Abstract
The National Science Foundation-funded Chlamydomonas reinhardtii genome project involves (a) construction and sequencing of cDNAs isolated from cells exposed to various environmental conditions, (b) construction of a high-density cDNA microarray, (c) generation of genomic contigs that are nucleated around specific physical and genetic markers, (d) generation of a complete chloroplast genome sequence and analyses of chloroplast gene expression, and (e) the creation of a Web-based resource that allows for easy access of the information in a format that can be readily queried. Phases of the project performed by the groups at the Carnegie Institution and Duke University involve the generation of normalized cDNA libraries, sequencing of cDNAs, analysis and assembly of these sequences to generate contigs and a set of predicted unique genes, and the use of this information to construct a high-density DNA microarray. In this paper, we discuss techniques involved in obtaining cDNA end-sequence information and the ways in which this information is assembled and analyzed. Descriptions of protocols for preparing cDNA libraries, assembling cDNA sequences and annotating the sequence information are provided (the reader is directed to Web sites for more detailed descriptions of these methods). We also discuss preliminary results in which the different cDNA libraries are used to identify genes that are potentially differentially expressed.
The unicellular, green alga Chlamydomonas reinhardtii has many characteristics that make it an ideal organism for elucidating the function, biosynthesis, and regulation of the photosynthetic apparatus (Harris, 1989). Photosynthetic mutants of C. reinhardtii are viable because this alga can be grown heterotrophically with acetate as a sole source of carbon, and because C. reinhardtii is haploid during vegetative growth, mutations are almost immediately expressed and specific mutant phenotypes can be readily observed as colonies on solid medium. Furthermore, C. reinhardtii lends itself to in vivo procedures that are difficult or impossible to perform with more complex systems, and the “molecular toolkit” with which investigators can manipulate genes and gene expression in C. reinhardtii is extensive and has become increasingly sophisticated in recent years (Rochaix, 1995; Lefebvre and Silflow, 1999; Grossman, 2000; Harris, 2001). Selectable markers are available for identification of nuclear and chloroplast transformants (Boynton et al., 1988; Kindle et al., 1989; Kindle, 1990; Goldschmidt-Clermont, 1991; Lumbreras et al., 1998), as are relatively simple procedures to introduce DNA into cells (Kindle, 1990; Shimogawara et al., 1998). Reporter genes, such as green fluorescent protein (Fuhrmann et al., 1999) and arylsulfatase (Davies et al., 1992), and anti-sense- and RNAi-based suppression of mRNA levels have been effectively used in C. reinhardtii (Schroda et al., 1999; Sineshchekov et al., 2002). The many advantageous features of C. reinhardtii have earned it the epithet “green yeast” (Goodenough, 1992; Rochaix, 1995).
Many types of physiological, genetic, and molecular manipulations of C. reinhardtii have become routine and have made this organism ripe for more extensive genetic studies. Genome-wide analyses of this alga will add considerably to our understanding of photosynthetic function and the ways in which photosynthetic activities are modulated as environmental conditions change. Such analyses will also yield information on many physiological, developmental, cellular, and molecular processes, including the biogenesis of flagella; acclimation of cells to their nutrient, temperature, and light environments; and gamete and zygote formation. cDNA sequences derived from RNA isolated from C. reinhardtii cells exposed to a number of different environmental conditions are providing the community with a wealth of interesting genes that are serving as substrates for functional genomics and for understanding some of the biological processes described above. Analysis of global gene expression using high-density cDNA microarrays has recently enabled researchers to view the landscape of mRNA changes as cells experience different environmental conditions (Im et al., 2002). Furthermore, mutants defective for a number of different putative regulators involved in acclimation processes have already been characterized (Schnell and Lefebvre, 1993; Davies et al., 1996; Wykoff et al., 1999; Fukuzawa et al., 2001; Xiang et al., 2001). These mutants can be immediately used to identify subsets of genes controlled by specific regulatory elements. Finally, generating a high-density physical map that is linked to the genetic map (Lefebvre and Silflow, 1999) will facilitate positional cloning of specific mutant alleles.
The goal of our work is to provide a functionally annotated set of unique genes through a process called “cDNA assembly,” which is described in detail in “Materials and Methods” and by Shrager et al. (2002). Assembly begins with short nucleotide sequences generated from the 3′ and 5′ ends of cDNA clones (each such sequence is called a “read”). The initial 3′-end assembly can be used to identify specific genes because all 3′-end sequences initiate at or near the very end of the cDNA clone. Overlapping reads are identified and assembled as “contigs,” and individual contigs composed of reads derived from the same clone are placed into an ACE. The resulting ACEs are assumed to represent unique genes. (The term “ACE” is derived historically from Phrap terminology and refers to a collection of contigs which, based on the results of the assembly process, are predicted to be part of the same gene.) ACEs are then annotated for proposed gene function, and all of the reads, contigs, ACEs, and annotations are registered in a public database.
