Abstract
S-adenosylhomocysteine hydrolase (SAH) is a key enzyme in the maintenance of methylation homeostasis in eukaryotes because it is needed to metabolize the by-product of transmethylation reactions, S-adenosylhomocysteine (AdoHcy), which causes by-product inhibition of methyltransferases (MTase's). Complete loss of SAH function is lethal. Partial loss of SAH function causes pleiotropic effects including developmental abnormalities and reduced cytosine methylation. Here we describe a novel partial-function missense allele of the Arabidopsis SAH1 gene that causes loss of cytosine methylation specifically in non-CG contexts controlled by the CMT3 DNA MTase and transcriptional reactivation of a silenced reporter gene, without conferring developmental abnormalities. The CMT3 pathway depends on histone H3 lysine 9 methylation (H3 mK9) to guide DNA methylation. Our results suggest that this pathway is uniquely sensitive to SAH impairment because of its requirement for two transmethylation reactions that can both be inhibited by AdoHcy. Our results further suggest that gene silencing pathways involving an interplay between histone and DNA methylation in other eukaryotes can be selectively impaired by controlled SAH downregulation.
S-ADENOSYLMETHIONINE (AdoMet) serves as the methyl-group donor for transmethylation reactions including modification of DNA, RNA, proteins, and lipids, generating S-adenosylhomocysteine (AdoHcy) as a by-product (reviewed in Chiang 1998). In eukaryotes, AdoHcy is further metabolized by S-adenosylhomocysteine hydrolase (SAH) to yield homocysteine and adenosine. The SAH-catalyzed reaction is critical to relieve by-product inhibition of methyltransferases (MTase's) by AdoHcy. Consequently, loss of SAH function can have pleiotropic effects. For example, null mutations in the major Arabidopsis gene encoding SAH (At4g13940, hereafter referred to as SAH1) are embryonic lethal (Rocha et al. 2005). Similarly, complete loss of SAH function in the mouse is embryonic lethal (Miller et al. 1994). Partial inactivation of plant SAH activity by missense mutations or anti-sense RNA expression confers developmental abnormalities including slow growth, reduced fertility, and reduced size (Tanaka et al. 1997; Rocha et al. 2005). In humans partial loss of SAH function confers severe defects, including a lack of myelin in the central nervous system, implicating perturbation of lipid biosynthesis as a key component of the SAH deficiency (Baric et al. 2004, 2005). Nonetheless, controlled downregulation of SAH has potential beneficial outcomes, including suppression of viral replication in plants and mammals (Masuta et al. 1995; Chiang 1998; De Clercq 2004) and inhibition of mammalian parasites (Bujnicki et al. 2003; Rapp et al. 2006).
Among the diverse transmethylation reactions controlled by SAH, cytosine methylation has received particular attention because this modification plays a key role in regulating gene expression and genome stability via heterochromatin formation in mammals and plants. In mammalian and plant cells, suppression of SAH by chemical inhibitors or anti-sense RNA results in reduced cytosine methylation (Wolfson et al. 1986; Tanaka et al. 1997; Fojtova et al. 1998). Similarly, hog1 missense mutations in the Arabidopsis SAH1 gene cause reduced cytosine methylation on a transgene reporter and ribosomal DNA (rDNA) repeats (Rocha et al. 2005). One mechanism for this reduction is AdoHcy inhibition of cytosine MTase activity. Such inhibition has been demonstrated in vitro for the mammalian Dnmt1 enzyme (Bacolla et al. 1999). However, in some cases cytosine methylation is targeted by histone protein methylation. For example, in the fungus Neurospora crassa, mutation of either the histone H3 lysine 9 (H3 K9) MTase DIM-5 or mutation of the K9 residue of H3 confers genomewide loss of cytosine methylation (Tamaru and Selker 2001). In Arabidopsis, mutation of the H3 K9 MTase SUVH4/KYP (hereafter referred to as SUVH4) confers reduced cytosine methylation in non-CG sequence contexts that are controlled by the DNA MTase CMT3 (Jackson et al. 2002; Malagnac et al. 2002). In mouse embryonic stem cells, double mutation of the related Suv39h1 and Suv39h2 H3 K9 MTase's, or mutation of the G9a H3 K9 MTase, results in loss of cytosine methylation from specific regions of the genome (Lehnertz et al. 2003; Xin et al. 2003). Thus, modulation of SAH function can potentially disrupt DNA methylation patterning through both cytosine MTase and histone MTase targets.
Here we describe a missense mutation in the Arabidopsis SAH1 gene, sah1L459F, that confers reduced genomic cytosine methylation specifically in non-CG sequences controlled by CMT3. In contrast to the hog1 missense mutants in SAH1 that display phenotypic abnormalities including slow growth and reduced fertility (Rocha et al. 2005), the sah1L459 mutant is morphologically normal. Nonetheless, this weak allele is sufficient to cause depletion of non-CG methylation and reactivation of a silenced reporter gene. Our results suggest that CMT3-dependent non-CG methylation is uniquely susceptible to even slight perturbations in SAH activity because it relies on a pathway involving two types of enzymes that can be inhibited by AdoHcy, the SUVH H3 K9 MTase's and the CMT3 cytosine MTase. Simultaneous inhibition of these enzymes, even at a very low level, can thus result in a synergistic effect on the DNA methylation output of the CMT3 pathway.
MATERIALS AND METHODS
Isolation of the sah1L459F mutation:
The sah11L459F mutation was isolated from a previously described screen for mutations that reduce PAI2 silencing in the Wassilewskija (Ws) pai1 background, resulting in a reduced blue fluorescence phenotype (Bartee et al. 2001). The sah1L459F allele segregated in crosses as a single recessive mutation. The mutation was mapped as previously described for cmt3 mutations isolated in the PAI2 silencing suppressor screen (Bartee et al. 2001). Briefly, the Ws pai1 sah1L459F mutant was crossed to the polymorphic strain Nd-0, which has a similar arrangement of methylated PAI genes to Ws. F2 progeny of this cross with the weak fluorescent phenotype diagnostic of individuals homozygous for both the pai1 mutation and the silencing suppressor mutation were isolated and used to identify the genomic region that cosegregated with the suppressor mutation. The mapping analysis localized the mutation to a region of ∼43 kb on the lower arm of chromosome 4. Within this region, we focused on the SAH1 gene as a likely candidate to alter methylation metabolism. Sequencing of SAH1 from the mutant identified a single C:G to T:A mutation that changed a leucine to a phenylalanine, consistent with ethyl methanesulfonate (EMS) mutagenesis used on the Ws pai1 strain. To facilitate genetic analysis, a PCR-based dCAPS (Neff et al. 1998) marker was designed, where the base change created by the sah1L459F mutation is combined with a mismatch at the end of a nearby PCR primer to create a restriction site polymorphism. For this marker, PCR products amplified with DRAF 5′ CAGCTCGAGCTCTGGAACGAGAAAGCAAGC 3′ and STUR 5′ GTCAGATTGGTCCTTTGACAGCTTTGTAG 3′ were cleaved with StuI: sah1L459F yields a 144-bp fragment and the wild type yields 117- and 27-bp fragments.
