 10.1101/gr.226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, Stein L, McCombie WR, Martienssen RA. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- Roe BA, Crabtree JS, Khan AS. DNA Isolation and Sequencing. Essential Techniques Series. New York: John Wiley & Sons; 1996. [Google Scholar]
- SanMiguel P, Bennetzen JL. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot. 1998;82:37–44. [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker: a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coliusing an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic N, Florea L, Riemer C, Gumucio D, Slightom J, Goodman M, Miller W, Hardison R. Comparison of five methods for finding conserved sequences in multiple alignments of gene regulatory regions. Nucleic Acids Res. 1999;27:3899–3910. doi: 10.1093/nar/27.19.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramova Z. Colinearity and its exceptions in orthologous adhregions of maize and sorghum. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze AE, Sorrells ME, Park WD, Ayres NM, Fu H, Cartinhour SW, Paul E, McCouch SR. Anchor probes for comparative mapping of grass genera. Theor Appl Genet. 1998;97:356–369. [Google Scholar]
- Ventelon M, Deu M, Garsmeur O, Doligez A, Ghesquiere A, Lorieux M, Rami JF, Glaszmann JC, Grivet L. A direct comparison between the genetic maps of sorghum and rice. Theor Appl Genet. 2001;102:379–386. [Google Scholar]
- Whitkus R, Doebley J, Lee M. Comparative genome mapping of sorghum and maize. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S-S, Jiang J, Gill BS, Paterson AH, Wing RA. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-W, Lai K-N, Tai P-Y, Li W-H. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassicaand other angiosperm lineages. J Mol Evol. 1999;48:597–604. doi: 10.1007/pl00006502. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK-S, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhang H-B, Choi S, Woo S-S, Li Z, Wing RA. Construction and characterization of two rice bacterial artificial chromosome libraries from the parents of a permanent recombinant inbred mapping population. Mol Breed. 1996;2:11–24. [Google Scholar]