RESULTS
The assembly that we call 20020630 (June 30, 2002, the date on which this assembly was initiated) was generated from 3′ and 5′ reads (sequences) from individual cDNA clones present in three composite cDNA libraries designated Core, Stress I, and Stress II Libraries. Other libraries for which sequences are currently being completed are the Stress III, Deflagellation, and Gametogenesis-Zygote Libraries. The growth conditions for cells used in the construction of the different libraries, the project number of each library, and the number of clones sequenced are presented in Table I. The cDNAs represented in the Core Libraries are those present under the various “standard conditions” of growth used by most investigators in the field. The Deflagellation and Gametogenesis-Zygote Libraries could be used to study the biogenesis of flagella and events important for the formation of gametes and zygotes. The three stress libraries are enriched for cDNAs generated from cells exposed to various stress conditions. The Stress I Library is enriched for cDNAs synthesized from mRNAs that appear during deprivation for the macronutrients nitrogen, sulfur, and phosphorus, as well as during the shift between growth on the two major nitrogen compounds used by C. reinhardtii, nitrate and ammonium. The Stress II and Stress III Libraries are enriched for cDNAs synthesized from mRNAs that appear during a host of other stress conditions including, anaerobiosis, oxidative stress, high-light conditions, high-activity osmotic conditions, heavy-metal exposure, and iron and copper deprivation. The multitude of conditions to which the cells were exposed before generating these libraries ensures a broad range of cDNA representation in the total cDNA population.
Table I.
cDNA libraries
| Library | Conditions | Strain | Normalization | Project No. | Clones |
|---|---|---|---|---|---|
| Core | TAP light, TAP dark, HS + CO2, HS | 21gr | Not Normalized | 874 | 768 |
| Core | As above | 21gr | Normalized | 894 | 10,080 |
| Core | As above | 21gr | Subtracted (894) | 1,024 | 12,096 |
| Stress I | NO3 to NH4 (30 min, 1, 4 h), NH4 to NO3 (30 min, 1, 4 h), TAP-N (30 min, 1, 4 h), TAP-S (30 min, 1, 4 h), TAP-P (4, 12, 24 h) | 21gr | Normalized | 963 | 12,000 |
| Stress II | NH4 to NO3 (24 h), H2 production (0, 12, 24 h), TAP + H2O2 (1, 12, 24 h), TAP + Sorbitol (1, 2, 6, 24 h), TAP + Cd (1, 2, 6, 24 h) | 21gr | Normalized | 1,031 | 10,752 |
| Stress III | TAP-Fe, TAP-Cu, TAP-O2, TAP high light, HS high light (0, 0.5, 1, 2, 4, 6, 12 h) | 21gr | Normalized | 3,510 | Ongoing |
| Deflagellation | 15, 30, 60 min | 21gr | Normalized | 1,030 | 12,480 |
| Gametogenesis-Zygote | Gamete (2, 8, 10, 12, 15, 17 h), Zygote (30, 60 min) | 21gr | Normalized | 3,511 | Ongoing |
| S1D2 | S1D2 | Normalized | 925 | 124 |
Conditions of cell growth before RNA preparation, project number, and the number of clones sequenced are presented. -N, -S and -P refer to medium devoid of a nitrogen, sulfur, and phosphorus source, respectively. The normalization procedure is described in the text.
The assembly of 3′ and 5′ reads into contigs, each of which is represented by a consensus sequence in the database, was achieved by the use of an iterative assembly process (see “Materials and Methods” and http://www.biology.duke.edu/chlamy/ejournal. html for additional details). The 20020630 assembly started with 80,286 combined 3′ and 5′ reads, of which 62,780 were used in the final assembly to generate 14,410 contigs in 8,628 ACEs. The 3′ reads are initially assembled to identify a unique gene set, and they are then assembled with their 5′ ends to obtain as much sequence information about the individual genes as possible. As indicated above, approximately 25% of the reads were not used in the final assembly, primarily because their inclusion in specific contigs was ambiguous. Of the 8,628 ACEs generated during this assembly, most (4,374) contain two contigs, with the next most common group containing a single contig (3,661). There are also 454 ACEs with three contigs, 94 ACEs with four contigs, and the remaining (45) have more than four contigs. Whereas 7,750 ACEs generated from the assembly are composed of both 3′ and 5′ reads, 1,074 contain only 3′ reads, and four contain only 5′ reads.
Size Distribution of Contigs and ACEs
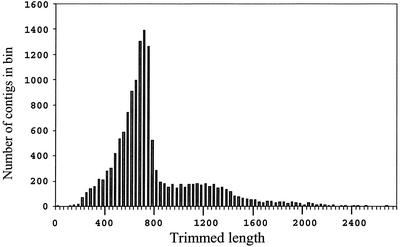
Figure 1 displays the vector-trimmed lengths of contigs that compose the ACEs. The longest contig was 3,321 nucleotides (composed solely from 5′-end reads), and the mean value was 791 nucleotides. The mean quality of the final pool of contigs was approximately 39 (38.9), which represents a very low probability of sequence error (approximately one error in approximately 103.9 or in about 8,000 bases). In 2,583 cases, the 3′ and 5′ reads assembled into a single contig; these contigs range in length from 164 to 3,062 nucleotides, with a mean value of 1,240 bases. The actual mean value of a C. reinhardtii mRNA is likely to be well over 1,240 because most ACEs consist of 3′ and 5′ contigs that do not overlap.
Figure 1.
Trimmed length (nt.) of all contigs; n = 14,410, min = 0, max = 3,321, mean = 791. Data has been placed into bins of 33 units in width. Data on the far right side of the graph, with two or fewer entries, are not shown.