The pai1 sah1L459F mutant was two times outcrossed with the wild-type Ws to segregate pai1 sah1L459F and sah1L459F strains used in this analysis. Other DNA methylation-deficient strains used for comparison to sah1L459F in Figures 2, 3, and 5 were previously described: cmt3i11a (Bartee et al. 2001), suvh4R302* (Malagnac et al. 2002), the met1-1 allele (Kankel et al. 2003) introgressed into the Ws background (Bartee and Bender 2001), and the Ws drm1 drm2 double mutant (Cao and Jacobsen 2002b). Southern blot assays for DNA methylation were performed as previously described (Melquist et al. 1999). Previously described Southern blot probes for the PAI genes (Melquist et al. 1999), the Mu1 DNA transposon (Lippman et al. 2003), the Ta3 retrotransposon (Johnson et al. 2002), the MEA-ISR repeat (Cao and Jacobsen 2002a), and the 180-bp centromere repeat (Vongs et al. 1993) were used.
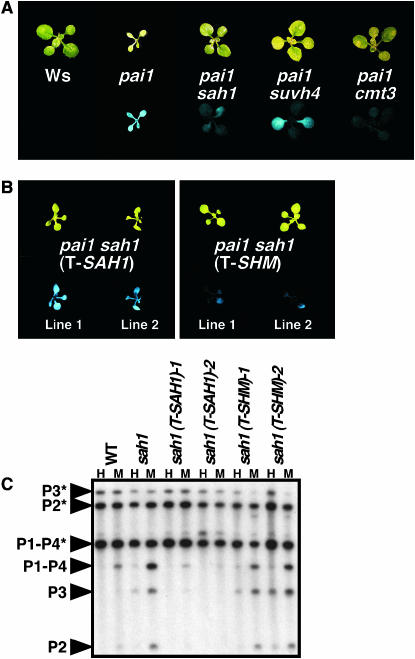
Figure 2.—
The sah1L459F mutation suppresses PAI2 silencing. (A and B) Two-week-old seedlings of the indicated genotypes grown on agar medium were photographed under visible (top) or UV (bottom) light, with sah1 indicating sah1L459F. (A) Representative seedlings of mutants isolated from the PAI2 silencing suppressor screen. (B) Representative T2 generation seedlings of Ws pai1 sah1L459F transformed with either an SAH1 (T-SAH1) or a control SHM (T-SHM) genomic transgene. (C) Genomic DNAs prepared from 4-week-old plants of the indicated genotypes were cleaved with HpaII (H) or MspI (M) and used for Southern blot analysis with a PAI probe. P1–P4 is PAI1–PAI4, P2 is PAI2, and P3 is PAI3, with asterisks indicating the positions of species methylated at PAI-internal sites. WT is Ws pai1, sah1 is Ws pai1 sah1L459F, and the remaining strains are Ws pai1 sah1L459F transformed with the indicated transgenes, corresponding to the transgenic lines shown in B.
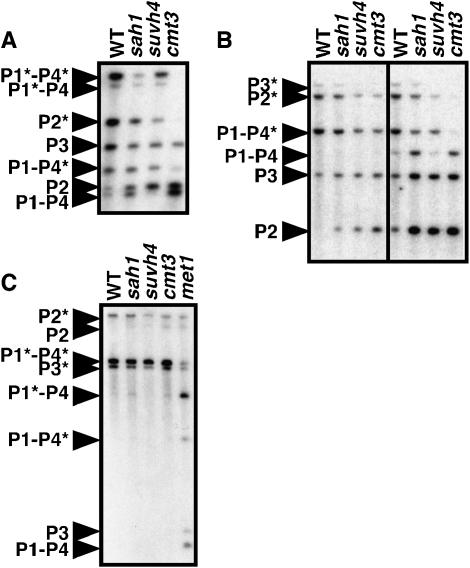
Figure 3.—
The sah1L459F mutation reduces PAI non-CG methylation. Genomic DNA prepared from 4-week-old plants of the indicated genotypes was cleaved with (A) HincII, (B) HpaII (left) or MspI (right), or (C) ClaI and used for Southern blot analysis with a PAI probe. P1–P4 is PAI1–PAI4, P2 is PAI2, and P3 is PAI3, with asterisks indicating the positions of species methylated at PAI-internal sites. All strain backgrounds carried the pai1 mutation. WT is Ws pai1 and sah1 is Ws pai1 sah1L459F.
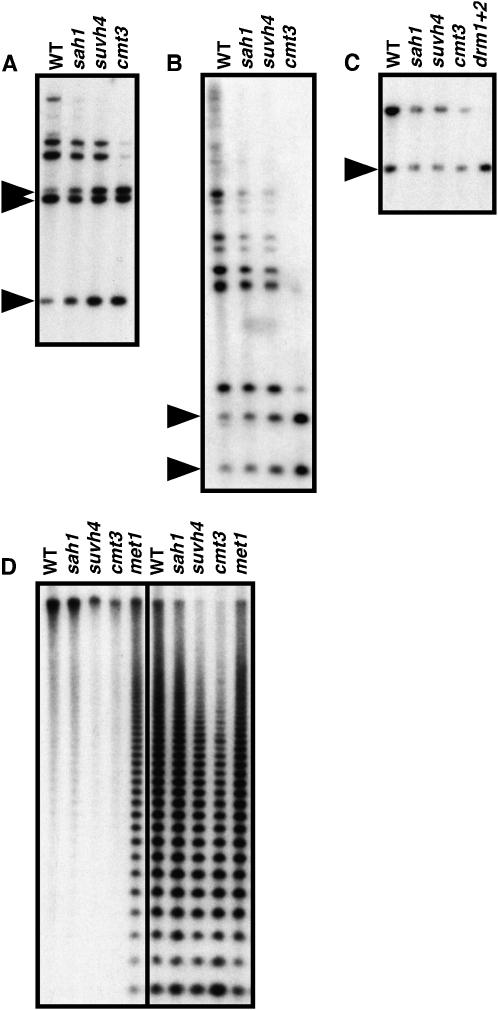
Figure 5.—
The sah1L459F mutation reduces non-CG methylation on transposon sequences, similarly to suvh4 and cmt3 mutations. Genomic DNA prepared from 4-week-old plants of the indicated genotypes was cleaved with the indicated restriction enzymes and used for Southern blot analysis with the indicated probes. (A) DNA digested with MspI and HindIII and probed for Mu1. (B) DNA digested with MspI and probed for Ta3. (C) DNA digested with MspI and probed for MEA-ISR. (D) DNA digested with HpaII (left) or MspI (right) and probed for the 180-bp centromere repeat. WT is the wild-type Ws, sah1 is sah1L459F, and drm1+2 is Ws drm1 drm2. Arrowheads in left margins indicate the positions of fully cleaved species.
Plant transformation with the T-SAH1 and T-SHM genomic clones:
A 5.5-kb SnaBI Ws genomic fragment carrying the SAH1 gene was cloned into the SmaI site of the pBIN19 plant transformation vector (Bevan 1984) to make the T-SAH1 construct (Figure 1B). A 7.4-kb XbaI-SmaI Ws genomic fragment carrying the neighboring SHM gene was cloned into the XbaI and SmaI sites of pBIN19 to make the T-SHM control construct. Both constructs were transformed into the pai1 sah1L459F strain using the floral dip method (Clough and Bent 1998).
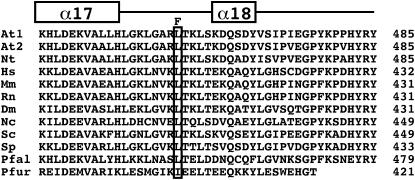
Figure 1.—
The sah1L459F mutation affects a conserved residue in the SAH carboxy-terminal domain. The aligned predicted carboxy-terminal amino acid sequences of SAH from a variety of organisms are shown. These sequences correspond to the region extending from α-helix α17 through α-helix α18 to the carboxy terminus of the protein in the crystal structure of the human SAH protein (Turner et al. 1998), as indicated over the alignment. The residue corresponding to L459 in the Arabidopsis SAH1 protein is boxed, with the mutation to F indicated over this position. The coordinate number of the carboxy-terminal residue for each amino acid sequence is shown in the right margin. At1 is Arabidopsis SAH1, At2 is Arabidopsis SAH2, Nt is Nicotiana tabacum, Hs is Homo sapiens, Mm is Mus musculus, Rn is Rattus norvegicus, Dm is Drosophila melanogaster, Nc is Neurospora crassa, Sc is Saccharomyces cerevisiae, Sp is Schizosaccharomyces pombe, Pfal is Plasmodium falciparum, and Pfur is Pyrococcus furiosus.