ACEs consisting of a single contig in which the 3′ and 5′ reads overlap may represent full-length or near full-length cDNAs or clones truncated at their 5′ ends. In the case of longer cDNAs, where the 3′ and 5′ contigs fail to overlap, two or more contigs will alternatively be created within a single ACE. For example, ACE 40 comprises three contigs: Contig 40.1 was constructed from eleven 5′ reads, and contig 40.2 from the 3′ reads of many of the same clones. Contig 40.3 consists of a single 5′ read from one of the clones included in contig 40.2. The 3′ reads (40.2) and 5′ reads (40.1 and 40.3) do not overlap with one another, but because all of these were derived from the same clones, they are de facto part of the same cDNA. In another example, ACE 3790 comprises two contigs, each of which includes several overlapping 3′ and 5′ reads, but the reads in the separate contigs do not overlap one another. Again, the two contigs are considered part of the same cDNA because the reads were derived from the same clones.
In cases for which the ACEs comprise only 3′ ends, the corresponding 5′ reads were either too short or of quality too poor to be included in the assembly. The very small number (four) of ACEs comprising only 5′ reads appear to be derived from 3′ read-based contigs formed during assembly; but the 3′ reads did not pass later, more stringent quality inspection, leaving the four associated 5′ reads as “orphans.”
In Silico Identification of Potential Stress-Specific Genes
We are interested in how C. reinhardtii acclimates to diverse environmental conditions, and the programs of gene regulation that accompany acclimation (Im and Grossman, 2002; Im et al., 2003). Because we know the specific cDNA project from which each read was derived and how the cells were treated before isolation of mRNA for library construction, we can perform in silico subtractions, sorting contigs according to those that are potentially specific for each project or library. For example, we present in Table II some genes that have been identified at least four times (different 3′ reads) in the Stress I (project 963), Stress II (project 1031), or both Stress I and Stress II libraries, but which where never isolated from the Core Library.
Table II.
Polypeptides encoded by genes sequenced in Stress I and Stress II Libraries, but not in the Core Library
| ACE | Contig | Stress I | Stress II | Annotation(s) |
|---|---|---|---|---|
| 1,176 | 1,176.2 | 8 | 5 | Periplasmic arylsulfatase (ns; 0.0) |
| 6,106 | 6,106.1 | 10 | 0 | Extracellular polypeptide Ecp76 (ns; 0.0) |
| 3,899 | 3,899.1 | 5 | 9 | l-Amino acid oxidase catalytic subunit M [a] (ns; 0.0) |
| 1,121 | 1,121.2 | 0 | 5 | Nitrite reductase structural locus (ns; 0.0) |
| 8,377 | 8,377.1 | 0 | 6 | Ser acetyl transferase (Sat1; ns; 0.0) |
| 6,817 | 6,817.1 | 2 | 6 | h43 gene for high-CO2-inducible, periplasmic protein (ns; 0.0) |
| 2,590 | 2,590.1 | 0 | 7 | Putative Ser/Thr phosphatase (PP2A alpha; r; 3.00E-61) |
| 6,281 | 6,281.1 | 5 | 0 | CAAT-box DNA-binding protein subunit B (NF-YB; r; 2.00E-45) |
| 3,632 | 3,632.2 | 0 | 6 | Translation initiation factor (r; 5.00E-92) |
| 6,172 | 6,172.1 | 0 | 12 | Degreening-related gene dee76 (r; 4.00E-80) |
| 6,971 | 6,971.1 | 2 | 12 | Ribosomal protein P1 (r) |
| 7,764 | 7,764.2 | 5 | 0 | GlsA, involved in asymmetric cell division in dividing Volvox embryos (r; 8.00E-22) |
| 2,964 | 2,964.1 | 2 | 3 | DnaK chaperone (bpb; 0.0) |
| 220 | 220.2 | 4 | 17 | Protease DegP (bpb; 1.00E-43) |
| 4,644 | 4,644.1 | 0 | 10 | Possible ubiquitin-conjugating enzyme (bpb; 7.00E-18) |
| 3,550 | 3,550.2 | 0 | 6 | Ubiquitin-conjugating enzyme (bpb; 3.00E-68) |
| 2,307 | 2,307.1 | 0 | 11 | 26S Protease regulatory subunit (bpb; 1.00E-136) |
| 4,236 | 4,236.1 | 5 | 0 | Ubiquitin-conjugating enzyme E2 isoform (UBCX; bpb; 0) |
| 4,207 | 4,207.2 | 2 | 4 | 26S Proteasome AAA-ATPase subunit (bpb; 3.00E-85) |
| 3,327 | 3,327.1 | 5 | 12 | NADPH:protochlorophyllide oxidoreductase (lpcr-1; o; 0.0) |
| 4,002 | 4,002.2 | 3 | 4 | Vacuolar membrane proton-translocating inorganic pyrophosphate (o; 4.00E-32) |
| 1,941 | 1,941.1 | 0 | 7 | γ-Subunit of the coatomer protein complex (secretory pathway; o; 6.00E-68) |
| 1,559 | 1,559.3 | 2 | 6 | GDP-mannose transporter (o; 4.00E-30) |
| 6,361 | 6,361.1 | 2 | 9 | Oxoglutarate dehydrogenase E2 subunit (TCA cycle; o; 3.00E-13) |
| 5,659 | 5,659.2 | 2 | 6 | Squalene mono-oxygenase (cholesterol biosynthesis; o; 1.00E-12) |
| 6,943 | 6,943.3 | 2 | 14 | Chloroplast membrane-associated protein (o; 9.00E-13) |
| 443 | 443.1 | 2 | 4 | Unknown function (o; 3.00E-29) |
| 6,170 | 6,170.1 | 0 | 6 | Unknown function (o; 8.00E-12) |
| 7,121 | 7,121.1 | 2 | 4 | Unknown function (o; 7.00E-15) |
Stress I and II columns represent the number of reads found in each of those libraries. No other libraries were represented in the contigs composing these ACEs. Possible functional categories are indicated in parentheses: ns, nutrient stress; r, regulatory; bpb, biogenesis/pigment biosynthesis; o, other. Values following the functional category indicate the Blast e-value of the target used to determine this annotation.
Identification of genes that are highly represented in the Stress libraries but not in the Core Library may provide information concerning processes important for acclimation to stress conditions. However, because the libraries were normalized, the validity of statistical examination of the data is questionable; therefore these results only reveal genes that are “potentially stress-related.”
Some genes determined from this analysis to be potentially stress-related have been shown, in previous studies, to be activated under specific stress or environmental conditions. For example, levels of transcripts encoding the extracellular arylsulfatase (ACE1176) and Ecp76 polypeptide (ACE6106) have been previously shown to dramatically increase during sulfur stress (Davies et al., 1996; Takahashi et al., 2001). Sequences encoding both of these genes are present in the Stress I Library (which contains cDNAs of sulfur-stressed C. reinhardtii cells). Interestingly, the arylsulfatase but not the Ecp76 cDNA is also found in the Stress II Library. A component of the Stress II Library is derived from sulfur-stressed cells, but during growth under anaerobic conditions. It is possible that the anaerobic environment suppresses expression of some genes that are normally activated during sulfur stress (e.g. ECP76). The stress libraries also contain a number of genes encoding proteins involved in nitrogen metabolism, including nitrite reductase (Quesada et al., 1998), and l-amino acid oxidase (Vallon et al., 1993). These findings are consistent with the fact that both stress libraries used in this study were constructed from RNA derived from cells that had been transferred from growth on ammonium to nitrate (the nitrogen source for the Core Library was strictly ammonium) and/or starved for nitrogen. Another gene detected exclusively in the two stress libraries is H43. This gene was previously shown to be associated with iron deprivation and heavy-metal exposure (Rubinelli et al., 2002) but may also have a more general function during stress conditions. The SAT1 gene may also be involved in tolerance to heavy metals. SAT1 cDNA, detected only in the Stress II Library, encodes a Ser acetyl transferase that is required for Cys biosynthesis (Saito, 2000). Cys is the precursor of glutathione and phytochelatins (Meister, 1994), which help organisms cope with exposure to heavy metals (Scheller et al., 1987; Grill et al., 1989); one component of the cDNAs in the Stress II Library was generated from cadmium-treated cells. Interestingly, a protein similar to a chloroplast membrane protein identified in rice (Oryza sativa) genome sequences (accession no. AP003407) was a dominant cDNA in the stress libraries, especially in the Stress II Library. The function of this putative protein in cellular metabolism is not known.
Several genes encoding regulatory proteins also appear in the Stress I and Stress II Libraries but not in the Core Library. Among these putative polypeptides are a Ser/Thr phosphatase, a CAAT box DNA-binding protein, a translation initiation factor, and the ribosomal protein P1. Also in this category may be the degreening protein Dee76 (Hortensteiner et al., 2000) and the Volvox polypeptide GlsA. The Dee76 mRNA accumulates during degreening of nitrogen-starved Chlorella protothecoides and resembles a cell division factor (Hortensteiner et al., 2000), although its exact function with respect to degreening is not known. The GlsA gene, only identified from sequences in the Stress I Library, encodes a chaperone-like protein that is involved in early zygote cell divisions in Volvox (Miller and Kirk, 1999).
One of the most prominent groups of genes represented in the stress libraries encodes polypeptides involved in proteolysis, including ubiquitin-conjugating enzymes (Bachmair et al., 2001), proteasome subunits (Koster et al., 1995), and the chaperone DnaK (Mayer et al., 2000; Marcario and de Marcario, 2001). Interestingly, there is potentially very high expression of a DegP-like protease in stressed C. reinhardtii cells (it was sequenced 21 times in the Stress libraries but not once in the Core Library). DegP, an essential protease in Escherichia coli, and DegP homologs degrade damaged or denatured periplasmic proteins generated during various stress conditions (Pallen and Wren, 1997), including oxidative stress (Pedersen et al., 2001). Because the C. reinhardtii cell wall is mostly protein and appears to be modified under specific stress conditions (Takahashi et al., 2001), the DegP-like protease may also be involved in restructuring the cell wall. Overall, proteases and chaperone activities may help the organism eliminate damaged proteins and tailor protein content and function to stress conditions. On the basis of these comparative analyses, a gene encoding an enzyme involved in chlorophyll biosynthesis, protochlorophyllide oxidoreductase (POR), may also be elevated under stress conditions. This result is not very surprising because, under conditions of low pigment synthesis, POR may bind and protect the cells from phototoxic pigment intermediates such as protochlorophyllide, which can accumulate. Overexpression of genes encoding POR was previously shown to be photoprotective, probably by decreasing the steady-state level of the photosensitizer protochlorophyllide (Sperling et al., 1997).