Complementation of a yeast sah1 mutant:
A Saccharomyces cerevisiae diploid strain heterozygous for an sah1 deletion was obtained from Open Biosystems (no. 20176, YSC1021-669867) in strain background BYB4743 (MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 met15Δ0/MET1 lys2Δ0/LYS2). A DNA fragment containing the S. cerevisiae SAH1 promoter and full-length coding sequence (ScSAH1) was amplified by PCR and cloned into the polylinker of pRS315 (LEU2 CEN), pRS316 (URA3 CEN), pRS325 (URA3 2μ), or pRS425 (LEU2 2μ) yeast vectors (Sikorski and Hieter 1989). The same ScSAH1 promoter sequence was fused to the start codon of the Arabidopsis SAH1 cDNA (AtSAH1, obtained from the Arabidopsis Biological Resource Center), the sahlL459F cDNA (Atsah1L459F, derived from the SAH1 cDNA by site-directed mutagenesis), or the SAH2 cDNA (AtSAH2, amplified from Ws RNA by reverse transcriptase–PCR). These expression cassettes were cloned into the polylinker of pRS315 or pRS425.
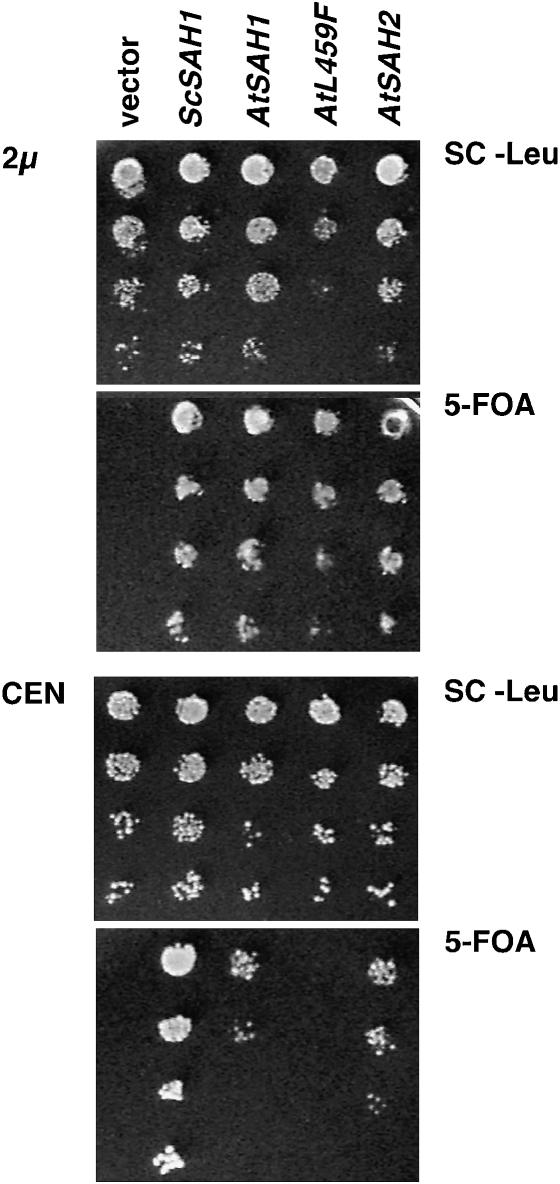
The heterozygous sah1/SAH1 yeast strain was transformed with either the ScSAH1 URA3 CEN plasmid or the ScSAH1 URA3 2μ plasmid. Tetrads were dissected from sporulated transformants and analyzed for genetic and molecular markers to identify Δsah1 (ScSAH1 URA3 CEN) and Δsah1 (ScSAH1 URA3 2μ) haploid strains. These strains were supertransformed with LEU2 CEN or LEU2 2μ plasmids carrying no insert or with ScSAH1, AtSAH1, Atsah1L459F, or AtSAH2 expression cassettes. Cultures were grown overnight in selective SC −Leu broth and density was determined by spectrophotometry at 600 nm. A series of 10-fold dilutions of each culture, starting at ∼10,000 cells per spot and ranging down to ∼10 cells per spot, were spotted on either SC −Leu medium or 0.1% 5-fluoroorotic Acid (5-FOA) (United States Biological F5050) medium. Plates were photographed after 2 days of growth at 30° (Figure 6).
Figure 6.—
Arabidopsis sah1L459F can complement a yeast SAH deficiency when overexpressed. Haploid yeast deleted for SAH1 and carrying a complementing yeast SAH1 (ScSAH1) gene on a URA3 vector were supertransformed with ScSAH1 or the Arabidopsis SAH1 (AtSAH1), sah1L459F (AtL459F), or SAH2 (AtSAH2) genes all expressed from the yeast SAH1 promoter carried on a LEU2 vector. As a negative control the strain was also transformed with the empty LEU2 vector. Both high-copy 2μ and low-copy CEN vectors were tested. Serial dilutions of cultures grown from each strain were spotted in parallel on medium lacking leucine to indicate the total number of supertransformed cells plated in each spot (SC −Leu) or on medium containing 5-FOA to select against URA3 cells.
Chromatin immunoprecipitation:
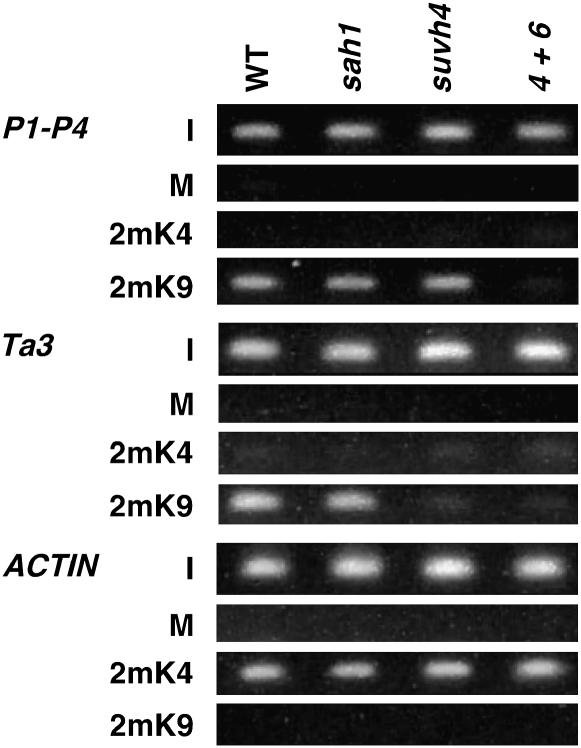
Chromatin immunoprecipitation (ChIP) assays were performed using a previously described method (Gendrel et al. 2002) starting with 0.7 g of leaf tissue from 3-week-old plants grown in soilless potting mix (Fafard mix no. 2) under continuous illumination. Chromatin was immunoprecipitated with anti-H3 dimethyl K4 antibodies (Upstate Biotechnologies, Lake Placid, NY) or with anti-H3 dimethyl K9 antibodies (gift of T. Jenuwein, Vienna Biocenter) or carried through the protocol with no antibody added as a control (mock precipitation). PCR primer sets for the center of the PAI1-PAI4 inverted repeat, Ta3, and ACTIN were previously described (Ebbs and Bender 2006). PCR-amplified products from ChIP template DNA were visualized on a 2.5% agarose gel stained with Gel-Star (Cambrex). Each ChIP assay was performed in three independent experiments, with results from a representative experiment shown in Figure 7.