A number of cDNAs encoding functionally diverse gene products are also identified as potentially specific to the stress libraries. In addition to those with unknown function, there are putative proteins with similarity to a vacuolar pyrophosphate transporter, the γ-subunit of the coatomer complex, a putative GDP Man transporter, the E2 subunit of oxoglutarate dehydrogenase, squalene mono-oxygenase, and a chloroplast membrane-associated protein. Although similarities between some of these putative polypeptides and their potential homologs are very high (inorganic pyrophosphate transporter, and γ-coatomer subunit), others are not so high (oxoglutarate dehydrogenase, squalene mono-oxygenase, and chloroplast membrane-associated protein). These analyses provide interesting directions for future research, however, validation of the results at the level of both gene expression and protein function are required.
Expressed Sequence Tag (EST) Database Details
An on-line database of EST sequences is accessible at http://www.biology.duke.edu/chlamy_genome/. Users can search the database by clone, read (EST), or annotation. This database includes the cDNA sequences defined from this study and other C. reinhardtii EST sequences reported by other groups and for other strains (for details, see “Materials and Methods”). Sequence data files and full blast results can be downloaded by anonymous ftp at: ftp://ftp.biology.duke.edu/pub/chlamy_genome/. These resources will eventually link the cDNA data with the information currently maintained within ChlamyDB, the database maintained by the Chlamydomonas Genetics Center, and the nuclear genome database under construction at the Joint Genome Institute of the Department of Energy (http://bahama.jgi-psf.org/prod/bin/chlamy/home.chlamy.cgi).
CONCLUSIONS
C. reinhardtii is an ideal organism for elucidating the mechanism of photosynthesis, the biogenesis of complexes of the photosynthetic apparatus, flagellar structure and function, and acclimation processes. The establishment of a high-quality C. reinhardtii uni-gene set is greatly facilitating the investigation of gene function, structure, and regulation and will help establish evolutionary relationships between C. reinhardtii and other photosynthetic eukaryotes. This paper describes and discusses the creation, normalization, and sequencing of C. reinhardtii cDNA libraries for the efficient formation of a high-quality uni-gene set from cDNA reads and for the annotation of that uni-gene set. Complete, detailed descriptions of the assembly protocol and all data generated in this project are available on the Web site http://www.biology.duke.edu/chlamy/ejournal.html. From the analyses of the different recombinant libraries used for these studies, we have identified genes that are potentially activated under stress conditions. Furthermore, the high quality uni-gene set identified by the assembly process has been used to build a first-generation microarray that represents over 2,700 unique genes (Im et al., 2003), and a second generation array is currently under construction. Finally, the cDNA sequence information is greatly facilitating an understanding of gene function and the control of physiological and developmental processes in C. reinhardtii and has served as a stimulus for the Joint Genome Institute to sequence the entire C. reinhardtii genome. The genome sequence should be completed in the fall/winter of 2002/2003.
MATERIALS AND METHODS
This section provides an overview of the complex methods used in library construction and assembly process. Much more detail is provided on-line at http://www.biology.duke.edu/chlamy/ejournal.html.
Growth Conditions Used to Generate RNA for cDNA Synthesis
To obtain sequence information from as many genes as possible, cDNA libraries were constructed using RNA isolated from cells exposed to a variety of conditions. These libraries were then normalized, and individual cDNAs were sequenced. Growth conditions for cells used in the construction of the cDNA libraries are given in Table I. The Core Library was generated from four separate cDNA libraries, each constructed from pooled RNA isolated from Chlamydomonas reinhardtii strain 21gr (CC-1690, Chlamydomonas Genetics Center, Duke University) grown under the following conditions: (a) Tris-acetate-phosphate medium (TAP) in moderate light (approximately 75 μmol photon m−2 s−1); (b) TAP medium in the dark; (c) high-salt minimal medium (HS) at ambient levels of CO2; (d) HS medium in air supplemented with 5% (v/v) CO2 (for composition of these media, see Harris, 1989). An equal number of plasmids from each library was combined, and this composite library was normalized (see normalization protocol below) to generate the Core Library.
In addition to the Core Library, five other composite libraries were constructed and normalized (see Table I). The Stress I Library was generated from pooled RNA isolated from C. reinhardtii strain 21gr cells at several time intervals after shifts to various conditions: (a) from nitrate- to ammonium-containing medium; (b) from ammonium- to nitrate-containing medium; (c) from ammonium-containing medium to medium devoid of nitrogen; (d) from sulfur-replete medium to medium devoid of sulfur; (e) from phosphate-replete medium to medium devoid of phosphorus.
The Stress II Library was similarly generated from C. reinhardtii strain 21gr cells exposed to different environmental conditions. RNA was isolated and pooled from cells that were: (a) shifted from ammonium- to nitrate-containing medium 24 h before harvesting the cells (long-term acclimation to growth on nitrate); (b) starved for sulfur under anaerobic conditions (conditions used for H2 production); (c) treated with 2 mm H2O2 (oxidative stress); (d) incubated with 0.3 m sorbitol (osmotic shock); or (e) treated with sublethal concentrations (100 μm) of the potentially toxic heavy-metal cadmium.