Figure 7.—
The sah1L459F mutation does not deplete H3 2mK9 or H3 2mK4 from modified regions. ChIP analysis of the indicated mutants is shown. WT is Ws pai1, sah1 is Ws pai1 sah1L459F, suvh4 is Ws pai1 suvh4, and 4+6 is pai1 suvh4 suvh6 (Ebbs et al. 2005). Primer sets specific for each locus were used to amplify PCR products from total-input chromatin (I), no antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (2mK9), or chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (2mK4) from the indicated mutants. GelStar-stained PCR products are shown. P1–P4 indicates the PAI1–PAI4 inverted repeat.
RESULTS
Isolation of the sah1L459F mutation as a suppressor of PAI2 transcriptional silencing:
The sah1L459F allele was identified in a genetic screen for mutations that reduce cytosine methylation and silencing of the Arabidopsis PAI2 gene, which encodes the tryptophan biosynthetic enzyme phosphoribosylanthranilate isomerase (PAI). In the Ws strain of Arabidopsis, duplicated PAI genes are arranged as an inverted repeat (PAI1–PAI4) at one locus and singlet genes (PAI2 and PAI3) at two other unlinked loci (Bender and Fink 1995). These four genes are densely methylated in both CG and non-CG contexts over their regions of shared sequence identity, which includes proximal promoter sequences of each gene (Luff et al. 1999). The proximal promoters are thus transcriptionally silenced. Moreover, PAI3 and PAI4 encode nonfunctional PAI enzyme (Melquist et al. 1999). Nonetheless, Ws expresses sufficient PAI enzyme for normal tryptophan synthesis due to expression of PAI1 from a promoter that lies upstream of the methylated region (Melquist and Bender 2003). Transcription from this upstream promoter is also required to maintain PAI cytosine methylation, indicating that the PAI1–PAI4 inverted repeat locus generates an RNA signal for PAI DNA methylation (Melquist and Bender 2003).
To exploit the functional but methylated and silenced PAI2 gene as a reporter for PAI cytosine methylation, we isolated a Ws pai1 missense mutation that inactivates PAI1 enzyme without affecting the RNA signal for PAI DNA methylation generated by transcription of PAI1–PAI4 (Bartee and Bender 2001). The pai1 mutant displays a number of PAI-deficient phenotypes including blue fluorescence under ultraviolet (UV) light due to accumulation of a fluorescent tryptophan precursor, yellow–green leaf color, pointed tips on juvenile leaves, reduced plant size, and reduced fertility. In the pai1 reporter background, mutations that decrease the transcriptional silencing of the functional PAI2 gene can be easily detected by suppression of blue fluorescence. Using this pai1 suppressor screening strategy, we previously isolated 7 loss-of-function alleles in the SUVH4 H3 K9 MTase (Malagnac et al. 2002) and 11 loss-of-function alleles in the CMT3 cytosine MTase (Bartee et al. 2001). Both suvh4 and cmt3 mutants display a strong loss of non-CG methylation from the PAI2 reporter. PAI2 is particularly responsive to loss of non-CG methylation because the majority of cytosines in its promoter are methylated non-CG residues (Luff et al. 1999).
In addition to multiple alleles of cmt3 and suvh4, the pai1 suppressor screen yielded a single sah1L459F allele of SAH1. This mutation was cloned on the basis of its map position (materials and methods). The mutated leucine residue is conserved in SAH enzymes from a variety of organisms including humans, Drosophila, and fungi, although thermophilic archaea maintain an isoleucine at the analogous position (Porcelli et al. 2005, Figure 1). This residue occurs near the carboxy terminus of the protein. In the three-dimensional structure of the human SAH tetramer solved by X-ray crystallography, the analogous leucine residue lies in a linker region between two α-helicies (α17 and α18, Figure 1) that reach across and make contacts with the nicotinamide adenine dinucleotide (NAD) cofactor-binding domain on the adjacent monomer (Turner et al. 1998).
In the pai1 background, the sah1L459F mutation conferred a partial suppression of fluorescence similar to suvh4, but not as strong as cmt3 (Figure 2A). Like suvh4 and cmt3 mutations (Bartee et al. 2001; Malagnac et al. 2002), the sah1L459F mutation did not confer any obvious morphological defects, either in the pai1 background or in the wild-type Ws background. In contrast, previously described hog1 missense alleles of SAH1 confer slow growth and reduced fertility (Rocha et al. 2005).
The sah1L459F mutation was confirmed as the PAI2 silencing suppressor mutation by complementation with a transgene carrying a genomic clone of the SAH1 locus (T-SAH1). Transgenic pai1 sah1L459F (T-SAH1) plants displayed strong fluorescence similar to that of the pai1 parental strain (Figure 2B). In contrast, pai1 sahlL459F mutant plants transformed with a control transgene carrying a genomic clone of a neighboring gene At4g13930 encoding a serine hydroxymethyltransferase (T-SHM) displayed weak fluorescence similar to that of the untransformed pai1 sah1L459F strain. The pai1 sah1L459F (T-SAH1) transgenic plants also displayed remethylation of the PAI2 reporter gene as assessed by HpaII and MspI Southern blot assays for DNA methylation (Figure 2C). These assays are described in detail below.
The sah1L459F mutation reduces PAI cytosine methylation in non-CG contexts:
The suppression of PAI-deficient phenotypes in the pai1 background by the sah1L459F mutation correlated with reduced methylation on the PAI2 reporter gene as well as the PAI3 and PAI1–PAI4 loci. The PAI demethylation conferred by this mutation occurred primarily in non-CG sequence contexts, as determined by Southern blot assays with methylation-sensitive restriction enzymes and by bisulfite genomic sequencing.
For Southern blot analysis, genomic DNA was cleaved with different enzymes that monitor methylation in non-CG or CG sequence contexts. For comparison, we included suvh4 and cmt3 mutants previously recovered from the PAI2 silencing suppressor screen (see above). The cmt3 mutation confers a strong reduction of non-CG methylation on all three PAI loci, whereas CG methylation patterning is only slightly reduced (Bartee et al. 2001). The suvh4 mutation also confers a loss of non-CG methylation on the singlet genes PAI2 and PAI3, but does not alter the CG plus non-CG methylation patterning on the PAI1–PAI4 inverted repeat (Malagnac et al. 2002) because histone H3 lysine 9 methylation (H3 mK9) at this locus is uniquely controlled by the combined action of SUVH4 and a related enzyme SUVH6 (Ebbs et al. 2005; Ebbs and Bender 2006).
To assess methylation in non-CG contexts, genomic DNA was cleaved with HincII, which has recognition sites at the translational start codons of PAI1, PAI4, and PAI2 (but not PAI3 due to a polymorphism) (Malagnac et al. 2002). HincII monitors methylation of cytosines in the contexts 5′ CAG 3′ and 5′ CAT 3′on each of the two DNA strands of its recognition site. In this assay, the sah1L459F mutant displayed cleavage of PAI1–PAI4 and PAI2 intermediate between the wild type and the cmt3 mutant, implying a partial loss of non-CG methylation (Figure 3A). As a second assay for non-CG contexts, genomic DNA was cleaved with MspI, which has recognition sites in the second introns of PAI2, PAI3, and PAI4 (but not PAI1 due to a polymorphism) (Malagnac et al. 2002) and monitors methylation of cytosines in the context 5′ CCG 3′. The sah1L459F mutant displayed MspI cleavage of all three PAI loci intermediate between the wild type and the cmt3 mutant (Figure 3B), similar to the patterns observed for HincII cleavage.