Pooled RNA isolated from cells exposed to high light (1,100 μmol photon m−2 s−1) or deprived of oxygen, iron, and copper for various times was used to construct the Stress III Library. A Deflagellation Library was constructed from RNA isolated from cells synthesizing or assembling flagella after deflagellation. Deflagellation was accomplished by adding 1 m acetic acid to the cultures until the pH declined to 4.5. The cultures were left undisturbed for 2 min and neutralized to pH 7.0 with 1 m KOH, and the cells harvested at various times after neutralization. A Gametogenesis-Zygote Formation Library was constructed using RNA from cells transferred to nitrogen-deficient medium and sampled at intervals during the process of gametogenesis and after mixing fully differentiated gametes of both mating types to permit initiation of the mating process. Together, these libraries provide a rich source of genes that are expressed during normal growth and as the cells acclimate to a number of different environmental conditions.
We have also constructed a cDNA library from the S1D2 strain (CC-2290, Chlamydomonas Genetics Center), a natural isolate of C. reinhardtii with extensive nucleotide sequence polymorphisms relative to the most commonly used laboratory strains (Harris, 2001; Vysotskaia et al., 2001). The S1D2 library was constructed from cells grown to mid-logarithmic phase in TAP medium in moderate light (70 μmol photon m−2 s−1) and bubbled with air.
Library Construction
RNA was isolated (see http://www.biology.duke.edu/chlamy/ejournal. html) from cells grown in 100 mL cultures (approximately 5 × 106 cells mL−1). cDNA libraries were constructed using a λZAP cDNA synthesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol, with two modifications during first-strand synthesis to accommodate the GC-rich content of C. reinhardtii nuclear genes. To reduce secondary structure in the mRNA template, the reverse transcription reaction was performed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) at 50°C. To facilitate oligonucleotide annealing for first-strand synthesis at the higher temperature, we used the primer GAGA-XhoI-(dT)30, which has a 3′ end extended by 12 oligo(dT) [from (dT)18 to (dT)30]. Libraries of phagemids containing cloned cDNA inserts were normalized or subtracted to reduce the abundance of cDNAs appearing in high copy number. The normalization procedure was modified from a re-association-based method developed by Bonaldo et al. (1996). A 10-fold excess of cDNA inserts (“Driver DNA”) generated by PCR amplification of a small fraction of the single-stranded library was hybridized to the single-stranded cDNA clones of the library until a COt value of 5 was reached. At that point, highly and moderately abundant cDNAs anneal to form double-stranded DNA. The remaining single-stranded phagemid DNA was purified by hydroxylapatite column chromatography and converted to double-stranded cDNA, yielding the normalized library.
Driver DNA used for normalization was mixed with the single-stranded library, and 5′-blocking oligonucleotide (5′-GAAT TCCT GCAG CCCG GGGG ATCC ACTA GTTC TAGA) and 3′-blocking oligonucleotide (5′-AATA CGAC TCAC TATA GGGC GAAT TGGG TACC GGGC CCCC CCTC GAG), and this mixture was heated at 80οC for 3 min before the addition of hybridization buffer. The resulting reaction mixture was incubated at 35οC for 20 h 30 min (calculated COt approximately 5). Salmon sperm DNA (50 μg denatured) was added to the hybridization reaction as a carrier, and the remaining single-stranded circles were purified by hydroxylapatite chromatography and then converted from single- to double-stranded DNA by amplification with four 20-mer oligonucleotides (JM1–JM4; see below) that anneal at positions approximately evenly spaced around the pBluescript SK. Eight microliters of single-stranded DNA circles was mixed with 3 μg of each of the primers JM1 (5′-GCTA TGTG GCGC GGTA TTAT), JM2 (5′-CTAC CAGC GGTG GTTT GTTT), JM3 (5′-CTGG CGTA ATAG CGAA GAGG), and JM4 (5′-TGTG GAAT TGTG AGCG GATA). Resulting double-stranded DNA circles were purified through a Chroma Spin-200 TE (pH 8.0) column (BD Biosciences Clontech, Palo Alto, CA) and precipitated. The effectiveness of the normalization procedure was assessed by plating the un-normalized and normalized phage libraries at a density of approximately 5,000 plaque-forming units per plate (100 × 15 mm) and hybridizing the plaque lifts to RBCS2 and ATS1 gene-specific probes. Normalization resulted in a considerable reduction in the frequency at which the clones encoding both of these polypeptides were represented in the library (Shrager et al., 2001).
The subtraction procedure used to reduce sequencing redundancy was the same as for normalization, except that the starting material was the normalized library, and the driver DNA was amplified inserts of all previously sequenced cDNA clones. One microliter of phagemid DNA from each sequenced clone from the normalized Core Library (894) was pooled to generate a mixture of cDNAs. The inserts from this mixture were amplified as above and used as driver DNA. The driver DNA was hybridized with the source library itself to subtract out previously sequenced clones.
Assembly of Contigs and Generation of Unique Gene Sets
More than 40,000 C. reinhardtii cDNAs were sequenced from both the 5′ and 3′ ends. Sequences generated from the Core, Stress I, and Stress II Libraries were assembled based on sequence similarity. Our computational protocol for sequence assembly is based upon the commonly used Phrap assembly program (http://www.phrap.org), but uses an iterative assembly cycle. Details of the computational methods and parameters that we used for the iterative assembly are given on the Web site http://www.biology.duke.edu/chlamy/ejournal.html.