We additionally tested cleavage with HpaII, an isoschizomer of MspI that is sensitive to methylation of either cytosine in the recognition site 5′ CCGG 3′ (CG and CCG methylation). In this assay a slight increase in PAI2 cleavage was evident in sah1L459F relative to the wild type, similarly to suvh4 and cmt3 (Figure 3B).
To monitor methylation specifically in a CG context we used the enzyme ClaI, which recognizes the sequence 5′ ATCGAT 3′ in the third intron of each of the four PAI genes. For comparison, we included a met1 mutant deficient in the major CG MTase (Bartee and Bender 2001; Kankel et al. 2003). The sah1L459F, suvh4, and cmt3 mutants displayed PAI ClaI cleavage patterns similar to the wild type (Figure 3C). The met1 mutant displayed partial ClaI cleavage diagnostic of reduced CG methylation at all three PAI loci. Together, the Southern blot analyses indicate that the sah1L459F mutation primarily reduces non-CG methylation patterning on the PAI genes.
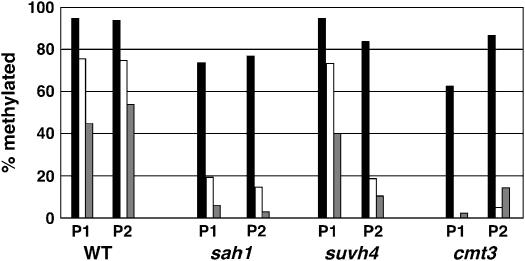
In addition to Southern blot assays, we monitored methylation patterning on the proximal promoter regions of PAI1 and PAI2 in the sah1L459F mutant background using genomic bisulfite sequencing. This analysis showed reduced methylation at both PAI1 and PAI2 relative to the wild type, with a stronger reduction in CNG and other non-CG contexts than in CG contexts (Figure 4). Overall, the methylation patterns at the PAI2 reporter locus were similar among sah1L459F, suvh4, and cmt3. However, the sah1L459F mutant had a slightly stronger depletion of CG methylation at PAI2 than either suvh4 or cmt3, suggesting additional weak inhibition of the MET1 CG methylation pathway specifically in this background.
Figure 4.—
Genomic bisulfite methylation sequencing data for PAI1 and PAI2 proximal promoter regions in the sah1L459F mutant. Eight independent top strand clones were sequenced for PAI1 or PAI2 from bisulfite-treated DNA of Ws pai1 sah1L459F (sah1) as previously described (Melquist and Bender 2003). The percentage of 5-methyl-cytosines detected out of total cytosines available within the region of PAI sequence identity (344 bp for PAI1 or 338 bp for PAI2) is shown, divided into the contexts CG (solid bars), CNG (open bars), and other contexts (shaded bars). For comparison, previously determined methylation patterns for wild-type Ws (WT) (Luff et al. 1999), Ws pai1 suvh4R302* (suvh4) (Malagnac et al. 2002), and Ws pai1 cmt3G456D (cmt3) (Bartee et al. 2001) are shown.
The sah1L459F mutation reduces non-CG methylation on transposon sequences:
Our finding that the sah1L459F allele causes demethylation of the PAI genes at non-CG residues, similarly to suvh4 and cmt3 mutations, suggested that the CMT3-controlled methylation pathway might be uniquely susceptible to SAH perturbations. To further investigate this issue, we monitored methylation patterning at known endogenous targets for the three cytosine methylation pathways characterized in Arabidopsis—SUVH4/CMT3, DRM, or MET1 (reviewed in Mathieu and Bender 2004)—in the sah1L459F mutant background.
Some Arabidopsis transposon sequences including the Mu1 mutator-related DNA transposon and the Ta3 retrotransposon are densely methylated at both CG and non-CG cytosines (Johnson et al. 2002; Lippman et al. 2003; Ebbs and Bender 2006). Like the PAI genes, these transposons are partially demethylated in a suvh4 mutant background and strongly demethylated in a cmt3 mutant background in CCG contexts monitored by MspI cleavage. Using MspI Southern blot analysis, we found that the sah1L459F mutation caused partially increased cleavage of both of these transposons relative to the wild-type parental strain (Figure 5). Therefore SUVH4/CMT3 target loci in addition to the PAI genes are partially demethylated in non-CG contexts by the sah1L459F mutation.
The Arabidopsis DRM1 and DRM2 cytosine MTase's control establishment of new methylation imprints, as well as maintenance of non-CG methylation at some loci (Cao and Jacobsen 2002a,b; Cao et al. 2003). In particular, the MEA-ISR direct repeat sequence is methylated and inhibited from cleavage by MspI in the wild type, but is demethylated and completely cleaved in the drm1 drm2 mutant (Cao and Jacobsen 2002a). We found that in the sah1L459F mutant there was no increase in MspI cleavage of MEA-ISR relative to the wild type (Figure 5C). This result suggests that DRM-mediated non-CG methylation is not affected by the sah1L459F mutation.
A major target for cytosine methylation in the Arabidopsis genome is the 180-bp centromere-associated repeat sequence (Vongs et al. 1993). This sequence carries mainly CG methylation and is demethylated by mutation of the MET1 CG-specific MTase, as indicated by increased cleavage with the CG-sensitive enzyme HpaII (Kankel et al. 2003, Figure 5D). HpaII Southern blot analysis showed no increase in centromere repeat cleavage for the sah1L459F mutant relative to the wild type (Figure 5D). Thus, the sah1 defect does not impair MET1-controlled CG methylation at the centromere repeats. The centromere repeats also carry non-CG methylation, as indicated by increased cleavage with MspI in suvh4 and cmt3 mutant backgrounds relative to the wild type (Figure 5D). However, MspI Southern blot analysis showed no obvious increase in centromere repeat cleavage for the sah1L459F mutant relative to the wild type. The lack of a centromere demethylation phenotype might reflect the limited sensitivity of the Southern blot assay for this highly repetitive sequence and an overall weaker effect on CCG methylation for sah1L459F relative to suvh4 or cmt3.
The sah1L459F mutation is a weak allele of SAH1:
To understand the degree to which the sah1L459F allele disrupts SAH1 function, we determined whether the Arabidopsis SAH1 vs. sah1L459F enzymes could complement the lethality of a yeast sah1 deletion mutant. We also included Arabidopsis SAH2 (At3g23810) in this analysis to determine whether it encodes a functional enzyme. The predicted SAH2 protein is 96% identical to SAH1 (Rocha et al. 2005). Both SAH1 and SAH2 are represented in databases by multiple cDNA and expressed sequence tag clones from a range of different tissue sources. Moreover, RNA gel blot analysis with gene-specific probes indicates that both SAH1 and SAH2 transcripts accumulate to readily detectable levels in total RNA prepared from adult plants (data not shown). However, an insertion mutation in SAH2 was previously reported to confer no morphological or DNA methylation phenotypes, in contrast to the lethal phenotype of SAH1 insertion mutations (Rocha et al. 2005). A possible explanation for this result is that SAH2 does not encode a functional enzyme. Alternatively, both SAH1 and SAH2 might be functional, but SAH1 might serve the dominant role in the plant due to its expression patterns or enzymatic properties.