Annotation
Analysis of contigs and gene products encoded by an ACE involves three phases: (a) placing the final assembled contigs into a nonredundant uni-gene set; (b) finding contigs encoding previously identified C. reinhardtii genes by cross-matching sequences of the set of final contigs to a non-EST Volvocales database with more than 2,000 sequences; and (c) identifying potential orthologs of the genes encoded by the remaining contigs. Those assembled contig sequences that did not match previously sequenced Volvocales genes were annotated by searching derived amino acid sequences in the GenBank database using the WU-BlastX program. A putative homolog is selected only if the E value of the matching high-scoring segment pair is less than 1 × 10−10, corresponding to proteins with a high degree of sequence similarity to the inferred C. reinhardtii translation products. The collected annotation data for all putative homologs is deposited in the ChlamyEST database (http://www.biology.duke.edu/chlamy_genome/; “Gene Search” link) and serves as a starting point for full annotation.
EST Grouping and Uni-Gene Set
Independent of Phrap-mediated assembly of contigs into ACE groups, we generated a set of unique EST groups by cross-matching each cDNA sequence read against a dataset of all C. reinhardtii EST reads using WU-blastn (S > 1,000; or high-scoring segment pair, >95% identity). In contrast to the ACEs, which were assembled only from sequences derived from strain 21gr (CC-1690), the EST groups include sequences from the other widely used strains, 137C (CC-125) and S1D2 (CC-2290), as well as clones sequenced by the Kazusa group, and strain C9, equivalent to CC-408 of the Chlamydomonas Genetics Center (Asamizu et al., 1999). This grouping permits identification of homologous cDNA sequences derived from all strains represented in the database and therefore can be used to find potential sequence polymorphisms among strains. The EST groups are included in the ChlamyDB database (http://www.biology.duke.edu/chlamydb/) as an aid to finding related sequences.
In most cases, we found that all reads in a given contig came from the same EST group. Exceptions to this rule were examined individually and led to discovery of a few chimeric sequences and other anomalies. The EST group thus serves as an additional means of quality control for the assembly process.
Web Access to Sequence and Annotation Data
We have created a comprehensive relational database using PostgreSQL (http://www.postgresql.org/) containing the sequence and annotation data generated by the cDNA project, which is maintained at the Chlamydomonas Genome Project Web site (http://www.biology.duke.edu/chlamy_genome/). The primary goal in creating the database is to provide the community with a maximal set of C. reinhardtii genes and to use this information as a platform for gene annotation, and construction of a cDNA-based microarray representative of a large assemblage of unique genes. The database may be queried for annotation data (“Gene search”) or for sequence information (“Contig/Clone search”). A search for a gene or gene product will return a list of all contigs annotated as such. For each contig returned, a set of links lead the user to detailed information on that contig, including the contig sequence, a table of the cDNA clones used to construct the contig, cDNA sequence information, quality data, library information (the library from which the sequence was derived), the position of the cDNA on the microarray, and links that facilitate blast analysis of the sequence using either the C. reinhardtii-specific database or GenBank for a more general analysis. In addition, all of the primary sequence and sequence quality data can be downloaded from the Chlamydomonas Genome FTP site (ftp://ftp.biology.duke.edu/pub/chlamy_genome).
ACKNOWLEDGMENTS
We thank people at the Stanford Genome Technology Center for providing both strong technical support and helpful discussions. We are grateful for the help provided by other members of the team of investigators, including David Stern, Pete Lefebvre, and Carolyn Silflow.
Footnotes
This work was supported by the National Science Foundation (Molecular and Cellular Biosciences grant no. 9975765). This is a Carnegie Institution of Washington publication no. 1,554.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.016899.