To test the activities of the Arabidopsis SAH enzymes in yeast, we used a plasmid shuffle strategy. For this strategy, we constructed a haploid yeast strain where a deletion of the genomic SAH1 coding region is complemented by the yeast SAH1 gene expressed from its own promoter carried on a URA3-marked plasmid. We then supertransformed this strain with LEU2-marked plasmids, where each of the Arabidopsis SAH cDNAs, or as a positive control the yeast SAH1 gene, is expressed from the yeast SAH1 promoter. Serial dilutions of cultures grown from each strain were plated on medium containing 5-FOA, which selects against cells expressing URA3. Thus, 5-FOA-resistant colonies recovered in this assay are those where the yeast SAH1 gene on the URA3 vector has segregated away and where the plant SAH gene on the LEU2 vector is sufficient to complement the genomic sah1 deletion; an absence of 5-FOA-resistant colonies indicates that the plant SAH gene is not able to complement the genomic sah1 deletion. We found that when the Arabidopsis SAH1, SAH2, or sah1L459F cDNAs were expressed from a high copy 2μ yeast vector, they were all able to support growth on 5-FOA medium (Figure 6). However, when the Arabidopsis SAH cDNAs were expressed from a low-copy centromeric (CEN) plasmid, only wild-type SAH1 and SAH2 were able to support growth on 5-FOA medium. The plasmid shuffle data indicate that the sah1L459F allele confers partial loss of function that can be compensated for by overexpression. The data also indicate that both SAH1 and SAH2 encode functional SAH enzyme. Thus, the dominant role of SAH1 in the plant is likely due to differences in expression patterns between SAH1 and SAH2.
Histone H3 methylation is maintained near wild-type levels in the sah1L459F mutant:
H3 dimethylated at K9 (H3 2mK9) catalyzed by SUVH histone MTase's is required for CMT3-mediated DNA methylation (Jackson et al. 2002, 2004; Malagnac et al. 2002; Ebbs et al. 2005; Ebbs and Bender 2006). The sah1L459F mutation could thus selectively impair the CMT3 pathway by combined inhibition of SUVH MTase's and the CMT3 MTase. To estimate the severity of the sah1L459F mutation toward H3 K9 MTase's, we performed ChIP analysis of H3 2mK9 levels on heterochromatic loci in sah1L459F relative to the wild type and suvh mutants. We also used ChIP analysis to monitor levels of H3 dimethylated at lysine 4 (H3 2mK4), a modification associated with transcriptionally active genes (Gendrel et al. 2002; Lippman et al. 2004).
H3 mK9 can be lost either through direct impairment of SUVH activity or as an indirect consequence of transcriptional activation (Johnson et al. 2002; Ebbs et al. 2005). To avoid this ambiguity, we assayed H3 2mK9 levels at two heterochromatic loci where transcription is not activated by reduced DNA methylation: the Ta3 retrotransposon and the PAI1–PAI4 constitutively transcribed inverted repeat. In the wild type, Ta3 is transcriptionally silenced and enriched for H3 2mK9; in suvh4, Ta3 is only partially demethylated and remains transcriptionally silenced, but is strongly depleted for H3 2mK9 (Johnson et al. 2002; Ebbs et al. 2005; Ebbs and Bender 2006). In the wild type, PAI1–PAI4 is both DNA methylated and transcriptionally active due to an unmethylated promoter that lies upstream of PAI1 (Melquist and Bender 2003). PAI1–PAI4 is enriched for H3 2mK9 despite its transcriptional activity (Ebbs et al. 2005; Ebbs and Bender 2006). In addition, the accumulation of transcripts from the locus is not significantly altered by reduced DNA methylation of the internal inverted repeat sequences. Interestingly, H3 2mK9 at PAI1–PAI4 is not reduced in the suvh4 mutant because a related H3 K9 MTase SUVH6 is also active at this locus; both SUVH4 and SUVH6 must be mutated to cause loss of PAI1–PAI4 H3 2mK9 and non-CG methylation (Ebbs et al. 2005; Ebbs and Bender 2006). However, the effects of the suvh6 mutation are locus specific: partial demethylation of Ta3 in suvh4 is not enhanced in suvh4 suvh6.
ChIP analysis showed that the sah1L459F mutant maintained similar levels of H3 2mK9 relative to the wild type at both Ta3 and PAI1-PAI4 (Figure 7). In contrast, H3 2mK9 was lost from Ta3 in suvh4 or suvh4 suvh6 mutant backgrounds and from PAI1–PAI4 in the suvh4 suvh6 mutant background. Neither Ta3 nor PAI1–PAI4 carried H3 2mK4 in any of the strains assayed, consistent with a lack of transcriptional activation. These results indicate that the sah1L459F mutation does not inhibit the CMT3 DNA methylation pathway by strongly impairing SUVH H3 K9 MTase activity. However, it should be noted that a subtle modulation in H3 2mK9 levels would not be detectable within the sensitivity of the ChIP assay.
The ACTIN gene is enriched for H3 2mK4 in the wild-type and DNA methylation-deficient backgrounds including suvh4 and cmt3 (Johnson et al. 2002; Ebbs et al. 2005; Ebbs and Bender 2006). The levels of ACTIN H3 2mK4 were similar in the wild-type, sahlL459F, and suvh mutant backgrounds (Figure 7). This result suggests that as for SUVH H3 K9 MTase's, Arabidopsis H3 K4 MTase activity is not strongly inhibited by the sah1L459F mutation.
DISCUSSION
The addition of a methyl group to the 5-position of cytosine is a key epigenetic modification in mammals and plants, signaling heterochromatin formation and transcriptional silencing. Cytosine methylation functions in genome defense against invasive sequences (Yoder et al. 1997; Walsh et al. 1998; Miura et al. 2001; Kato et al. 2003; Lippman et al. 2003, 2004; Bourc'his and Bestor 2004), in regulation of gene dosage through parental imprinting (da Rocha and Ferguson-Smith 2004; Gehring et al. 2004), and in centromere compaction (Xu et al. 1999; Soppe et al. 2002). However, aberrant methylation and silencing of tumor suppressor genes is a common event in the progression of human cancers (Jones and Baylin 2002). In addition, methylation and silencing of newly integrated transgenes often presents a technical barrier to the expression of novel traits in transgenic plants and animals. Here we show that weak impairment of the Arabidopsis SAH enzyme that controls overall methylation homeostasis can specifically impair cytosine methylation and silencing without other phenotypic consequences.
We recovered the sah1L459F missense allele of the Arabidopsis SAH1 gene through a genetic screen designed to monitor transcriptional silencing of the PAI2 promoter sequence, which contains mainly methylated non-CG cytosines (Luff et al. 1999). In the non-CG methylation maintenance pathway H3 mK9 mediated by SUVH4 is required for DNA methylation mediated by CMT3 (Jackson et al. 2002; Malagnac et al. 2002). The sahlL459F mutation confers similar partial non-CG demethylation of PAI2 and transposon sequences to a suvh4 null mutation, but both sah1L459F and suvh4 confer weaker non-CG demethylation than a cmt3 null mutation (Figures 3 and 5). The sah1L459F mutation does not cause demethylation of sequences controlled by the DRM and MET1 MTase's (Figures 3 and 5). Together these results indicate that the SUVH4/CMT3 cytosine methylation pathway is preferentially inhibited by impairment of SAH1 relative to the DRM or MET1 cytosine methylation pathways.
The sah1L459F mutant protein can complement a yeast sah1 deletion mutant when expressed from a high-copy plasmid, demonstrating that the protein retains partial enzymatic activity (Figure 6). Consistent with this weak defect in enzyme activity, the sah1L459F mutation does not confer developmental abnormalities. In contrast, previously described hog1 missense mutations in SAH1 cause slow growth and decreased fertility (Rocha et al. 2005). The hog1 morphological defects might be caused by partial impairment of MTase's that catalyze essential metabolic functions and/or by dysregulation of gene expression due to loss of cytosine methylation. For example, Southern blot and thin layer chromatography-based assays show that the hog1-1 mutant has reductions in both CG and non-CG methylation (Rocha et al. 2005). Loss of methylation in CG contexts, such as in a met1 mutant, is associated with developmental abnormalities including reduced size, abnormal flower development, and delayed time to flowering (Kankel et al. 2003; Saze et al. 2003; Xiao et al. 2006). Furthermore, loss of methylation in both CG and non-CG contexts, such as in a met1 cmt3 double mutant, is associated with transposon reactivation (Kato et al. 2003).
The methylated transgene reporter used to isolate the hog1 alleles can be reactivated by met1 or cmt3 mutations, but not by a suvh4 mutation (Rocha et al. 2005). Since the sahlL459F mutation causes similar or weaker loss of non-CG methylation compared to suvh4 and has minimal effects on CG methylation, weak sah1 alleles would perhaps not be recovered from the hog1 mutant screen. Conversely, the pai1 mutant reporter background used to isolate the sah1L459F allele might have selected against stronger hog1-like sah1 mutations due to pleiotropic effects of combined tryptophan deficiency and SAH1 impairment. Selection against strong sah1 alleles, together with a limited spectrum of EMS-induced mutations, could account for why we recovered only one sah1 allele from a screen that also yielded multiple loss-of-function alleles for suvh4 and cmt3 (Bartee et al. 2001; Malagnac et al. 2002). In addition, SAH is highly conserved across eukaryotes (Figure 1), indicating that there are strong structural constraints on SAH function.
In the crystal structure of human SAH, the L459-analogous residue (L406) lies in the center of a linker region connecting two α-helicies (Figure 1) that make contacts with the NAD cofactor-binding domain of the adjacent monomer (Turner et al. 1998). The substitution of the bulky phenylalanine side chain at this position might thus impair optimal intersubunit contacts. In comparison, the strongest of the hog1 alleles, hog1-1 (T414I) (Rocha et al. 2005), mutates a conserved threonine residue (T363 in the human SAH) to isoleucine at a position adjacent to a conserved phenylalanine (F362 in human SAH) that forms the base of a hydrophobic binding pocket for the substrate adenine ring in the human SAH structure (Turner et al. 1998).
The CMT3 pathway for maintenance of non-CG methylation presents at least two types of MTase enzymes that can be inhibited by accumulation of AdoHcy in an sah mutant background: SUVH H3 K9 MTase's and the CMT3 cytosine MTase. In particular, SUVH-catalyzed dimethylation of H3 K9 is the epigenetic mark associated with CMT3-mediated cytosine methylation (Jackson et al. 2004). However, ChIP analysis at the Ta3 retrotransposon and the PAI1–PAI4 transcribed inverted repeat targets of SUVH4 and the related enzyme SUVH6 showed that these targets maintain similar levels of H3 2mK9 between the wild type and the sah1L459F mutant (Figure 7). Thus, SAH impairment does not preferentially inhibit SUVH MTase's to cause strong depletion of H3 2mK9, with loss of CMT3-mediated cytosine methylation occurring as a secondary effect.
Another possible explanation for preferential inhibition of the CMT3 pathway by SAH impairment is that the CMT3 cytosine MTase is uniquely susceptible to inhibition by AdoHcy. However, this possibility would require that CMT3 has a significantly different active site architecture than the DRM or MET1 cytosine MTase's, which are not strongly inhibited in the sah1L459F background (Figures 3 and 5). The chromomethylase family to which CMT3 belongs is defined by the insertion of a chromodomain between conserved cytosine MTase catalytic motifs (Goll and Bestor 2005). Thus, this extra domain has the potential to alter the catalytic properties of CMT3 relative to other cytosine MTase's. Future elucidation of the three-dimensional structures of plant cytosine MTase's will clarify this possibility.
Similarly, a histone MTase other than the SUVH enzymes required specifically in the CMT3 pathway could be uniquely susceptible to inhibition by AdoHcy. For example, in vitro studies have implicated H3 mK27 as well as H3 mK9 in CMT3 binding (Lindroth et al. 2004). However, this possibility would require that the putative susceptible histone MTase has a significantly different active site architecture than the SUVH H3 K9 MTase's or the MTase's that maintain H3 2mK4, which are not strongly inhibited in the sah1L459F background (Figure 7).
The simplest explanation for the preferential inhibition of the SUVH/CMT3 DNA methylation pathway by weak impairment of SAH1 is that simultaneous weak inhibition of the SUVH- and CMT3-mediated transmethylation reactions results in a synergistic strong inhibition of the output of the pathway. Even though SUVH activity is not impaired at the limit of detection in a ChIP assay in the sah1L459F background (Figure 7), a slight defect in this step of the pathway could combine with a slight defect in the CMT3-catalyzed step of the pathway to create a substantial pathway bottleneck. If this explanation indeed accounts for the preferential inhibition of SUVH/CMT3 vs. the DRM or MET1 DNA methylation pathways by sah1L459F, it also suggests that the DRM and MET1 pathways do not require histone methylation signals.
In Neurospora H3 mK9 mediated by the DIM-5 histone MTase is required for DNA methylation mediated by the DIM-2 cytosine MTase (Tamaru and Selker 2001). It would thus be interesting to determine whether weak mutations in the Neurospora SAH gene (such as a leucine-to-phenylalanine mutation at the residue analogous to the Arabidopsis SAH L459 residue, Figure 1) would cause substantial genomic demethylation without other phenotypic consequences. This result would support the view that weak SAH impairment causes preferential inhibition of histone methylation-dependent cytosine methylation because the pathway depends on sequential transmethylation reactions, rather than because one of the enzymes in the pathway is uniquely susceptible to inhibition by AdoHcy.
In the mouse, cells deleted for the related Suv39h1 and Suv39h2 H3 K9 MTase's display reduced cytosine methylation at major satellite repeats (Lehnertz et al. 2003), and cells deleted for the G9a H3 K9 MTase display a loss of methylation at an imprinted locus (Xin et al. 2003). SAH impairment might thus be a useful tool in further dissecting which regions of the mammalian genome are under control of histone methylation-dependent DNA methylation pathways. Controlled SAH impairment by chemical inhibitors or RNA interference could also serve as an alternative to inhibition of cytosine MTase's by cytidine analogs in probing the relationship between chromatin structure and gene silencing.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the SAH1 cDNA clone and Thomas Jenuwein (Vienna Biocenter) for H3 anti-dimethyl K9 antibodies. We also thank Eric Grote and Martin Romeo (Johns Hopkins University) for assistance with yeast strain constructions. This work was supported by National Institutes of Health grant GM61148 to J.B., by National Cancer Institute (NCI) training grant T32 CA09110 to L.M., and by National Institute of Environmental Health Sciences training grant T32 ES07141 and NCI training grant T32 CA09110 to M.L.E.