LITERATURE CITED
- Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. A large scale structural analysis of cDNAs in a unicellular green alga Chlamydomonas reinhardtii: generation of 3:433 non-redundant expressed sequence tags. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci. 2001;6:463–470. doi: 10.1016/s1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Davies J, Weeks DP, Grossman AR. Expression of the arylsulfatase gene from the β2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 1992;20:2959–2965. doi: 10.1093/nar/20.12.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Yildiz F, Grossman AR. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 1996;15:2150–2159. [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho KI, Saito T, Kohinata T, Ohyama K. Ccm1, a regulatory gene controlling the induction of a carbon concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc Natl Acad Sci USA. 2001;98:5347–5352. doi: 10.1073/pnas.081593498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenyl transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW. Green yeast. Cell. 1992;70:533–538. doi: 10.1016/0092-8674(92)90424-b. [DOI] [PubMed] [Google Scholar]
- Grill E, Loffler S, Winnacker EL, Zenk MH. Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific y-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthetase) Proc Natl Acad Sci USA. 1989;86:6838–6842. doi: 10.1073/pnas.86.18.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR. Chlamydomonas reinhardtii and photosynthesis: genetics to genomics. Curr Opin Plant Biol. 2000;3:132–137. doi: 10.1016/s1369-5266(99)00053-9. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Hortensteiner S, Chinner J, Matile P, Thomas H, Donnison IS. Chlorophyll breakdown in Chlorella protothecoides: characterization of degreening and cloning of degreening-related genes. Plant Mol Biol. 2000;42:439–450. doi: 10.1023/a:1006380125438. [DOI] [PubMed] [Google Scholar]
- Im CS, Grossman AR. Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. Plant J. 2002;30:301–313. doi: 10.1046/j.1365-313x.2001.01287.x. [DOI] [PubMed] [Google Scholar]
- Im CS, Zhang Z, Shrager J, Chang CW, Grossman AR (2003) Analysis of light and CO2 regulation in Chlamydomonas reinhardtii using genome-wide approaches. Photosynth Res (in press) [DOI] [PubMed]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Schnell RA, Fernández E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109:2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster AJ, Walz J, Lupas A, Baumeister W. Structural features of archaebacterial and eukaryotic proteasomes. Mol Biol Rep. 1995;21:11–20. doi: 10.1007/BF00990965. [DOI] [PubMed] [Google Scholar]
- Lefebvre PA, Silflow CD. Chlamydomonas: the cell and its genomes. Genetics. 1999;151:9–14. doi: 10.1093/genetics/151.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. [Google Scholar]
- Marcario AJL, de Marcario EC. The molecular chaperone system and other anti-stress mechanisms in archaea. Front Biosci. 2001;6:262–283. doi: 10.2741/macario. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Rudiger S, Bukau B. Molecular basis for interaction of the DnaK chaperones with substrates. J Biol Chem. 2000;381:875–885. doi: 10.1515/BC.2000.109. [DOI] [PubMed] [Google Scholar]
- Meister A. Glutathione-ascorbic acid antioxidant system in animals. J Biol Chem. 1994;269:9397–9400. [PubMed] [Google Scholar]
- Miller SM, Kirk DL. glsA, a Volvox gene required for asymmetric division and germ cell specification encodes a chaperone-like protein. Development. 1999;126:649–658. doi: 10.1242/dev.126.4.649. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- Pedersen LL, Radulic M, Doric M, Abu Kwaik Y. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect Immunol. 2001;69:2569–2579. doi: 10.1128/IAI.69.4.2569-2579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada A, Gomez I, Fernandez E. Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta. 1998;206:259–265. doi: 10.1007/s004250050398. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D. Chlamydomonas reinhardtii as the photosynthetic yeast. Annu Rev Genet. 1995;29:209–230. doi: 10.1146/annurev.ge.29.120195.001233. [DOI] [PubMed] [Google Scholar]
- Rubinelli P, Siripornadulsil S, Gao-Rubinelli F, Sayre RT. Cadmium- and iron-stress-inducible gene expression in the green alga Chlamydomonas reinhardtii: evidence for H43 protein function in iron assimilation. Planta. 2002;215:1–13. doi: 10.1007/s00425-001-0711-3. [DOI] [PubMed] [Google Scholar]
- Saito K. Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol. 2000;3:188–195. [PubMed] [Google Scholar]
- Scheller HV, Huang B, Hatch E, Goldsbrough PB. Phytochelatin synthesis and glutathione levels in response to heavy metals in tomato cells. Plant Physiol. 1987;85:1031–1035. doi: 10.1104/pp.85.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell RA, Lefebvre PA. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genet. 1993;134:737–747. doi: 10.1093/genetics/134.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell. 1999;11:1165–1178. doi: 10.1105/tpc.11.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman AR, Usuda H. High efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genet. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager J, Chang C-W, Davies J, Harris EH, Hauser C, Tamse R, Surzycki R, Gurjal M, Zhang Z, Grossman AR. Proceedings of the 12th International Congress on Photosynthesis, Aug 18–23, Australia: Brisbane; 2001. Chlamydomonas cDNAs: assembly and potential role in understanding metabolic processes.http://www.publish.csiro.au/ps2001 , Article no. 541–001. [Google Scholar]
- Shrager J, Hauser C, Chang C-W, Harris EH, Davies J, McDermott J, Tamse R, Zhang Z, Grossman AR (2002) The generation and organization of C. reinhardtii cDNA information. Chlamydomonas Articles. http://www.biology.duke.edu/chlamy/ejournal.html (Nov 18, 2002) [DOI] [PMC free article] [PubMed]
- Sineshchekov OA, Jung KH, Spudich JL. The rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:225–230. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling U, van Cleve B, Frick G, Apel K, Armstrong GA. Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 1997;12:649–658. doi: 10.1046/j.1365-313x.1997.00649.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Braby CE, Grossman AR. Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol. 2001;127:665–673. [PMC free article] [PubMed] [Google Scholar]
- Vallon O, Bulte L, Kuras R, Olive J, Wollman F-A. Extensive accumulation of an extracellular L-amino-acid oxidase during gametogenesis of Chlamydomonas reinhardtii. Eur J Biochem. 1993;215:351–360. doi: 10.1111/j.1432-1033.1993.tb18041.x. [DOI] [PubMed] [Google Scholar]
- Vysotskaia VS, Curtis DE, Voinov AV, Kathir P, Silflow CD, Lefebvre PA. Development and characterization of genome-wide single nucleotide polymorphism markers in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2001;127:386–389. [PMC free article] [PubMed] [Google Scholar]
- Wykoff D, Grossman A, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA. 1999;96:15336–15341. doi: 10.1073/pnas.96.26.15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang J, Weeks DP. The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2001;98:541–546. doi: 10.1073/pnas.101534498. [DOI] [PMC free article] [PubMed] [Google Scholar]