References
- Bacolla, A., S. Pradhan, R. J. Roberts and R. D. Wells, 1999. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated dna. J. Biol. Chem. 274: 33011–33019. [DOI] [PubMed] [Google Scholar]
- Baric, I., K. Fumic, B. Glenn, M. Cuk, A. Schulze et al., 2004. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc. Natl. Acad. Sci. USA 101: 4234–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric, I., M. Cuk, K. Fumic, O. Vugrek, R. H. Allen et al., 2005. S-adenosylhomocysteine hydrolase deficiency: a second patient, the younger brother of the index patient, and outcomes during therapy. J. Inherit. Metab. Dis. 28: 885–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, L., and J. Bender, 2001. Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 29: 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, L., F. Malagnac and J. Bender, 2001. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15: 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, J., and G. R. Fink, 1995. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83: 725–734. [DOI] [PubMed] [Google Scholar]
- Bevan, M., 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12: 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his, D., and T. H. Bestor, 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. [DOI] [PubMed] [Google Scholar]
- Bujnicki, J. M., S. T. Prigge, D. Caridha and P. K. Chiang, 2003. Structure, evolution, and inhibitor interaction of S-adenosyl-L-homocysteine hydrolase from Plasmodium falciparum. Proteins 52: 624–632. [DOI] [PubMed] [Google Scholar]
- Cao, X., and S. E. Jacobsen, 2002. a Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99(Suppl. 4): 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., and S. E. Jacobsen, 2002. b Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- Cao, X., W. Aufsatz, D. Zilberman, M. F. Mette, M. S. Huang et al., 2003. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13: 2212–2217. [DOI] [PubMed] [Google Scholar]
- Chiang, P. K., 1998. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol. Ther. 77: 115–134. [DOI] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- da Rocha, S. T., and A. C. Ferguson-Smith, 2004. Genomic imprinting. Curr. Biol. 14: R646–R649. [DOI] [PubMed] [Google Scholar]
- De Clercq, E., 2004. Antivirals and antiviral strategies. Nat. Rev. Microbiol. 2: 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M. L., and J. Bender, 2006. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M. L., L. Bartee and J. Bender, 2005. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 25: 10507–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojtova, M., A. Kovarik, I. Votruba and A. Holy, 1998. Evaluation of the impact of S-adenosylhomocysteine metabolic pools on cytosine methylation of the tobacco genome. Eur. J. Biochem. 252: 347–352. [DOI] [PubMed] [Google Scholar]
- Gehring, M., Y. Choi and R. L. Fischer, 2004. Imprinting and seed development. Plant Cell 16(Suppl.): S203–S213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A. V., Z. Lippman, C. Yordan, V. Colot and R. A. Martienssen, 2002. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873. [DOI] [PubMed] [Google Scholar]
- Goll, M. G., and T. H. Bestor, 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74: 481–514. [DOI] [PubMed] [Google Scholar]
- Jackson, J. P., A. M. Lindroth, X. Cao and S. E. Jacobsen, 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Jackson, J. P., L. Johnson, Z. Jasencakova, X. Zhang, L. PerezBurgos et al., 2004. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112: 308–315. [DOI] [PubMed] [Google Scholar]
- Johnson, L., X. Cao and S. Jacobsen, 2002. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367. [DOI] [PubMed] [Google Scholar]
- Jones, P. A., and S. B. Baylin, 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3: 415–428. [DOI] [PubMed] [Google Scholar]
- Kankel, M. W., D. E. Ramsey, T. L. Stokes, S. K. Flowers, J. R. Haag et al., 2003. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., A. Miura, J. Bender, S. E. Jacobsen and T. Kakutani, 2003. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13: 421–426. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos et al., 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13: 1192–1200. [DOI] [PubMed] [Google Scholar]
- Lindroth, A. M., D. Shultis, Z. Jasencakova, J. Fuchs, L. Johnson et al., 2004. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23: 4146–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z., B. May, C. Yordan, T. Singer and R. Martienssen, 2003. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z., A. V. Gendrel, M. Black, M. W. Vaughn, N. Dedhia et al., 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476. [DOI] [PubMed] [Google Scholar]
- Luff, B., L. Pawlowski and J. Bender, 1999. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3: 505–511. [DOI] [PubMed] [Google Scholar]
- Malagnac, F., L. Bartee and J. Bender, 2002. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21: 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuta, C., H. Tanaka, K. Uehara, S. Kuwata, A. Koiwai et al., 1995. Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethionine-dependent transmethylation reactions. Proc. Natl. Acad. Sci. USA 92: 6117–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, O., and J. Bender, 2004. RNA-directed DNA methylation. J. Cell Sci. 117: 4881–4888. [DOI] [PubMed] [Google Scholar]
- Melquist, S., and J. Bender, 2003. Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes Dev. 17: 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melquist, S., B. Luff and J. Bender, 1999. Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics 153: 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. W., D. M. Duhl, B. M. Winkes, F. Arredondo-Vega, P. J. Saxon et al., 1994. The mouse lethal nonagouti (a(x)) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. EMBO J. 13: 1806–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada et al., 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214. [DOI] [PubMed] [Google Scholar]
- Neff, M. M., J. D. Neff, J. Chory and A. E. Pepper, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392. [DOI] [PubMed] [Google Scholar]
- Porcelli, M., M. A. Moretti, L. Concilio, S. Forte, A. Merlino et al., 2005. S-adenosylhomocysteine hydrolase from the archaeon Pyrococcus furiosus: biochemical characterization and analysis of protein structure by comparative molecular modeling. Proteins 58: 815–825. [DOI] [PubMed] [Google Scholar]
- Rapp, M., T. A. Haubrich, J. Perrault, Z. B. Mackey, J. H. McKerrow et al., 2006. Antitrypanosomal activity of 5′-deoxy-5′-(iodomethylene)adenosine and related 6-N-cyclopropyladenosine analogues. J. Med. Chem. 49: 2096–2102. [DOI] [PubMed] [Google Scholar]
- Rocha, P. S., M. Sheikh, R. Melchiorre, M. Fagard, S. Boutet et al., 2005. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 17: 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze, H., O. M. Scheid and J. Paszkowski, 2003. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34: 65–69. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe, W. J., Z. Jasencakova, A. Houben, T. Kakutani, A. Meister et al., 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru, H., and E. U. Selker, 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., C. Masuta, K. Uehara, J. Kataoka, A. Koiwai et al., 1997. Morphological changes and hypomethylation of DNA in transgenic tobacco expressing antisense RNA of the S-adenosyl-L-homocysteine hydrolase gene. Plant Mol. Biol. 35: 981–986. [DOI] [PubMed] [Google Scholar]
- Turner, M. A., C. S. Yuan, R. T. Borchardt, M. S. Hershfield, G. D. Smith et al., 1998. Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength. Nat. Struct. Biol. 5: 369–376. [DOI] [PubMed] [Google Scholar]
- Vongs, A., T. Kakutani, R. A. Martienssen and E. J. Richards, 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. [DOI] [PubMed] [Google Scholar]
- Walsh, C. P., J. R. Chaillet and T. H. Bestor, 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20: 116–117. [DOI] [PubMed] [Google Scholar]
- Wolfson, G., J. Chisholm, A. H. Tashjian, Jr., S. Fish and R. H. Abeles, 1986. Neplanocin A. Actions on S-adenosylhomocysteine hydrolase and on hormone synthesis by GH4C1 cells. J. Biol. Chem. 261: 4492–4498. [PubMed] [Google Scholar]
- Xiao, W., K. D. Custard, R. C. Brown, B. E. Lemmon, J. J. Harada et al., 2006. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z., M. Tachibana, M. Guggiari, E. Heard, Y. Shinkai et al., 2003. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 278: 14996–15000. [DOI] [PubMed] [Google Scholar]
- Xu, G. L., T. H. Bestor, D. Bourc'his, C. L. Hsieh, N. Tommerup et al., 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402: 187–191. [DOI] [PubMed] [Google Scholar]
- Yoder, J. A., C. P. Walsh and T. H. Bestor, 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335–340. [DOI] [PubMed] [Google Scholar]